Significance
Epithelial cells undergo remarkable changes in their intracellular organization during wound healing, morphogenesis, and metastasis. Among these changes, the relocation of the Golgi apparatus from an apical to a basal position is key to epithelial migration, but a mechanistic understanding of this process remains elusive. To this end, we use an interdisciplinary approach, which includes super-resolution imaging, live cell dynamics, cell stretching, and molecular perturbations. Using these techniques, we show a migration-associated Golgi-remodeling process involving equatorial Golgi dispersion around the nucleus. This dispersion stems from a dynamic Golgi–actin interaction, is independent of Golgi–microtubule association, and differs from the well-known mitotic Golgi fragmentation. Finally, we demonstrate a critical role of this Golgi remodeling in persistent cell migration.
Keywords: collective cell migration, Golgi, cell polarity, actin cytoskeleton, epithelial tissue
Abstract
As a sedentary epithelium turns motile during wound healing, morphogenesis, and metastasis, the Golgi apparatus moves from an apical position, above the nucleus, to a basal position. This apical-to-basal repositioning of Golgi is critical for epithelial cell migration. Yet the molecular mechanism underlying it remains elusive, although microtubules are believed to play a role. Using live-cell and super-resolution imaging, we show that at the onset of collective migration of epithelial cells, Golgi stacks get dispersed to create an unpolarized transitional structure, and surprisingly, this dispersal process depends not on microtubules but on actin cytoskeleton. Golgi–actin interaction involves Arp2/3-driven actin projections emanating from the actin cortex, and a Golgi-localized actin elongation factor, MENA. While in sedentary epithelial cells, actin projections intermittently interact with the apically located Golgi, and the frequency of this event increases before the dispersion of Golgi stacks, at the onset of cell migration. Preventing Golgi–actin interaction with MENA-mutants eliminates Golgi dispersion and reduces the persistence of cell migration. Taken together, we show a process of actin-driven Golgi dispersion that is mechanistically different from the well-known Golgi apparatus fragmentation during mitosis and is essential for collective migration of epithelial cells.
In 1970, Robert Trelstad observed an apical-to-basal shift in the intracellular positioning of Golgi apparatus during the development of corneal epithelium in embryonic chicks and proposed that there must be an active change in cellular organization driving this shift in Golgi positioning (1). Over the next 50 y, multiple groups have observed apical-to-basal shift of Golgi in diverse tissue and organism contexts (2–6). Yet, the mechanism that the cells employ to change the intracellular position of the Golgi apparatus during epithelial morphogenesis and wound healing, remains elusive. Broadly, cell polarity in the epithelium involves the asymmetric organization of cellular elements, including the cell membrane composition, positioning of intracellular organelles, and arrangement of the cytoskeleton (3, 4, 7–13). In this respect, the apical positioning of Golgi between the nucleus and the apical membrane is a well-known signature of the inherent asymmetry and apico-basal polarity of epithelium (14–18). However, during epithelial morphogenesis, wound healing, and epithelial to mesenchymal transition (EMT), a new front-to-rear Golgi polarity appears in migrating cells (5, 19–21). In these cells, Golgi moves to a basal position, closer to the basal membrane. This apical-to-basal shift in Golgi positioning is critical for sustaining the cell migration, as it aids the transport of cell-matrix adhesion proteins to the basal membrane and facilitates the basal secretion of extracellular matrix proteins (21). Despite its importance, the underlying molecular mechanism that moves Golgi from an apical to a basal position and thus causes a fundamental shift in the cell function, remains unknown (14).
Nevertheless, it is possible that cytoskeletal elements such as microtubules and actin microfilaments might influence Golgi structure and positioning during epithelial migration. Since microtubules serve as the transport-track for Golgi vesicles and since Golgi is one of the major microtubule nucleation sites, researchers have long speculated that Golgi–microtubule association might play a role in Golgi repositioning (14, 22–24). However, several recent reports have shown that Golgi apparatus can be assembled and maintained as a compact organelle in absence of centrosomal microtubules (10, 19, 25). Do noncentrosomal microtubules play a role? In fact, the majority of microtubules in epithelial cells are of noncentrosomal origin, and in these cells, apically located calmodulin regulated spectrin-associated protein (CAMSAP)-family proteins seem to have critical roles in both organization of noncentrosomal microtubules and apical positioning of Golgi (10, 26). These reports, therefore, indicate that at least noncentrosomal microtubules might influence Golgi positioning. Yet, how these microtubules might drive the apical-to-basal shift in Golgi positioning during epithelial migration remains unknown.
In recent times, Golgi–actin association has also emerged to be critical for Golgi dynamics, mechanics, and morphology (14, 27, 28). For example, Golgi-membrane protein GOLPH3 binds to an unconventional myosin MYO18A and thus connects the Golgi to actin cytoskeleton (29). Overexpression of GOLPH3 in some cancer types leads to aberrant Golgi organization (30). In addition, an actin elongation factor, MENA, promotes local actin polymerization and facilitates linking of Golgi stacks into a ribbon. While actin cytoskeleton does undergo massive reorganization at the onset of epithelial cell migration during morphogenesis, wound healing, and EMT (31), whether it influences the apical-to-basal repositioning of Golgi remains an open question. In addition to these missing mechanistic details, we do not also know the structural changes associated with Golgi repositioning. For example, does Golgi undergo full disintegration like what happens during mitosis or does the hierarchical structure of Golgi, in the form of stacks and ribbons, remain intact during repositioning? Last, are these structural changes in Golgi connected to the dynamics of a specific cytoskeletal element during epithelial cell migration?
Results
Migration-Induced Golgi Apparatus Remodeling.
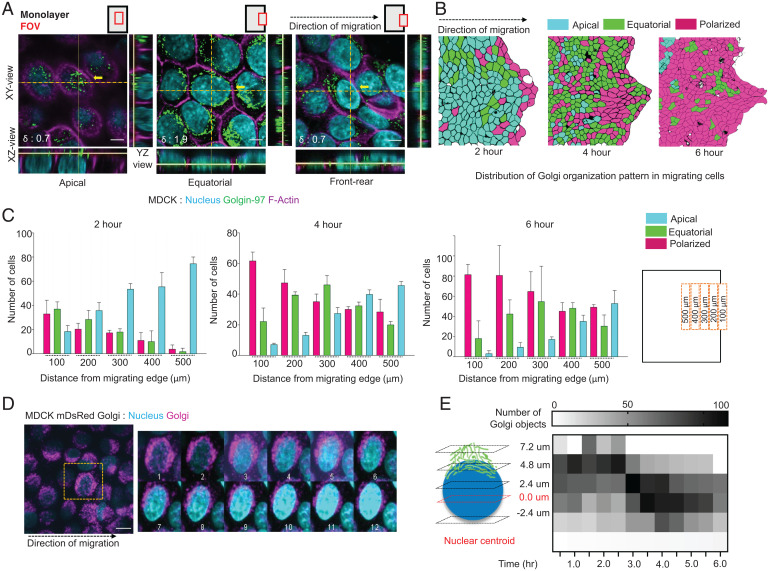
To answer these questions, we first grew a confluent monolayer of Madin-Darby canine kidney (MDCK) epithelial cells within a confined area and then lifted off the confinement, prompting collective cell migration (32). Existing studies on collective cell migration suggest that the change in Golgi positioning at the experimental wound edge could be observed as early as 2 h, and with time, the number of cells with basally positioned Golgi would increase. To understand the positional and structural changes in Golgi as sedentary epithelial cells start migrating, we fixed the cells at 0, 2, 4, and 6 h after the confinement removal and imaged Golgi with an antibody against a well-known Golgi structural protein, Golgin-97, which localized to trans-Golgi compartments (33) (Fig. 1A and SI Appendix, Fig. S1 A–C). Results from fixed samples hinted at the existence of a unique repositioning and remodeling route of Golgi apparatus during epithelial migration. In the samples fixed at 4 h after the confinement removal, we observed a compact Golgi apparatus localized apically—on top of the nucleus—within the cells that were away from the wound edge (Fig. 1A). On the other hand, cells very close to wound edge (within five cell rows) showed a basally located polarized and compact Golgi. Interestingly, cells in between (six to ten cell rows from the wound edge) showed a unique dispersed Golgi, which located at the basal region and distributed equatorially all around the nucleus (Fig. 1A and SI Appendix, Fig. S1D). Given that cell migration propagates inward, these images indicated that between apically located compact Golgi in sedentary cells and basally located compact Golgi in fully migrating cells, there exists an intermediate transition state of dispersed Golgi, which possibly arises at the onset of epithelial cell migration (Fig. 1A and SI Appendix, Fig. S1E). While apical-to-basal transition of Golgi was long known, here, our experiment revealed a previously unknown dispersed and relatively unpolarized transitional Golgi organization. Moreover, looking at images taken at 2, 4, and 6 h post confinement removal, we noticed that this transitional dispersed Golgi continued to appear but their abundance progressively got shifted away from the wound edge (Fig. 1 B and C). These results further indicated that the observed Golgi dispersion might be connected to the initiation of cell migration. To understand how this basally localized dispersed state arose dynamically from an apically localized compact Golgi apparatus, we created an epithelial cell line that stably expressed Golgi-localized mDsRed protein (SI Appendix, Fig. S1F). This system enabled us to image the transition process in live cells (Fig. 1D and Movie S1). From live imaging, we found that Golgi height with respect to cell nucleus reduced gradually, along z axis, until it reached a plateau and then Golgi reorganization occurred in XY-plane, which led to the establishment of front-rear polarity (Fig. 1 A and D). Next, to quantitatively capture the Golgi reorganization in three dimensions, we used two complementary methods. First, we measured the distance of individual Golgi objects from the centroid of the nucleus along z axis and plotted a heat-map representing the distribution of this Z-distance of the Golgi objects (Fig. 1E). This representation elucidated a migration time-dependent decrease in the number of apically located Golgi objects with respect to the nucleus, where the majority of Golgi objects moved on to or below the nuclear centroid plane with time (Fig. 1E). Next, to capture the distribution of Golgi objects on the transverse plane (XY-plane), we represented each Golgi object as a point (SI Appendix, Fig. S1 G and H), measured the distance (d) of each point from center of mass of Golgi, and finally, computed the root mean square of the distances, normalized to the nuclear radius (R). We term this quantity as dispersion index or δ, which is computed as , where N is the number of Golgi objects. A higher δ value (>1.5) indicated an equatorially dispersed Golgi where the Golgi objects are dispersed around the nucleus, whereas a polarized Golgi with aggregated Golgi objects showed a lower δ value (<1.5) (SI Appendix, Fig. S1H). Specifically focusing on a fixed area (edge of monolayer to ∼250 µm deep into the monolayer) of the wound edge every time, we observed that dispersion index reached its peak at 1.5 h after the confinement lift-off as the Golgi distributed around the cell nucleus (SI Appendix, Fig. S1I). From there on, dispersion index started decreasing due to the aggregation of Golgi in one end of the migrating cells (SI Appendix, Fig. S1I). Interestingly, migration speed became significant only after the dispersal and aggregation phases had completed (SI Appendix, Fig. S1J), showing that this Golgi dispersal and apical-to-basal transition preceded the cell movement. Next, we tested the generality of our observation in another epithelial cell line, namely Caco-2. Caco-2 is of human origin and was originally derived from a colon carcinoma. However, given that these cells develop very prominent apico-basal polarity and form tight epithelial barriers, researchers have used it extensively for studying epithelial differentiation and barrier function (34, 35). Relevantly, the epithelial differentiation process includes apical localization of the Golgi apparatus. Here, we differentiated Caco-2 cells by growing these cells for 21 d in cell culture medium containing 1% NEAA. Once the cells established the polarized structure, we performed the wound-healing assay for 4 and 8 h and compared these samples against the samples fixed at 0 h at the migration edge (spanning from edge of monolayer to ∼250 µm deep into the monolayer) (SI Appendix, Fig. S2 A–C). As expected, in migrating Caco-2 cells, Golgi apparatus undergoes very conspicuous dispersion while switching from an apical localization to a basal localization (SI Appendix, Fig. S2 D and E).
Fig. 1.
A Golgi remodeling process observed during epithelial collective cell migration. (A) Immunostained images of migrating MDCK monolayer displaying the three Golgi organizations. Left to Right, apical Golgi polarity, equatorially dispersed Golgi around the nucleus, and front–rear polarized Golgi. The orthogonal views illustrate the change in Golgi localization with respect to nucleus in the three scenarios. FOV: field of view. Black dotted arrow represents the direction of migration. The δ values for the selected cells (crosshair) shown at the Bottom Left corner. (Scale bar: 5 μm.) (B) Distribution pattern of the Golgi organization types after 2, 4, and 6 h of migration. Black dotted arrow represents the direction of migration. (C) Bar plot displaying the distribution of Golgi organization as a function of distance from the migrating edge after 2, 4, and 6 h of migration. (D) A time-lapse montage (Right) of mDsRed Golgi MDCK cells stained with Hoechst depicting the change in Golgi organization during migration. Images were acquired at 30-min interval for 4 h. The yellow square (Left) shows the cell used in the montage. Black dotted arrow represents the direction of migration. (Scale bar: 5 μm.) (Movie S1). (E) A heatmap showing the change in number of Golgi objects with respect to nuclear centroid with time.
We next addressed the question whether migration-associated Golgi dispersion and apical-to-basal repositioning might be a passive consequence of cell flattening, which occurs as the sedentary cells start migrating. Researchers have long debated over the passive versus active nature of Golgi repositioning (1). To check for the aforementioned possibility, we tested whether simple cell flattening by monolayer stretching could lead to a similar apical-to-basal transition of Golgi (SI Appendix, Fig. S3A). Subsequently, we found that while stretching-induced cell flattening indeed led to apical-to-basal relocalization of Golgi, it did not induce any dispersion (SI Appendix, Fig. S3A). Upon stretching-induced cell flattening, Golgi moved to a basal position as a compact organelle (SI Appendix, Fig. S3A). These evidences suggested that Golgi dispersion was cell migration-associated and possibly involved an active process, and we named it migration-induced Golgi apparatus remodeling (MIGAR). Since apico-basal to front-rear polarity change is a property of collective epithelial systems, we looked at how the process got affected if the collectivity of the epithelial monolayer was abolished. To this end, migration in presence of E-cadherin blocking antibody, DECMA1, showed a clear loss of the Golgi reorganization (SI Appendix, Fig. S3B). Live cell imaging of DECMA1-treated cells showed polarization of Golgi without an intermittent equatorial distribution, along with single-cell-like migration out of the monolayer (SI Appendix, Fig. S3C and Movie S2). Finally, we elucidated migration-induced changes in Golgi dispersion using multiple Golgi markers, including cis-Golgi matrix protein GM130 and GRASP65 for cis-Golgi, medial-Golgi matrix protein Mannosidase 2A, mDsRed fluorophore tagged with the N-terminal signal sequence from trans-Golgi membrane localizing beta-1,4-galactosyltransferase 1 enzyme, and Golgin-97 for trans-Golgi (SI Appendix, Fig. S3D). Taken together, here we discovered a migration-induced Golgi remodeling process in epithelial cells, which involves a previously unknown dispersed and unpolarized Golgi structure.
MIGAR Is Different from Golgi Fragmentation during Mitosis.
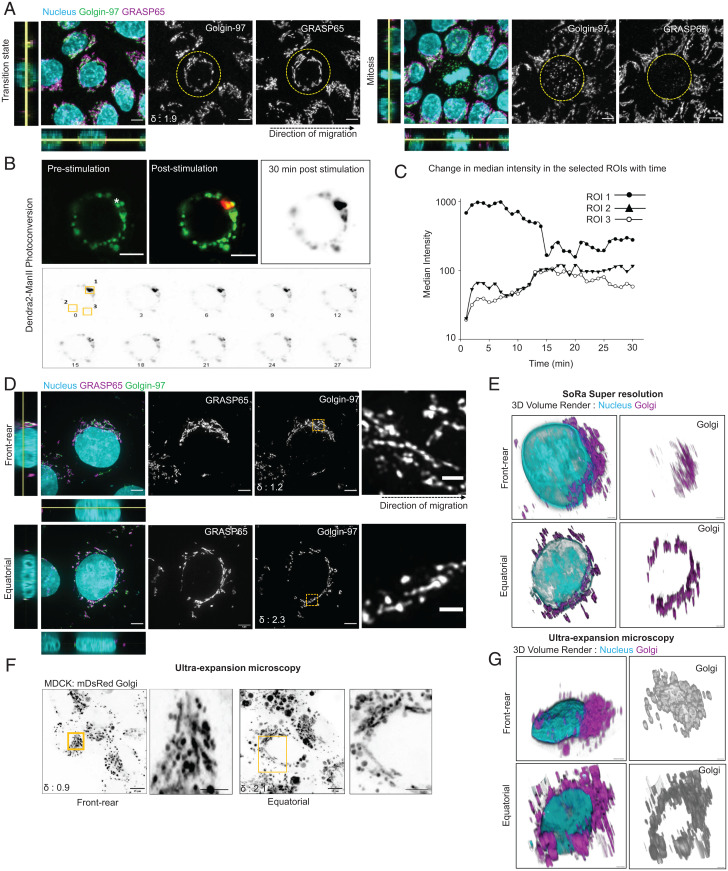
We next attempted to investigate the molecular mechanism underlying MIGAR. In this respect, Golgi fragmentation and dispersion during mitosis is a relatively well-characterized event (36, 37). During mitosis, Golgi reassembly stacking proteins (GRASPs), especially GRASP65 and GRASP55, undergo a series of phosphorylation events (38). This chemical modification abolishes the homophilic interactions between GRASP molecules localized at neighboring stacks and leads to a progressive structural fragmentation of Golgi from a fully connected ribbon to individual stacks to isolated cisternae and finally, to a cluster of vesicles (36, 37, 39). Golgi fragmentation during mitosis facilitates equipartitioning of Golgi components into two daughter cells, which a compact Golgi would fail to achieve. Given this precedence, we asked whether Golgi follows the same mechanistic route during its dispersion in MIGAR. To this end, at first, staining for the localization of endogenous GRASP65, we observed intact and elongated filament-like organization of GRASP65 during MIGAR, as opposed to a haze-like signal in mitotic cells (Fig. 2A). Second, we over-expressed two GRASP55 mutants, namely constitutively phosphorylated GRASP55 T-E and phosphorylation-inactive GRASP55 T-A. These mutants have a dominant negative effect on Golgi fragmentation during mitosis. In contrast, during cell migration, both of them failed to eliminate the dispersed organization of Golgi (SI Appendix, Fig. S4A). Finally, we over-expressed three dominant negative mutants of BARS protein. BARS links adjacent stacks into ribbons and gets phosphorylated during mitosis (40, 41). We, therefore, expressed constitutively phosphorylated BARS-S147D mutants and phosphorylation inactive BARS- S147A and BARS-D355A mutants, which prevent Golgi fragmentation during mitosis in a dominant negative manner (42, 43). Again, these mutants also failed to eliminate the dispersed organization of Golgi during cell migration (SI Appendix, Fig. S4B). Taken together, these experiments showed that the signaling axis involved in MIGAR is different from the one involved in mitosis. Next, given that during mitosis, Golgi apparatus fragments into disconnected vesicles, we probed whether dispersed Golgi units in MIGAR were also disconnected. To this end, we first expressed a photo-switchable fluorescent protein, Dendra2, in Golgi (Dendra2-ManII). Then, as the fluorescently labeled Golgi became dispersed during migration, we switched the color of Dendra2, from green-to-red in spectra, in one segment and checked whether the photo-switched red Dendra2 molecules moved to other segments (Fig. 2B). Here, we hypothesized that if the dispersed Golgi units were really disconnected, the red Dendra2 would not spread to other segments. To our surprise, the photoconverted signal appeared in distal segments within 5 min and it gradually spread to other segments reaching a nearly homogenous distribution and making the entire Golgi apparatus visible in the red channel within 15 min (Fig. 2 B and C and Movie S3). These results indicated that dispersed Golgi units were still retaining mutual connections. Finally, we sought to uncover the structural nature of these units. Researchers have conventionally used transmission electron microscopy (TEM) to study Golgi structure in detail. However, TEM suffers from the narrow field-of-view and insensitivity to dynamic changes. Given that Golgi structure during migration was dynamic and sensitive to the location of a cell within the collective (Fig. 1B), it would be difficult to reveal Golgi structure as a function of the cell and organelle polarity with TEM. Therefore, we used two super-resolution techniques, namely super-resolution via optical reassignment (SoRa) and expansion microscopy (44), that offered a large enough field-of-view to pick cells with the target Golgi polarization as well as sufficient resolution to study organelle structure (∼200 nm). In both methods, cells with polarized Golgi showed parallel-arranged interconnected Golgi units or stacks which formed a Golgi network (Fig. 2 D–G). In contrast, cells with dispersed Golgi organization showed Golgi stacks arranged end-to-end around the nucleus in an equatorial fashion with significantly reduced network organization (Fig. 2 D–G). Taken together, these results indicated that although Golgi underwent dispersion at the onset of epithelial migration, this process was structurally and mechanistically different from the Golgi fragmentation process that takes place during mitosis. MIGAR, therefore, is a signature of epithelial collective migration.
Fig. 2.
Migration induced Golgi remodeling is different from mitotic Golgi remodeling. (A) Fluorescence images comparing the Golgin-97 and GRASP65 organization in a cell with migration induced transition state (Left) and a mitotic cell (Right). Left shows nonfragmented Golgin-97 and ribbon organization of GRASP65. Right displays a fragmented Golgin-97 and significantly diminished signal for GRASP65. Yellow dotted circles mark the cells in focus; the black dotted arrow shows the direction of migration. The δ value for the selected cell shown at the Bottom Left corner in Golgin-97 image, does not apply to mitotic cell. (Scale bar, 5 μm.) (B) Photoconversion of Dendra2-ManII displaying emission upon excitation with 488 nm (for green emission) and 561 nm (for red emission) wavelengths. Prestimulation image shows signal only in the 488 channel; the white asterisk marks the site of stimulation. Poststimulation image shows signals in both 488 and 561 channels. The time-lapse montage (Bottom) of the poststimulation red signal shows the diffusion of the photoconverted signal across Golgi. (Scale bar: 5 μm.) (Movie S3). (C) Scatter line plot of median intensity changes in the selected ROIs. (D) SoRa super-resolution images showing Golgin-97 and GRASP65 organization in polarized (Top) and equatorially dispersed Golgi (Bottom). Zoomed-in area from the yellow box shows Golgi retains ribbon organization in both scenarios. The black dotted arrow shows the direction of migration. The δ values for the cells in focus shown at the Bottom Left corner in Golgin-97 image. (Scale bars: 4 μm, Top; 6 μm, Bottom; 1 μm-zoomed in Insets.) (E) 3D volume render of the SoRa images. (F) Ultra-expansion images of mDsRed Golgi cells with polarized (Left) and equatorially dispersed Golgi (Right). The zoomed-in area in the yellow box shows long ribbons in both the cases. The δ values for the cells in focus shown at the Bottom Left corner. (Scale bar: 15 μm; 5 μm zoomed-in Insets.) (G) 3D volume render of the expansion images.
Golgi-Associated Microtubules Are Dispensable for MIGAR.
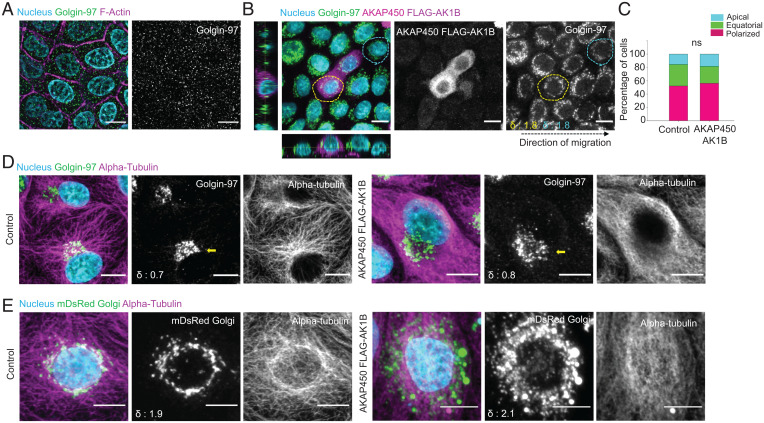
Since stretching experiments indicated that MIGAR might not be a passive consequence of cell flattening, we next asked what cellular element might be actively enabling it. To this end, Golgi-associated microtubules are known to be critical for the establishment and maintenance of Golgi polarity during single cell as well as collective migration (25, 45, 46). We, therefore, first investigated the role of microtubules in MIGAR through the classical approach, by carrying out epithelial migration in presence of a microtubule-destabilizing drug, nocodazole. Microtubule disruption following nocodazole treatment results in fragmentation of Golgi. We were able to recapitulate the fragmented Golgi morphology reported in literature (Fig. 3A), however, the global fragmentation of Golgi in this approach was unable to address the precise role of microtubules in Golgi distribution and repositioning during migration. Therefore, to understand the specific involvement of microtubules better, we sought to unlink microtubules from Golgi membrane. To this end, previous works had reported that AKAP450 is critical for nucleation and polymerization of microtubules on Golgi membranes (47). Over-expression of an AKAP450 mutant, namely AKAP450-AK1B, prevents localization of Golgi-associated microtubules on Golgi membrane by displacing the endogenous AKAP450 from Golgi (48). We hypothesized that if Golgi-associated microtubules were involved in MIGAR, then overexpressing AKAP450-AK1B mutant would prevent Golgi dispersion during migration. However, contrary to our expectation, during migration, Golgi showed very similar dispersed stage in AK1B-expressing cells as in surrounding wild-type cells (Fig. 3 B and C), although staining for microtubules with an antibody against α-tubulin showed disruption of Golgi–microtubule association in these cells (Fig. 3D). During the dispersion phase, while the wild-type cells exhibited a clear localization of microtubules to Golgi forming a prominent ring around the nucleus, the cells expressing AKAP450 mutant lacked Golgi localization of the microtubules (Fig. 3E). However, this reduction in Golgi–microtubule interaction did not affect MIGAR (Fig. 3E). Together, these results demonstrated that Golgi-associated microtubules were dispensable for MIGAR, in contrast to the conventional belief that Golgi-associated microtubules should be involved in defining Golgi polarity and structure.
Fig. 3.
Golgi-associated microtubules are dispensable for MIGAR. (A) Immunostained images of nocodazole-treated migrating MDCK monolayer showing a fragmented Golgi dispersed across cytoplasm. (Scale bar: 5 μm.) (B) Fluorescence images of migrating mosaic monolayer of MDCK cells and MDCK-AKAP450-FLAG-AK1B overexpressing cell (yellow dotted area), both control (cyan dotted area) and mutant show equatorial Golgi dispersion. The δ values for the selected cells shown at the Bottom Left corner in Golgin-97 image. (Scale bar: 10 μm.) (C) A Stacked vertical bar plot showing no significant change in percentage of cells with equatorial Golgi in control versus AKAP450-AK1B overexpressing cells. n = 81 cells. (D) Microtubule immunostaining pattern in control MDCK cell and AKAP450-AK1B overexpressing cell. Left shows the microtubules originating from Golgi in control cells. Right shows the AKAP450-AK1B overexpressing cells with a clear hollow for microtubules around Golgi. The δ values for the selected cell (yellow arrow) shown at the Bottom Left corner in Golgin-97 image. (Scale bar: 10 μm.) (E) Immunofluorescence images showing the perinuclear microtubule organization pattern during equatorial Golgi dispersion in control cells (Left), and undiscernible microtubule organization in AKAP450-AK1B overexpressing cells (Right). The δ values shown at the bottom-left corner in mDsRed Golgi image. (Scale bar 5 μm.)
Golgi–Actin Interaction Is Critical for MIGAR.
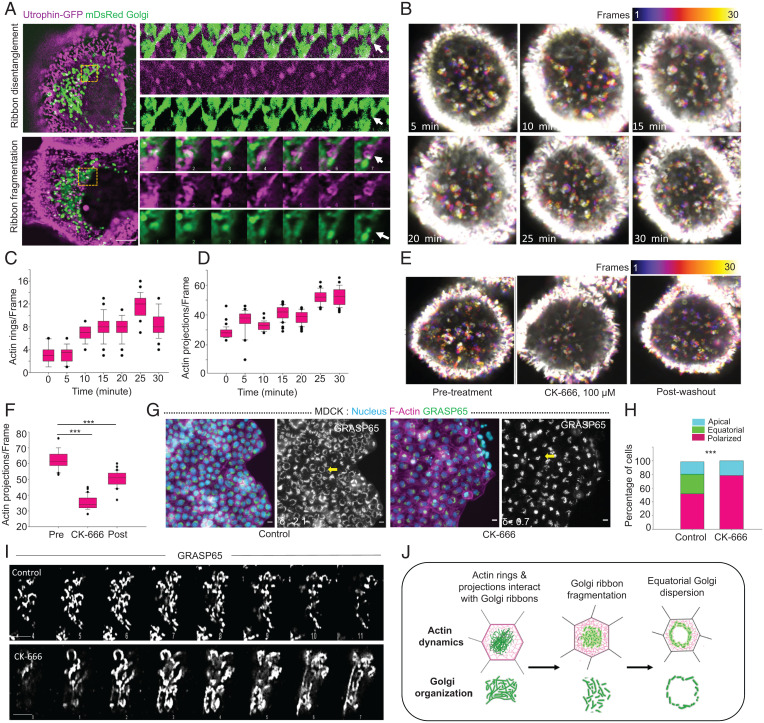
We then asked whether another cytoskeletal element, actin, might play important roles in MIGAR. In this respect, the linkage between Golgi and contractile actin cytoskeleton has emerged to be critical in defining Golgi structure and function. For example, there are reports of oncogenic elevation of actomyosin contractility leading to unusual Golgi distributions within cancer cells (29, 49). To perturb actin dynamics, we used a suboptimal concentration (500 nM) of an actin filament polymerizing and stabilizing drug, jasplakinolide (Jasp), or a potent inhibitor of actin polymerization, cytochalasin D (Cyt D) (SI Appendix, Fig. S5A). These drug concentrations did not affect the collective cell migration, and we were able to capture polarized Golgi in drug-treated cells in the wound area spanning from the edge of the monolayer to ∼250 µm deep into the monolayer. However, no equatorial Golgi organization was found in the cells (SI Appendix, Fig. S5B), indicating that a dynamic Golgi–actin interaction was indeed critical for MIGAR. To be specific, treating the cells with Jasp abolished the equatorial dispersion of Golgi (SI Appendix, Fig. S5A). Along with the loss of dispersed Golgi organization, in Jasp-treated cells, we observed formation of an actin aggresome closely encased by Golgi (SI Appendix, Fig. S5 A, Top). However, unlike actin aggresomes reported in the literature, we observed that these aggresomes displayed a twisted ring structure in each cell, hinting at possible deformation of an existing actin structure within the cell. This led us to question whether an existing actin structure in the cell could be playing a part in MIGAR. Relevantly, epithelial cells are known to form an apical circumferential actin belt, which is important for epithelial barrier function. At the onset of migration, the circumferential actin belt undergoes fragmentation and rearrangement to facilitate cell migration. This information implied that the twisted actin ring structures upon Jasp treatment (SI Appendix, Fig. 5 A, Top) were possibly the Jasp-stabilized circumferential actin belts, which otherwise would have rearranged during migration. On the other hand, treating the cells with Cyt D led to the loss of equatorial dispersion of Golgi (SI Appendix, Fig. S5 A, Bottom). In addition, a quantification of cells for the three types of Golgi organization showed a complete absence of equatorial Golgi dispersion in both Jasp and Cyt D-treated cells (SI Appendix, Fig. S5C). These results from actin-perturbation experiments indicated that Golgi dispersion in MIGAR possibly depends on the integrity and dynamics of actin cytoskeleton. Next, to uncover the precise role of actin dynamics and the exact involvement of circumferential actin belt in MIGAR, we imaged actin and Golgi dynamics together by expressing actin-marker utrophin-GFP and Golgi-marker mDsRed-Golgi7 (Fig. 4A and SI Appendix, Fig. S5D and Movies S4–S7). Subsequently, we detected small actin projections (0.5–1.0 µm) emanating from the circumferential actin belt and over time evolving into small actin rings in sedentary cells (Fig. 4A and SI Appendix, Fig. S5D and Movies S4–S7). These actin rings or projections were then loaded onto Golgi ribbon. Interestingly, post confinement lift-off, we observed a progressively increased number of actin rings and projections emanating from the circumferential actin belt (Fig. 4 B–D). We then asked whether Cdc42-formin-mediated actin bundling or Arp2/3-mediated branched actin polymerization created these actin projections. Treating the cells with a formin inhibitor (SMIFH2) as well as a Cdc42 inhibitor (ML141) did not change the frequency of Golgi-interacting actin projections (SI Appendix, Fig. S5E), but an Arp2/3 inhibitor CK-666 reduced it significantly (Fig. 4 E and F). These results indicated that Arp2/3-mediated actin polymerization drives these actin projections. Consequently, we asked whether these projections might be important for Golgi-dispersion. To this end, treating the cells with a suboptimal level of CK-666, which did not perturb cell migration but reduced the frequency of Golgi-interacting actin projections, eliminated migration-associated Golgi dispersion (Figs. 4 G and H). Golgi under CK-666 treated conditions showed comparatively longer ribbons (Fig. 4I and SI Appendix, Fig. S5F). To test the generality of our observation-related actin dynamics and MIGAR, we finally probed the role of actin dynamics in Golgi dispersion in Caco-2 cells by treating the differentiated Caco-2 monolayer with actin polymerizing or destabilizing drugs, including Jasp, Cyt D, and CK-666. As compared to MDCK cells, here, we used still suboptimal but higher concentrations of the drugs since differentiated Caco-2 cells formed more tightly sealed epithelial barrier than MDCK cells. To our expectation, we observed treatment with these drugs resulted in the loss of equatorial Golgi dispersion in the migrating Caco-2 cells (SI Appendix, Fig. S6). As compared to control cells where Golgi dispersed around the nucleus, Golgi in drug-treated Caco-2 cells showed no preference for perinuclear localization and was rather distributed all over the cytoplasm (SI Appendix, Fig. S6). Together these results provide the preliminary and yet robust evidence that MIGAR required a dynamic interaction between actin projections and Golgi, and the increased Golgi–actin interaction at the onset of migration is critical for Golgi dispersion (Fig. 4J). However, the molecular players involved in this interaction remained unknown.
Fig. 4.
Interaction of Actin rings and projections with Golgi is required for controlled ribbon dynamics and organization during MIGAR. (A) Live cell time-lapse images of cells expressing Utrophin GFP and mDsRed Golgi showing actin-Golgi interaction, Top shows ribbon disentanglement following actin projection interaction with Golgi ribbon (Movie S4). Bottom shows ribbon fragmentation following actin ring sliding over the Golgi ribbon (Movie S5). A montage of the boxed areas shows both Golgi and actin dynamics with time, white arrowhead marks the fate of the interaction. Images were taken at 1-s intervals for 1 min. (Scale bar: 2.5 μm.) (B) Temporal color projected images of MDCK cells expressing utrophin GFP following initiation of the migration at 5-min intervals. The number of actin rings and projections increases with time as migration ensues. Color key shows the frame numbers. Image dimension, 16.69 × 16.69 μm. (C and D) Box-and-whiskers plot of the number of actin rings (Left) and projections (Right) depicting gradual increase in the number with migration. n = 30 frames. (E) Temporal color projected images displaying the change in number of actin projections following Arp2/3 inhibition by CK-666 (100 μM) and post drug washout. Color key shows the frame numbers. Image dimension, 25.56 × 25.56 μm. (F) The box-and-whiskers representation showing the change in number of actin projections under conditions of pretreatment, CK-666, and post washout, (P < 0.001). n = 30 frames. (G) Fluorescence images of MDCK monolayer showing normal MIGAR distribution pattern in control cells (Left) and loss of equatorial Golgi dispersion in CK-666-treated sample (Right). The δ values for the selected cell (yellow arrow) shown at the Bottom Left corner in GRASP65 image. (Scale bar: 10 μm.) (H) A stacked vertical bar plot showing the percentage of cells with polarized, equatorial, and apical Golgi in control and CK-666-treated condition (P < 0.001). n = 95 cells. (I) Zoomed in Z-stack of Golgi ribbons showing the difference in Golgi ribbon in control cells (Top) and CK-666-treated cells (Bottom). (Scale bar: 5 μm.) (J) Schematic representing the mechanism of Golgi ribbon fragmentation following interaction with actin.
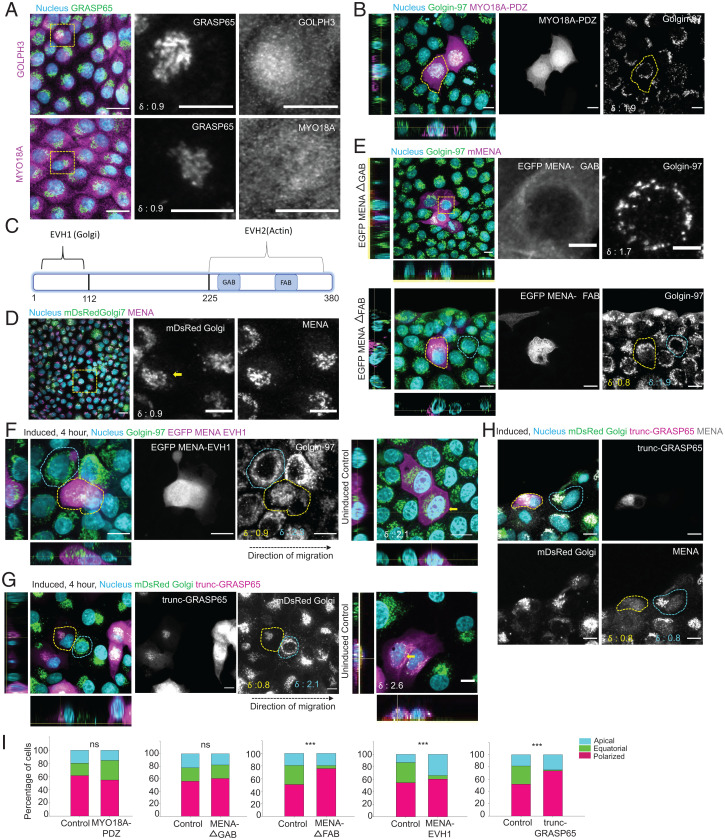
Golgi-Localized Actin Elongation Factor MENA Is Essential for MIGAR.
Next, searching for the prospective molecular candidates that might mediate Golgi–actin interaction during epithelial migration, we explored two previously elucidated signaling axes—one involving GOLPH3-MYO18A (29) and another involving MENA (50). The GPP34 domain (aa 30–293) of GOLPH3 binds to phosphatidylinositol-4-phosphate or PtdIns(4)P on the Golgi membrane and its N-terminal interacts with an unconventional myosin MYO18A (29). MYO18A in turn binds to the actin filament, thus connecting Golgi and actin cytoskeleton. We first looked at the localization of these proteins during MIGAR and found both GOLPH3 and MYO18A localized to Golgi, though significant non-Golgi fractions were also present (Fig. 5A). Next, to block the GOLPH3-MYO18A pathway, we over-expressed the MYO18A-PDZ domain in migrating cells. This mutant acts as dominant negative disrupter of GOLPH3-MYO18A pathway by saturating the MYO18A binding domain of GOLPH3 (51). Therefore, if GOLPH3-MYO18A axis were important for Golgi–actin interaction during MIGAR, we would not observe Golgi dispersion in MYO18A-PDZ-expressing cells. However, even in the cells overexpressing MYO18A-PDZ, we observed dispersed Golgi around the nucleus, and MIGAR was seemingly unperturbed (Fig. 5B, quantification: 5I). This result showed that GOLPH3-MYO18A axis might not be relevant for MIGAR. We then investigated the role of actin elongation factor MENA in MIGAR. Localization of MENA to Golgi happens through its interaction with GRASP65 via the EVH1 domain (Fig. 5C), while it binds to actin via its EVH2 domain (50). Interestingly, MENA can bind to both G-actin and F-actin via its G-actin and F-actin binding domains (GAB and FAB; Fig. 5C), respectively (50). On immunostaining for the endogenous protein, MENA showed a strong localization to Golgi (Fig. 5D). Interestingly, over-expression of a MENA mutant lacking the F-actin binding domain (MENA-ΔFAB) resulted in the loss of equatorial Golgi dispersion, while over-expression of another mutant lacking the G-actin binding domain (MENA-ΔGAB) did not perturb it (Fig. 5E, quantification: 5I). However, since MENA also exhibits focal adhesion localization, MENA-ΔFAB could also be perturbing the focal adhesion-associated MENA. Hence, we sought for further Golgi-specific perturbation and hypothesized that overexpressing EVH1 domain of MENA (MENA-EVH1) in a mosaic manner would block Golgi-MENA interaction in MENA-EVH1 overexpressing cells. If MENA were important for Golgi–actin interaction during MIGAR, we would not observe Golgi dispersion in these cells, while surrounding nonexpressing cells would continue displaying Golgi dispersion. To this end, we created a doxycycline-inducible MENA-EVH1 expression system, where cells transfected with MENA-EVH1 construct expressed the transgene only to a very basal level (due to leaky expression) until we added doxycycline to the medium. Further, to perturb the Golgi repositioning in a controlled way, 40 min before the confinement lift off, 5 μg mL−1 doxycycline was added to the cells to induce transgene expression. Subsequently, over-expression of MENA-EVH1 led to complete failure of Golgi dispersion during migration, as the mutant cells showed apical localization of Golgi even during migration, and equatorial Golgi distribution was conspicuously absent (Fig. 5F, quantification: 5I). While these results indicated that MENA is critical for Golgi–actin interaction during MIGAR, it is plausible that MENA-EVH1 constructs caused additional actin changes in the migrating cells and could be directly controlling the overall front-back actin architecture, not only at the Golgi but throughout the cell body. To address if MENA-EVH1 affects the front-rear actin organization, we used use SiR-actin, which is a better probe for staining actin stress fibers in live cells than utrophin (52). We used very low concentration of the dye (50 nM for 12–16 h) to stain the actin fibers, allowing us to capture well-resolved actin stress fibers. The quantification and comparison of actin fibers in wild-type and MENA-EVH1-expressing cells showed no significant change in length and numbers (SI Appendix, Fig. S7A). In addition, we used actin and myosin markers, including Arp3, phosphorylated myosin light chain (pMLC), and SiR-actin, to compare the front-back actin architecture with and without over-expression of MENA-EVH1 (SI Appendix, Fig. S7B). In this case, we carried out migration experiments for 4 h and then stained the cells with antibodies (against Arp3 and pMLC) or SiR-actin. Comparison between localization of these proteins in MENA-EVH1 expressing and the surrounding wild-type cells did not reveal any perceptible difference in the front-to-back distribution of these proteins (SI Appendix, Fig. S7B).
Fig. 5.
Actin-Golgi interaction mediated by MENA-GRASP65 is critical for MIGAR. (A) Fluorescence images displaying the localization of GOLPH3, MYO18A during migration. GOLPH3 and MYO18A show significant non-Golgi localization. The δ values for the cells in focus shown at the Bottom Left corner in GRASP65 panel. (Scale bar: 5 μm.) (B) Cells overexpressing MYO18A-PDZ show normal progression through MIGAR. The yellow dotted area marks the mutant cell. The δ value for the cell in focus shown at the Bottom Left corner in Golgin-97 panel. (Scale bar: 5 μm.) (C) The schematic illustrates the domain structure of MENA. EVH1 domain binds to Golgi and EVH2 interacts with actin via GAB and FAB subdomains. (D) Immunofluorescence images showing MENA exhibits specific Golgi localization. The yellow square marks the zoomed in cells on right showing the Golgi and MENA signal. The δ values for the selected cell (yellow arrow) shown at the Bottom Left corner in mDsRed Golgi image. (Scale bar: 10 μm in the images; 5 μm in the zoomed in panel.) (E) Immunofluorescence images showing equatorial Golgi dispersion in MENA ΔGAB overexpressing cells (Top), whereas MENA ΔFAB overexpressing cell (yellow dotted area) shows loss of equatorial dispersion. The control cell (cyan dotted area) shows equatorial Golgi dispersion. The δ values for the selected cells shown at the Bottom Left corner in Golgin-97 panel. (Scale bar: 10 μm; 5 μm in the zoomed in box.) (F) Expression of MENA EVH1 domain results in a failed MIGAR and loss of equatorial Golgi dispersion. The fluorescent image panel on the left displays the apical localization of Golgi in MENA EVH1 overexpressing cell (yellow dotted area). (Scale bar: 10 μm.) The orthogonal view of uninduced MENA EVH1 cells (Right) shows equatorial localization in uninduced control cells. Contrast adjusted to show the mutant cell. Black dotted arrow represents the direction of migration. The δ values for the selected cells shown at the Bottom Left corner in Golgin-97 image and uninduced panel. (Scale bar: 10 μm.) (G) Immunofluorescence image showing the polarized Golgi in trunc-GRASP65 overexpressing cell (yellow dotted area) versus the equatorial Golgi in control MDCK cell (cyan dotted area). The uninduced control on right shows equatorial Golgi. Contrast adjusted to show the mutant cell. Black dotted arrow represents the direction of migration. The δ values for the selected cells shown at the Bottom Left corner in mDsRed Golgi image and uninduced panel. (Scale bar: 10 μm.) (H) Immunostained images showing the displacement of endogenous MENA protein form Golgi in trunc-GRASP65 overexpressing cell (yellow dotted area), compared to the Golgi localized MENA signal in control cell (cyan dotted area). The δ values for the selected cells shown at the Bottom Left corner. (Scale bar: 10 μm.) (I) A quantification of percentage of cells with apical, equatorial, and polarized Golgi in control versus cells overexpressing MYO18A-PDZ (n = 82 cells), MENA ΔGAB (n = 95 cells), MENA ΔFAB (n = 95 cells), MENA-EVH1 (n = 98 cells), and trunc-GRASP65 (n = 90 cells). A significant reduction in cells with equatorial Golgi is seen in MENA ΔFAB (P < 0.001), MENA-EVH1 (P < 0.001), and trunc-GRASP65 (P < 0.001).
We next targeted the MENA-actin linkage by an alternative means. MENA localizes to Golgi through its interaction with GRASP65 (aa 236–241) (53). We, therefore, generated a construct that enabled us to express a GRASP65 truncation mutant (GRASP65 aa 1–201 or trunc-GRASP65), lacking the MENA-binding domain, in tetracycline-inducible manner. We envisaged that the over-expression of this mutant would saturate GRASP65 binding to Golgi, and because of its inability to interact with the endogenous MENA, it would disconnect the downstream linkage with actin cytoskeleton and act as a dominant negative mutant. Following transfection of the monolayer with trunc-GRASP65 construct, we induced its expression 40 min before the confinement lift-off. We then allowed the cells to migrate for 4 h and analyzed the Golgi organization in the trunc-GRASP5-expressing vis-à-vis the surrounding nonexpressing cells. As expected, the cells expressing trunc-GRASP5 lacked the migration-induced Golgi dispersion (Fig. 5G, quantification: 5I), and at the same time, endogenous MENA dislocated from Golgi in these cells (Fig. 5H). Since beyond the MENA-actin axis, GRASP65 is not known to control the actin dynamics, results obtained from these experiments robustly established the role MENA-GRASP65 pathway in MIGAR. Finally, to test whether the MENA-GRASP65 pathway might be involved in Golgi dispersion in Caco-2 model as well, we carried out transient transfection of the differentiated Caco-2 cells with either MENA-EVH1 construct or trunc-GRASP65 construct. As expected, Caco-2 cells expressing either of two constructs lacked Golgi dispersion (SI Appendix, Fig. S8 A and B). We quantified the percentage of cells with the three types of Golgi organization in control Caco-2 cells and cells overexpressing the two constructs. In this case, to obtain significant fractions of all three types of Golgi organization in control and thereof, to study effect of the mutant construct expression unbiasedly, we allowed Caco-2 cells to migrate for 12 h, since they migrated at a slower speed than MDCK cells. However, for quantification, we again focused on cells in an area spanning from the edge of the monolayer to ∼250 µm deep into the monolayer. A comparison of these showed a significant reduction in equatorial Golgi organization in cells expressing either of the two mutant constructs (SI Appendix, Fig. S8 C and D). Together, these experiments elucidated that Golgi-localized actin elongation factor MENA is essential for Golgi–actin interaction during epithelial migration.
Preventing Golgi Dispersion Disrupts Coherent Cell Polarization and Migration Persistence.
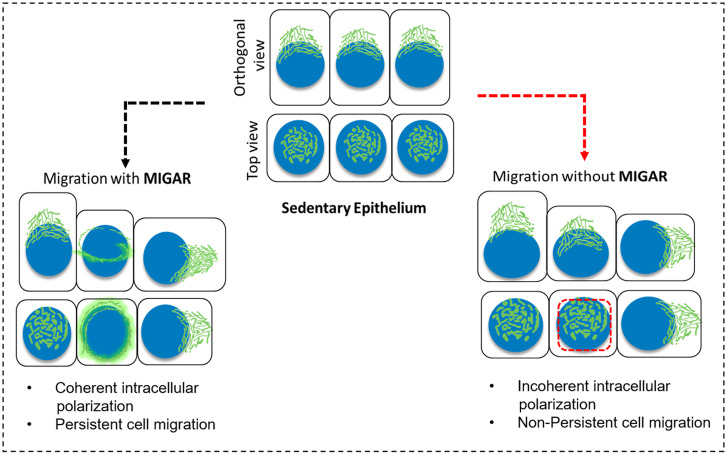
We next asked what might be the importance of Golgi dispersal in epithelial migration. To this end, we utilized the diminished Golgi remodeling upon disruption of MENA-GRASP65 axis. First, we looked at the state of global intracellular polarization in MENA-EVH1 overexpressing cells vis-à-vis surrounding nonexpressing cells by examining the centrosome position. We observed an increase (∼0.4 μm) in the distance between Golgi and centrosome as well as increased variability in the distance, in MENA-EVH1 overexpressing MDCK cells (SI Appendix, Fig. S9 A and B). This observation emphasized the abnormal intracellular polarization in absence of normal MIGAR. To understand the effect of the incoherent intracellular polarization on cell migration, we dissected and compared the cell migration attributes of MENA-EVH1 and trunc-GRASP65 overexpressing cells vis-à-vis control cells. In this regard, we first segregated collective cell migration from single-cell migration in terms of the change in Golgi polarity with time. Isolated MDCK did not move significantly. However, tracking the Golgi apparatus showed that it changed its intracellular position frequently (SI Appendix, Fig. S10 A, Left). On contrary, intracellular positioning of Golgi changed a little within a collective, and overall Golgi movement essentially mirrored the cell movement (SI Appendix, Fig. S10 A, Right). Given these results, we then examined the movement of control cells and of cells expressing MENA-EVH1 or trunc-GRASP65 constructs, embedded within cell collectives. As expected, the control cells within collectives showed a persistent directional migration (SI Appendix, Fig. S10 B, Left), exhibiting a similar pattern as the Golgi tracks in collective cell migration. However, cells overexpressing either of the two mutant constructs lacked a specific directionality in cell movement (SI Appendix, Fig. S10 B, Middle and Right). Further, to understand the loss of persistent directional cell migration in cells expressing the mutant constructs, we looked at other important aspects of the migration in these cells. First, we found no discernable change in cell migration speed between control MDCK cells and MENA-EVH1-expressing cells. However, trunc-GRASP65 expressing cells displayed a large variation in cell migration speed (SI Appendix, Fig. S10C). We then measured the change in angle at each turn during migration (SI Appendix, Fig. S10D). As expected, the control cells displayed a minimum change in direction at each turn (SI Appendix, Fig. S10E), however, cells expressing the mutant constructs MENA-EVH1 (SI Appendix, Fig. S10F) and trunc-GRASP65 (SI Appendix, Fig. S10G) showed larger changes in direction at each turn (SI Appendix, Fig. S10H) and higher cell-to-cell variation. As a result of large change in direction at each turn, the net displacement turned out to be significantly lower in case of both MENA-EVH1 and trunc-GRASP65 (SI Appendix, Fig. S10I). Together, we have established previously unknown role GRASP65-MENA molecular pathway in regulating MIGAR during collective cell migration (Fig. 6).
Fig. 6.
Schematic describing the process of migration-induced Golgi apparatus remodeling (MIGAR). Normal MIGAR goes through equatorial Golgi dispersion and results in coherent intracellular polarization and persistence cell migration. In absence of MIGAR, incoherent intracellular polarization results in nonpersistent cell migration.
Discussion
Among the results that we reported above, perhaps the linkage between the shift in Golgi positioning and actin dynamics is the most surprising one. Specifically, the microscopic evidence of interaction between Golgi and the projections coming of the actin cortex is a unique discovery. Of three types of cytoskeletal elements, it is not actin but microtubules that are conventionally known to be important in Golgi positioning and integrity, although actin-associated proteins seem to influence the Golgi polarity in migrating fibroblasts (54). Following the discovery that microtubule disruption upon nocodazole treatment leads to Golgi fragmentation in fibroblasts cells (55), the field quickly explored the role of microtubules in Golgi integrity and positioning. Golgi emerged to act as a major microtubule nucleation site. In addition, Golgi membrane appeared to promote microtubule nucleation in vitro (56). In due course, researchers identified several Golgi-residing proteins, and their role in microtubule nucleation on the Golgi membrane was also identified. For example, CLASPs nucleate microtubule on trans-Golgi membrane by their interaction with GCC185, and knockdown of CLASPs resulted in loss of Golgi-derived microtubules and disrupted Golgi ribbon organization in RPE1 cells (22, 57). Similarly, expressing AKAP450 mutants that were unable to nucleate microtubule on Golgi membrane, resulted in dramatic change in Golgi organization. Golgi lost the long-extended ribbon like organization and collapsed into a sphere-like organization around the centrosome and led to consequent loss of Golgi reorientation during migration (48). However, recent works with epithelial cells have suggested that a compact Golgi can exist in the absence of both centrosomal and Golgi-associated microtubules, although uncoupling Golgi from microtubule perturbs the vesicular trafficking required for migration-invasion (25). Another report showed that the loss of centrosome had little effect on the ability of endothelial cells to polarize Golgi and move in 2D and 3D environments (19). These recent developments hint that perhaps it is necessary to think beyond microtubules as the main mediator of Golgi organization everywhere, and consider that actin filaments might play as critical roles as microtubules in some context. However, what then links actin and Golgi?
To this end, there is evidence that at least two proteins, GOLPH3 and MENA, which mediate Golgi–actin cytoskeleton connection and are critical for Golgi morphology and cell migration of cancer cells. GOLPH3 is a proven oncogenic protein and is known for its interaction with an unconventional myosin, myosin 18A. Perturbing this interaction by gene silencing of myosin18A caused significant reduction in Golgi reorientation in migrating MDA-MB-231 cells (58). Another key molecule reported to bring together actin dynamics and Golgi structure is mammalian homolog of Enabled, MENA. MENA localizes to cis Golgi membrane through its interaction with the Golgi structural protein GRASP65 mediated by MENA EVH1 domain. MENA EVH2 domain binds to both the G-actin and F-actin form of actin via G-actin binding domain (GAB) and F-actin binding domain (FAB). Loss of endogenous MENA by gene knockdown led to Golgi ribbon fragmentation in HeLa cells (50). From these discoveries, while it seemed that actin dynamics and acto-myosin contractility should play critical roles in Golgi morphology and function in cancer cells, it was not known whether these connections play any role in the apical-to-basal shift in Golgi positioning in migrating epithelial cells. Pertinently, although MENA activity decreases from embryonal to adult life, it gets reactivated in some breast tumors (59). In fact, studies have indicated that MENA expression increases during malignant transformation, possibly playing an important role in tumor invasiveness (60). We can now speculate that hyper-activated Golgi–actin interaction due to MENA over-expression may lead to fragmented Golgi morphology, which nevertheless is a characteristic cell phenotypical marker of cancer cells. One caveat here could be that although two mutants, MENA-EVH1 and trunc-GRASP65, demonstrated a connection between MIGAR and the persistence of cell migration, both of these mutants nevertheless targeted the same signaling axis (MENA-GRASP65). Ideally it would have been more confirmatory if it were possible to perturb different axes targeting the same event, thus robustly proving a causal connection between MIGAR and the persistence of migration. However, given that connection between the Golgi apparatus and actin cytoskeleton is specific, the state-of-the-art does not suggest any other alternatives.
It is important to note that while our study is restricted to mammalian epithelial cells, a previous study has reported dispersal of Golgi apparatus into several puncta or ministacks containing cis- and trans-Golgi complex proteins during early zebrafish embryogenesis (16). Considering this evidence, what could be the functional significance of a fragmented or dispersed Golgi? During mitosis, Golgi fragmentations help in equipartitioning of the organelle in two daughter cells. While Golgi does not undergo such fragmentation and the stacks remain connected during migration, dispersion does generate a momentary loss of polarity. Hence, it is tempting to speculate that during migration, this dispersed Golgi around the nucleus creates a transition state from which a stable polarization can be flexibly created, depending on the intercellular cues that the cells receive from their neighbors. The formation of the transition state thus provides a temporal interlude for the reorientation of other intracellular organelles and facilitates global intracellular reorganization during the switch from apico-basal to front-to-rear polarity. However, it needs to be investigated next how the basally located dispersed Golgi stacks reassemble on one side of the nucleus and what chemical and mechanical cues determine the specific site of reassembly. Inhibiting Golgi dispersion using MENA-EVH1 or trunc-GRASP65 mutant prevents the active actin-mediated Golgi dispersion around the nucleus, and Golgi in the mutant cell remains on top of nucleus. However, as the mutant cells migrate, the concomitant decrease in cell height leads to Golgi getting squeezed out to a random position with respect to the nucleus. We presume that this random polarization may not follow the local cues experienced by a cell within a collective, which leads to erratic cell movements (SI Appendix, Fig. S10). It is known that dynamics of individual cells within a collective follows local biochemical and biophysical field (32). It is possible that ultimate Golgi polarization too follows such fields, and a mutant cell with uncoordinated Golgi organization amid wild-type cells may not respond to these collective fields in the same way as the wild-type cells do. Supported by the increased Golgi-centrosome distance in MIGAR-deficient cells (SI Appendix, Fig. S9 A and B), one can speculate that these cells exhibit a compromised polarity axis, which results in an inefficient collective migration. Future work addressing the selection of cell polarity and Golgi polarity in cells of a collective should uncover further interaction between the two processes in greater detail.
Finally, it needs to be highlighted that the present work focuses specifically on how actin cytoskeleton influences the dispersal of Golgi. It does not, however, elucidate the molecular and mechanistic details of how the Golgi reassembly and polarization regulate actin organization during cell migration. Golgi polarity within a cell is an established marker of the cell polarity under many physiological contexts. It is, therefore, possible that the molecular pathways regulating cell polarity and cell migration intersect with the pathways regulating Golgi remodeling and polarity. We can also speculate that for an efficient collective cell migration, the existence of a parallel or sequential control of Golgi polarization and cytoskeletal remodeling is very likely. It is possible that Golgi polarity of a migrating cell controls the directional transport of the molecular factors that are related to actin remodeling at cell-cell junctions, at lamellipodial protrusions, and at cell–matrix adhesions. Further works need to be done to reveal the mechanisms connecting Golgi polarization and actin cytoskeleton remodeling during collective cell migration. Nevertheless, our current discovery reveals an actin-driven process and a previously unknown transition state during apical-to-basal repositioning of Golgi apparatus in epithelial cells and resolves many questions related to Golgi dynamics in epithelial tissue that have been looking for answers, perhaps for many decades.
Methods
All the experiments were done using the Madin-Darby canine kidney (MDCK) epithelial cell line and Caco-2 cells (ATCC-HTB-37) derived from colon tissue. Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with GlutaMax (Gibco) with 10% fetal bovine serum (tetracycline-free FBS, Takara Bio) and 10 U mL−1 penicillin and 10 μg mL−1 streptomycin (Pen-Strep, Invitrogen) in an incubator maintained at 37 °C and 5% CO2. To carry out cell migration experiments as before (32), adhesive biocompatible silicone culture-insert (ibidi) was press-bonded to glass-bottom culture dish (ibidi). The two wells in the culture insert was seeded with 5 × 104 cells, suspended in 80 μL cell culture medium, and were allowed to adhere and form cell monolayer for 18 h at 37 °C in a 5% CO2 humidified incubator. Following an incubation period, the culture-insert was lifted off to initiate cell migration, and 2 mL media was added to the dish. In case of tetracycline-inducible plasmids, expression was induced with 5 μg mL−1 doxycycline in culture media 30 min prior to removal of culture inserts, and migration was carried out in the induction medium. Confocal fluorescence images were acquired using 60× oil objective mounted on an Olympus IX83 inverted microscope equipped with a scanning laser confocal head (Olympus FV3000). Time-lapse images of live samples were done in the live-cell chamber provided with the microscopy setup. 20 mM Hepes (Gibco) was used to maintain CO2 levels. Super-resolution images were acquired with a 60× oil objective mounted on an Olympus IX83 inverted microscope supplied with Yokogawa CSU-W1 (SoRa Disk) scanner. To calculate dispersion index, first Golgi channel images were segmented to find Golgi objects in 3D Suite-FIJI. The distance between center of mass of Golgi and centroid of Golgi objects were measured, and root mean square (RMS) value was calculated. RMS values were then divided by nuclear radius. All statistical analysis was carried out in SigmaPlot 10.0. Depending on the experiment, either χ2 analysis or unpaired t test with Welch’s correction was used for determining the statistical significance. Please see SI Appendix text for the detailed materials and methods.
Supplementary Material
Acknowledgments
We thank Mahendra Sonawane, Aprotim Mazumder, Mustansir Barma, and the Collective Cellular Dynamics laboratory members for critical discussion. T.D. is a Department of Biotechnology (DBT)/Wellcome Trust India Alliance intermediate fellow and partner group leader of the Max Planck Society, Germany. R.M. is a DBT/Wellcome Trust India Alliance early career fellow. The authors sincerely acknowledge generous funding from the DBT/Wellcome Trust India Alliance (Grant Nos. IA/E/19/1/504967 to R.M. and IA/I/17/1/503095 to T.D.), a partner group grant from the Max Planck Society, and intramural funds at Tata Institute of Fundamental Research, Hyderabad from the Department of Atomic Energy Energy, India (under Project Identification No. RTI 4007).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2204808119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Trelstad R. L., The Golgi apparatus in chick corneal epithelium: Changes in intracellular position during development. J. Cell Biol. 45, 34–42 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carney P. R., Couve E., Cell polarity changes and migration during early development of the avian peripheral auditory system. Anat. Rec. 225, 156–164 (1989). [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Boulan E., Macara I. G., Organization and execution of the epithelial polarity programme. Nat. Rev. Mol. Cell Biol. 15, 225–242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson W. J., Remodeling epithelial cell organization: Transitions between front-rear and apical-basal polarity. Cold Spring Harb. Perspect. Biol. 1, a000513 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burute M., et al. , Polarity reversal by centrosome repositioning primes cell scattering during epithelial-to-mesenchymal transition. Dev. Cell 40, 168–184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu W., et al. , Hepatocyte growth factor switches orientation of polarity and mode of movement during morphogenesis of multicellular epithelial structures. Mol. Biol. Cell 14, 748–763 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayor R., Etienne-Manneville S., The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 17, 97–109 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Bacallao R., et al. , The subcellular organization of Madin-Darby canine kidney cells during the formation of a polarized epithelium. J. Cell Biol. 109, 2817–2832 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apodaca G., Gallo L. I., Bryant D. M., Role of membrane traffic in the generation of epithelial cell asymmetry. Nat. Cell Biol. 14, 1235–1243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toya M., et al. , CAMSAP3 orients the apical-to-basal polarity of microtubule arrays in epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 113, 332–337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugioka K., Sawa H., Formation and functions of asymmetric microtubule organization in polarized cells. Curr. Opin. Cell Biol. 24, 517–525 (2012). [DOI] [PubMed] [Google Scholar]
- 12.van Bergeijk P., Hoogenraad C. C., Kapitein L. C., Right time, right place: Probing the functions of organelle positioning. Trends Cell Biol. 26, 121–134 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Mishra R., Grzybek M., Niki T., Hirashima M., Simons K., Galectin-9 trafficking regulates apical-basal polarity in Madin-Darby canine kidney epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 107, 17633–17638 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravichandran Y., Goud B., Manneville J. B., The Golgi apparatus and cell polarity: Roles of the cytoskeleton, the Golgi matrix, and Golgi membranes. Curr. Opin. Cell Biol. 62, 104–113 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Grindstaff K. K., Bacallao R. L., Nelson W. J., Apiconuclear organization of microtubules does not specify protein delivery from the trans-Golgi network to different membrane domains in polarized epithelial cells. Mol. Biol. Cell 9, 685–699 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sepich D. S., Solnica-Krezel L., Intracellular Golgi complex organization reveals tissue specific polarity during zebrafish embryogenesis. Dev. Dyn. 245, 678–691 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W., Gulvady A. C., Goreczny G. J., Olson E. C., Turner C. E., Paxillin-dependent regulation of apical-basal polarity in mammary gland morphogenesis. Development 146, dev174367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terasaki M., Dynamics of the endoplasmic reticulum and Golgi apparatus during early sea urchin development. Mol. Biol. Cell 11, 897–914 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin M., Veloso A., Wu J., Katrukha E. A., Akhmanova A., Control of endothelial cell polarity and sprouting angiogenesis by non-centrosomal microtubules. eLife 7, 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bisel B., et al. , ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65. J. Cell Biol. 182, 837–843 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yadav S., Puri S., Linstedt A. D., A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol. Biol. Cell 20, 1728–1736 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller P. M., et al. , Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat. Cell Biol. 11, 1069–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vinogradova T., Miller P. M., Kaverina I., Microtubule network asymmetry in motile cells: Role of Golgi-derived array. Cell Cycle 8, 2168–2174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sütterlin C., Colanzi A., The Golgi and the centrosome: Building a functional partnership. J. Cell Biol. 188, 621–628 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J., et al. , Molecular pathway of microtubule organization at the Golgi apparatus. Dev. Cell 39, 44–60 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Tanaka N., Meng W., Nagae S., Takeichi M., Nezha/CAMSAP3 and CAMSAP2 cooperate in epithelial-specific organization of noncentrosomal microtubules. Proc. Natl. Acad. Sci. U.S.A. 109, 20029–20034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guet D., et al. , Mechanical role of actin dynamics in the rheology of the Golgi complex and in Golgi-associated trafficking events. Curr. Biol. 24, 1700–1711 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Gurel P. S., Hatch A. L., Higgs H. N., Connecting the cytoskeleton to the endoplasmic reticulum and Golgi. Curr. Biol. 24, R660–R672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dippold H. C., et al. , GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell 139, 337–351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott K. L., et al. , GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature 459, 1085–1090 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haynes J., Srivastava J., Madson N., Wittmann T., Barber D. L., Dynamic actin remodeling during epithelial-mesenchymal transition depends on increased moesin expression. Mol. Biol. Cell 22, 4750–4764 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das T., et al. , A molecular mechanotransduction pathway regulates collective migration of epithelial cells. Nat. Cell Biol. 17, 276–287 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Alzhanova D., Hruby D. E., A trans-Golgi network resident protein, golgin-97, accumulates in viral factories and incorporates into virions during poxvirus infection. J. Virol. 80, 11520–11527 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu S. K., et al. , Cortical F-actin stabilization generates apical-lateral patterns of junctional contractility that integrate cells into epithelia. Nat. Cell Biol. 16, 167–178 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Baas A. F., et al. , Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell 116, 457–466 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Carlton J. G., Jones H., Eggert U. S., Membrane and organelle dynamics during cell division. Nat. Rev. Mol. Cell Biol. 21, 151–166 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Sütterlin C., Hsu P., Mallabiabarrena A., Malhotra V., Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell 109, 359–369 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Tang D., Yuan H., Vielemeyer O., Perez F., Wang Y., Sequential phosphorylation of GRASP65 during mitotic Golgi disassembly. Biol. Open 1, 1204–1214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang Y., Wang Y., GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J. Cell Biol. 188, 237–251 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hidalgo Carcedo C., et al. , Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science 305, 93–96 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Liberali P., et al. , The closure of Pak1-dependent macropinosomes requires the phosphorylation of CtBP1/BARS. EMBO J. 27, 970–981 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonazzi M., et al. , CtBP3/BARS drives membrane fission in dynamin-independent transport pathways. Nat. Cell Biol. 7, 570–580 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Valente C., et al. , A 14-3-3γ dimer-based scaffold bridges CtBP1-S/BARS to PI(4)KIIIβ to regulate post-Golgi carrier formation. Nat. Cell Biol. 14, 343–354 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Gambarotto D., Hamel V., Guichard P., Ultrastructure expansion microscopy (U-ExM). Methods Cell Biol. 161, 57–81 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Hao H., et al. , Golgi-associated microtubules are fast cargo tracks and required for persistent cell migration. EMBO Rep. 21, e48385 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aoki T., et al. , Intraflagellar transport 20 promotes collective cancer cell invasion by regulating polarized organization of Golgi-associated microtubules. Cancer Sci. 110, 1306–1316 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rivero S., Cardenas J., Bornens M., Rios R. M., Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 28, 1016–1028 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hurtado L., et al. , Disconnecting the Golgi ribbon from the centrosome prevents directional cell migration and ciliogenesis. J. Cell Biol. 193, 917–933 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zilberman Y., et al. , Involvement of the Rho-mDia1 pathway in the regulation of Golgi complex architecture and dynamics. Mol. Biol. Cell 22, 2900–2911 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang D., et al. , Mena-GRASP65 interaction couples actin polymerization to Golgi ribbon linking. Mol. Biol. Cell 27, 137–152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buschman M. D., Field S. J., MYO18A: An unusual myosin. Adv. Biol. Regul. 67, 84–92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lukinavičius G., et al. , Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat. Methods 11, 731–733 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Ahat E., Li J., Wang Y., New insights into the Golgi stacking proteins. Front. Cell Dev. Biol. 7, 131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Magdalena J., et al. , Involvement of the Arp2/3 complex and Scar2 in Golgi polarity in scratch wound models. Mol. Biol. Cell 14, 670–684 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Minin A. A., Dispersal of Golgi apparatus in nocodazole-treated fibroblasts is a kinesin-driven process. J. Cell Sci. 110, 2495–2505 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Chabin-Brion K., et al. , The Golgi complex is a microtubule-organizing organelle. Mol. Biol. Cell 12, 2047–2060 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Efimov A., et al. , Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell 12, 917–930 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xing M., et al. , GOLPH3 drives cell migration by promoting Golgi reorientation and directional trafficking to the leading edge. Mol. Biol. Cell 27, 3828–3840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gurzu S., Ciortea D., Ember I., Jung I., The possible role of Mena protein and its splicing-derived variants in embryogenesis, carcinogenesis, and tumor invasion: A systematic review of the literature. BioMed Res. Int. 2013, 365192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Di Modugno F., et al. , Splicing program of human MENA produces a previously undescribed isoform associated with invasive, mesenchymal-like breast tumors. Proc. Natl. Acad. Sci. U.S.A. 109, 19280–19285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.