Abstract
Soil contaminated with C5+, which contained benzene (45%, wt/wt), dicyclopentadiene (DCPD) plus cyclopentadiene (together 20%), toluene (6%), styrene (3%), xylenes (2%), naphthalene (2%), and smaller quantities of other compounds, served as the source for isolation of 55 genomically distinct bacteria (standards). Use of benzene as a substrate by these bacteria was most widespread (31 of 44 standards tested), followed by toluene (23 of 44), xylenes (14 of 44), styrene (10 of 44), and naphthalene (10 of 44). Master filters containing denatured genomic DNAs of all 55 standards were used to analyze the community compositions of C5+ enrichment cultures by reverse sample genome probing (RSGP). The communities enriched from three contaminated soils were similar to those enriched from three uncontaminated soils from the same site. The compositions of these communities were time dependent and showed a succession of Pseudomonas and Rhodococcus spp. before convergence on a composition dominated by Alcaligenes spp. The dominant community members detected by RSGP were capable of benzene degradation at all stages of succession. The enrichments effectively degraded all C5+ components except DCPD. Overall, degradation of individual C5+ hydrocarbons followed first-order kinetics, with the highest rates of removal for benzene.
Although degradation of single aromatic compounds by pure strains of hydrocarbon-degrading bacteria is generally well understood, bioremediation processes in the environment usually involve degradation of mixtures of compounds by communities of microorganisms. The biodegradation kinetics for benzene-toluene-ethylbenzene-xylene (BTEX) components by an enrichment culture from gasoline-contaminated soil were recently reported (14). The rates of biodegradation of benzene and toluene were 1 to 2 orders of magnitude lower when the compounds were degraded as gasoline components than when the pure compounds were degraded (22). No attempts were made to evaluate the nature of the microorganisms in the enrichment. Deeb and Alvarez-Cohen (1) also reported that degradation of individual BTEX components differed significantly from degradation of the BTEX mixture. This was demonstrated both for a consortium from gasoline-contaminated soil and for a pure culture of Rhodococcus rhodochrous. When added individually, BTEX components were degraded in the order toluene-benzene-ethylbenzene-xylenes, whereas in BTEX mixtures the ethylbenzene was degraded most rapidly, followed by toluene, benzene, and the xylenes.
We have previously reported on degradation of C5+, a complex mixture of aromatic hydrocarbons that is formed during ethane pyrolysis to ethylene during polyethylene production (4, 13, 16). We used reverse sample genome probing (RSGP) in initial experiments to define the communities involved in this process. RSGP involves hybridization of labeled total community DNA with a master filter containing the denatured chromosomal DNAs of genomically distinct bacteria (referred to as standards) isolated from the target environment (20). Initially, a set of 35 standards, isolated primarily on rich media, was used (13). This set was augmented with six standards isolated with toluene as the sole carbon and energy source (4). In the present work we used the C5+ components benzene, styrene, xylenes, and naphthalene to isolate another 14 genomically distinct hydrocarbon degraders. The resulting master filter, representing 55 standards, was used to monitor the community response to hydrocarbons in the present study.
MATERIALS AND METHODS
Soil samples.
Soil samples C1, C2, and C3 (0.5 to 1 kg) were obtained from depths of 0.9 to 1.2 m in a C5+-contaminated area close to an existing polyethylene plant in Alberta, Canada. Benzene, toluene, xylene, styrene, dicyclopentadiene (DCPD), and naphthalene were all detected in this soil. Samples U1, U2, and U3, consisting of 1 to 2 kg of uncontaminated sediment, were obtained from different locations at the same site. Sample U1 consisted of clay till and sandy material taken from a depth of 12.0 to 12.5 m. Sample U2 consisted of sandy material taken from a depth of 12.5 m. Sample U3 was a composite of wet clay material taken from a depth of 5.5 m and sandy material collected at a depth of 14 m from the same borehole. Uncontaminated sediment samples U1 and U3 were taken from new boreholes made beyond the edges of a known C5+ contaminant plume. Uncontaminated sediment sample U2 was taken from a location where a new ethylene plant was under construction. Samples from a contaminated soil pile that was constructed for a bioremediation project (16) and has been described previously (4, 13) were used for isolation of master filter organisms.
Isolation and characterization of C5+-degrading bacteria.
The single C5+ components benzene, styrene, xylenes (a mixture of o-, m-, and p-xylenes), and naphthalene were used as sole carbon and energy sources for isolation of strains. Appropriate dilutions of suspensions or enrichments of contaminated soil samples were spread on minimal salts medium (MSM) agar plates (13). These preparations were incubated at room temperature in desiccators containing specific hydrocarbon substrates at a concentration of 1 or 10% (wt/wt) in vacuum pump oil (vpo), which provided a constant concentration of each hydrocarbon substrate for microbial growth (4). Pure cultures were obtained by successive culturing of colonies picked from these plates on MSM. Purity was checked by plating colonies on rich tryptone-yeast extract (TY) agar (21). Pure cultures were stored as glycerol stocks at −70°C. The genomic relatedness of the cultures was determined by dot blot assays. Chromosomal DNA of a given isolate was labeled by the random hexamer procedure using [α-32P]dCTP and was used to probe a dot blot preparation containing known amounts of denatured genomic DNAs of all isolated strains, as well as a master filter containing denatured genomic DNAs of 41 standards isolated previously (Table 1, standards 1 to 41). The dot blot assay was repeated for all newly obtained isolates. Genomically distinct isolates were characterized by partial 16S rRNA gene sequencing of PCR products obtained with primers f10 (11) and r1406 (3), as described previously (13, 19). The 16S rRNA sequences were compared to nucleotide sequences obtained from GenBank by using the advanced basic local alignment search tool (BLASTN) and to nucleotide sequences obtained from the Ribosomal Database Project (RDP) by using the SIMILARITY_RANK tool (7).
TABLE 1.
Composition of the soil master filter
| Positiona | Nameb | Mediumc | Sabd | Nearest homologe | Hydrocarbons degradedf |
|---|---|---|---|---|---|
| 1 | LQ1 | PTYG | 0.802 | Bordetella bronchiseptica | — |
| 2 | LQ5 | PTYG | 0.375 | Pseudomonas syringae | ben |
| 3 | LQ6 | PTYG | 0.396 | Azospirillum sp. | ben, tol, sty, nap |
| 4 | LQ10 | PTYG | 0.598 | Sphingomonas parapaucimobilis | — |
| 5 | LQ11 | PTYG | 0.769 | Bacillus macroides | ben, tol, xyl |
| 6 | LQ14 | PTYG | 0.615 | Xanthomonas campestris | xyl |
| 7 | LQ15 | PTYG | 0.580 | Bacillus pseudomegaterium | tol, nap |
| 8 | LQ16 | PTYG | 0.861 | Pseudomonas syringae | ben, tol, sty, xyl, nap |
| 9 | LQ17 | PTYG | 0.931 | Agrobacterium rubi | ben, tol, nap |
| 10 | LQ19 | PTYG | 0.349 | “Flavobacterium lutescens” | — |
| 11 | LQ20 | PTYG | 0.816 | Pseudomonas syringae | ben, tol, xyl |
| 12 | LQ21 | PTYG | 0.791 | Bordetella parapertussis | ben |
| 13 | LQ26 | PTYG | 0.865 | Bordetella parapertussis | — |
| 14 | LQ27 | PTYG | Bordetella sp.g | tol | |
| 15 | LQ29 | PTYG | 0.843 | Bordetella parapertussis | — |
| 16 | LQ30 | PTYG | 0.935 | Sphingomonas yanoikuyae | ben, tol |
| 17 | LQ33 | PTYG | NDh | — | |
| 18 | LQ34 | PTYG | 0.969 | Pseudomonas flavescens | ben, tol |
| 19 | LQ35 | PTYG | 0.972 | Pseudomonas flavescens | — |
| 20 | LQ36 | PTYG | 0.965 | Pseudomonas flavescens | ben, tol |
| 21 | Q1 | HDM, benzene | 0.851 | Rhodococcus sp. | ben, tol, sty, xyl |
| 22 | Q2 | PTYG | 0.879 | Bacillus cereus-Bacillus thuringiensis | ND |
| 23 | Q3 | PTYG | 0.763 | Nocardioides luteus | ND |
| 24 | Q4 | PTYG | 0.683 | Flavobacterium ferrugineum | ND |
| 25 | Q5 | HDM, naphthalene | 0.779 | Pseudomonas syringae | ben, nap |
| 26 | Q6 | HDM, styrene | 0.836 | Rhodococcus globerulus | ben, tol, sty, xyl, nap |
| 27 | Q7 | TY | 0.741 | Pseudomonas syringae | ben, tol, sty |
| 28 | Q8 | TY | 0.879 | Bacillus benzeovorans | ND |
| 29 | Q9 | TY | 0.809 | Bacillus polymyxa | ND |
| 30 | Q10 | TY, anoxic | 0.504 | Bacteroides distasonis | ND |
| 31 | Q11 | TY, anoxic | 0.498 | Bacteroides heparinolyticus | ND |
| 32 | Q12 | TY, anoxic | 0.846 | Clostridium xylanolyticum | ND |
| 33 | Q13 | TY, anoxic | 0.337 | Clostridium sp. | ND |
| 34 | Q14 | Medium C, anoxic | 0.582 | Desulfovibrio longus | ND |
| 35 | Q15 | Medium C, anoxic | 0.571 | Desulfovibrio desulfuricans | ND |
| 36 | Cstd1 | MSM, toluene | 0.715 | Pseudomonas putida | ben, tol |
| 37 | Cstd2 | MSM, toluene | 0.816 | Unidentified bacterium isolate LX1 | ben, tol |
| 38 | Cstd5 | MSM, toluene | 0.761 | Pseudomonas stutzeri | ben, tol, xyl |
| 39 | Cstd6 | MSM, toluene | 0.796 | Microbacterium lacticum | ben, tol |
| 40 | Cstd7 | MSM, toluene | 0.823 | Pseudomonas fluorescens | ben, tol |
| 41 | Cstd8 | MSM, toluene | 0.857 | Pseudomonas putida | ben, tol |
| 42 | S1 | MSM, styrene | 0.889 | Pseudomonas sp. | ben, sty, xyl, nap |
| 43 | S2 | MSM, styrene | 0.872 | Pseudomonas corrugata | ben, sty |
| 44 | S3 | MSM, styrene | 0.920 | Pseudomonas syringae | sty |
| 45 | S6 | MSM, styrene | 0.935 | Pseudomonas aureofaciens | sty |
| 46 | Bsc30 | MSM, benzene | 0.870 | Pseudomonas agarici | ben, tol, xyl |
| 47 | B4 | MSM, benzene | 0.884 | Alcaligenes sp. | ben, tol, sty, xyl |
| 48 | X1 | MSM, xylene | 0.946 | Alcaligenes sp. | ben, tol, xyl |
| 49 | X2 | MSM, xylene | 0.872 | Pseudomonas sp. | ben, tol, xyl |
| 50 | X5p | MSM, xylene | 0.895 | Rhodococcus marinoascens | ben, tol, sty, xyl |
| 51 | X7p | MSM, xylene | 0.839 | Microbacterium laevaniformans | ben, tol, sty, xyl |
| 52 | N1 | MSM, naphthalene | 0.935 | Alcaligenes sp. | ben, nap |
| 53 | N2 | MSM, naphthalene | 0.658 | Arthrobacter oxydans | ben, tol, nap |
| 54 | N3 | MSM, naphthalene | 0.825 | Pseudomonas pseudoalcaligenes | ben, nap |
| 55 | N5 | MSM, salicylate | 1.000 | Pseudomonas sp. strain BI*7 | — |
Position of denatured chromosomal DNA on the master filter.
Name assigned at the time of isolation.
Medium used for isolation. Standards 30 to 35 were isolated under anaerobic conditions.
Similarity coefficient for query and matching sequences (7).
Nearest homolog in the RDP database as determined by the program SIMILARITY_RANK (7).
ben, benzene; tol, toluene; sty, styrene; xyl, mixture of o-, p-, and m-xylenes; nap, naphthalene;— no growth.
Inferred from cross-hybridization with other Bordetella spp. genomes on the filter.
ND, not determined.
Community analysis of soil enrichment cultures on C5+.
Aerobic enrichment cultures were started with 2 g of soil and 10 ml of MSM in test tubes (2 by 15 cm) that were closed with loosely fitting metal caps to allow exchange of gases and hydrocarbon vapor. These tubes were incubated in a desiccator equilibrated with 1% synthetic C5+ (148.5 g of vpo and 1.5 g of a mixture containing 56% benzene, 25% DCPD, 7.5% toluene, 4% styrene, 4% m-xylene, and 4% naphthalene). The cultures were transferred every second or third week by using 10% inocula. Following two transfers the medium was changed to B+NP medium (2) because this medium provided trace elements which were considered necessary in the absence of soil. At the time of each transfer, plate counts were obtained by using 0.25× Luria-Bertani agar (12); the remainder of the culture was used for isolation of total community DNA for RSGP analysis. DNA was isolated by a modified procedure of Marmur (8), as described by Shen et al. (13). Humic acids were removed with Sepharose 4B spin columns (5). For RSGP analysis 100 ng of total community DNA was combined with 0.1 ng of bacteriophage λ DNA, which served as an internal standard. Following random hexamer labeling with [α-32P]dCTP, the probe was hybridized with a master filter containing known amounts (typically 100 ng) of the denatured genomic DNAs of all 55 standards (Table 1). Following washing and drying the filters were exposed to a BASIII imaging plate. Images were analyzed with a Fuji BAS1000 bioimaging analyzer (13, 19). Calculated fx values (20) for all 55 standards were normalized by setting the Σfx value at 1. Dendrograms were generated with the cluster analysis program SYSTAT.
Hydrocarbon degradation kinetics.
The kinetics of C5+ degradation were studied with enrichment cultures and synthetic consortia. Triplicate enrichment cultures were maintained by biweekly transfer for 23 weeks by inoculating 2 ml of each enrichment into 8 ml of B+NP medium. Following 2 weeks of growth in a desiccator equilibrated with a 1% C5+ atmosphere, the cells were harvested by centrifugation and then washed twice with and resuspended in 3.5 ml of B+NP medium. For preparation of synthetic consortia, 41 bacterial standards were grown separately in 5 ml of TY medium for 5 days at room temperature. Portions (1 ml) of selected cultures were then combined, centrifuged, washed twice with B+NP medium, and finally resuspended in 3 ml of B+NP medium. Synthetic consortia SC-5, SC-9, and SC-41 consisted of, respectively, standards 3, 11, 25, 38, and 52, standards 11, 25, 26, 37, 38, 39, 41, 47, and 52, and all standards except standards 4, 21, 22, 23, 24, 28, 30, 31, 32, 33, 34, 35, 50, and 53. To determine C5+ degradation rates, 1-ml portions of resuspended enrichments or 0.5-ml portions of resuspended synthetic consortia were injected into 158-ml serum bottles containing 60 ml of B+NP medium and 2 μl of C5+ hydrocarbon (final concentration, approximately 300 μM). Sterile controls were prepared by autoclaving preparations on three consecutive days and then injecting mercuric chloride (final concentration, 2.5 mM). The amounts of individual C5+ components were determined by culture headspace analysis by using a Hewlett-Packard HP5190 gas chromatograph equipped with a flame ionization detector. The column used was a 0.5 μ ID RTX-5 column (30 m by 0.2 mm; Restek, Brockville, Ontario, Canada). The flow rate of the He carrier gas was 1.23 ml/min; the flow rate of air was 400 ml/min; and the flow rate of H2 was 30 ml/min. The injector and detector temperatures were 210 and 250°C, respectively. The oven temperature program was as follows: 60°C for 3 min, followed by a 10°C/min increase to 200°C and then 200°C for 5 min. Plate counts were obtained on 0.25× Luria-Bertani agar at time zero and at the end of incubation; cell densities were measured by determining the optical densities at 600 nm at the same times with a Shimadzu UV-265 spectrophotometer.
Biodegradation rate constants were determined by the method described by Tchobanoglous (18). Substrate removal data was plotted to determine whether it was best described by zero-order or first-order kinetics.
Chemical and biochemical reagents.
[α-32P]dCTP (10 mCi/ml; 3,000 Ci/mmol) was purchased from ICN; Taq polymerase, the Klenow fragment of DNA polymerase I, and bacteriophage λ DNA (0.5 mg/ml) were obtained from Pharmacia. Reagent grade chemicals were obtained from BDH (Toronto, Ontario, Canada), Fisher (Fair Lawn, N.J.), or Sigma/Aldrich (St. Louis, Mo.). Agar and rich medium components were obtained from Difco (Detroit, Mich.). The C5+ components were obtained as follows: benzene (high-performance liquid chromatography grade) and DCPD (reagent grade) were obtained from Aldrich (Milwaukee, Wis.); toluene (99.5% pure), mixed xylenes, and styrene (99%) were obtained from BDH (Toronto, Ontario, Canada); m-xylene was obtained from BDH (Poole, England); and naphthalene (99%) was obtained from Sigma (St. Louis, Mo.). Vacuum pump oil 19, a 100% parafinnic oil with a density of 0.85 g cm−3, was obtained from VWR Scientific (West Chester, Pa.). Compressed gases were obtained from Praxair Products Inc. (Calgary, Alberta, Canada).
RESULTS
Isolation and characterization of novel bacterial standards.
Soil samples from a contaminated soil pile (13, 16) were used to obtain 30 new isolates. Single colonies were picked from MSM plates containing soil suspensions or enrichments incubated in desiccators equilibrated with either 1 or 10% (wt/wt) benzene, styrene, xylenes, or naphthalene in vpo. DNAs from all 30 isolates were tested for genomic cross-hybridization with each other and with the 41 standards obtained previously; this resulted in identification of 14 new genomically distinct isolates (standards). Four of these strains were isolated on styrene (Table 1, standards 42 to 45), two were isolated on benzene (Table 1, standards 46 and 47), four were isolated on xylenes (Table 1, standards 48 to 51), and four were isolated on naphthalene (Table 1, standards 52 to 55). Following subculturing, standard 55 grew poorly on naphthalene but grew well on salicylate.
Standards were identified by 16S rRNA gene sequencing as members of Pseudomonas spp. (Table 1, standards 42 to 46, 49, 54, and 55), Alcaligenes spp. (Table 1, standards 47, 48, and 52), Rhodococcus sp. (Table 1, standard 50), Microbacterium sp. (Table 1, standard 51), or Arthrobacter sp. (Table 1, standard 53). The Sab values indicating the degrees of similarity of these standards to their nearest RDP homologs were high (Sab > 0.82) except for standard 53 (Sab = 0.66). The ratio of hybridization constants for each standard and bacteriophage λ DNA was determined. These data were required to calculate fx values from experimentally determined hybridization intensities (19).
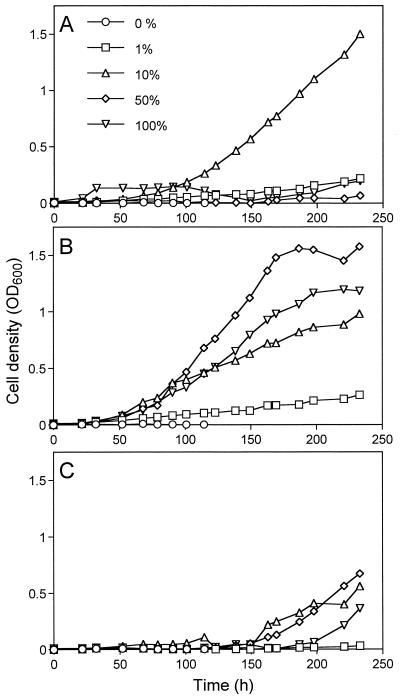
Like previously isolated toluene-degrading standards (Table 1, standards 36 to 41) (4), many of the new isolates grew optimally at a preferred hydrocarbon concentration. When liquid MSM cultures of Pseudomonas spp. strains S1, S2, and S3 (Table 1, standards 42 to 44), which originally were isolated on styrene, were grown in desiccators equilibrated with 0, 1, 10, 50, or 100% (wt/wt) styrene in vpo, S1 grew optimally in the presence of 10% styrene and S2 grew optimally in the presence of 50% styrene. S3 grew equally well in atmospheres equilibrated with 10 and 50% styrene, although it grew considerably more slowly than S1 and S2 (Fig. 1). All aerobically growing standards (except standards 22 to 24 and 27 to 29) were tested for growth on MSM plates with individual C5+ hydrocarbons as the sole carbon and energy sources. The plates were incubated in desiccators equilibrated with a C5+ hydrocarbon at a concentration of either 1 or 10% (wt/wt) in vpo. The results are summarized in Table 1. Of the 44 standards tested, 36 were capable of degrading one or more C5+ compounds. Of these, 31 were able to use benzene, 26 were able to use toluene, 14 were able to use xylenes, 12 were able to use styrene, and 10 were able to use naphthalene as sole carbon and energy sources (Table 1). Only two standards (standard 8 [Pseudomonas syringae strain LQ16] and standard 26 [Rhodococcus globerulus strain Q6]) were able to grow well on all five of the C5+ hydrocarbons tested. Strain LQ16 had been isolated on rich PTYG medium, whereas strain Q6 had been isolated with styrene as the sole carbon and energy source. It is noteworthy that standards 1 to 20 were all initially isolated on PTYG medium and 13 of these standards grew with one or more C5+ components. This indicates that the ability to degrade hydrocarbons is a common trait among the culturable heterotrophs in the contaminated soil tested.
FIG. 1.
Growth of Pseudomonas spp. in MSM equilibrated with vpo containing either 0, 1, 10, 50, or 100% (wt/wt) styrene. The cell densities of the cultures were determined by determining optical densities at 600 nm (OD600). (A) Pseudomonas sp. strain S1 (Table 1, standard 42). (B) Pseudomonas sp. strain S2 (Table 1, standard 43). (C) Pseudomonas sp. strain S3 (Table 1, standard 44).
Enrichment of soil microbial communities on C5+.
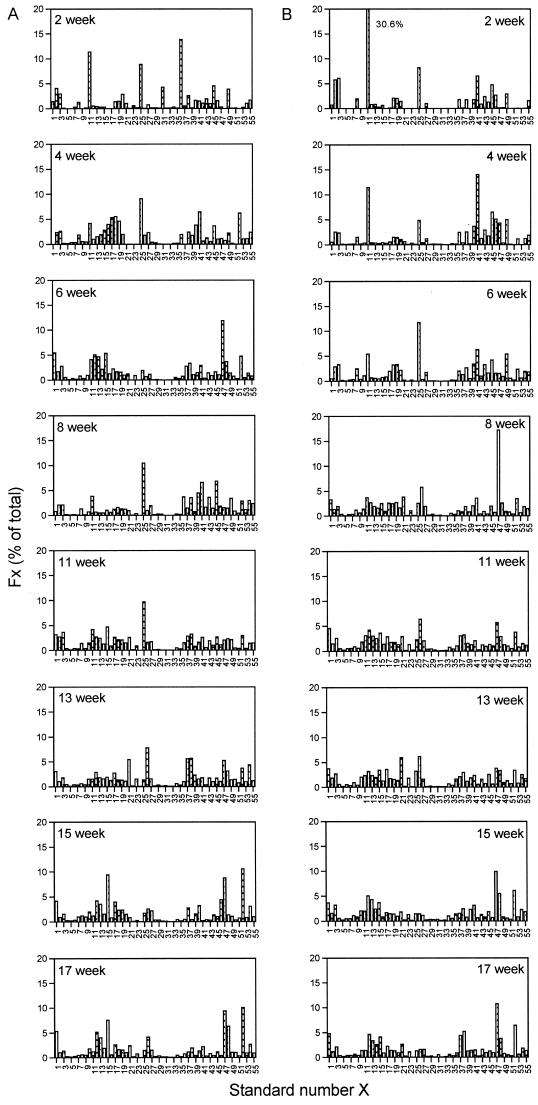
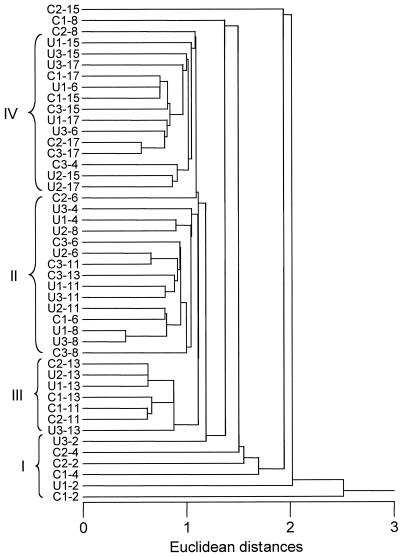
Microbial communities were serially enriched from three C5+-contaminated soils (C1, C2, and C3) and from three uncontaminated sediments (U1, U2, and U3) on media in which C5+ was the sole carbon and energy source. Cultures were transferred every 2 or 3 weeks. The enrichments obtained at the different time points were identified by the accumulated time of incubation as 2-, 4-, 6-, 8-, 11-, 13-, 15-, and 17-week enrichments. The evolution of the microbial community composition, as monitored by RSGP, indicated that the compositions of all six communities, as defined by RSGP, were strongly time dependent (Fig. 2). The RSGP profiles for enrichments obtained from uncontaminated soil U1 were dominated by standards 36, 11, and 25 (all Pseudomonas spp.) at 2 weeks, by standard 25 (Pseudomonas syringae strain Q5) at 4 weeks, by standard 47 (Alcaligenes sp. strain B4) at 6 weeks, by standard 25 (Pseudomonas syringae strain Q5) at 8 and 11 weeks, by standard 26 (Rhodococcus globerulus strain Q6) at 13 weeks, and by standards 52, 47, and 15 (Alcaligenes sp. strains N1 and B4, Bordetella parapertussis strain LQ29) at 15 and 17 weeks. The enrichments obtained from contaminated soil C3 were dominated by standards 11 and 25 (both Pseudomonas spp.) at 2 weeks, by standards 41 and 11 (both Pseudomonas spp.) at 4 weeks, by standard 25 (Pseudomonas syringae strain Q5) at 6 weeks, by standard 47 (Alcaligenes sp. strain B4) at 8 weeks, by standard 26 (Rhodococcus globerulus strain Q6) at 11 and 13 weeks, and by standards 47 and 52 (Alcaligenes sp. strains B4 and N1) at 15 and 17 weeks. The community profiles of these two enrichments over time were remarkably similar even though the soil samples were obtained from different depths and have distinct contamination histories. The dominant organisms in the four other enrichments (C1, C2, U2, and U3) were also similar. All the community compositions obtained over time are compared in a dendrogram in Fig. 3. Early enrichments did not form a distinct clade, but many of the enrichment communities occurred together in the bottom part of the dendrogram (Fig. 3, cluster I). RSGP analysis of these early communities demonstrated that standard 11 (Pseudomonas syringae strain LQ20) (Fig. 2) was frequently enriched to a greater extent than other community members detectable by RSGP. All six enrichments converged to have similar community compositions, and they were dominated by Alcaligenes sp. strains B4 and N1 (standards 47 and 52). The communities at 15 and 17 weeks formed a distinct clade (Fig. 3, cluster IV), in which only three enrichments obtained at intermediate times (Fig. 3, enrichments C3-4, U1-6, and U3-6) were present and from which only one late community (enrichment C1-15) was missing. These late communities were preceded by those in clusters II and III (Fig. 3), which represented compositions dominated by Pseudomonas syringae strain Q5 (standard 25) and by Rhodococcus globerulus strain Q6 (standard 26). The latter organism was not identified as a major community component in any of the enrichments obtained from contaminated soil sample C3. Taken together, the data indicate that there was a succession of organisms in the enrichment rather than continuous enrichment for a few strains. Key organisms in this succession were Pseudomonas syringae strain LQ20 (standard 11) at 2 to 4 weeks, Pseudomonas syringae strain Q5 (standard 25) at 6 to 8 weeks, Rhodococcus globerulus strain Q6 at 11 to 13 weeks, and Alcaligenes sp. strains B4 and N1 at 15 to 17 weeks.
FIG. 2.
Community compositions of enrichment cultures derived from uncontaminated soil sample U1 (A) and contaminated soil sample C3 (B). Enrichments were transferred to fresh medium every 2 to 3 weeks and were identified on the basis of their cumulative growth times (2, 4, 6, 8, 11, 13, 15, and 17 weeks). The compositions of these enrichments, as determined by RSGP, are shown as the calculated fraction of each standard fx compared with the standard number.
FIG. 3.
Dendrogram for all community compositions of enrichment cultures. The compositions of enrichments from three contaminated (C1, C2, and C3) and three uncontaminated (U1, U2, and U3) soils were compared by using the cluster analysis program SYSTAT. The tree was constructed by using pairwise Euclidean distance comparisons of nearest neighbors (horizontal scale). Clusters I, II, III, and IV are dominated by selected standards, as explained in the text.
Degradation of C5+ by microbial enrichments and consortia.
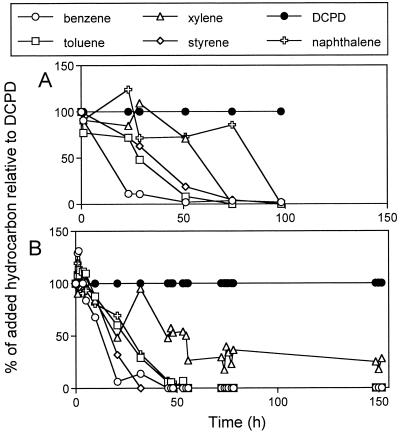
Aerobic C5+ degradation by six enrichments and three synthetic consortia was monitored. Synthetic consortia SC-5 and SC-9 consisted of isolates that appeared frequently as major community components, whereas synthetic consortium SC-41 consisted of all 41 standards that readily grew aerobically on rich medium. Sufficient oxygen for complete mineralization of the C5+ added was present in each of the serum bottles. Typical results for degradation of C5+ hydrocarbons by one of the enrichments and one of the synthetic consortia are shown in Fig. 4. DCPD was not removed from any of the cultures; all the other compounds were partially or completely removed during incubation. DCPD was therefore used as an internal standard.
FIG. 4.
Aerobic degradation of C5+ components as a function of time by enrichment culture U1 (A) and synthetic consortium SC-9 (B), as determined by gas chromatographic headspace analysis.
Analysis of the rates of removal of individual hydrocarbons from the C5+ mixture by the method of Tchobanoglous (18) showed that most compounds obeyed first-order kinetics better than they obeyed zero-order kinetics. Lag times and first-order rate constants for the degradation kinetics of all C5+ components except DCPD are listed in Table 2. Benzene was degraded almost immediately by all enrichments except the enrichment obtained from soil C3; toluene degradation began immediately with enrichments U1, U3, and C2. Degradation of the other C5+ components began within 1 day of addition of the C5+ mixture to the cultures. Only enrichments U2 and U3 showed lag times greater than 1 day for m-xylene (enrichment U2) and m-xylene plus naphthalene (enrichment U3). The synthetic consortia all started degradation of C5+ compounds within 1 day after C5+ was added (Table 2).
TABLE 2.
Lag times, first-order rate constants, and order of degradation of C5+ components by enrichment cultures or synthetic consortia
| Culture | Lag time (day)
|
First-order rate constant (days−1)
|
Order of degradation
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bena | tol | xyl | sty | nap | ben | tol | xyl | sty | nap | ben | tol | xyl | sty | nap | |
| SC-5b | 0.16 | 0.25 | 0.54 | 0.17 | 0.11 | 3.37 | 1.66 | 0.35 | 2.25 | 2.57 | 1 | 4 | 5 | 2 | 3 |
| SC-9b | 0.06 | 0.15 | 0.18 | 0.09 | 0.13 | 3.40 | 1.63 | 1.02 | 2.22 | 1.21 | 1 | 3 | 4 | 2 | 3 |
| SC-41b | 0.10 | 0.14 | 0.88 | 0.14 | 0.10 | 3.78 | 2.47 | 0.58 | 2.83 | 0.81 | 1 | 3 | 5 | 2 | 4 |
| U1c | 0.00 | 0.00 | 0.62 | 0.02 | 0.48 | 1.92 | 1.03 | 0.38 | 0.84 | 0.30 | 1 | 2 | 4 | 3 | 5 |
| U2c | 0.02 | 0.65 | 1.25 | 0.53 | 0.65 | 1.09 | 0.79 | 2.30 | 0.16 | 0.83 | 1 | 4 | 3 | 2 | 5 |
| U3c | 0.02 | 0.02 | 1.55 | 0.00 | 1.44 | 2.71 | 1.16 | 2.40 | 0.55 | 0.40 | 1 | 3 | 4 | 2 | 5 |
| C1c | 0.02 | 0.43 | 0.52 | 0.00 | 0.41 | 3.67 | 3.02 | 1.19 | 2.47 | 0.92 | 1 | 3 | 5 | 2 | 4 |
| C2c | 0.00 | 0.00 | 0.57 | 0.00 | 0.14 | 1.74 | 1.91 | 0.94 | 3.25 | 1.46 | 2 | 2 | 4 | 1 | 3 |
| C3c | 0.38 | 0.38 | 0.38 | 0.38 | 0.19 | 4.88 | 3.55 | 2.30 | 2.38 | 1.49 | 1 | 2 | 3 | 2 | 4 |
ben, benzene; tol, toluene; xyl, m-xylene; sty, styrene; nap, naphthalene.
ynthetic consortia SC-5, SC-9, and SC-41 contained 5, 9, and 41 standards, respectively. Data are averages based on six cultures.
Enrichments from uncontaminated (U1 to U3) or contaminated (C1 to C3) soil samples were maintained for 23 weeks by biweekly transfer. Data are averages based on two cultures.
The rate constants for degradation of benzene, toluene, styrene, and naphthalene were on average greater for enrichments from contaminated soils than for enrichments from uncontaminated soils (Table 2). Degradation of m-xylene was the only exception, because enrichments U2 and U3 had substantially higher degradation rate constants (2.3 and 2.4 days−1, respectively) than most of the enrichments from contaminated soils. The rate constants obtained for the synthetic consortia were similar to those obtained for the enrichment cultures (Table 2). Averaging all the rate constants for enrichments yielded values of 2.67, 1.91, 1.59, 1.61, and 0.90 days−1 for benzene, toluene, xylene, styrene, and naphthalene, respectively. For the synthetic consortia the corresponding average rate constants were 3.51, 1.92, 0.65, 2.43, and 1.53 days−1, respectively. In all cases except enrichment C2, benzene degradation proceeded most rapidly (Table 2).
The order of degradation of the various C5+ components for each culture is also shown in Table 2. This order was a function of both the rate constant and the lag time and was based on the average results for two cultures for each of the enrichments and six cultures for each artificial consortium. By adding all the average rank orders, the overall order of degradation of all of the C5+ components was established as benzene-styrene-toluene-naphthalene-xylene. Benzene and styrene were clearly removed from all of the cultures first and second, respectively, while the order of removal for toluene, naphthalene, and xylene was more culture dependent (Table 2). Synthetic consortium SC-41 degraded C5+ components in the order benzene-styrene-toluene-naphthalene-xylene. Interestingly, synthetic consortium SC-9 degraded C5+ in the following order: benzene, styrene, naphthalene and toluene simultaneously, and then xylene. Synthetic consortium SC-5 degraded these components in the order benzene-styrene-naphthalene-toluene-xylene.
DISCUSSION
One of the great challenges of microbiology is to find efficient ways to monitor and thereby understand environmental microbial diversity. If environmental microbial diversity can be adequately monitored, then the direction and rate of processes catalyzed by environmental microbial communities may become better understood. This applies both to in situ processes and to processes in controlled environments (e.g., reactors).
With respect to in situ processes, Shi et al. (14) defined the differences between microbial community structures in pristine and fuel-contaminated aquifers by using 16S rRNA probes. The fuel-contaminated areas were enriched in members of the β subclass of the class Proteobacteria (β-proteobacteria) and γ-proteobacteria compared to α-proteobacteria (14). Addition of toluene in microcosm studies enriched microorganisms that were only minor constituents of the fuel-contaminated community in situ. These results suggested that characterization of the microbial community change when toluene is added might help identify key toluene degraders in situ. In another recent study the in situ microbial population changes during oil spills were characterized by phospholipid fatty acid analysis and PCR-amplified 16S ribosomal DNA denaturing gradient gel electrophoresis (DGGE) (6). DGGE indicated that oil enhanced the presence of gram-negative α-proteobacteria. Although phospholipid fatty acid analysis indicated that the microbial communities of oil-contaminated and noncontaminated plots were similar after 14 weeks, DGGE still revealed major population differences.
With respect to microbial communities in controlled environments, Stoffels et al. (17) extensively characterized degradation of Solvesso 100, a complex mixture of aromatic hydrocarbons used in industrial painting. Inoculation of a fermentor with a sample from a Solvesso 100-laden waste stream resulted in a community in which γ-proteobacteria, especially Pseudomonas spp., were dominant, even though the original inoculum was dominated by α- and β-proteobacteria. Interestingly, the original diversity did not appear to be lost during fermentor enrichment; use of the fermentor culture to inoculate a trickle-bed bioreactor resulted in a community in which α- and β-proteobacteria were dominant. These transitions were established by direct fluorescent in situ hybridization of samples with 16S and 23S rRNA-targeted probes and by performing probe analysis of colonies obtained following plating on different media. Both methods indicated that distinctly different populations derived from the same environmental sample were able to degrade Solvesso 100. Stoffels et al. (17) concluded that establishment of an enrichment culture prior to inoculation of a reactor may result in enrichment of bacteria that do not effectively colonize the reactor. However, this would be a problem only if some of the original diversity in the sample was permanently lost during enrichment. Communities may be merely dynamic (i.e., capable of reestablishing dominance depending on the growth conditions).
Our results suggest that contaminated and uncontaminated soils at depths ranging from 0 to 14 m at the ethylene plant site investigated have populations in which specific C5+-degrading bacterial strains are typically community members. The RSGP results for community DNA without enrichment on hydrocarbons revealed community profiles (data not shown) that were more evenly distributed than those obtained following enrichment on hydrocarbons (Fig. 2). Enrichment of similar organisms from diverse backgrounds occurs after exposure to C5+, suggesting that evolution of a single dynamic microbial community occurs during growth on hydrocarbons. Different conditions result in differential expression of community members. Enrichment under identical conditions leads to development of similar community compositions, irrespective of contamination history (Fig. 2 and 3). These results are similar to those of Shi et al. (14), who demonstrated that similar microbial communities developed in soils exposed to toluene regardless of whether the soils were initially contaminated. The soil environment targeted in our study supports a variety of different culturable bacteria. The ability to degrade aromatic hydrocarbons is widely distributed in this community, and the community includes γ-proteobacteria. (Pseudomonas spp.), β-proteobacteria (Alcaligenes spp.), α-proteobacteria (Sphingomonas spp.), high-G+C-content gram-positive bacteria, such as actinobacteria (Rhodococcus spp.) and Microbacterium, and low-G+C-content gram-positive bacteria (Bacillus spp.). We used RSGP rather than rRNA-targeted probes to monitor the dynamics of this community. The main advantage of this technique is that multiple cultured bacteria can be tracked in a single hybridization step. This has not been accomplished yet with rRNA targeted probes. Some disadvantages of the technique are the fact that the microbial community is described solely in terms of its culturable component, although this is likely to be a significant fraction of the total bacteria in enrichment cultures, the fact that genomically similar microorganisms cannot be distinguished, and the fact that calculated fx values contain cross-hybridization contributions for which we cannot correct (19, 20). Despite these shortcomings, the RSGP method demonstrated that enrichments from contaminated and uncontaminated soils from the same site developed along similar paths when they were exposed to similar conditions. The samples were initially dominated by γ-proteobacteria (Pseudomonas spp.) but converged to a community dominated by β-proteobacteria (Alcaligenes and Bordetella spp.), as shown in Fig. 2 and 3. The reasons for the observed succession are not clear. In our experimental system the supply of C5+ was continuous at a constant concentration, whereas other required nutrients were replenished every 2 weeks when the culture was transferred. The increases and decreases in some of the early major community components (for instance, standard 11 [Pseudomonas syringae LQ 20]) may be related to the gradual removal of soil from the enrichment.
Spills of C5+ have occurred occasionally at the ethylene plant site investigated, leading to exposure of microbial communities to nearly pure C5+ at the center of the spill and to lower C5+ concentrations away from the center of the spill. We demonstrated that communities derived from an uncontaminated location where a new ethylene plant is under construction can develop the ability to degrade most C5+ hydrocarbons, which is important for the plant operators in view of the potential for accidental contamination of this location. Benzene, toluene, and xylene are major constituents of C5+. Degradation of mixtures of BTEX compounds has been investigated by numerous authors. Oh et al. (10) demonstrated that p-xylene was cometabolically removed by benzene and toluene degraders. The presence of p-xylene decreased the rates of degradation of these primary substrates. Similarly, Deeb and Alvarez-Cohen (1) found that the rates of degradation of BTEX compounds in mixtures were lower than rates of degradation of the pure compounds. Also, the order of degradation of compounds in mixtures was different than the order of degradation of the pure compounds. The presence of o-xylene enhanced benzene and toluene removal, but the latter two compounds inhibited removal of xylene. We determined the degradation kinetics only for the C5+ components present in the mixture, as exposure to pure components is unlikely to occur at sites where this mixture is generated. Benzene is the main component of C5+ (45%, wt/wt), followed by poorly degradable DCPD and cyclopentadiene (20%, wt/wt). Toluene, styrene, xylenes, and naphthalene, the other components that we monitored, are present at much smaller concentrations (6, 3, 2, and 2% [wt/wt], respectively). A large number of the standards isolated (31 of the 44 standards tested) were capable of benzene degradation, whereas degradation of the minor C5+ components was less widespread (Table 1). The main community members identified in enrichment cultures (Fig. 2) are all capable of benzene degradation (Table 1). The degradation curves for all C5+ components suggested that there was an acclimation period (lag time), followed by degradation (9). The lag time may involve adaptation to the C5+ concentration used in the degradation experiments; hydrocarbon degradation genes should have been induced in the inocula, which were all grown in desiccators with 1% C5+ in vpo as the sole carbon and energy source. Despite its much higher concentration, benzene was generally removed first, due to a shorter lag time and/or a higher first-order rate constant than those observed for the other degradable C5+ components (Table 2). This was true for enrichment cultures derived from contaminated and uncontaminated soils and for synthetic consortia. Inhibition of xylene removal by benzene, as described by Deeb and Alvarez-Cohen (1), may have contributed to the slower removal of xylene by some of our cultures. The fact that the benzene concentration was highest and the benzene supply was unlimited during growth of enrichments or synthetic consortia on 1% C5+ in vpo in the desiccators prior to the rate studies may have forced the communities to make benzene removal a priority, because benzene metabolism can reduce its toxicity (15). The communities at field sites contaminated with C5+ may be similarly adapted to benzene removal. Our enrichment studies suggest that a variety of communities with different compositions are able to metabolize the benzene-dominated C5+ mixture (Fig. 2). Hence, just as communities with different compositions have been shown to effectively degrade Solvesso 100 (17), our results indicate that a variety of communities with different compositions are active in C5+ degradation. A single community structure that is effective in C5+ degradation therefore cannot be defined.
ACKNOWLEDGMENTS
This work was supported by a strategic grant from the Natural Science and Engineering Research Council of Canada to G.V. and by a financial contribution from NOVA Research & Technology Corporation.
REFERENCES
- 1.Deeb R A, Alvarez-Cohen L. Temperature effects and substrate interactions during the aerobic biotransformation of BTEX mixtures by toluene-enriched consortia and Rhodococcus rhodochrous. Biotechnol Bioeng. 1999;62:526–536. [PubMed] [Google Scholar]
- 2.Fedorak P M, Gbrić-Gallić D. Aerobic microbial cometabolism of benzothiophene and 3-methylbenzothiophene. Appl Environ Microbiol. 1991;57:932–940. doi: 10.1128/aem.57.4.932-940.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hicks R, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hubert C, Shen Y, Voordouw G. Composition of toluene-degrading microbial communities from soil at different concentrations of toluene. Appl Environ Microbiol. 1999;65:3064–3070. doi: 10.1128/aem.65.7.3064-3070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson C R, Harper J P, Willoughby D, Roden E E, Churchill P F. A simple and efficient method for the separation of humic substances and DNA from environmental samples. Appl Environ Microbiol. 1997;63:4993–4995. doi: 10.1128/aem.63.12.4993-4995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacNaughton S J, Stephen J R, Venosa A D, Davis G A, Chang Y-J, White D C. Microbial population changes during bioremediation of an experimental oil spill. Appl Environ Microbiol. 1999;65:3566–3574. doi: 10.1128/aem.65.8.3566-3574.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 9.Odormatt J R. Simulations of intrinsic biodegradation using a non-linear modification of first-order reaction kinetics. J Soil Contam. 1997;6:495–508. [Google Scholar]
- 10.Oh Y-S, Shareefdeen Z, Baltzis B C, Bartha R. Interactions between benzene, toluene and p-xylene (BTX) during their biodegradation. Biotechnol Bioeng. 1994;44:533–538. doi: 10.1002/bit.260440417. [DOI] [PubMed] [Google Scholar]
- 11.Olsen G J, Lane D J, Giovannoni S J, Pace N R, Stahl D A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 13.Shen Y, Stehmeier L G, Voordouw G. Identification of hydrocarbon-degrading bacteria in soil by reverse sample genome probing. Appl Environ Microbiol. 1998;64:637–645. doi: 10.1128/aem.64.2.637-645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y, Zwolinski M D, Schreiber M E, Bahr J M, Sewell G W, Hickey W J. Molecular analysis of microbiol community structures in pristine and contaminated aquifers: field and laboratory microcosm experiments. Appl Environ Microbiol. 1999;65:2143–2150. doi: 10.1128/aem.65.5.2143-2150.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sikkema J, de Bont J A M, Poolmans B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev. 1995;59:201–222. doi: 10.1128/mr.59.2.201-222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stehmeier L G, Francis M M, Jack T R, Voordouw G. Biodegradation of dicyclopentadiene in the field. Biodegradation. 1999;10:135–148. doi: 10.1023/a:1008301631122. [DOI] [PubMed] [Google Scholar]
- 17.Stoffels M, Amann R, Ludwig W, Hekmat D, Schleifer K-H. Bacterial community dynamics during start-up of a trickle-bed bioreactor degrading aromatic compounds. Appl Environ Microbiol. 1998;64:930–939. doi: 10.1128/aem.64.3.930-939.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tchobanoglous G. Wastewater engineering treatment, disposal, reuse. 2nd ed. New York, N.Y: McGraw-Hill; 1979. pp. 146–148. . 146–148. [Google Scholar]
- 19.Telang A J, Ebert S, Foght J M, Westlake D W S, Jenneman G E, Gevertz D, Voordouw G. The effect of nitrate injection on the microbial community in an oil field as monitored by reverse sample genome probing. Appl Environ Microbiol. 1997;63:1785–1793. doi: 10.1128/aem.63.5.1785-1793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voordouw G. Reverse sample genome probing of microbial community dynamics. ASM News. 1998;64:627–633. [Google Scholar]
- 21.Voordouw G, Strang J D, Wilson F R. Organization of the genes encoding [Fe] hydrogenase in Desulfovibrio vulgaris subsp. oxamicus Monticello. J Bacteriol. 1989;171:3881–3889. doi: 10.1128/jb.171.7.3881-3889.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yerushalmi L, Guiot S R. Kinetics of degradation of gasoline and its hydrocarbon constituents. Appl Microbiol Biotechnol. 1998;49:475–481. doi: 10.1007/s002530051201. [DOI] [PubMed] [Google Scholar]