Abstract
Background
Global longitudinal shortening (GL‐Shortening) and the mitral annular plane systolic excursion (MAPSE) are known markers in heart failure patients, but measurement may be subjective and less frequently reported because of the lack of automated analysis. Therefore, a validated, automated artificial intelligence (AI) solution can be of strong clinical interest.
Methods and Results
The model was implemented on cardiac magnetic resonance scanners with automated in‐line processing. Reproducibility was evaluated in a scan–rescan data set (n=160 patients). The prognostic association with adverse events (death or hospitalization for heart failure) was evaluated in a large patient cohort (n=1572) and compared with feature tracking global longitudinal strain measured manually by experts. Automated processing took ≈1.1 seconds for a typical case. On the scan–rescan data set, the model exceeded the precision of human expert (coefficient of variation 7.2% versus 11.1% for GL‐Shortening, P=0.0024; 6.5% versus 9.1% for MAPSE, P=0.0124). The minimal detectable change at 90% power was 2.53 percentage points for GL‐Shortening and 1.84 mm for MAPSE. AI GL‐Shortening correlated well with manual global longitudinal strain (R 2=0.85). AI MAPSE had the strongest association with outcomes (χ2, 255; hazard ratio [HR], 2.5 [95% CI, 2.2–2.8]), compared with AI GL‐Shortening (χ2, 197; HR, 2.1 [95% CI,1.9–2.4]), manual global longitudinal strain (χ2, 192; HR, 2.1 [95% CI, 1.9–2.3]), and left ventricular ejection fraction (χ2, 147; HR, 1.8 [95% CI, 1.6–1.9]), with P<0.001 for all.
Conclusions
Automated in‐line AI‐measured MAPSE and GL‐Shortening can deliver immediate and highly reproducible results during cardiac magnetic resonance scanning. These results have strong associations with adverse outcomes that exceed those of global longitudinal strain and left ventricular ejection fraction.
Keywords: artificial intelligence; cardiac magnetic resonance imaging; global longitudinal shortening, reproducibility; image processing; prognosis
Subject Categories: Magnetic Resonance Imaging (MRI)
Nonstandard Abbreviations and Acronyms
- CNN
convolutional neural network
- GL‐Shortening
global longitudinal shortening
- GL‐Strain
global longitudinal strain
- HCMR
Hypertrophic Cardiomyopathy Registry
- HHF
hospitalization for heart failure
- MAPSE
mitral annular plane systolic excursion
- MR
magnetic resonance
Clinical Perspective
What Is New?
We developed an artificial intelligence deep neural network model to measure indices of left ventricular systolic function from cardiac magnetic resonance cine images: global longitudinal shortening and mitral annular plane systolic excursion (MAPSE).
The artificial intelligence model was deployed directly on magnetic resonance scanners to generate immediate global longitudinal shortening and MAPSE values without any supervision, which provided better scan–rescan reproducibility than human experts.
Artificial intelligence global longitudinal shortening and MAPSE measurements associated with adverse outcomes in a large prospective cohort of 1572 patients with MAPSE being the strongest predictor, better than manual measures, of global longitudinal strain and left ventricular ejection fraction.
What Are the Clinical Implications?
Automated artificial intelligence–derived global longitudinal shortening and MAPSE measures provide immediate, reproducible, robust, clinically relevant measures to assess left ventricular contractile function.
Cardiac contractile dysfunction is associated with adverse cardiovascular outcomes and mortality. 1 , 2 Cardiac magnetic resonance (CMR) imaging is considered the gold standard for measuring left ventricular (LV) volumes and LV ejection fraction (LVEF). However, LVEF changes are frequently late in disease development, so earlier and more sensitive predictors are needed, particularly where linked to therapeutic choices such as chemotherapy‐related decisions.
Contractile function can be globally assessed by measuring the longitudinal ventricular shortening between end‐diastole and end‐systole, 3 which predicts outcomes in a variety of scenarios, such as dilated cardiomyopathy 4 , 5 or heart failure with preserved ejection fraction (EF). 6 The term global longitudinal shortening (GL‐Shortening) has been proposed as a positive percentage ratio to quantify LV contraction, 7 , 8 where a larger number implies better contractile ability. Changes in GL‐Shortening occur earlier than the LVEF reduction and complement in heart failure with preserved EF. Longitudinal function is also tied to the atrioventricular plane displacement, also referred to as mitral annular plane systolic excursion (MAPSE), which is a key contributor to pumping 9 and is prognostic for patients with heart failure. 10 , 11
GL‐Shortening can be assessed using CMR cine imaging. One method relies on segmenting the myocardium and estimating tissue strain using either feature tracking 12 or image registration, 13 from one or more long‐axis (LAX) views. The per‐pixel or sector strain values are averaged for the entire myocardium to get the global longitudinal strain (GLS or GL‐Strain), as a measure of longitudinal shortening. Other methods directly measure left ventricle lengths and compute longitudinal shortening from cine images by tracking myocardium. 14 , 15 These processes require manual delineation. 16 , 17 , 18 , 19 MAPSE can be measured from CMR by manually tracking the movement of the lateral annulus position. 3 , 9 Although useful as a prognostic marker, 20 all methods are in part subjective, time consuming, and require a dedicated workstation and software packages. 9 , 16 , 21 Consequently, GL‐Shortening and MAPSE are less embedded in guidelines and treatment‐decision algorithms.
We developed a fully automated solution to measure GL‐Shortening and MAPSE using an artificial intelligence (AI) deep neural network model, implemented directly on magnetic resonance (MR) scanners. This study sought to evaluate the reproducibility and prognostic performance of the AI measures in 2 existing data sets. First, we used a multicenter, multidisease, scan–rescan data set 22 (n=160 patients) to assess model precision and reproducibility compared with manual labeling by a human expert. Second, we explored the prognostic association of AI‐derived GL‐Shortening and MAPSE in a large cohort (n=1572) 21 and compared the prognostic performance with that of manually measured GL‐Strain and LVEF. We hypothesized that AI‐derived metrics could be reproducible, accurate, and prognostic.
Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request. The deep‐learning source code is available at https://github.com/xueh2/CMR_LandMark_Detection.git.
Patient Population
Two data sets were each acquired with the corresponding required ethical and/or secondary audit use approvals or guidelines (as per each center) that permitted retrospective analysis of anonymized data for the purpose of technical development, protocol optimization, and quality control by the institutional review committee of each center. The scan‐rescan study was approved by the institutional review board at the Barts Heart Center and the outcome study was approved by the IRB at UPMC CMR center. Written informed consent was obtained for all particpants for both studies. All data were anonymized and delinked for analysis with approval by the local Office of Human Subjects Research (exemption approval for collaboration number 13156).
Scan–Rescan
Patients (n=160) were scanned twice within a short time interval (median interval 1 day, 82% scanned on the same day) to minimize changes in cardiac function. 22 , 23 This patient cohort included a representative clinical spectrum, including health, dilatation, hypertrophy, and regional cardiac diseases. More details of this data set and subject characteristics are published at https://thevolumesresource.com (Validation of Left Ventricular Myocardial and Endocardial Segmentation Resource). Scan–rescan data sets were collected across 5 institutions in the United Kingdom with 5 different magnetic resonance imaging scanners (3 Siemens Avanto 1.5T scanners and 1 Siemens Aera 1.5T scanner; Siemens AG Healthcare, Erlangen, Germany; 1 Philips Achieva 3T scanner; Philips HealthCare, Best, the Netherlands). The standard cine imaging sequence was used with comparable imaging parameters. Standard 4‐chamber, 2‐chamber, and 3‐chamber cine images were acquired for all participants. For rescans on the same day, patients were taken off the imaging table after the first scan before returning and re‐isocentering and re‐planned for the second scan. Cine imaging was performed either before or after the administration of a gadolinium‐based contrast agent.
Images were analyzed by an expert cardiologist (R.H.D.; level 3 European Association of Cardiovascular Imaging accreditation and 5 years of experience in CMR). To help for better throughput, a trainee (R.A.) prelabeled the images for the expert who reviewed the labels and performed corrections if necessary. For every cine scan, the end‐diastolic and end‐systolic phases were picked by the human operator, and valve and apical points were manually marked using the VGG Image Annotator software (Oxford, UK; http://www.robots.ox.ac.uk/~vgg/software/via/), from which the GL‐Shortening and MAPSE were computed.
Outcome Study
Patients (n=1578) were scanned at the University of Pittsburgh Medical Center Cardiovascular Magnetic Resonance Center (Pittsburgh, PA) from June 2010 to March 2016. Clinical outcomes were collected until October 2018 (median follow‐up 5.6 years), 21 with follow‐up data available for n=1572 patients whose data were included for AI analysis. This patient cohort was curated from 2368 consecutive patients who underwent clinical CMR at the University of Pittsburgh Medical Center, with exclusion criteria including hypertrophic cardiomyopathy, adult congenital disease, myocardial edema, amyloidosis, iron overload, Anderson‐Fabry disease, and inadequate image quality. This cohort was curated around measuring myocardial fibrosis with T1 and extracellular volume measures for comparison with global longitudinal strain measured with commercial software that we have published previously. 21 Myocardial edema, amyloidosis, iron overload, and Anderson‐Fabry disease confound T1 and extracellular volume fraction measures of myocardial fibrosis and thus were excluded. Hypertrophic cardiomyopathy was excluded from the cohort because this unique genetic condition is typically characterized by hypercontractile systolic function and low event rates in young people, which requires an adequately powered large sample size cohort (currently being investigated by the National Institutes of Health–funded multicenter HCMR [Hypertrophic Cardiomyopathy Registry]). To foster the comparison of the proposed AI‐based measures with our prior work, the same exclusions were applied to the cohort as before. Further information about patient population and characteristics have been described. 21 All patients were scanned with a clinical 1.5T scanner (Espree; Siemens AG Healthcare). Standard balanced steady state‐free precession cine imaging 21 was performed to acquire 2‐chamber, 3‐chamber, and 4‐chamber imaging planes. Recorded events included all‐cause death and hospitalization for heart failure (HHF), extracted from a REDCap (Research Electronic Data Capture) database. 24 A total of 335 events were recorded, including deaths and HHF.
AI Image Analysis
A convolutional neural network (CNN) model 25 was used for the automated measurement of GL‐Shortening and MAPSE in the 2‐chamber, 3‐chamber, and 4‐chamber cine images, collectively termed LAX. This model was trained on a separate, large data set (2329 patients with 34 019 images) curated from 2 hospitals to output landmark points from LAX cine images. Among them, 6323 images were acquired after contrast. The inferoseptal and anterolateral mitral annular hinge points were detected from the 4‐chamber view. From the 2‐chamber view, the anterior and inferior points were detected. The inferolateral and anteroseptal points were detected from the 3‐chamber view. The apex was marked for all views. The detection was performed for every cine phase covering the entire cardiac cycle. All detections were also checked visually to find failures, which were defined as: (1) any landmarks missed in any phase in the cine series or (2) any landmark detected at the wrong locations such as outside the left ventricle. There was no overlap between the images used for training the AI landmarking algorithm25 and either the scan–rescan or outcome data sets reported in this study. The previously described25 training data set consisted of consecutive patients who successfully completed a routine CMR study, without intentionally excluding structural abnormalities.
As an example, Figure 1 demonstrates the landmark points detected on a 4‐chamber cine series and the measurement of GL‐Shortening and MAPSE. Videos [Link], [Link], [Link] give examples of this dynamic landmark detection scheme for the 3 LAX views. The length of the left ventricle was measured from the apical point to the midpoint of valve plane. For each LAX view, GL‐Shortening was computed as the percentage of LV length shortening between end‐diastole and end‐systole, reported as a positive value, as proposed in 7 :
Figure 1. Example of artificial intelligence global longitudinal shortening (GL‐Shortening) and mitral annular plane systolic excursion (MAPSE) measurements.
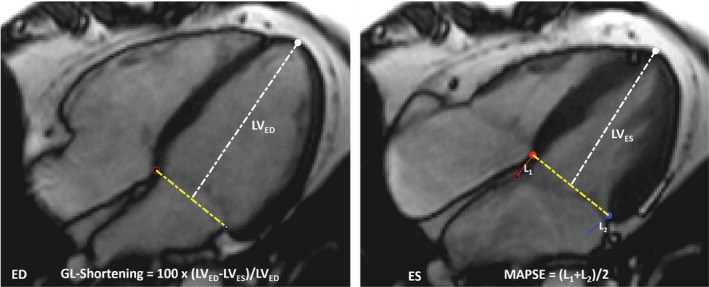
A cine 4‐chamber scan is displayed for end‐diastolic (ED) and end‐systolic (ES) phases with detected landmarks overlaid. GL‐Shortening is computed as the percentage shortening of left ventricle (LV) length (from apical to midpoint of 2 valve landmarks). MAPSE is computed as the mean moved distance in millimeters for 2 valve points in each long‐axis image. L is the moved distance of valve point between the ED and ES.
MAPSE in millimeters was computed as the mean distance for 2 valve points between end‐diastole and end‐systole. 20 The CNN model was applied to all acquired LAX views. For every patient, the mean values across the different LAX views were computed and reported. Additionally, a comparison was made between MAPSE and lateral MAPSE. Lateral MAPSE was computed from the lateral mitral valve point only using the 4‐chamber view, whereas MAPSE is the average of both valve points from all 3 LAX views (6 measurements in total).
The CNN model was integrated on MR scanners using the open‐source Gadgetron InlineAI toolbox. 26 As demonstrated in Figure 2, where an MR scanner ran the proposed AI solution, the reconstructed cine images, the images overlaid with detected landmarks, LV length curves, GL‐Shortening, and MAPSE measurements are displayed on the scanner console. Images and results are seamlessly returned into the digital imaging and communications in medicine workflow and thereby saved to picture archiving and communication system, and available for reporting by clinicians. The data for the scan–rescan and outcome cohorts were previously acquired, whereas the in‐line measurement of MAPSE and GL‐Shortening is otherwise normally prospectively performed on the MR scanner at the time of imaging. Video S4 illustrates the cine acquisition, image reconstruction, and AI processing happening in‐line on an MR scanner. The AI solution automates the whole process, eliminating the need for postprocessing and providing these measures directly to reporting clinicians. It is timesaving and improves reproducibility compared with manual measurement.
Figure 2. The proposed artificial intelligence solution was deployed to the magnetic resonance (MR) scanners in the in‐line fashion illustrated by this screen snapshot of the MR scanner console, where a 4‐chamber cine series was processed with the model.
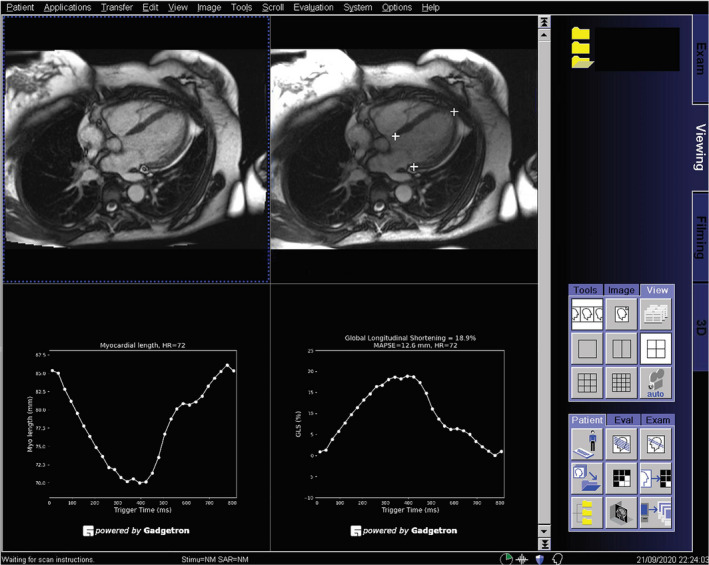
The cine images are displayed at the top left, and detected landmarks are overlaid on the images at the top right. This allows a convenient check of performance of landmark detection. The global longitudinal shortening (GL‐Shortening) and mitral annular plane systolic excursion (MAPSE) were measured automatically and displayed as signal curves. In this way, a fully automated solution was achieved, without requiring any user interaction for processing.
For comparison, GL‐Strain was manually measured for the outcome data set as reported previously 21 using commercially available software (Circle cvi42; Calgary, AB, Canada). The endocardial and epicardial boundaries were manually traced in cine images, and feature tracking was adopted to compute deformation across cardiac phases. GL‐Strain was computed as the mean of the longitudinal strain component across the myocardium. The automated AI MAPSE and GL‐Shortening and the manually measured feature tracked GL‐Strain and LVEF were compared on prognostic power. Note the standard definition of (Lagrangian) strain is (L1–L0)/L0, where L0 is end‐diastolic length and L1 is end‐systole length. This generates negative percentages with healthier myocardium being more negative and disease being less negative, causing confusion. 7 As recently suggested, 7 GL‐Shortening is herein referred to as a positive percentage ratio; thus, lower values indicate reduced cardiac contractility. LVEF was measured manually from the short‐axis stack of cine images for every patient using standard methods. 27
CNN Model and Training
As a summary of the CNN model and training process previously presented, 25 a variation of U‐Net architecture was implemented for the landmark detection. The network was organized as layers for different spatial resolution. The input to the model was a 2‐dimensional image series (ie, to detect the landmarks from a time series), and the model was applied to each 2‐dimensional image using the current model configuration. Each layer can contain several blocks. Each block had 2 convolution layers with batch normalization and LeakyReLu activation functions. The network can be made deeper by inserting more resolution layers or by inserting more blocks. Going down the downsampling branch, the image spatial resolution was reduced by 2× for every layer with the number of filters increased. Going up the upsampling branch, the spatial resolution was restored with a reduced number of filters. All convolution layers had filter size 3×3 with stride 1 and padding 1. The final convolution layer outputs a per‐pixel score tensor, which is converted to a probability tensor using a SoftMax.
The landmark detection problem was formulated as a heat map detection problem. Every landmark point was convolved with a Gaussian kernel (σ was 4.0 pixels), and the resulting blurred distribution represents the spatial probability of this landmark. Detecting 3 landmarks was equivalent to a semantic segmentation problem for 4 classes (background class and 1 object class for each landmark). Class labels for different landmarks were represented as channels in probability maps; thus, if there are 3 landmarks to be detected, it will have 4 heat maps (3 maps for 3 landmarks and 1 for background). The heat maps were normalized to be probability maps. That is, for every pixel in the field of view, the sum over all classes including the background is 1.0. The pixel value in a heat map is the probability that this pixel belongs to corresponding landmark class or background. With the probability heat maps as the target, the training process will optimize the network parameters to minimize the distance between network outputs (after SoftMax) to the per‐pixel probability distribution. For comparison, in binary segmentation, the mask is either 0 or 1 (hard map). But in the current heat map setting, it uses soft probability. The entropy‐based cost function (such as Kullback–Leibler divergence, which computes distance between 2 probability distributions) can be applied to both scenarios. In this way, landmark detection is formulated as a semantic segmentation problem.
In the data preparation step, all images were resampled and cropped to 400×400 pixels square. The CNN output score tensor had dimensions 400×400×4. To train the network, the Kullback–Leibler divergence was computed between ground‐truth heat map and SoftMax tensor of scores. In addition to this entropy‐based loss, the shape loss was further computed as the soft Dice ratio. The soft Dice ratio was computed as the production of 2 probability maps over their sum. The final loss was a sum of entropy‐based loss and soft Dice ratio, which used both entropy‐based information and region costs. This strategy to use a combined loss has been previously used in deep‐learning segmentation and found to improve segmentation robustness.
For the LAX, all views were trained together as a multitask learning task. Because the number of images for each LAX view was roughly equal, no extra data rebalancing strategy was applied. Instead, every minibatch was randomly selected from 2‐chamber, 3‐chamber, or 4‐chamber images and refined network weights.
Images were acquired using both 1.5T (4 MAGNETOM Aera; Siemens AG Healthcare) and 3T (1 MAGNETOM Prisma; Siemens AG Healthcare) MR scanners. In the training set, 1790 patients were scanned with 1.5T scanners and 539 were scanned with 3T. In the test set, 462 patients were scanned at 1.5T magnetic resonance imaging and 69 scanned at 3T. Typically, 30 cardiac phases were reconstructed for each heartbeat for every cine scan. For training, a total of 34 019 images were included from 2329 patients (mean age, 54.1 years; 1471 men).
The data for training were split, with 90% of all patients for training and 10% for validation. The training and validation data sets were split on a per‐study basis, such that there was no mixing of patients between the 2 data sets. The Adam optimizer was used, with an initial learning rate of 0.001, βs were 0.9 and 0.999, and ε was 1e–8. The learning rate was reduced by 2 whenever the cost function plateaued. Training lasted 50 epochs (≈4 hours), and the model was selected as the one giving the highest performance on the validation set. The CNN model was implemented using PyTorch, and training was performed on an Ubuntu 20.04 personal computer with 4 NVIDIA GTX 2080Ti graphics processing unit cards, each with 11 GB random‐access memory. Data parallelization was used across multiple graphics processing unit cards to speed up training. More information about the CNN model is provided in a previous publication. 25
After the model was trained, the model was applied to the scan–rescan and outcome data sets. No data from these 2 data sets were used in any way during the training process and was a completely held‐out data set.
In‐Line Deployment
The trained model was integrated onto MR scanners using the Gadgetron InlineAI toolbox. After training, the model weights were saved as the Open Neural Network Exchange format, using the Pytorch onnx (Open Neural Network Exchange) functionalities. The saved model was loaded when the imaging started, and after the cine images were reconstructed, it was applied to the images as part of the image reconstruction workflow (in‐line processing) at the time of scan. The resulting detection and measurements are displayed on the scanner console, adding only a few seconds to the reconstruction. This process was fully automated.
Statistical Analysis
The scan–rescan data were analyzed using MATLAB (R2017b; MathWorks, Natick, MA). The outcome data were analyzed using SAS (version 9.4; SAS Institute, Cary, NC). Continuous variables are presented as mean±SD or median (interquartile range) for normal and nonnormal distribution, respectively. Reproducibility was measured with absolute differences of measurements between 2 scans. Bland‐Altman limits of agreement and plots were generated, together with the within‐subject coefficient of variation. The 2‐sided t test was performed for group comparisons. The F test was used to compare measures of variability. A P value <0.05 was deemed statistically significant. To illustrate clinical usefulness, the sample size required to detect a 1% change of GL‐Shortening or 1 mm change of MAPSE was computed with a power of 90% and a significance level of 0.05. The minimal detectable change 28 for AI measurements was estimated at the significant level of 0.90: , where . was computed with 1‐way ANOVA as . is the total sum of squares in 1‐way ANOVA and is the 1‐way intraclass correlation coefficient (form ). 28 , 29 For the interoperator variation, Lin concordance correlation coefficient 30 was computed, where higher value means better agreement between operators.
For the outcome data set, survival analyses examined a combined end point of time from CMR scanning to either first HHF or death (all‐cause mortality). Univariable and multivariable Cox regression models quantified associations between variables and outcomes, whereby χ2 values tested the strength of these associations and permitted benchmark comparisons between MAPSE, GL‐Shortening, GL‐Strain, and LVEF. To illustrate associations visually, Kaplan‐Meier curves used the log‐rank test dividing MAPSE and GL‐Shortening into 5 equally spaced strata, with 1 SD intervals (2 strata above the median and 3 strata below given skewed distributions). We preferred this approach over using quantiles because the clinician wishes to estimate risk with progressive deviation from normal, regardless of whether strata contain equal numbers of patients. The multivariable Cox model was adjusted for a set of clinical variables according to patient condition, hospitalization status, and other imaging findings (details in Table 1). The hazard ratio was scaled to 1 SD intervals for comparison purpose.
Table 1.
Reproducibility Measured on the Scan–Rescan Data Set for GL‐Shortening and MAPSE
| Median | Interquartile range | Difference between the 2 scans | R 2 | P value | CV | N | MDC90 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Scan 1 | Scan 2 | Scan 1 | Scan 2 | ||||||||
| GL‐Shortening | AI | 14.6% | 14.7% | 11.8%–18.0% | 12.0%–17.3% | 0.23%±1.49% | 0.86 | 0.59 | 7.2% | 14 | 2.53% |
| Expert | 14.2% | 14.4% | 10.9%–18.3% | 11.6%–18.0% | 0.11%±2.26% | 0.76 | 0.83 | 11.1% | 29 | 3.85% | |
| MAPSE | AI | 12.4 mm | 11.8 mm | 9.5–13.9 mm | 9.9–13.5 mm | 0.20±1.08 mm | 0.87 | 0.54 | 6.5% | 9 | 1.84 mm |
| Expert | 12.5 mm | 12.1 mm | 10.1–14.4 mm | 10.6–14.0 mm | 0.20±1.57 mm | 0.76 | 0.57 | 9.1% | 16 | 2.70 mm | |
AI was treated as an independent operator and compared with the expert. The median and percentile values are given. Interscan and intrasubject differences between the 2 scans are reported as mean±SD, together with the R 2 ratio and within‐subject CV. The number of samples (N) required to detect 1 mm or 1% change in MAPSE and GL‐Shortening was computed. Minimal detectable changes are reported with 90% power of significance. AI indicates artificial intelligence; CV, coefficient of variation; GL‐Shortening, global longitudinal shortening; MAPSE, mitral annular plane systolic excursion; and MDC90, minimal detectable changes with 90% power of significance.
To evaluate whether MAPSE and GL‐Shortening provide added value for prognostic prediction over GL‐Strain and LVEF, further analysis was performed to evaluate the net reclassification index and the integrated discriminant improvement by comparing the MAPSE and GL‐Shortening against LVEF and GL‐Strain.
Results
AI Inference
The trained model was applied to all cine scans, and GL‐Shortening and MAPSE were computed. Visual manual inspection of all results for plausibility found no implausible or failed detections in the scan–rescan cohort and 1 in the outcome cohort because of suboptimal imaging slice planning. This suggests the model was robust to slice planning, anatomical changes, and diseases. Figure 3 shows examples of AI detection in health, global disease (dilated cardiomyopathy), and regional disease (chronic myocardial infarction), demonstrating the typical performance.
Figure 3. Examples of artificial intelligence detection on long‐axis cine images.
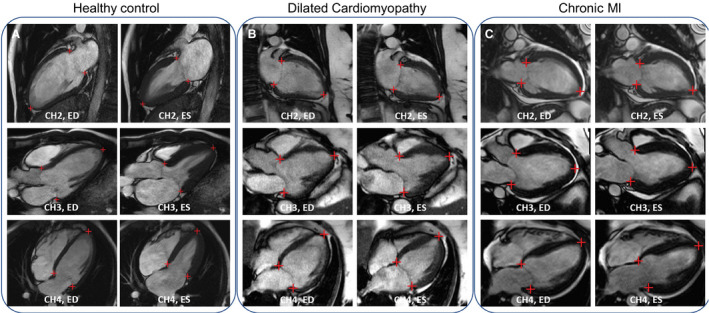
The detected landmarks were overlaid on the image to indicate the accuracy of the model. A, A healthy 31‐year‐old man from the scan–rescan cohort was scanned to acquire 3 long‐axis cine views. B, A 61‐year‐old woman was diagnosed with dilated cardiomyopathy. C, A 30‐year‐old man with myocardial infarction (MI) was scanned and found to have impaired cardiac function with the ejection fraction being 22%. CH2 indicates 2‐chamber; CH3, 3‐chamber; CH4, 4‐chamber; ED, end‐diastolic; and ES, end‐systolic.
Running on the scanners with a dedicated Linux server, the AI measurement and image analysis took 1.1 seconds to process for a typical cine series (30 phases, Ubuntu 20.04, NVIDIA GTX 2080Ti graphics processing unit with 11 GB random‐access memory), in addition to usual MR image reconstruction that took ≈5 seconds. The human measurement of GL‐Strain using the Circle cvi42 commercial software took 261±36 seconds (minimum: 233 seconds; maximum: 342 seconds) for 3 LAX views, timed by an expert (E.B.S., with 20 years of CMR experience) on 8 consecutive patients. Another timing test was performed by the second expert (R.H.D., with 5 years of CMR experience) to measure MAPSE and GL‐Shortening using the Circle cvi42 commercial software, which took 3 to 3.5 minutes per patient to process 3 LAX views, timed on 8 consecutive patients. By comparison, the in‐line AI processing to measure and report MAPSE and GL‐Shortening was fully automated. It did not require any manual interaction or guidance from the MR scanner operator. As shown in Video S4, the cine imaging is acquired in the conventional manner, and the optimized AI analysis is completely integrated into the clinical workflow, with no additional burden of manual interaction. The MR scanner operator can review these results immediately following cine imaging.
Scan–Rescan
Table 1 lists results for the scan–rescan experiment for the AI and human expert.
Accuracy and Interoperator Variation
There was good agreement between AI and expert manual analysis with no bias for either GL‐Shortening or MAPSE (GL‐Shortening, P=0.49 for scan 1 and P=0.65 for scan 2; MAPSE, P=0.26 for scan 1 and P=0.24 for scan 2). For GL‐Shortening, the differences between 2 scans were 0.23% for the model and 0.11% for the manual measurement, whereas the model showed improved reproducibility with a lower standard variation compared with manual measurement by a human expert (1.49% versus 2.26%). Similar results were found for AI MAPSE compared with manual MAPSE (SD, 1.1 mm versus 1.6 mm). Lin concordance correlation coefficient was 0.91 to 0.94 between AI and manual for all metrics. The interoperator coefficient of variation of GL‐Shortening and MAPSE for AI versus manual was 8.4% and 6.8%.
Reproducibility and Intraoperator Variation
Reproducibility was used as an estimate of precision. GL‐Shortening and MAPSE measured from 2 scans were in good agreement for AI and manual (P>0.5 for all measurements). Compared with manual, AI had a lower within‐subject coefficient of variation (GL‐Shortening: 7.2% versus 11.1%, P=0.002; MAPSE: 6.5% versus 9.1%, P=0.012), and a numerically smaller minimal detectable change (GL‐Shortening MDC90, 2.53 versus 3.85 percentage points; MAPSE MDC90, 1.84 mm versus 2.70 mm).
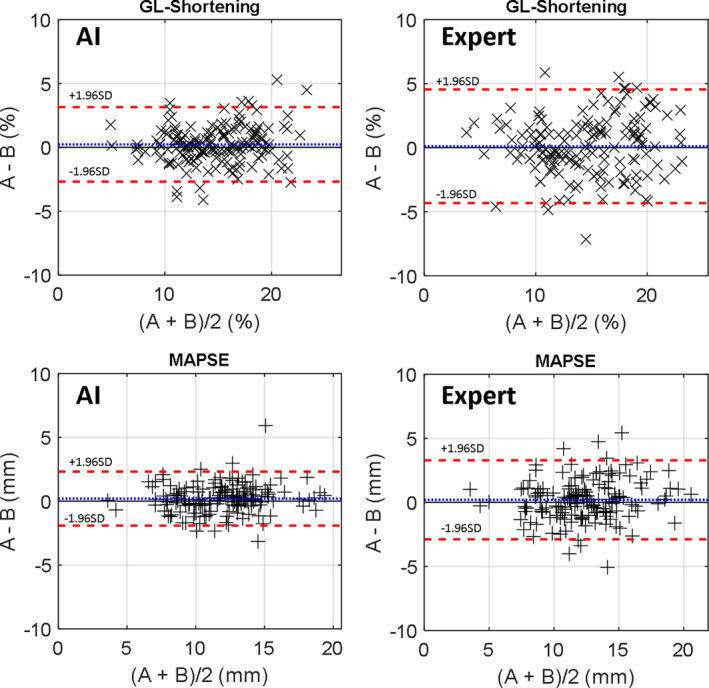
Figure 4 shows the Bland‐Altman plots for intraoperator variation, where AI had the better reproducibility. The 95% CI for GL‐Shortening was −2.69 to 3.15 mm for AI and −4.32 to 4.54 mm for manual. For MAPSE, the confidence range was −1.92 to 2.32 mm for AI and −2.88 to 3.28 mm for manual.
Figure 4. Bland‐Altman plots of scan–rescan data sets for global longitudinal shortening (GL‐Shortening) (top) and mitral annular plane systolic excursion (MAPSE) (bottom) for the artificial intelligence (AI) and expert.
Outcome Analysis
Across the outcomes cohort, GL‐Shortening was 14.7 (10.7%–17.4%) and the range was 2.5% to 25.0%; MAPSE was 11.5 mm (9.1–13.9 mm) and the range was 2.6 to 21.9 mm. MAPSE was moderately correlated with GL‐Shortening and manual GL‐Strain (R 2=0.71 and 0.63, respectively), whereas AI GL‐Shortening was strongly correlated with the manual GL‐Strain (R 2=0.85). AI MAPSE and AI GL‐Shortening were associated with adverse events, demonstrated in the Cox regression analysis (Table 1). Notably, MAPSE was the strongest predictor of adverse events. In the univariable Cox regression model for HHF or death, the χ2 values of AI MAPSE, AI GL‐Shortening, and manual GL‐Strain were 255.2, 197.3, and 191.6, all of which exceeded manually measured EF 146.7. The same trend remained when testing for only death or HHF, respectively, as shown in Table 2. For all testing categories (death, HHF, or both), AI MAPSE had the highest χ2 values, and LVEF had the lowest. In death, HHF, or both AI GL‐Shortening and manual GL‐Strain had close χ2 values (111.2 versus 111.0 for death, 137.6 versus 126.5 for HHF, 197.3 versus 191.6 for both;). Lateral MAPSE was a strong predictor of HHF (χ2, 130.9; P<0.0001), death (χ2, 105.1; P<0.0001), or both (χ2, 180.6; P<0.0001), which was lower than MAPSE averaged over 6 measurements in 3 LAX views (χ2 HHF, 160.8; death, 163.6; and both, 255.2; P<0.0001 for all).
Table 2.
AI MAPSE Is Associated With Outcomes (Composite of HHF or Death [n=335]) More Strongly Than AI GL‐Shortening and Manual GL‐Strain Based on χ2 Values in Univariable and Multivariable Cox Regression Models Among 1572 Participants
| Variables | Univariable Cox regression | Multivariable Cox regression* | |||||
|---|---|---|---|---|---|---|---|
| χ2 |
HR (95% CI) |
P value | χ2 |
HR (95% CI) |
P value | ||
| HHF or death | MAPSE, per mm | 255.2* | 2.5 (2.2–2.8) | <0.001 | 52.1* | 1.7 (1.5–2.0) | <0.001 |
| GL‐Shortening, % | 197.3 | 2.1 (1.9–2.4) | <0.001 | 26.9 | 1.5 (1.3–1.8) | <0.001 | |
| GL‐Strain, % | 191.6 | 2.1 (1.9–2.3) | <0.001 | 24.9 | 1.5 (1.3–1.7) | <0.001 | |
| LVEF, % | 146.7 | 1.8 (1.6–2.0) | <0.001 | 16.8 | 1.4 (1.2–1.6) | <0.001 | |
| Death | MAPSE, per mm | 163.6* | 2.3 (2.0–2.6) | <0.001 | 29.3* | 1.6 (1.4–1.9) | <0.001 |
| GL‐Shortening, % | 111.2 | 1.9 (1.7–2.2) | <0.001 | 7.7 | 1.3 (1.1–1.6) | 0.0056 | |
| GL‐Strain, % | 111.0 | 1.9 (1.7–2.1) | <0.001 | 7.1 | 1.3 (1.1–1.5) | 0.0078 | |
| LVEF, % | 79.1 | 1.6 (1.5–1.8) | <0.001 | 4.7 | 1.2 (1.0–1.4) | 0.0307 | |
| HHF | MAPSE, per mm | 160.8* | 3.1 (2.6–3.7) | <0.001 | 39.2* | 2.1 (1.7–2.7) | <0.001 |
| GL‐Shortening, % | 137.6 | 2.7 (2.3–3.1) | <0.001 | 30.5 | 2.0 (1.6–2.6) | <0.001 | |
| GL‐Strain, % | 126.5 | 2.5 (2.1–2.9) | <0.001 | 25.6 | 1.8 (1.4–2.3) | <0.001 | |
| LVEF, % | 106.9 | 2.1 (1.8–2.4) | <0.001 | 20.1 | 1.7 (1.3–2.1) | <0.001 | |
GL‐Shortening indicates global longitudinal shortening; GL‐Strain, global longitudinal strain; HHF, hospitalization for heart failure; HR, hazard ratio; LVEF, left ventricular ejection fraction; and MAPSE, mitral annular plane systolic excursion.
*Adjusted for: age, sex, race, diabetes, hypertension, hyperlipidemia, smoking status, glomerular filtration rate, prior percutaneous intervention, prior coronary bypass surgery, moderate or severe aortic stenosis, moderate or severe mitral regurgitation, myocardial infarction by late gadolinium enhancement, nonischemic scar, extracellular volume fraction, left ventricular mass index, end diastolic volume index, and stratified by hospitalization status.
Multivariable Cox regression was performed for HHF, death, or both. Table 2 shows the outcomes of the analysis adjusted for multiple factors and including hazard ratios. For HHF or death, the multivariable χ2 values of AI MAPSE, AI GL‐Shortening, GL‐Strain, and LVEF were 52.1, 26.9, 24.9, and 16.8, respectively. AI MAPSE had the highest χ2 values if counting only death or HHF, indicating it had the strongest association. AI GL‐Shortening had the second strongest association with outcomes, higher than manual GL‐Strain. LVEF remained as the measure with the weakest association with outcomes in this test.
Figure 5 plots the Kaplan‐Meier survival curves for AI MAPSE and AI GL‐Shortening for death, HHF, or both, illustrating the dose‐response relationships. Reduced MAPSE or GL‐Shortening was associated with lower survival rates and higher risk of adverse events. Compared with GL‐Shortening, MAPSE was a stronger predictor with higher log‐rank (176 versus 118 for death, 187 versus 182 for HHF, 277 versus 222 for both). This is consistent with the results of the Cox regression analysis.
Figure 5. Kaplan‐Meier survival curves for artificial intelligence (AI)‐measured global longitudinal shortening (GL‐Shortening) and mitral annular plane systolic excursion (MAPSE) for the outcome data set.
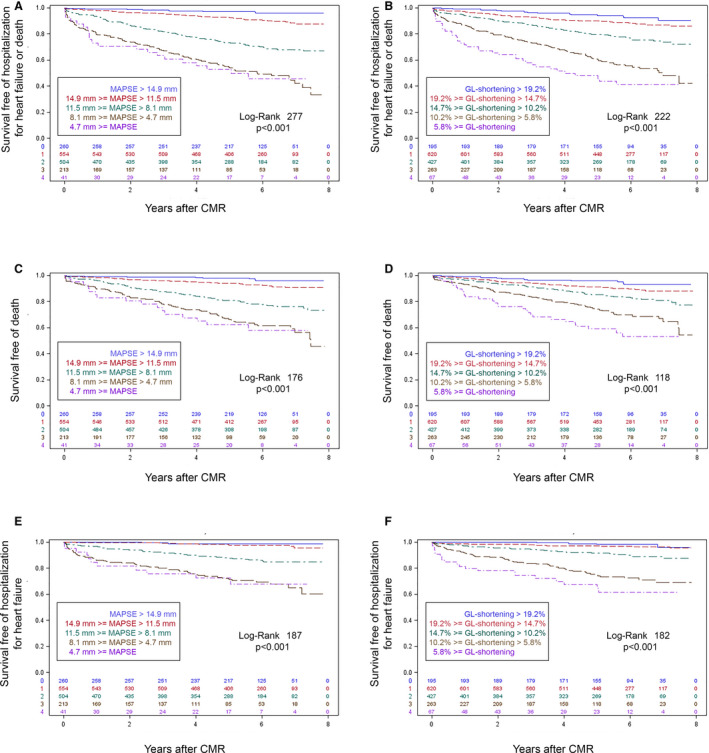
Based on the log‐rank statistic, MAPSE (A, C, and E) associated more strongly with outcomes of the composite of hospitalization for heart failure (HHF) or death (top row, n=335), death only (middle row, n=250), and HHF in survivors (bottom row, n=147) than the GL‐Shortening (B, D, and F) among participants (n=1572). To illustrate dose‐response relationships in both MAPSE and GL‐Shortening, Kaplan‐Meier curves relative to the median, standard deviation increments separated the strata, with the top 2 strata above the median and the lower 3 strata below the median (given the skewed distributions visible on histograms). CMR indicates cardiac magnetic resonance.
For LVEF, the patients were divided into 2 groups (Group A: EF >55%, 899 patients with 118 events; Group B: EF <55%, 673 patients with 217 events). For Group A, MAPSE and GL‐Shortening had χ2 values of 89.3 (P<0.0001) and 36.3 (P<0.0001), respectively. For Group B, MAPSE and GL‐Shortening had χ2 values of 83.6 (P<0.0001) and 69.1 (P<0.0001), respectively. Analysis was also conducted for multivariable Cox regression, with the adjustments listed in the footnotes of Table 2. Both MAPSE and GL‐Shortening remained strong predictors for prognosis (χ2 values, MAPSE: 40.3, P<0.0001 for Group A and 16.7, P<0.0001 for Group B; GL‐Shortening: 7.7, P=0.0055 for Group A and 11.4, P=0.0007 for Group B). For GL‐Strain among patients with GL‐Strain higher than 15.5 (849 patients with 88 events), MAPSE and GL‐Shortening had χ2 values of 55.0 (P<0.0001) and 18.8 (P<0.0001), respectively. For patients with higher GL‐Strain (<15.5, 723 patients with 247 events), χ2 values were 78.0 (P<0.0001) for MAPSE and 46.9 (P<0.0001) for GL‐Shortening. For the multivariable analysis, MAPSE had χ2 values of 34.9 (P<0.0001) and 21.7 (P<0.0001) for patients with higher and lower GL‐Strain, respectively. For GL‐Shortening, these values were 9.3 (P=0.0023) and 9.1 (P=0.0026), respectively.
Table S1 summarizes the correlation between MAPSE, GL‐Shortening, GL‐Strain, EF and end‐diastolic volume, end‐systolic volume, and late gadolinium enhancement findings. Results show that MAPSE was weakly to modestly correlated with end‐diastolic volume, end‐systolic volume, and late gadolinium enhancement mass (r=−0.28 to −0.51), whereas more strongly correlated with GL‐Shortening (r=0.84) and GL‐Strain (r=0.79).
The net reclassification index and integrated discriminant improvement results are given in Table S2 by comparing MAPSE and GL‐Shortening against LVEF and GL‐Strain. Net reclassification index and integrated discriminant improvement parameters were all significant for all comparisons except for GL‐Shortening versus GL‐Strain, which may be related to their high correlation (r=0.79, P<0.001). Fully adjusted multivariable models yielded significant continuous net reclassification index parameters, but not categorical net reclassification index or integrated discriminant improvement parameters.
Figure S1 further provides the multivariable Cox modeled curves for HHF or death.
Discussion
This study proposed and implemented an AI approach for the automated measurement of GL‐Shortening and MAPSE from LAX CMR cine images. Validation in healthy and diseased subjects across multiple scanners and hospitals showed automated AI measurements are reliable and have prognostic value to predict adverse cardiovascular events, whereas AI MAPSE was found to be the strongest predictor of all‐cause‐mortality, HHF, or both. AI GL‐Shortening is equivalent to manual GL‐Strain and exceeds LVEF for predicting adverse events. These findings are consistent across the univariable and multivariable Cox regression analysis. The AI solution has been integrated into MR scanners and is capable of fast computation to produce analysis results in a few seconds after the conclusion of cine imaging, with the resultant landmarks and plot of LV length displayed to allow for manual quality assurance oversight.
Four key aspects related to clinical applicability were investigated. First, the model was fast (additional computing time ≈1.1 seconds) and runs on MR scanners. The results are presented to clinicians because the generated landmarks are available for visual inspection. It is noted this visualization only servers as a check to capture failed detection and further discard incorrect measurement results. Second, the AI‐measured parameters, GL‐Shortening and MAPSE, were in excellent agreement with the human expert. Third, the reproducibility was higher than for a human expert in the multicenter, multidisease scan–rescan data set. Finally, the results of AI were prognostic, with AI MAPSE being the strongest predictor over manual GL‐Strain and LVEF. Taken together, these results suggest the proposed solution saves processing time and can be further deployed and evaluated, ideally in multicenter trials, for its usefulness as a component in clinical workflows where currently manual measurements are used.
Inspecting the Kaplan‐Meier curves qualitatively, GL‐Shortening showed a more graded dose‐response relationship with outcomes than MAPSE, whereas the relationship between MAPSE and outcomes was not entirely linear in those MAPSE categories that most deviated from normal. This observation may indicate a potential threshold or plateau effect for MAPSE.
Results of the scan–rescan test showed the model surpassed human operators achieving better reproducibility. This is consistent with previous studies, where the variations introduced by manual measurements were found to be equivalent or greater than a model in measuring LVEF. 22 Better precision could lead to broader and faster adoption of automated analysis and reporting provided by deep‐learning models, 31 , 32 which is encouraged, because the AI processing shown in this study can be much faster than manual measurement. 33 , 34 Precision improvement can help disease discrimination because with more precise measurement, normal reference ranges will likely narrow, helping detect early disease. Models have started showing such advantages for LVEF. 17 , 22 , 34 , 35 We predict models trained with large and representative patient data sets will be more objective, reproducible, and much faster than manual measurements.
The proposed solution was deployed in‐line on MR scanners. This way of model deployment can be used for other imaging applications. We anticipate the success of deep learning may open new opportunities to build more in‐line solutions to streamline imaging, analysis, and reporting workflows. The potential benefit of in‐line versus off‐line processing is that by providing immediate results, the user has more information to optimize the ongoing image acquisition as it happens. For example, the user will be able to make an immediate assessment about whether LV function is normal or not, which might drive additional imaging that is not in the original protocol. In the case of poor LV longitudinal function that might be indicative of infiltrative disease such as amyloidosis, the user might add multiparametric (T1 and T2) tissue characterization that otherwise might not be in the standard protocol because of time constraints. Furthermore, in instances of poor LV function, the user might choose to adjust protocols such as first pass perfusion to increase the number of measurements to capture the first pass, or they might add additional assessment of the valves such as phase contrast based on AI landmark measurement of valve diameters. The AI analysis output figure plots as well as a summary report (see Video S4) may also be used for quality control. When having access to in‐line measurement results, the reporting physician has more information for a preliminary assessment at the conclusion of the study, which might be used to refer the patient for immediate care, if warranted. The in‐line reporting with the prompt feedback during the imaging study can drive additional imaging protocols to be added. For example, a reduced LAX function indicated by the reduced GL‐Strain may prompt a T1/extracellular volume fraction map to look for fibrosis. In patients who have repeat imaging (eg, patients on cardiotoxic chemotherapy), a newly found abnormal GL‐Strain may provide guidance to further acquire contrast‐enhanced scans. Finally, having the analysis complete at the conclusion of the scan shortens the offline analysis, because off‐line analysis of longitudinal function will not be needed. The scanner image reconstruction computer is powerful, and the in‐line implementation is rapid. Some off‐line tools are incompatible with deployment on slower computers such as laptops. In‐line analysis may potentially eventually lead to better standardization of analysis, which is currently an issue using multiple analysis tools.
Although CMR image analysis is conventionally performed on an offline high‐performance workstation with significant manual interaction, deep learning could permit far simpler and faster ways of reviewing and reporting studies with increased reliability. Further comparison and standardization may be worthwhile to facilitate more widespread adoption of deep learning. This study evaluated the prognostic value and reproducibility of AI derived GL‐Shortening and MAPSE from LAX cine imaging. The benefits of in‐line processing include freeing reporting cardiologists from manual measurement and providing fast, in‐session feedback to operators while the scan is ongoing. The in‐line AI analysis can be extended to enable other measurements (eg, EF, stroke volume, LV mass) to further increase the level of automation. A set of thoroughly evaluated deep‐learning solutions can serve as a standardized benchmark and be available on imaging devices, with the potential for broader deployment.
The outcome study presented in this article was performed retrospectively, because the data set has been previously curated with approximately a 5‐year follow‐up time period. It should be noted that a more ideal prospective study is the next step to further evaluate the clinical usefulness of the in‐line AI solution on the imaging device. As demonstrated in the Video S4, the in‐line analysis runs on the MR imaging device and completed the reconstruction and analysis within a few seconds after the end of cine data acquisition, where the AI‐generated report was saved to the picture archiving and communication system and available for reviewing by the reporting cardiologists. This study evaluated the prognostic value and quantified reproducibility. Based on these results, identical models are being used prospectively.
The usefulness of this AI solution is an add‐on to the existing imaging protocol and does not lengthen imaging acquisition. The AI analysis adds only a few seconds to the imaging reconstruction time. This added computing time is often overlapped with the succeeding acquisition or the operator preparing the next scan. For a typical CMR study to exam the structure and function of the heart, one or a few LAX views are typically acquired to assess longitudinal contractility. In this case, the added automation can free cardiologists from downstream manual measurement. If the operator decides not to acquire the LAX views, the AI analysis will not be applied.
Previous studies have established the prognostic value of MAPSE and GL‐Strain. Both measurements have been found to be associated with adverse events, 10 , 36 , 37 with few direct comparisons. The current study compared their prognostic association in a large patient data set, showing MAPSE was a stronger predictor than GL‐Strain and GL‐Shortening. This could be attributable to the fact that atrioventricular plane displacement is the major contributor to LV systolic function. 9 The measurement of apex motion can add extra confounding factors such as imperfect imaging planning and difficulties in visualizing the apex because of high intensity epicardial fat or balanced steady state‐free precession banding artifacts. Some CMR studies report only lateral wall atrioventricular plane displacement as a measurement of MAPSE, 15 whereas the current study averaged 2 valve plane excursions per view across all 3 LAX views. With the detected landmarks, either or both ways of reporting are possible.
This study evaluated the prognostic value of AI‐measured GL‐Shortening and MAPSE, but did not measure their direct clinical impact, which is out of the scope of this study. The current study did deploy the model in MR scanners (Video S4) to enable automated measurement and reporting. This deployment will facilitate future studies to measure direct impact on clinical care.
The current study showed MAPSE was a stronger predictor for outcomes than GL‐Strain. A recent study 38 compared the predictive power of echo MAPSE and GL‐Strain among patients with ischemic heart failure (n=1277) and reduced LVEF (<50%). That study reported similar prediction power for all‐cause mortality between MAPSE and GL‐Strain . A possible reason for the difference in outcome associations is likely related to the differences in the tested patient cohorts. In the ischemic heart failure study, patients had much lower LVEF (median [full range] 39% [32%–45%] versus 57% [7%–79%] in the current study). Importantly, the current study included patients without amyloidosis who were enrolled consecutively and had greater variability in LVEF, which is associated with a statistically greater challenge in predicting outcomes. These results warrant the need to conduct further outcome studies comparing MAPSE and GL‐Strain. Echo MAPSE has the disadvantage of the dependence on the angle image acquisition. Best practices suggests that measurements should be conducted bilaterally and from multiple cardiac views. 10 Echo GL‐Strain measurements using speckle tracking have grown in popularity as an alternative for the evaluation of global LV function. The more complicated processing algorithm used for echo GL‐Strain has shown higher vendor dependence. Differences in GL‐Strain between vendors can be up to 3.7% (≈20% relative difference). 39 A test–retest study in patients (n=30) 40 reported the relative smallest detectable change to be 10.7% for MAPSE and 14.7% for echo GL‐Strain, which are comparable to the current study.
In the outcome study, the Kaplan‐Meier survival curves was computed with the median, and the SD increments separated the strata, with the top 2 strata above the median and the lower 3 strata being below the median. The purpose is to illustrate the dose‐response relationships to death, HHF, or both. Another good method is to fit the survivor data with spline function and generate more continuous response with ability to extrapolate beyond the observed data. Although this study exploited the SD method, the flexible parametric spline model is a good alternative for further investigation.
Compared with echo measurement of MAPSE and GL‐Strain, the proposed method for CMR measurement has the benefits of being automated, and AI analysis eliminates operator‐introduced variation, as shown in the test–retest results. The processing time is most likely reduced because of the in‐line processing, whereas extra measurements are often needed off‐line for echo. 10 Head‐to‐head comparison between 2 imaging modalities showed good intermodality agreement, 41 with CMR GL‐Strain having better test–retest repeatability. This finding was further confirmed by another study 42 reporting the good agreement in CMR and echo‐measured MAPSE in adults (n=111).
Limitations
First, this study was conducted retrospectively on 2 previously curated data sets, because the CMR cine imaging is standardized across vendors, and the objective of this study was to evaluate the performance of an AI image analysis solution. Further prospective studies are required to test model performance in multicenter trials. Second, for the outcome data set, which was previously curated to study the relationship of GL‐Strain and extracellular volume to the patient outcome, the subjects with hypertrophic cardiomyopathy were excluded. The trained CNN model was tested on the hypertrophic cardiomyopathy subjects in the scan–rescan data set and was found to correctly detect landmarks (as demonstrated in Figure S2). However, lacking this patient cohort represents a clear limitation for this study. Third, only the reproducibility of trained models was evaluated on the scan–rescan data sets. A recent study on short‐axis cine image analysis 22 shows deep‐learning models provide precision comparable to experts, with significant savings in processing time. The inter‐ and intraobserver variation in manual LAX cine measurements could be further evaluated and compared with AI performance. Fourth, outcome data are from a single center with exclusion for specific disease groups. However, the disease groups excluded, especially cardiac amyloidosis, are those where longitudinal shortening and strain are most likely to be prognostic, and therefore, inclusion of these patients would likely increase rather than decrease the prognostic power of the tool. Model performance should be further validated for individual diseases, which would require collection of more training data. Fifth, global contractility was the focus of this study, whereas regional measurement will require development of new deep‐learning models. Although regional strain has been measured using feature‐tracking tools for assessing cardiac function, it is generally the global measures that are more likely to have widespread application. Sixth, because the manual MAPSE and GL‐Shortening results were not available for the outcome data set, they were not included in this study for further analysis. In the outcome study, the AI‐derived measurements were compared with manually measured EF. A better comparison is against the AI LVEF, which is not available for this study. This comparison is to be conducted in a future study. Last, AI performance was compared with 1 expert. Future studies will be needed to conduct multicenter clinical trials, establish normal reference ranges for the AI measures, and evaluate diagnostic performance for different pathological conditions.
The source code for the model and training is available at the github repository (https://github.com/xueh2/CMR_LandMark_Detection.git).
Conclusions
An AI approach was proposed, evaluated, and deployed to provide automated measurement of GL‐Shortening and MAPSE from CMR cine imaging. Reproducibility of the model was better than the human operators. The outcome study showed both AI measurements were associated with adverse outcomes, with MAPSE being the strongest predictor, being better than manual measures of GL‐Strain and LVEF.
Sources of Funding
This work was supported by the National Heart, Lung, and Blood Institute, National Institutes of Health by the Division of Intramural Research (Z1A‐HL006214‐05, Z1A‐HL006242‐02).
Disclosures
None.
Supporting information
Table S1‐S2
Figure S1‐S2
Supplemental Video Legends
Video S1‐S4
Supplemental Material for this article is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.121.023849
For Sources of Funding and Disclosures, see page 14.
References
- 1. Russo C, Jin Z, Elkind MSV, Rundek T, Homma S, Sacco RL, Di Tullio MR. Prevalence and prognostic value of subclinical left ventricular systolic dysfunction by global longitudinal strain in a community‐based cohort. Eur J Heart Fail. 2014;16:1301–1309. doi: 10.1002/ejhf.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Solomon SD, Anavekar N, Skali H, McMurray JJV, Swedberg K, Yusuf S, Granger CB, Michelson EL, Wang D, Pocock S, et al. Influence of ejection fraction on cardiovascular outcomes in a broad spectrum of heart failure patients. Circulation. 2005;112:3738–3744. doi: 10.1161/CIRCULATIONAHA.105.561423 [DOI] [PubMed] [Google Scholar]
- 3. Asgeirsson D, Hedström E, Jögi J, Pahlm U, Steding‐Ehrenborg K, Engblom H, Arheden H, Carlsson M. Longitudinal shortening remains the principal component of left ventricular pumping in patients with chronic myocardial infarction even when the absolute atrioventricular plane displacement is decreased. BMC Cardiovasc Disord. 2017;17:1–9. doi: 10.1186/s12872-017-0641-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buss SJ, Breuninger K, Lehrke S, Voss A, Galuschky C, Lossnitzer D, Andre F, Ehlermann P, Franke J, Taeger T, et al. Assessment of myocardial deformation with cardiac magnetic resonance strain imaging improves risk stratification in patients with dilated cardiomyopathy. Eur Heart J Cardiovasc Imaging. 2015;16:307–315. doi: 10.1093/ehjci/jeu181 [DOI] [PubMed] [Google Scholar]
- 5. Jung IH, Park JH, Lee JA, Kim GS, Lee HY, Byun YS, Kim BO. Left ventricular global longitudinal strain as a predictor for left ventricular reverse remodeling in dilated cardiomyopathy. J Cardiovasc Imaging. 2020;28:137–149. doi: 10.4250/jcvi.2019.0111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Romano S, Judd RM, Kim RJ, Heitner JF, Shah DJ, Shenoy C, Evans K, Romer B, Salazar P, Farzaneh‐Far A. Feature‐tracking global longitudinal strain predicts mortality in patients with preserved ejection fraction: a multicenter study. JACC Cardiovasc Imaging. 2020;13:940–947. doi: 10.1016/j.jcmg.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Flachskampf FA, Blankstein R, Grayburn PA, Kramer CM, Kwong RYK, Marwick TH, Nagel E, Sengupta PP, Zoghbi WA, Chandrashekhar Y. Global longitudinal shortening: a positive step towards reducing confusion surrounding global longitudinal strain. JACC Cardiovasc Imaging. 2019;12:1566–1567. doi: 10.1016/j.jcmg.2019.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schuster A, Backhaus SJ, Stiermaier T, Kowallick JT, Stulle A, Koschalka A, Lotz J, Kutty S, Bigalke B, Gutberlet M, et al. Fast manual long‐axis strain assessment provides optimized cardiovascular event prediction following myocardial infarction. Eur Heart J Cardiovasc Imaging. 2019;20:1262–1270. doi: 10.1093/ehjci/jez077 [DOI] [PubMed] [Google Scholar]
- 9. Carlsson M, Ugander M, Mosén H, Buhre T, Arheden H. Atrioventricular plane displacement is the major contributor to left ventricular pumping in healthy adults, athletes, and patients with dilated cardiomyopathy. Am J Physiol‐Hear Circ Physiol. 2007;292:1452–1459. doi: 10.1152/ajpheart.01148.2006 [DOI] [PubMed] [Google Scholar]
- 10. Hu K, Liu D, Herrmann S, Niemann M, Gaudron PD, Voelker W, Ertl G, Bijnens B, Weidemann F. Clinical implication of mitral annular plane systolic excursion for patients with cardiovascular disease. Eur Heart J Cardiovasc Imaging. 2013;14:205–212. doi: 10.1093/ehjci/jes240 [DOI] [PubMed] [Google Scholar]
- 11. Willenheimer R, Cline C, Erhardt L, Israelsson B. Left ventricular atrioventricular plane displacement: an echocardiographic technique for rapid assessment of prognosis in heart failure. Heart. 1997;78:230–236. doi: 10.1136/hrt.78.3.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morton G, Schuster A, Jogiya R, Kutty S, Beerbaum P, Nagel E. Inter‐study reproducibility of cardiovascular magnetic resonance myocardial feature tracking. J Cardiovasc Magn Reson. 2012;14:1–8. doi: 10.1186/1532-429X-14-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jolly MP, Jordan JH, Meléndez GC, McNeal GR, D’Agostino RB, Hundley WG. Automated assessments of circumferential strain from cine CMR correlate with LVEF declines in cancer patients early after receipt of cardio‐toxic chemotherapy. J Cardiovasc Magn Reson. 2017;19:1–12. doi: 10.1186/s12968-017-0373-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saba SG, Chung S, Bhagavatula S, Donnino R, Srichai MB, Saric M, Katz SD, Axel L. A novel and practical cardiovascular magnetic resonance method to quantify mitral annular excursion and recoil applied to hypertrophic cardiomyopathy. J Cardiovasc Magn Reson. 2014;16:1–9. doi: 10.1186/1532-429X-16-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bulluck H, Ngamkasem H, Sado D, Treibel TA, Fontana M, Maestrini V, Moon J, Hausenloy DJ, Herrey AS, White SK. A simple technique to measure TAPSE and MAPSE on CMR and normal values. J Cardiovasc Magn Reson. 2014;16:1–2. doi: 10.1186/1532-429X-16-S1-P22 24387349 [DOI] [Google Scholar]
- 16. Tantiongco J‐P, Grover S, Perry R, Bradbrook C, Leong D, Selvanayagam J. Comparison of GLS via echocardiography and tissue tracking by Cardiac Magnetic Resonance imaging. J Cardiovasc Magn Reson. 2015;17:1–2. doi: 10.1186/1532-429X-17-S1-P77 25589308 [DOI] [Google Scholar]
- 17. Ito H, Ishida M, Makino W, Goto Y, Ichikawa Y, Kitagawa K, Omori T, Dohi K, Ito M, Sakuma H. Cardiovascular magnetic resonance feature tracking for characterization of patients with heart failure with preserved ejection fraction: correlation of global longitudinal strain with invasive diastolic functional indices. J Cardiovasc Magn Reson. 2020;22:1–11. doi: 10.1186/s12968-020-00636-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Erley J, Genovese D, Tapaskar N, Alvi N, Rashedi N, Besser SA, Kawaji K, Goyal N, Kelle S, Lang RM, et al. Echocardiography and cardiovascular magnetic resonance based evaluation of myocardial strain and relationship with late gadolinium enhancement. J Cardiovasc Magn Reson. 2019;21:1–11. doi: 10.1186/s12968-019-0559-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Onishi T, Saha SK, Delgado‐Montero A, Ludwig DR, Onishi T, Schelbert EB, Schwartzman D, Gorcsan J III. Global longitudinal strain and global circumferential strain by speckle‐tracking echocardiography and feature‐tracking cardiac magnetic resonance imaging: comparison with left ventricular ejection fraction. J Am Soc Echocardiogr. 2015;28:587–596. doi: 10.1016/j.echo.2014.11.018 [DOI] [PubMed] [Google Scholar]
- 20. Rangarajan V, Chacko SJ, Romano S, Jue J, Jariwala N, Chung J, Farzaneh‐Far A. Left ventricular long axis function assessed during cine‐cardiovascular magnetic resonance is an independent predictor of adverse cardiac events. J Cardiovasc Magn Reson. 2016;18:1–10. doi: 10.1186/s12968-016-0257-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fröjdh F, Fridman Y, Bering P, Sayeed A, Maanja M, Niklasson L, Olausson E, Pi H, Azeem A, Wong TC, et al. Extracellular volume and global longitudinal strain both associate with outcomes but correlate minimally. JACC Cardiovasc Imaging. 2020;13:2343–2354. doi: 10.1016/j.jcmg.2020.04.026 [DOI] [PubMed] [Google Scholar]
- 22. Bhuva AN, Bai W, Lau C, Davies RH, Ye Y, Bulluck H, McAlindon E, Culotta V, Swoboda PP, Captur G, et al. A multicenter, scan‐rescan, human and machine learning CMR study to test generalizability and precision in imaging biomarker analysis. Circ Cardiovasc Imaging. 2019;12:1–11. doi: 10.1161/CIRCIMAGING.119.009214 [DOI] [PubMed] [Google Scholar]
- 23. Lau C, Gerber B, Hamilton‐craig C. Diagnosis and risk stratification in hypertrophic cardiomyopathy using machine learning wall thickness measurement: a comparison with human test‐retest performance. Lancet Digit Heal. 2021;3:e20–e28. doi: 10.1016/S2589-7500(20)30267-3 [DOI] [PubMed] [Google Scholar]
- 24. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)‐A metadata‐driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xue H, Artico J, Fontana M, Moon JC, Davies RH, Kellman P. Landmark detection in Cardiac Magnetic Resonance Imaging using a convolutional neural network. Radiol Artif Intell. 2021;3:e200197. doi: 10.1148/ryai.2021200197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xue H, Davies R, Hansen D, Tseng E, Fontana M, Moon JC. Kellman P. Gadgetron inline AI: effective model inference on MR scanner. ISMRM. 2019:4837. [Google Scholar]
- 27. Puntmann VO, Valbuena S, Hinojar R, Petersen SE, Greenwood JP, Kramer CM, Kwong RY, McCann GP, Berry C, Nagel E. Society for Cardiovascular Magnetic Resonance (SCMR) expert consensus for CMR imaging endpoints in clinical research: part I—Analytical validation and clinical qualification. J Cardiovasc Magn Reson. 2018;20:1–23. doi: 10.1186/s12968-018-0484-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weir JP. Quantifying test‐retest reliability using the intraclass correlation coefficient and the SEM. J Strength Cond Res. 2005;19:231–240. [DOI] [PubMed] [Google Scholar]
- 29. Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 1979;86:420–428. doi: 10.1037/0033-2909.86.2.420 [DOI] [PubMed] [Google Scholar]
- 30. Lin LI. A Concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–268. doi: 10.2307/2532051 [DOI] [PubMed] [Google Scholar]
- 31. Johnson KW, Torres Soto J, Glicksberg BS, Shameer K, Miotto R, Ali M, Ashley E, Dudley JT. Artificial intelligence in cardiology. J Am Coll Cardiol. 2018;71:2668–2679. doi: 10.1016/j.jacc.2018.03.521 [DOI] [PubMed] [Google Scholar]
- 32. Petersen SE, Abdulkareem M, Leiner T. Artificial intelligence will transform cardiac imaging—opportunities and challenges. Front Cardiovasc Med. 2019;6:1–6. doi: 10.3389/fcvm.2019.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xue H, Davies R, Brown LA, Knott KD, Kotecha T, Fontana M, Plein S, Moon JC, Kellman P. Automated inline analysis of myocardial perfusion MRI with deep learning. Radiol Artif Intell. 2020;2:e200009. doi: 10.1148/ryai.2020200009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bai W, Sinclair M, Tarroni G, Oktay O, Rajchl M, Vaillant G, Lee AM, Aung N, Lukaschuk E, Sanghvi MM, et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J Cardiovasc Magn Reson. 2018;20:65. doi: 10.1186/s12968-018-0471-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suinesiaputra A, Bluemke DA, Cowan BR, Friedrich MG, Kramer CM, Kwong R, Plein S, Schulz‐Menger J, Westenberg JJM, Young AA, et al. Quantification of LV function and mass by cardiovascular magnetic resonance: multi‐center variability and consensus contours. J Cardiovasc Magn Reson. 2015;17:1–8. doi: 10.1186/s12968-015-0170-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mayr A, Pamminger M, Reindl M, Greulich S, Reinstadler SJ, Tiller C, Holzknecht M, Nalbach T, Plappert D, Kranewitter C, et al. Mitral annular plane systolic excursion by cardiac MR is an easy tool for optimized prognosis assessment in ST‐elevation myocardial infarction. Eur Radiol. 2020;30:620–629. doi: 10.1007/s00330-019-06393-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reindl M, Tiller C, Holzknecht M, Lechner I, Beck A, Plappert D, Gorzala M, Pamminger M, Mayr A, Klug G, et al. Prognostic implications of global longitudinal strain by feature‐tracking cardiac magnetic resonance in ST‐elevation myocardial infarction. Circ Cardiovasc Imaging. 2019;12:e009404. doi: 10.1161/CIRCIMAGING.119.009404 [DOI] [PubMed] [Google Scholar]
- 38. Liu D, Wagner C, Hu K, Lengenfelder B, Ertl G, Frantz S, Nordbeck P. Prognostic value of global longitudinal strain versus mitral annular plane systolic excursion in patients with ischemic heart failure. Eur Heart J. 2020;41 (Supplement_2):. doi: 10.1093/ehjci/ehaa946.1011 [DOI] [Google Scholar]
- 39. Støylen A, Mølmen HE, Dalen H. Relation between mitral annular plane systolic excursion and global longitudinal strain in normal subjects: the HUNT study. Echocardiography. 2018;35:603–610. doi: 10.1111/echo.13825 [DOI] [PubMed] [Google Scholar]
- 40. Baron T, Berglund L, Hedin EM, Flachskampf FA. Test–retest reliability of new and conventional echocardiographic parameters of left ventricular systolic function. Clin Res Cardiol. 2019;108:355–365. doi: 10.1007/s00392-018-1363-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pryds K, Larsen AH, Hansen MS, Grøndal AYK, Tougaard RS, Hansson NH, Clemmensen TS, Løgstrup BB, Wiggers H, Kim WY, et al. Myocardial strain assessed by feature tracking cardiac magnetic resonance in patients with a variety of cardiovascular diseases—A comparison with echocardiography. Sci Rep. 2019;9:11296. doi: 10.1038/s41598-019-47775-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sepúlveda‐Martínez A, Steding‐Ehrenborg K, Rodríguez‐López M, Ostenfeld E, Valenzuela‐Alcaráz B, Heiberg E, Gratacós E, Prat‐González S, Crispi F, Hedström E. Atrioventricular plane displacement versus mitral and tricuspid annular plane systolic excursion: a comparison between cardiac magnetic resonance and M‐mode echocardiography. Clin Physiol Funct Imaging. 2021;41:262–270. doi: 10.1111/cpf.12693 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1‐S2
Figure S1‐S2
Supplemental Video Legends
Video S1‐S4


