Abstract
We have constructed a genetic linkage map of the edible basidiomycete Pleurotus ostreatus (var. Florida). The map is based on the segregation of 178 random amplified polymorphic DNA and 23 restriction fragment length polymorphism markers; four hydrophobin, two laccase, and two manganese peroxidase genes; both mating type loci; one isozyme locus (est1); the rRNA gene sequence; and a repetitive DNA sequence in a population of 80 sibling monokaryons. The map identifies 11 linkage groups corresponding to the chromosomes of P. ostreatus, and it has a total length of 1,000.7 centimorgans (cM) with an average of 35.1 kbp/cM. The map shows a high correlation (0.76) between physical and genetic chromosome sizes. The number of crossovers observed per chromosome per individual cell is 0.89. This map covers nearly the whole genome of P. ostreatus.
Pleurotus ostreatus (oyster mushroom) is an edible mushroom that occupies the second most important position in the world mushroom market, led by the button mushroom Agaricus bisporus (5, 49). Besides its importance for food production, P. ostreatus is interesting for applications such as paper pulp bleaching, cosmetics, and the pharmaceutical industry. These different applications have fueled research on specific aspects of Pleurotus biochemistry and molecular biology (4, 8, 20, 31–34, 42).
Despite its economic importance, only a limited number of genetic studies of P. ostreatus have been done because of the difficulty in performing directed crosses between strains, contradictory data about the size and organization of its genetic material, and the lack of a genetic linkage map for it. Moreover, breeding of new P. ostreatus strains with a higher agricultural or industrial value has been traditionally carried out by trial and error because of the aforementioned reasons (2).
In order to facilitate the design of programs aimed to improve the strains currently available, it is important to increase our knowledge of the genome organization of this fungus. However, the study of the organization of the P. ostreatus genome has been hampered by the small size of fungal chromosomes and by the occurrence of intranuclear mitosis (15). Different authors have reported different chromosome numbers for this species (15, 41, 51), and only recently has this number been determined using pulsed-field gel electrophoresis (32). This species contains 11 chromosomes that account for a total genomic size of about 35.1 Mbp per haploid genome. Furthermore, chromosome length polymorphisms occur between the homologous chromosomes present in each of the two nuclei in the dikaryon. Electrophoretic separation of P. ostreatus chromosomes allowed the physical mapping of some genes or phenotypic markers on specific chromosomes (for instance, the A mating locus physically mapped on chromosome III and the B locus was on chromosome IX) (32).
The use of molecular markers combined with the construction of linkage maps is a potent strategy for designing breeding strategies and for attempting positional cloning of genes of interest. Linkage maps are available for some filamentous fungi such as Bremia lactucae (24), Cochliobolus heterostrophus (58), Aspergillus niger (16), Cladosporium fulvum (3), Magnaporthe grisea (54), Phytophthora sojae (62), and Fusarium moniliforme (Gibberella fujikuroi) (64). Among basidiomycetes, however, genetic linkage maps are scarce: a restriction fragment length polymorphism (RFLP)-based map of the white rot fungus Phanerochaete chrysosporium strain ME446 (44) and two linkage maps of the button mushroom A. bisporus have been published (13, 28). The linkage relationship between 19 allozyme-encoding loci in P. ostreatus was described by May et al. (35).
Random amplification of polymorphic DNA (RAPD) is a PCR-based strategy for the generation of molecular markers (RAPD markers) (63) suitable for the construction of linkage maps. RAPD markers show a dominant genetic behavior and are prone to be highly influenced by the reaction's environmental conditions. To overcome these difficulties, they can be converted into more robust markers, such as RFLPs, which uncover the occurrence of heterozygotes in a segregating population. The bulked segregant analysis strategy (37) can be used as a complementary tool because it allows the generation of RAPD markers genetically linked to characters of interest. This approach has allowed the identification of markers linked to the A and B mating factors of P. ostreatus and the study of their flanking regions (31, 45).
In this paper, we present a genetic linkage map of the edible and white rot fungus P. ostreatus based on RAPD and RFLP markers, phenotypic characters, and cloned genes. The number of genetic linkage groups obtained agrees with the number of chromosomes described by our group in this fungus (32). This is, to our knowledge, the first linkage map constructed for this species.
MATERIALS AND METHODS
Fungal strain and culture conditions.
P. ostreatus strain N001 has been previously described (31, 32, 42) and corresponds to the commercial variety Florida. The two nuclei present in it have been previously separated by de-dikaryotization (32), and the two corresponding protoclones (monokaryons carrying only one of the nuclei present in dikaryon N001) are in the Spanish Type Culture Collection (PC9 [CECT20311] and PC15 [CECT20312]). Culture techniques were performed as previously described by Larraya et al. (31).
Molecular techniques.
For the generation of RAPD markers, 10-mer oligonucleotides belonging to the L, P, R, and S Operon series (Operon Technologies Inc., Alameda, Calif.) were used as primers. Amplification reactions were carried out in a PTC-200 (Peltier Thermal Cycler; MJ Research, Watertown, Mass.) using the following program: 4 min of denaturation at 94°C, followed by 39 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 37°C, and 1.5 min of extension at 72°C. After the 39 cycles, an additional extension step of 2 min at 72°C was performed. A total of 80 different oligonucleotides were initially evaluated as primers for the generation of RAPD markers using (as the template) genomic DNA purified from dikaryon N001 and from 10 monokaryons belonging to the mapping population, and a set of 59 was finally used to carry out the RAPD reactions with the 80 members of the mapping population because they provided good-quality, reproducible polymorphic profiles. Faint bands were discarded even though they showed polymorphism. For the RFLP analysis, different restriction enzymes were used to digest genomic DNA (see Table 1). In order to identify the enzymes yielding polymorphic restriction patterns, DNAs purified from the dikaryon and from protoclones PC9 and PC15 were digested with the restriction enzymes and probed with the corresponding digoxigenin-labeled probe (Boehringer Mannheim, Mannheim, Germany) (23). Only those enzyme-probe combinations detecting polymorphisms were used for segregation analysis of the population of 80 monokaryons. Isolation of genomic DNA suitable for chromosome separation by pulsed-field gel electrophoresis (PFGE) was performed as described by Sonnenberg et al. (55). Preparation of protoplasts of N001, PC9, and PC15; PFGE conditions appropriate for the separation of P. ostreatus chromosomes; and blotting of PFGE-separated chromosomes onto the appropriate membranes were performed as previously described by Larraya et al. (32). Other molecular protocols were performed as described elsewhere (31, 52).
TABLE 1.
Summary of the genetic linkage map of P. ostreatus
| Marker type | Primera | Marker size (bp) or nameb (linkage group)c | Enzymed | Copy no.e | Chromosomef |
|---|---|---|---|---|---|
| RAPD | L1 | 1,075 (VI) | XhoI | Single | VI |
| 2,600 (X) | |||||
| RAPD | L2 | 1,850 (IV) | XhoI | Single | IV |
| 2,475 (XI) | |||||
| RAPD | L3 | 1,350 (IX) | PstI | Single | IX |
| RAPD | L4 | 2,400 (I) | |||
| RAPD | L5 | 875 (I) | Multiple | ||
| 1,475 (X) | XhoI | Single | X | ||
| 2,525 (IV) | |||||
| RAPD | L6 | 1,800 (IX) | PstI | Single | |
| RAPD | L7 | 825 (VII), 900 (I), 2,025 (I) | |||
| RAPD | L8 | 575 (IV) | Any oneg | Single | IV |
| RAPD | L10 | 2,175 (III), 2,925 (III) | |||
| RAPD | L11 | 1,325 (V), 1,450 (I) | |||
| RAPD | L13 | 2,575 (I) | |||
| RAPD | L14 | 1,525 (IX) | Multiple | ||
| 1,800 (II), 2,375 (III) | |||||
| RAPD | L15 | 625 (V) | XhoI | Single | V |
| 1,125 (I) | Multiple | ||||
| 1,275 (XI) | Multiple | ||||
| 1,525 (VI) | |||||
| RAPD | L16 | 875 (VII) | EcoRI | Single | VII |
| 1,100 (I) | PstI | Single | I | ||
| RAPD | L18 | 2,175 (I) | EcoRI | Single | I |
| 2,900 (II) | |||||
| RAPD | L19 | 1,750 (VII), 2,525 (IV) | |||
| RAPD | P1 | 750 (I), 1,600 (IX), 1,825 (VII) | |||
| RAPD | P2 | 725 (IX) | XhoI | Single | |
| 1,150 (IV), 2,100 (IX), 2,650 (IX) | |||||
| RAPD | P3 | 1,375 (IX), 1,550 (V) | |||
| RAPD | P4 | 600 (VI), 1,450 (III), 2,550 (VII) | |||
| RAPD | P5 | 1,225 (III) | |||
| RAPD | P6 | 950 (VI), 1,100 (X), 1,350 (N/A), 2,300 (XI) | |||
| RAPD | P7 | 1,275 (N/A), 1,525 (I) | |||
| RAPD | P8 | 1,875 (IV), 2,775 (X) | |||
| RAPD | P9 | 300 (N/A), 400 (VII), 875 (IV), 950 (II), 1,400 (VII), 1,500 (VII) | |||
| RAPD | P10 | 2,325 (VIII) | |||
| RAPD | P11 | 800 (V), 850 (III), 950 (IV) | |||
| RAPD | P12 | 950 (IX) | PstI | Single | IX |
| 1,350 (I) | |||||
| 1,525 (III) | Multiple | ||||
| 2,025 (II), 2,150 (VII), 3,525 (III) | |||||
| RAPD | P13 | 675 (IV), 1,250 (VI), 1,550 (V) | |||
| 1,625 (II) | |||||
| RAPD | P14 | 650 (VI) | |||
| RAPD | P15 | 950 (III), 1,050 (I), 1,225 (VIII) | |||
| RAPD | P16 | 400 (X), 1,525 (IX), 2,175 (II) | |||
| RAPD | P17 | 425 (II), 1,325 (VII), 1,425 (III) | |||
| RAPD | P19 | 525 (IX), 550 (III), 950 (II), 1,125 (II), 1,425 (VIII), 2,000 (VII), 2,100 (VI) | |||
| RAPD | R1 | 775 (I), 1,475 (VI) | |||
| RAPD | R2 | 1,600 (IX), 1,675 (V) | |||
| RAPD | R3 | 850 (III) | PstI | Single | |
| 925 (VI), 975 (III), 2,050 (VIII), 2,275 (IX), 2,475 (II), 3,625 (III) | |||||
| RAPD | R4 | 425 (VII), 450 (N/A), 925 (III), 1,175 (X), 2,425 (VIII), 3,150 (N/A) | |||
| RAPD | R6 | 400 (I), 950 (VI), 1,525 (II), 1,550 (II) | |||
| RAPD | R7 | 1,450 (X), 2,225 (IV), 2,400 (III), 2,700 (VII) | |||
| RAPD | R8 | 300 (VIII) | EcoRI | Single | VIII |
| 575 (III), 775 (VI), 1,475 (VI) | |||||
| RAPD | R9 | 1,300 (IV), 2,450 (X) | |||
| RAPD | R10 | 1,375 (V), 2,100 (VIII), 2,225 (VIII) | |||
| RAPD | R11 | 575 (VIII) | Multiple | ||
| 675 (III), 775 (I), 1,875 (VI) | |||||
| RAPD | R12 | 400 (I), 925 (VI), 1,475 (II), 1,500 (II) | |||
| RAPD | R13 | 525 (VII), 1,425 (IV), 2,175 (II) | |||
| RAPD | R14 | 700 (XI), 1,750 (VII) | |||
| RAPD | R15 | 575 (XI), 675 (IX), 1,025 (I), 1,200 (V), 1,750 (V), 3,100 (V) | |||
| RAPD | R16 | 775 (III), 975 (N/A), 1,025 (N/A), 1,200 (XI), 1,250 (II) | |||
| RAPD | R17 | 850 (VI), 1,500 (III) | |||
| RAPD | R19 | 1,300 (VII), 2,100 (III), 3,300 (XI) | |||
| RAPD | R20 | 650 (VIII) | EcoRI | Single | VIII |
| 1,400 (II), 1,500 (III), 1,950 (VIII), 2,700 (II) | |||||
| RAPD | S7 | 400 (VI) | |||
| 1,200 (V) | PstI | Single | V | ||
| 1,725 (II) | |||||
| RAPD | S11 | 975 (III) | PstI | Single | III |
| 2,325 (VIII) | |||||
| RAPD | S12 | 800 (XI), 1,550 (VIII), 2,075 (I), 2,425 (IV) | |||
| RAPD | S16 | 1,250 (X) | |||
| 1,275 (X) | Multiple | ||||
| RAPD | S17 | 775 (I), 825 (X), 2,375 (I) | |||
| RAPD | S18 | 1,175 (I) | |||
| 1,275 (III) | SmaI | Single | III | ||
| 1,925 (VII) | |||||
| RAPD | S19 | 1,250 (VII) | XhoI | Single | VII |
| 1,475 (VI), 2,600 (V) | |||||
| RFLP | O3 (X) | PstI | Single | X | |
| RFLP | O17 (II) | PstI | Single | ||
| RFLP | R3 (II) | PstI | Single | II | |
| RFLP | Hon2 (VI) | EcoRI | Single | ||
| RFLP | Rib (II) | XhoI | Single | II | |
| RFLP | fbh1 (XI) | EcoRI | Single | XI | |
| RFLP | vmh1 (I) | XhoI | Single | I | |
| RFLP | vmh2 (XI) | XhoI | Single | XI | |
| RFLP | vmh3 (X) | EcoRI | Single | X | |
| RFLP | pox1 (VI) | XhoI | Single | VI | |
| RFLP | poxC (VI) | XhoI | Single | VI | |
| RFLP | mnp1 (IV) | PstI | Single | IV | |
| RFLP | mnp3 (V) | HindIII | Single | V | |
| Mating | matA (III) | ||||
| Mating | matBα (IX) | ||||
| Mating | matBβ (IX) | ||||
| Isozyme | est1 (VIII) | ||||
| Repetitive DNA | rXhoI (II) | XhoI |
Operon oligonucleotide used as primer. RAPD markers are named after the primer oligonucleotide with the marker size as a subscript.
RAPD marker sizes are shown. RAPD markers transformed into RFLP markers are underlined. The names of RFLP, mating, isozyme, and repetitive DNA markers are those of the corresponding genes.
Indicates the linkage group where each marker maps. N/A, nonassigned markers.
Restriction enzyme used to reveal the RFLP markers used for linkage mapping.
Copy number of the corresponding RFLP marker. Only single-copy markers were used to construct the linkage map.
Physical mapping of the RFLP marker, confirmed by hybridization on PFGE, resolved the chromosomes of P. ostreatus N001.
Hemizygotic marker.
Markers used for construction of the linkage map. (i) DNA-based markers.
Four different types of DNA-based molecular markers were used as input data: RAPD markers, selected RAPD markers converted into RFLP markers, other cloned DNA sequences used as RFLP markers, and a repetitive DNA sequence. The DNA sequences used as RFLP markers included anonymous sequences from P. ostreatus strain N001 (markers R3 and Hon2) and from Somycel strain 3200 (O3 and O17, kindly provided by the Mushroom Experimental Station, Horst, The Netherlands), a sequence corresponding to a portion of the rRNA gene (rDNA) from Saccharomyces carlsbergensis (probe Rib) (60), and eight coding genes (four hydrophobin [42], two laccase [19, 20], and two manganese peroxidase [G. Sannia, unpublished data] genes). An additional DNA-based marker (rXhoI) suitable for mapping was identified when the genomic DNA was fully digested with restriction enzyme XhoI; it appeared as a pair of repetitive DNA bands showing different sizes and an allelic behavior.
(ii) Isozyme markers.
Several isozyme systems were studied for the identification of segregating alleles suitable as entries in the construction of the linkage map. Most of them (peroxidase [E.C.1.11.1.7], alcohol dehydrogenase [E.C.1.1.1.1], malate dehydrogenase [E.C.1.1.1.37], isocitrate dehydrogenase [E.C.1.1.1.42], 6-phosphogluconate dehydrogenase [E.C.1.1.1.44], and aspartate aminotransferase [E.C.2.6.1.1]) showed no polymorphism, and only one (esterase [E.C.3.1.1.2]) revealed polymorphism and was finally used. Total protein fractions were prepared as described by Roux and Labarère (48). Protein fractionation was performed using nondenaturing polyacrylamide gel electrophoresis, and the different enzymatic systems were developed using specific staining protocols described elsewhere (53, 59).
(iii) Phenotypic markers (mating factors).
The mating type of each one of the monokaryons forming the mapping population was determined using four mating testers specific for P. ostreatus N001 as previously described (31).
Data analysis and linkage mapping.
The mapping population consisted of a haploid progeny of 80 monokaryons derived from P. ostreatus N001 spores. These monokaryons correspond to single spore colonies developed during 5 days after the spores were placed under germinating conditions. The status of each one of the markers (molecular, isozyme, and phenotypic) used to construct the map was scored in each one of the members of the mapping population. The monokaryotic (haploid) nature of the members of this population allows the application of a backcross model for handling data. This is especially relevant considering the dominant behavior of the RAPD markers that, under these conditions, can be used directly for mapping. Analysis of linkage between markers, estimation of recombination frequencies, determination of the linear order of loci, including multipoint linkage analysis and the expectation maximization algorithm used for handling missing data, were performed as described by Ritter et al. (46) and by Ritter and Salamini (47) using the MAPRF program (22).
The mean likelihood odds ratio per marker interval was 17.2. In a first step of the linkage grouping, a maximum recombination frequency of 20% between any two or more markers was used as the threshold for the establishment of linkage groups. This value corresponds to a likelihood odds ratio of 6.7. In a second step, linkage subgroups were connected and far distant markers were assigned to linkage groups as described by Ritter and Salamini (47). In these last cases, linkage always was determined with an α error smaller than 5% to at least one of the markers present in an already existing χ2-based linkage group. The linkage groups were numbered in accordance with the numbers previously assigned to the chromosomes resolved by contour-clamped homogeneous electric field analysis (32). Chromosomes were numbered in accordance with their molecular sizes in protoclone PC9, from the largest (chromosome I) to the smallest (chromosome XI).
RESULTS
Polymorphism analysis.
The genetic polymorphisms detected by the different markers in the P. ostreatus mapping population are summarized in Table 1. When 59 selected oligonucleotide primers (see Materials and Methods) were used, an average of 12.1 RAPD bands per primer were amplified and about 25% of them displayed segregation in the mapping population. In a second selection step for mapping-suitable markers, between one and seven (average of three) RAPD bands per primer were finally used for the mapping analysis. This produced a total of 178 RAPD markers for linkage analysis. Most of them displayed presence-absence polymorphism in the progeny, although 12 codominant markers were also detected (L102175 and L102925, P91400 and P91500, R61525 and R61550, R102100 and R10225, R121475 and R121500, and S161250 and S161275).
Twenty-six RAPD markers preliminarily placed in different linkage groups were cloned to be used as probes to detect RFLP polymorphisms associated with them and to determine the correlation between the linkage map and the P. ostreatus molecular karyotype by hybridization on PFGE-separated chromosomes (Table 1). All of the DNA probes revealed polymorphisms with at least one of the restriction enzymes used. Out of the 26 probes tested, 7 highlighted high-copy-number DNA sequences and were discarded as RFLP markers for linkage analysis (Table 1). These probes hybridized to several P. ostreatus PFGE-resolved chromosomes (data not shown). Restriction polymorphisms were also detected when probes corresponding to cloned genes were used on DNA digested with the enzymes indicated above. In every case, the probes detected single-copy DNA sequences and some of the probes were also used for hybridization on chromosomes separated by PFGE (Table 1).
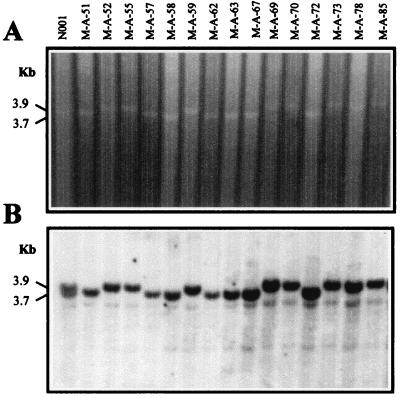
When the products of a complete digestion of genomic DNA with some restriction enzymes are separated by electrophoresis, a pattern of stronger DNA bands can be distinguished against the background corresponding to the digestion products in the electrophoresis lane. These stronger bands correspond to the high concentration of specific restriction fragments produced by digestion of DNA sequences present in high copy number in the genome. When P. ostreatus genomic DNA purified from dikaryon N001 and from the monokaryons of the mapping population were digested with restriction enzyme XhoI, it was observed that two stronger bands present in dikaryon N001 behaved as alleles in the mapping population (Fig. 1). These bands were considered an additional marker (marker rXhoI) and were used as input in the linkage analysis (Table 1).
FIG. 1.
(A) XhoI digestion of DNA purified from dikaryotic strain N001 and some monokaryons of the mapping population. Two polymorphic bands (3.7 and 3.9 kbp) of repetitive DNA segregating as alleles can be observed (rXhoI marker). (B) RFLP pattern obtained using a portion of the rDNA of S. carlsbergensis as a probe for Southern hybridization of the digested DNA shown in panel A. Two polymorphic bands segregating as alleles can be observed (Rib marker). These bands are coincident with the two bands of repetitive DNA in panel A.
The mating type genes of each one of the monokaryons were determined by crossings with the corresponding mating testers. For the A mating type gene, two allelic forms were segregating in the population (A1 and A2), whereas for mating type B genes, four haplotypes were obtained (B1, B2, B3, and B4). Two of them (B1 and B2) corresponded to those present in dikaryon N001, and the other two (B3 and B4) appeared as a result of recombination between the two linked mating subloci (matBα and matBβ).
Linkage mapping.
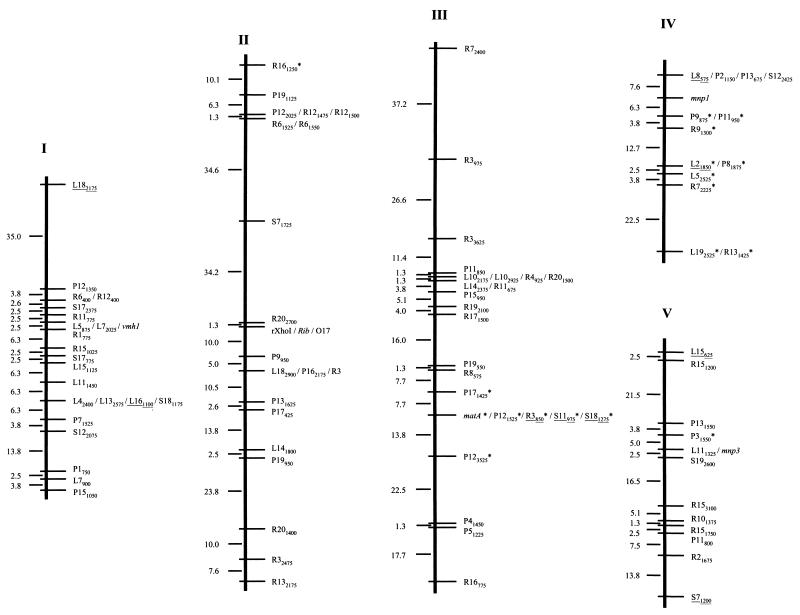
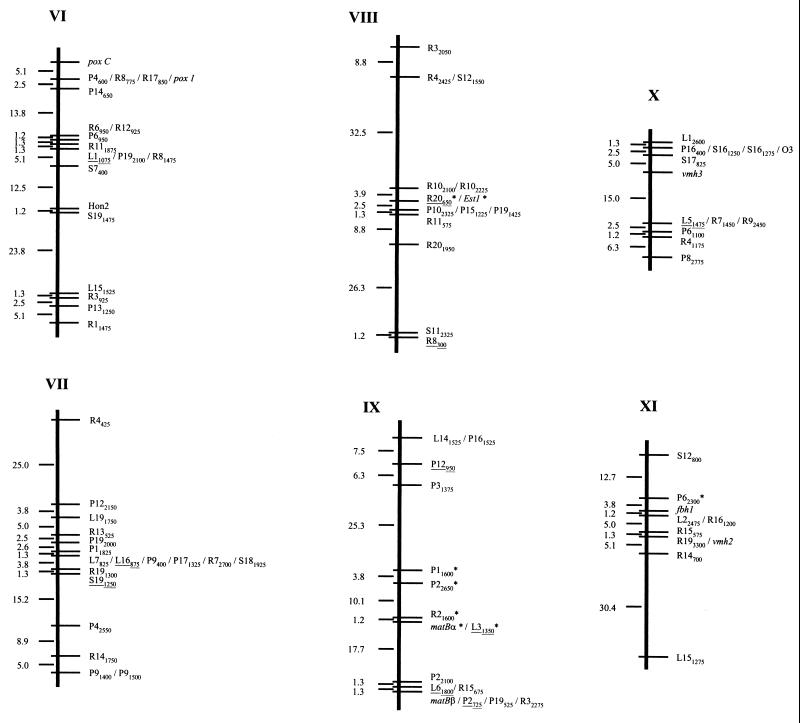
In order to carry out the linkage analysis of the 196 markers, the MAPRF (22) software was used as described by Ritter et al. (46) and by Ritter and Salamini (47). Only 7 out of the 196 markers described in the previous section could not be assigned to any linkage group (Table 1). The remaining 189 markers were assigned at a confidence level of at least 95% to 11 linkage groups (Table 1; Fig. 2) which span a total of 1,000.7 centimorgans (cM) (Kosambi units; 30) with an average marker distance between them of 5.3 cM. On average, 0.89 crossover event per chromosome per individual was found in the mapping population.
FIG. 2.
Linkage map of P. ostreatus. Linkage groups are numbered in accordance with the molecular size of the corresponding chromosome (in protoclone PC9) revealed by PFGE (32). Polymorphic RAPD markers were named by the primer designation, followed by the approximate size of the amplified fragment in base pairs. RAPD markers converted into RFLP markers are underlined. Genes are in italics. Markers that deviated from the expected 1:1 segregation (P < 0.05) appear with an asterisk to the right of the marker name. The linkage distances (centimorgans) between consecutive genetic markers are indicated on the left of each linkage group.
The characteristics of the linkage groups obtained are summarized in Table 2. Linkage group length varied between 33.8 (LG X) and 178.7 (LG III) cM, with an average of 91 cM per group. They clustered between 10 (LG XI) and 25 (LGIII) markers per group, with an average of 17.2 markers per linkage group. The average marker interval was also quite variable; a nearly threefold difference between the maximum (7.5 cM in LG III) and minimum (2.6 cM in LG X) intervals was observed. A total of 26 mapped markers exhibited distorted segregation ratios; 21 of them mapped in contiguous regions located on linkage groups III, IV, and IX.
TABLE 2.
Characteristics of the linkage and physical map of P. ostreatus
| Chromosome | Size in:
|
No. of markers | Size ratio (kbp/cM) | Avg marker interval (cM) | No. of crossover events | |
|---|---|---|---|---|---|---|
| Mbpa | cM | |||||
| I | 4.70 | 103.0 | 23 | 45.6 | 4.5 | 0.98 |
| II | 4.35 | 173.6 | 23 | 25.1 | 7.5 | 1.71 |
| III | 4.55 | 178.7 | 25 | 25.5 | 7.1 | 1.75 |
| IV | 3.55 | 59.2 | 14 | 60.0 | 4.2 | 0.59 |
| V | 3.45 | 82.0 | 13 | 42.1 | 6.3 | 0.81 |
| VI | 3.10 | 76.7 | 20 | 40.4 | 3.8 | 0.76 |
| VII | 3.15 | 74.4 | 18 | 42.3 | 4.1 | 0.74 |
| VIII | 2.95 | 85.3 | 14 | 34.6 | 6.1 | 0.84 |
| IX | 2.10 | 74.5 | 16 | 28.2 | 4.7 | 0.74 |
| X | 1.75 | 33.8 | 13 | 51.8 | 2.6 | 0.34 |
| XI | 1.45 | 59.5 | 10 | 24.4 | 5.9 | 0.59 |
| Avg | 3.19 | 91 | 17.2 | 35.1 | 5.3 | 0.89 |
| Total | 35.1 | 1,000.7 | 189 | |||
Physical size is the average of the two protoclones (32).
Map locations of the different markers used.
The characteristics and map locations of the markers are summarized in Table 1. RFLP markers derived from cloned RAPD markers cosegregated with the corresponding RAPD markers, with the exception of marker P2725, which showed an RFLP pattern different from that expected in 1 out of the 80 monokaryons of the mapping population. On the other hand, RAPD marker L8575, used as an RFLP probe, revealed hemizygosis for this locus. This RFLP marker was present in only protoclone PC9 and gave a hybridization signal in only one-half of the monokaryotic progeny of dikaryon N001. The DNA probe corresponding to marker Rib cosegregated with and had the same molecular size as marker rXhoI, which was identified as a high-copy DNA fragment evident upon digestion of P. ostreatus genomic DNA with XhoI. Both A and B incompatibility mating type genes were also mapped. They were located on different chromosomes. The A mating type locus was on linkage group III, and the B locus was on linkage group IX. Furthermore, the two subloci (matBα and matBβ) of the B locus were 19 cM apart, the matBβ sublocus being proximal to the closest telomere (Fig. 2).
Hybridization of selected RFLP probes on P. ostreatus PFGE-resolved chromosomes.
Most of the RFLP markers derived from RAPD markers and some of those corresponding to cloned genes were used as probes for Southern hybridization on P. ostreatus PFGE-separated chromosomes in order to determine their physical locations and to correlate the linkage map described here and the molecular karyotype of P. ostreatus (32). A total of 27 probes were used to assign unequivocally each linkage group to each one of the 11 chromosomes of this fungus (Table 1). In any case, two or three markers per linkage group were tested and no translocations were detected. In some cases, it was possible to assign one group's distal end to a chromosome (L182175 on chromosome I, L8575 on chromosome IV, poxC on chromosome VI, and R8300 on chromosome VIII) and, in some cases, RAPD markers corresponding to the linkage group's two ends were hybridized (markers L15626 and S71200 for linkage group V). Finally, molecular markers genetically linked to the mating factors were also physically mapped on PFGE-separated chromosomes.
DISCUSSION
In this paper, the construction of a linkage map based on the segregation analysis of a population of monokaryons is presented. The haploid nature of fungal monokaryons makes the analysis of segregation patterns similar to that obtained for the study of a backcross population in a diploid organism. The use of haploid populations as starting points in the construction of linkage maps has been previously reported for conifers (57) using megagametophytes and for the honeybee (Apis mellifera) (25) using drones as mapping populations. In the case of fungi, monokaryons have been occasionally used as tools for the mapping of genes of interest in P. chrysosporium (18).
The map described in this paper is based mainly on RAPD markers. Problems related to the reproducibility of this type of molecular marker, as well as the comigration of equally sized DNA fragments, have been frequently reported in the literature (10). However, the RAPD method has been proven to be simple, fast, and reliable for the identification of polymorphisms in many organisms and for the construction of linkage maps (7, 9, 12, 14, 21). Furthermore, it was found that error rates were similar when mapping was based on RAPD and on simple sequence repeats as markers (25). The reliability of RAPD segregation has been tested by different authors who converted the resulting markers into more stable and usually codominant markers such as SCAR (40) or RFLP (11). Some RAPD markers were used as probes for RFLP analysis in the construction of a P. ostreatus linkage map, and the corresponding RFLP markers cosegregated, in all cases but one, with the RAPD markers they proceeded from. The exception was marker P2725, which cosegregated with the corresponding RAPD marker, keeping the coupling phase in all but one of the monokaryons in the segregating population. This result can be explained by the occurrence of a crossover event between the DNA region amplified in the RAPD marker and one of the restriction sites flanking the RFLP marker in this specific monokaryon.
The level of polymorphism, as estimated by the average number of segregating markers per RAPD primer, was 3.0 in P. ostreatus. This value is higher than those found in other organisms, such as the honeybee A. mellifera (polymorphism level, 2.8) (25) and Tribolium castaneum (polymorphism level, 1.5) (7), whose linkage maps were based on markers of this type. Kerrigan et al. (28) reported a mean number of useful polymorphisms per primer of 4.2 in a linkage map of the button mushroom A. bisporus, based partially on RAPD markers. In order to compare this value with the polymorphism reported here for P. ostreatus, it should be taken into account that most of the oligonucleotide primers used by Kerrigan et al. did not reveal polymorphisms (23 out of 28) and only a limited number of them (5 out of 28) generated useful polymorphic RAPD markers.
Most of the markers used in the construction of this map showed normal segregation. However, distorted segregation was observed in 14% of them (markers labeled with an asterisk in Fig. 2). This percentage is similar to that found in the pathogenic fungus C. heterostrophus (16.4%) (58), the melon Cucumis melo (14.5%) (61), or the rainbow trout Oncorhynchus mykiss (13.3%) (65); higher than the value found in the lettuce plant Lactuca sativa (9.0%) (29); and considerably lower than that found in the button mushroom A. bisporus (32.8%) (28). Skewed markers mapped primarily to chromosomes III, IV, and IX, and the most prominent distorted group was that mapping to chromosome IV, where all of the markers but those mapping to one extreme of the linkage group showed skewed segregation. It is noteworthy that chromosome IV shows, on average, less than one crossover event per chromosome (Table 2), and this fact can be related to the distortion observed (see below). Distortions associated with linkage groups III and IX, on the other hand, have special interest because A and B mating type genes map to these two chromosomes, respectively. In order to explain the distorted segregation, the following hypotheses can be put forward: (i) the nonrandom segregation of mating type genes can drive a skewed segregation of the markers linked to them; (ii) differences in viability, germination, or vegetative growth rate associated with different mating haplotypes may have caused preferred selection of some genotypes when the mapping population was established; and (iii) balancing selection on mating types can at least transiently counteract some negative selection on loci linked to the mating type (while the disequilibrium persists). The effect of negative selection against slowly germinating spores has been previously discussed by Eger (17) in P. ostreatus var. Florida, by Kerrigan et al. (28) in A. bisporus, and by Mitchell-Olds (38) in Arabidopsis thaliana.
Although recombination rates would vary in different crosses, we can estimate the average ratios of physical to genetic distances in P. ostreatus. The importance of these data resides in the possibility of undertaking map-based strategies for the cloning of genes in this species. The haploid genome size of P. ostreatus has been estimated to be 35.1 Mbp (32), and the total recombination size estimated in this study is 1,000.7 cM. The ratio of these two measurements is 35.1 kbp/cM, with values ranging from 24.4 to 60 kbp/cM (chromosomes XI and IV, respectively; Table 2). The average ratio is similar to that found in the filamentous fungus F. moniliforme (32 kbp/cM) (64), although higher (B. lactucae, 25 kbp/cM [24]; C. heterostrophus, 23 kbp/cM [58]) and lower (P. chrysosporium, 59 kbp/cM [44]) ratios were found. We have used the procedure described by Hunt and Page (25) to estimate the theoretical number of crossovers per chromosome as the result of the ratio of the genome's total linkage size to the product of the chromosome number multiplied by a constant factor of 50 cM. According to this estimation, an average of 1.8 crossovers per chromosome would be expected in P. ostreatus. We have found, however, that the actual average number of crossovers (calculated by the MAPRF program), 0.89, is much lower in our linkage analysis (range, 0.34 [chromosome X] to 1.75 [chromosome III]) (Table 2). This value agrees with the 0.96 previously reported for F. moniliforme (64) but is higher than that found by Kerrigan et al. (28) in A. bisporus. Control of the number of crossovers per chromosome has been studied in different fungi, and in some cases, the existence of a mechanism by which small chromosomes undergo reciprocal recombination at rates (expressed in centimorgans per kilobase pair) higher than those of large chromosomes has been described (27). Our data do not support this type of control in P. ostreatus. Control of the number of crossovers per chromosome has been considered a method by which to ensure the occurrence of at least one crossover event per chromosome per cell, and this event is required for proper chromosome disjunction during meiosis (6). The data presented in Table 2 indicate that basidia in which less than one crossover per chromosome occurs are frequent, and how this fact affects the accuracy of meiotic product sorting has not been studied in P. ostreatus up to now.
The amount of repetitive DNA present in the P. ostreatus genome has not been accurately estimated. The short oligonucleotide primers used in RAPD analysis have a general tendency to amplify segments of repetitive DNA because palindromic sequences are more highly represented in such regions (63). Therefore, Kesseli et al. (29), Antolin et al. (1), and others have observed a nonrandom clustering of RAPD loci amplified by the same primer. Most of the RAPD markers used in this mapping corresponded to nonrepetitive DNA sequences. Out of 26 randomly selected RAPD markers converted into RFLP markers, 19 behaved as single-copy sequences. This suggests that the amount of repetitive DNA in the P. ostreatus genome should be small. An interesting polymorphism based on repetitive DNA was found when the restriction enzyme XhoI was used to digest total genomic DNA: marker rXhoI cosegregated with, and had the same size as, RFLP marker Rib, which corresponds to a highly conserved sequence coding for an rDNA fragment of S. carlsbergensis. Taking into account the high evolutionary conservation of these sequences, we believe that probe Rib highlights an rDNA region in the genome of P. ostreatus. Markers Rib and rXhoI mapped to chromosome II, and this chromosome showed 15% length polymorphism when the two homologous chromosomes present in protoclones PC9 and PC15 were compared (32). Length polymorphisms in chromosomes carrying rDNA genes have also been reported in other organisms, such as Ustilago hordei (36), Cladosporium fulvum (56), Candida albicans (26, 50), Leptosphaeria maculans (39), S. cerevisiae (50), and A. bisporus (55), and they have been associated with differences in rDNA copy number (43, 55). The difference in size between the two bands of repetitive DNA revealed by marker rXhoI is approximately 200 bp (Fig. 1), and the total length difference in chromosome II between PC9 and PC15 is 0.7 Mbp. In this context, 3,500 copies of this repetitive sequence would account for the chromosome size difference observed. This copy number is too high for rDNA genes, and consequently, size differences in marker rXhoI can be only partially responsible for the chromosome size difference.
The linkage map presented here shows a good correlation with the molecular karyotype of P. ostreatus (32). Different RAPD probes converted into RFLP probes hybridized in the corresponding chromosomes, indicating that no translocation events have occurred. The high correlation (r = 0.76) between physical size (megabase pairs) and recombinational size (centimorgans), an even higher correlation (r = 0.81) between physical size and the number of markers per chromosome, and the reduced number of unassigned markers suggest that this linkage map covers nearly the whole genome of P. ostreatus.
The availability of a genetic linkage map for P. ostreatus opens the possibility of addressing basic and applied questions such as those related to the syntheny of markers between P. ostreatus strains and between different species, analysis of the molecular basis for the chromosome length polymorphisms that exist and their fate through the meiotic cycle (66), mapping of quantitative trait loci and study of the genetic control of these polygenic characters. Moreover, mapping of quantitative trait loci would help in marker-assisted selection of economically important aspects, such as growth rate; different components of yield, such as number of flushes; number and average weight of mushrooms; and tolerance of pathogens, among others. The relatively small genome of P. ostreatus and the possibilities of classical genetic manipulation of this fungus make it an interesting model organism for breeding studies of edible basidiomycetes.
ACKNOWLEDGMENTS
We thank G. Sannia (University of Naples, Italy) for providing the probes for ligninolytic enzymes.
This work was supported by research projects BIO94-0443 and BIO99-0278 of the Comisión Nacional de Ciencia y Tecnología and by funds from the Universidad Pública de Navarra (Pamplona, Spain). L.M.L. and G.P. hold grants from the Departamento de Educación del Gobierno de Navarra and the Departamento de Industria del Gobierno de Navarra, respectively.
REFERENCES
- 1.Antolin M F, Bosio C F, Cotton J, Sweeney W, Strand M R, Black W C. Intensive linkage mapping in a wasp (Bracon hebetor) and a mosquito (Aedes aegypti) with single-strand conformation polymorphism analysis of random amplified polymorphic DNA markers. Genetics. 1996;143:1727–1738. doi: 10.1093/genetics/143.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias A, Ramírez R, Leal H. Cultivation of Pleurotus ostreatus hybrids resistant to 2DG obtained by pairings of neaohaplonts from selected dikaryons. In: van Griensven L J L D, editor. Science and cultivation of edible fungi. Vol. 1. Rotterdam, The Netherlands: A. A. Balkema; 2000. pp. 305–309. [Google Scholar]
- 3.Arnau J, Housego A P, Oliver R P. The use of RAPD markers in the genetic analysis of the plant pathogenic fungus Cladosporium fulvum. Curr Genet. 1994;25:438–444. doi: 10.1007/BF00351783. [DOI] [PubMed] [Google Scholar]
- 4.Asgeirsdóttir S A, de Vries O M H, Wessels J G H. Identification of three differentially expressed hydrophobins in Pleurotus ostreatus (oyster mushroom) Microbiology. 1998;144:2961–2969. doi: 10.1099/00221287-144-11-2961. [DOI] [PubMed] [Google Scholar]
- 5.Baars J J P, Sonnenberg A S M, Mikosch T S P, Griensven L J L D V. Development of a sporeless strain of oyster mushroom Pleurotus sotreatus. In: van Griensven L J L D, editor. Science and cultivation of edible fungi. Vol. 1. Rotterdam, The Netherlands: A. A. Balkema; 2000. pp. 317–323. [Google Scholar]
- 6.Baker B S, Carpenter A T, Esposito M S, Esposito R E, Sandler L. The genetic control of meiosis. Annu Rev Genet. 1976;10:53–134. doi: 10.1146/annurev.ge.10.120176.000413. [DOI] [PubMed] [Google Scholar]
- 7.Beeman R W, Brown S J. RAPD-based genetic linkage maps of Tribolium castaneum. Genetics. 1999;153:333–338. doi: 10.1093/genetics/153.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bezalel L, Hadar Y, Cerniglia C E. Enzymatic mechanisms involved in phenanthrene degradation by the white rot fungus Pleurotus ostreatus. Appl Environ Microbiol. 1997;63:2495–2501. doi: 10.1128/aem.63.7.2495-2501.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binelli G, Bucci G. A genetic linkage map of Picea abies Karst., based on RAPD makers, as a tool in population genetics. Theor Appl Genet. 1994;88:283–288. doi: 10.1007/BF00223633. [DOI] [PubMed] [Google Scholar]
- 10.Black W C. PCR with arbitrary primers: approach with care. Insect Mol Biol. 1993;2:1–6. doi: 10.1111/j.1365-2583.1993.tb00118.x. [DOI] [PubMed] [Google Scholar]
- 11.Botstein D, White R L, Skolnick M H, Davis R W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- 12.Brickner J H, Lynch T J, Zeilinger D, Orias E. Identification, mapping and linkage analysis of randomly amplified DNA polymorphisms in Tetrahymena thermophila. Genetics. 1996;143:811–821. doi: 10.1093/genetics/143.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Callac P, Desmerger C, Kerrigan R W, Imbernon M. Conservation of genetic linkage with map expansion in distantly related crosses of Agaricus bisporus. FEMS Microbiol Lett. 1997;146:235–240. doi: 10.1111/j.1574-6968.1997.tb10199.x. [DOI] [PubMed] [Google Scholar]
- 14.Chaparro J X, Werner D J, O'Malley D, Sederoff R R. Targeted mapping and linkage analysis of morphological, isozyme, and RAPD markers in peach. Theor Appl Genet. 1994;87:805–815. doi: 10.1007/BF00221132. [DOI] [PubMed] [Google Scholar]
- 15.Chiu S-W. Nuclear changes during fungal development. In: Chiu S-W, Moore D, editors. Patterns in fungal development. Cambridge, United Kingdom: Cambridge University Press; 1996. pp. 105–125. [Google Scholar]
- 16.Debets F, Swart K, Hoekstra R F, Bos C J. Genetic maps of eight linkage groups of Aspergillus niger based on mitotic mapping. Curr Genet. 1993;23:47–53. doi: 10.1007/BF00336749. [DOI] [PubMed] [Google Scholar]
- 17.Eger G. Rapid method for breeding Pleurotus ostreatus. Mushroom Sci. 1974;9:567–573. [Google Scholar]
- 18.Gaskell J, Dieperink E, Cullen D. Genomic organization of lignin peroxidase genes of Phanerochaete chrysosporium. Nucleic Acids Res. 1991;19:599–603. doi: 10.1093/nar/19.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giardina P, Aurilia V, Cannio R, Marzullo L, Amoresano A, Siciliano R, Pucci P, Sannia G. The gene, protein and glycan structures of laccase from Pleurotus ostreatus. Eur J Biochem. 1996;235:508–515. doi: 10.1111/j.1432-1033.1996.00508.x. [DOI] [PubMed] [Google Scholar]
- 20.Giardina P, Cannio R, Martirani L, Marzullo L, Palmieri G, Sannia G. Cloning and sequencing of a laccase gene from the lignin-degrading basidiomycete Pleurotus ostreatus. Appl Environ Microbiol. 1995;61:2408–2413. doi: 10.1128/aem.61.6.2408-2413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grattapaglia D, Sederoff R. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics. 1994;137:1121–1137. doi: 10.1093/genetics/137.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrán A, Estioko L, Becker D, Rodríguez M J B, Rohde W, Ritter E. Linkage mapping and QTL analysis in coconut (Cocos nucifera L.) Theor Appl Genet. 2000;101:292–300. [Google Scholar]
- 23.Holtke H J, Sagner G, Kessler C, Schmitz G. Sensitive chemiluminiscent detection of digoxigenin-labeled nucleic acids: a fast and simple protocol and its applications. BioTechniques. 1992;12:104–113. [PubMed] [Google Scholar]
- 24.Hulbert S H, Ilott T W, Legg E J, Lincoln S E, Lander E S, Michelmore R W. Genetic analysis of the fungus, Bremia lactucae, using restriction fragment length polymorphisms. Genetics. 1988;120:947–958. doi: 10.1093/genetics/120.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt G J, Page R E. Linkage map of the honey bee, Apis mellifera, based on RAPD markers. Genetics. 1995;139:1371–1382. doi: 10.1093/genetics/139.3.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwaguchi S, Homma M, Tanaka K. Clonal variation of chromosome size derived from the rDNA cluster in Candida albicans. J Gen Microbiol. 1992;138:1177–1184. doi: 10.1099/00221287-138-6-1177. [DOI] [PubMed] [Google Scholar]
- 27.Kaback D B, Steensma H Y, de Jonge P. Enhanced meiotic recombination on the smallest chromosome of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1989;86:3694–3698. doi: 10.1073/pnas.86.10.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerrigan R W, Royer J C, Baller L M, Kohli Y, Horgen P A, Anderson J B. Meiotic behavior and linkage relationships in the secondarily homothallic fungus Agaricus bisporus. Genetics. 1993;133:225–236. doi: 10.1093/genetics/133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kesseli R V, Paran I, Michelmore R W. Analysis of a detailed genetic linkage map of Lactuca sativa (lettuce) constructed from RFLP and RAPD markers. Genetics. 1994;136:1435–1446. doi: 10.1093/genetics/136.4.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosambi D D. The estimation of map distance from recombination values. Ann Eugenics. 1944;12:172–175. [Google Scholar]
- 31.Larraya L, Peñas M M, Pérez G, Santos C, Ritter E, Pisabarro A G, Ramírez L. Identification of incompatibility alleles and characterisation of molecular markers genetically linked to the A incompatibility locus in the white rot fungus Pleurotus ostreatus. Curr Genet. 1999;34:486–493. doi: 10.1007/s002940050424. [DOI] [PubMed] [Google Scholar]
- 32.Larraya L M, Pérez G, Peñas M M, Baars J P, Mikosch T S P, Pisabarro A G, Ramírez L. Molecular karyotype of the white rot fungus Pleurotus ostreatus. Appl Environ Microbiol. 1999;65:3413–3417. doi: 10.1128/aem.65.8.3413-3417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martínez A T, Camarero S, Guillén F, Gutiérrez A, Muñoz C, Varela E, Martínez M J, Barrasa J M, Ruel K, Pelayo J M. Progress in biopulping of non-woody materials: chemical, enzymatic and ultrastructural aspects of wheat straw delignification with ligninolytic fungi from the genus Pleurotus. FEMS Microbiol Rev. 1994;13:265–274. [Google Scholar]
- 34.Marzullo L, Cannio R, Giardina P, Santini M T, Sannia G. Veratryl alcohol oxidase from Pleurotus ostreatus participates in lignin biodegradation and prevents polymerization of laccase-oxidized substrates. J Biol Chem. 1995;270:3823–3827. doi: 10.1074/jbc.270.8.3823. [DOI] [PubMed] [Google Scholar]
- 35.May B, Henley C J, Fisher C G, Royse D J. Linkage relationships of 19 allozyme encoding loci within the commercial mushroom genus Pleurotus. Genome. 1988;30:888–895. [Google Scholar]
- 36.McCluskey K, Mills D. Identification and characterization of chromosome length polymorphisms among strains representing fourteen races of Ustilago hordei. Mol Plant-Microbe Interact. 1990;3:366–373. [Google Scholar]
- 37.Michelmore R W, Paran I, Kesseli R V. Identification of markers linked to resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genome regions by using segregating populations. Proc Natl Acad Sci USA. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell-Olds T. Interval mapping of viability loci causing heterosis in Arabidopsis. Genetics. 1995;140:1105–1109. doi: 10.1093/genetics/140.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morales V M, Séguin-Swartz G, Taylor J L. Chromosome size polymorphisms in Leptosphaeria maculans. Phytopathology. 1993;83:503–509. [Google Scholar]
- 40.Paran I, Michelmore R W. Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor Appl Genet. 1993;85:985–993. doi: 10.1007/BF00215038. [DOI] [PubMed] [Google Scholar]
- 41.Peberdy J F, Hanifah A H, Jia J-H. New perspectives on the genetics of Pleurotus. In: Chang S-T, Buswell J A, Chiu S W, editors. Mushroom biology and mushroom products. Hong Kong: The Chinese University Press; 1993. pp. 55–62. [Google Scholar]
- 42.Peñas M M, Asgeirsdóttir S A, Lasa I, Culiañez-Macià F A, Pisabarro A G, Wessels J G H, Ramírez L. Identification, characterization, and in situ detection of a fruit-body-specific hydrophobin of Pleurotus ostreatus. Appl Environ Microbiol. 1998;64:4028–4034. doi: 10.1128/aem.64.10.4028-4034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pukkila P J, Skrzynia C. Frequent changes in the number of reiterated ribosomal RNA genes throughout the life cycle of the basidiomycete Coprinus cinereus. Genetics. 1993;133:203–211. doi: 10.1093/genetics/133.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raeder U, Thompson W, Broda P. RFLP-based genetic map of Phanerochaete chrysosporium ME446: lignin peroxidase genes occur in clusters. Mol Microbiol. 1989;3:911–918. doi: 10.1111/j.1365-2958.1989.tb00240.x. [DOI] [PubMed] [Google Scholar]
- 45.Ramírez L, Larraya L M, Peñas M M, Pérez G, Eizmendi A, Agós I, Arana D, Aranguren J, Iribarren I, Olaberría N, Palacios E, Ugarte B E, Pisabarro A G. Molecular techniques for the breeding of Pleurotus ostreatus. In: van Griensven L J L D, editor. Science and cultivation of edible fungi. Vol. 1. Rotterdam, The Netherlands: A. A. Balkema; 2000. pp. 157–163. [Google Scholar]
- 46.Ritter E, Gebhardt C, Salamini F. Estimation of recombination frequencies and construction of RFLP linkage maps in plants from crosses between heterozygous parents. Genetics. 1990;125:645–654. doi: 10.1093/genetics/125.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritter E, Salamini F. The calculation of recombination frequencies in crosses of allogamous plant species with applications for linkage mapping. Genet Res. 1996;67:55–65. [Google Scholar]
- 48.Roux P, Labarère J. Isozyme characterization of dikaryotic strains of the edible basidiomycete Agaricus bitorquis (Quel.) Sacc. (Syn. Agaricus edulis) Exp Mycol. 1990;14:101–112. [Google Scholar]
- 49.Royse D J. Speciality mushrooms. In: Janick J, editor. Progress in new crops. Arlington, Va: ASHS Press; 1996. pp. 464–475. [Google Scholar]
- 50.Rustchenko E P, Curran T M, Sherman F. Variation in the number of ribosomal DNA units in morphological mutants and normal strains of Candida albicans and in normal strains of Saccharomyces cerevisiae. J Bacteriol. 1993;174:7189–7199. doi: 10.1128/jb.175.22.7189-7199.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sagawa I, Nagata Y. Analysis of chromosomal DNA of mushrooms in genus Pleurotus by pulsed field gel electrophoresis. J Gen Appl Microbiol. 1992;38:47–52. [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 53.Shaw C R, Prassad R. Starch gel electrophoresis of enzymes. A compilation of recipes. Biochem Genet. 1970;4:297–332. doi: 10.1007/BF00485780. [DOI] [PubMed] [Google Scholar]
- 54.Skinner D Z, Budde A D, Farman M L, Smith J R, Leung H. Genome organization of Magnaporthe grisea: genetic map, electrophoretic karyotype, and occurrence of repeated DNAs. Theor Appl Genet. 1993;87:545–557. doi: 10.1007/BF00221877. [DOI] [PubMed] [Google Scholar]
- 55.Sonnenberg A M, de Groot P W J, Schaap P J, Baars J J P, Visser J, van Griensven L L J L D. Isolation of expressed sequence tags of Agaricus bisporus and their assignment to chromosomes. Appl Environ Microbiol. 1996;62:4542–4547. doi: 10.1128/aem.62.12.4542-4547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Talbot N J, Oliver R P, Coddington A. Pulse field gel electrophoresis reveals chromosome length differences between strains of Cladosporium fulvum (syn. Fulvia fulva) Mol Gen Genet. 1991;229:267–272. doi: 10.1007/BF00272165. [DOI] [PubMed] [Google Scholar]
- 57.Tulsieram L K, Glaubitz J C, Kiss G, Carlson J E. Single tree genetic linkage mapping in conifers using haploid DNA from megagametophytes. Bio/Technology. 1992;10:686–690. doi: 10.1038/nbt0692-686. [DOI] [PubMed] [Google Scholar]
- 58.Tzeng T H, Lyngholm L K, Ford C F, Bronson C R. A restriction fragment length polymorphism map and electrophoretic karyotype of the fungal maize pathogen Cochliobolus heterostrophus. Genetics. 1992;130:81–96. doi: 10.1093/genetics/130.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vallejos C E. Enzyme activity staining. In: Tanksley S D, Orton T J, editors. Isozymes in plant genetics and breeding. Amsterdam, The Netherlands: Elsevier; 1983. pp. 469–516. [Google Scholar]
- 60.Verbeet M P, Klootwijk J, van Heerikhuizen H, Fontijn R, Vreugdenhil E, Planta R J. Molecular cloning of the rDNA of Saccharomyces rosei and comparison of its transcription initiation region with that of Saccharomyces carlsbergensis. Gene. 1983;23:53–63. doi: 10.1016/0378-1119(83)90216-0. [DOI] [PubMed] [Google Scholar]
- 61.Wang I-H, Thomas C E, Dean R A. A genetic map of melon (Cucumis melo L.) based on amplified fragment length polymorphism (AFLP) markers. Theor Appl Genet. 1997;95:791–798. [Google Scholar]
- 62.Whisson S C, Drenth A, Maclean D J, Irwin J A. Phytophthora sojae avirulence genes, RAPD, and RFLP markers used to construct a detailed genetic linkage map. Mol Plant-Microbe Interact. 1995;8:988–995. doi: 10.1094/mpmi-8-0988. [DOI] [PubMed] [Google Scholar]
- 63.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu J, Horgen P A, Anderson J B. Somatic recombination in the cultivated mushroom Agaricus bisporus. Mycol Res. 1996;100:188–192. [Google Scholar]
- 65.Young W P, Wheeler P A, Coryell V H, Keim P, Thorgaard G H. A detailed linkage map of rainbow trout produced using doubled haploids. Genetics. 1998;148:839–850. doi: 10.1093/genetics/148.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zolan M E. Chromosome-length polymorphism in fungi. Microbiol Rev. 1995;59:686–698. doi: 10.1128/mr.59.4.686-698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]