Abstract
Purpose
Cutaneous leishmaniasis (CL) is a major vector-borne disease that affects people globally, including Iran. Different factors are associated with leishmaniasis pathogenicity; recently, a link of the possible relationship between Leishmania RNA Virus (LRV) and disease severity was proposed, especially in the New World leishmaniasis (NWL). This study was aimed to investigate the presence of LRV2 in Leishmania isolates in Aran o Bidgol, Isfahan province.
Methods
Samples were collected from 110 CL-suspected patients referred to the health center. In this study, we aimed to investigate CL cases (parasitologically and clinically), identify Leishmania species (by ITS1-PCR–RFLP), and finally detection of LRV2 (by RdRp-semi-nested PCR).
Results
Parasitological methods showed 60 positive cases, based on the HaeIII enzyme restriction profile, 59 cases were caused by L. major and 1 case by L. tropica. Our project is the first study on LRV2 isolation in Aran o Bidgol city and the LRV was successfully detected from a single L. major isolated in a women’s hand lesion. Using BLAST, 94.8–100% similarity was observed in the RdRp sequence of current LRV isolate with those available in GenBank from Iran or overseas.
Conclusion
L. major was the main cause of CL in Aran o Bidgol, although L. tropica is also present in a much lower proportion in the area. This is the first report on the presence of LRV2 in Aran o Bidgol and the fifth in Iran.
Keywords: Cutaneous Leishmaniasis, Leishmania RNA virus, RdRp, Kashan, Iran
Introduction
Several species of Leishmania genus are transmitted by the bite of Phlebotomine sandflies that cause different clinical manifestations of leishmaniasis varies from cutaneous form to fatal form, with an annual estimation of 700,000 to 1 million new cases [1]. Cutaneous leishmaniasis (CL) is the most prevalent form of the disease, which produces skin lesions on exposed parts of the body that leave life-long scars. According to the World Health Organization’s most recent data, over 87% of new CL cases occurred in ten countries, including Iran and its 3 bordering nations (Afghanistan, Iraq, and Pakistan). CL is widely distributed in Iran, distributed in 25 provinces out of 31 [2–6]. Isfahan province is located in the central part of Iran and is one of the known foci for zoonotic cutaneous leishmaniasis (ZCL), with the highest reported prevalence (66%) [2, 6, 7]. One of the most important ZCL areas in Isfahan is Aran o Bidgol city [8], which is located in the northern part. Collected data from the health system showed an increasing trend in CL cases in recent years, which motivated us to conduct the project in this area. Fortunately, the number of cases decreased last year in terms of some interventional control methods, especially rodent control (http://healthab.kaums.ac.ir/Default.aspx?PageID=219). Moreover, the decrease in detected CL cases may be due to the fear of coronavirus, which made suspected patients refuse to refer to Health Medical Centers.
Naturally, leishmaniasis has a complex cycle involving vertebrate hosts, Phlebotominae sand flies and Leishmania parasites, consequently Leishmania pathogenicity is affected by several factors associated with these elements [9, 10]. Recently, the role of endosymbiotic organisms was highlighted in the pathogenicity of Leishmania parasites especially in the New World, with an increasing interest in Leishmania RNA viruses (LRV) as endosymbiotic organisms of Leishmania parasites [11, 12]. Leishmania RNA viruses belong to Totiviridae family; viral particle is composed of a dsRNA genome (4–8 kb in length), that encodes a non-enveloped capsid and an essential protein for the virus replication named RNA-dependent RNA polymerase (RDRP) [13]. LRVs showed significant sequence differences in Leishmania parasite of new and old world leishmaniasis which were categorized as LRV1 and LRV2, respectively. It seems that the divergence of LRV had occurred prior to their associated parasites and coevolution happened later due to the geographical separation of Leishmania parasites. Hence, the genetic distances between LRV1 and LRV2 are similar to differences among Leishmania parasites in the new and old world leishmaniasis [14, 15].
A link of the possible relationship of LRV and leishmaniasis severity or failure of treatment was made for the first time between the presence of LRV1 and muco-cutaneous form of the disease in the new world and has since been reported by several other researchers [11, 13, 16, 17]. Recently, some controversial published articles questioned the relationship between the virus and the severity of the disease [18, 19]; this discrepancy needed more comprehensive studies to be clarified.
Many researches were conducted across the globe to isolate the virus from various species of Leishmania parasites and to better understand its role in the pathogenesis of leishmaniasis [14, 17, 19]. The story of LRV in the old world began by its first isolation from L. major in Turkmenistan [14], and continued with reports from Ethiopia [20], Iran [21, 22], Turkey [23], and Uzbekistan [24]. The current study was performed with the aim of the first LRV isolation from Leishmania parasites in Aran o Bidgol city, Isfahan province.
Materials and Methods
Ethical Statements
The ethics committee of the Shahid Beheshti University of Medical Sciences approved the research (IR.SBMU.MSP.REC.1398.697). The project was financially supported by the research department of the school of medicine, Shahid Beheshti University of Medical Sciences (Grant no.: 20101). All human strains of Leishmania were isolated from patients as part of normal diagnosis and treatment, with no unnecessary invasive procedures, and with written informed consent recorded at the time of clinical examination. Data on human isolates were recorded in a questionnaire.
Study Area
The study was conducted in Aran o Bidgol city located in the northern part of Isfahan province (Fig. 1), with a population of about 468,000 people in 2016. The town is surrounded by desert from the north and east; thus, it has a typical climate of hot and dry in summer, cold and dry in winter and very little rainfall during the year. This area is one of the old ZCL foci in Iran with an increasing trend in CL cases [8, 25].
Fig. 1.
Map showing the geographical location of Aran o Bidgol (marked with black circle) in Isfahan province, Iran
Sampling Collection and Lesion Analysis
Individuals with clinical suspicion of CL who resided in the study area and were forwarded to the laboratory health center between the years 2018 and 2020 were included in the study. Before including a patient in the study, written informed consent was obtained from each adult participant or from the child’s parents. All information collected were kept under confidential cover.
The samples were taken from any patients that were diagnosed as CL, after careful clinical examination (about 110 cases). Maintaining aseptic conditions (sterilization of the lesions with 70% ethanol), sample collection was done by scrapping the swollen edge of the lesions.
Material Analysis and Leishmania Culture
Exudate materials were air-dried, fixed in methanol, Giemsa stained, and examined for amastigotes under oil immersion. Aspirates were inoculated in Novy–McNeal–Nicolle (NNN) culture medium. After 3–5 days, promastigotes were then transferred to RPMI 1640 medium (Gibco, Life technologies GmbH, Germany) with 10% heat-inactivated fetal bovine serum (Gibco, Germany), 100 U/ml penicillin, and 100 µg/ml streptomycin (Gibco, Germany), and incubated at 25º C. The cultures were examined every 3 days for a maximum period of 30 days to detect promastigotes under optic microscopy.
Leishmania Species Identification by ITS1-PCR–RFLP
All positive parasitological cases were subjected to molecular methods for species identification. DNA was extracted from the culture medium of each individual exudate material, if a sample was lost, extraction was performed from its corresponding stained smear. Bioneer kit (Bioneer Company, Korea) was used for DNA extraction from both materials, according to the manufacturer’s instruction. A fragment of approximately 350 bp of the Internal Transcribed Spacer 1 (ITS1) gene was submitted to amplification in all samples using the specific primer forward (LITSR: 5ʹ-CTGGATCATTTTCCGATG-3) and reverse (L5.8S: 5ʹ-TGATACCACTTATCGCACTT-3) based on the details previously described [26]. Amplification reactions were set for a total volume of 25 µl, containing 12.5 µl master mix (Ampliqon, Denmark), 1 µl forward and reverse primers (10 pmol), 3 µl DNA templates (70 ng), and 8.5 µl nuclease-free water. PCR amplification was done with an initial denaturing step at 94 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, 49 C for 30 s, and 72 °C for 45 s. This process was followed by a final extension at 72 °C for 5 min. According to the manufacturer’s instructions, PCR products (10 µl) were digested with Hae III enzyme. The restriction fragments were analyzed on 1.5% agarose gels and visualized. Standard strain, L. major (MRHO/IR/75/ER), and L. tropica (MHOM/IR/02/Mash10), obtained from the School of Public Health, Tehran University of Medical Sciences, were used to monitor all reactions.
RNA Extraction and cDNA Synthesis
RNA was extracted from samples containing 1 × 106 Stationary-phase promastigotes using a Favorgen RNA extraction kit (Favorgen Biotech) according to the manufacturer’s instructions. Re-suspension was done in 60 µl of nuclease-free water. RNA quantity was checked by the NanoDrop spectrometer (Thermo Scientific Fisher, USA), and based on the results, 1–2 µL was used for cDNA synthesis. The cDNA was then synthesized with a combination of oligo (dT) 18 and random hexamer primers using the cDNA Synthesis Kit (YTA, Iran, Cat No: YT4500) according to the manufacturer’s recommendation.
LRV2 Detection by RdRP-Semi-nested PCR
Hajjaran et al. [21] described a method that was used to detect LRV2. The amplification of about 526 bp of RdRp gene was done in the first run of PCR by LRV F1 (5ʹ TGT AAC CCA CAT AAA CAG TGT GC-3ʹ) and LRV R (5ʹ ATT TCA TCC AGC TTG ACT GGG-3). The forward primer for the second run of semi-nested PCR was LRV F2 (5ʹ AGG ACA ATC CAA TAG GTC GTG-3ʹ), that with LRV R as reverse primer amplify 315 bp of gene. PCR amplification was carried out in a total volume of 25 µl consisting 12.5 µl of master mix, 1 µl of each forward and reverse primers (10 pmol), 1 µl of cDNA and nuclease-free water up to final volume. The PCR conditions for the semi-nested PCR in both stages consisted of initial denaturation of DNA strands at 94 ºC for 5 min followed by 35 cycles of denaturation at 94 ºC for 35 s, primer annealing at 60 ºC for 35 s and strand elongation at 72 ºC for 45 s. A final extension of the strands was made at 72 ºC for 5 min [21]. Amplification product Visualizing was performed at each step on 1.2% Sybr safe gel-stained agarose gel and UV transilluminator. Standard reference strains of LRV 2 [LRV2/IR/2014/HM-2 (Acc. No. KP054245)] were used as positive controls to monitor all steps.
Sequencing and Phylogenetic Analysis
PCR products of the ITS related to 10 Leishmania isolates and PCR product of RdRp gene were purified and sequenced by the Sanger sequencing method in both directions, by Pouya Gostar Gene Company (www.pggene.com) in a Genetic Analyzer ABI 3730 (Bioneer, Daejeon, South Korea). Different software used for genetic data analysis as follows. BioEdit, version 7.0.5 used for multiple alignments, BLAST (available at: http://blast.ncbi.nlm.nih.gov) for finding sequence similarity with GenBank sequences, ClustalW for multiple sequence alignment, and Mega 7 0.0.7 to retrieve phylogenetic trees for the LRV data sets with the maximum likelihood methods using Kimura 2-parameter models, the sequence of Trichomonas vaginalis virus (TVV) used as out group. In addition, bootstrap resampling analysis (1,000 replications) was used to assess the value for a clade as the proportion of the replicate trees that recovered that particular clade [27].
Results
Totally, 110 samples were collected from the patients who were suspected of CL. But, based on parasitological observation, Leishmania amastigotes were found only in 60 cases out of 110 total ones (54%), regarded as positive samples for further work. As described above, in all cases, in addition to slide preparation, parasite culture was performed. Unfortunately, in most cases, the cultivation of the parasite was not successful and failed; as a result, successful cultivation was achieved just in 13 cases out of 60 (21%). The results on Leishmania species identification in 60 samples and LRV detection in 13 cultivations will be presented in the following.
Leishmania Species Identification by ITS1-PCR–RFLP
All parasitological positive samples (60 cases) were subjected to ITS1 PCR–RFLP for Leishmania species identification. PCR has given an amplified fragment of approximately 350 bp in all samples as a positive indicator for the presence of Leishmania species. Digestion pattern of Hae III restriction enzyme has revealed that at least two Leishmania species cause CL with the overwhelming majority of L. major. The restriction fragment profiles for 59 samples (98.3%) have revealed 2 bands (220 and 140 bp) as expected for L. major, and just in 1 sample (1.6%), the profile had 2 visible bands (200 and 60 bp (the third one (50 bp) one normally is invisible)) identified as L. tropica (Fig. 2). For more confidence, 10 samples were sequenced and deposited in GenBank (Accession numbers: MW843610–MW843619); they showed 97–99% similarity with sequences of L. major available in GenBank. Alignment of the sequences, at the minimum similar length, showed 100% intra-species similarity in sequences of nine specimens, just in 1 sequence a substitution of Guanine instead of Adenine was observed at 85 bp position.
Fig. 2.
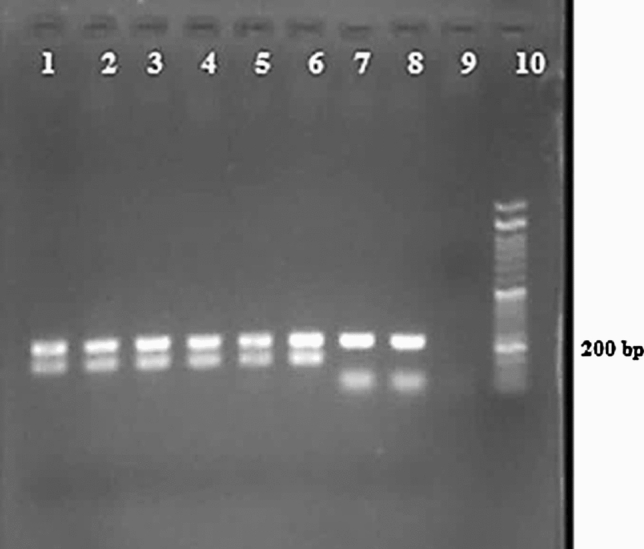
Hae III restriction enzyme digestion profile of amplified ITS 1 region from Leishmania species isolated from CL cases in Aran o Bidgol (on 1.5% gel agarose). Line 1–5: L. major (current isolation), line 6: positive control for L. major (MRHO/IR/75/ER), line 7: L. tropica (current isolation), line 8: positive control for L. tropica (MHOM/IR/02/Mash10), line 9: negative control, line 10: molecular marker (50 bp, Cinnagen Company)
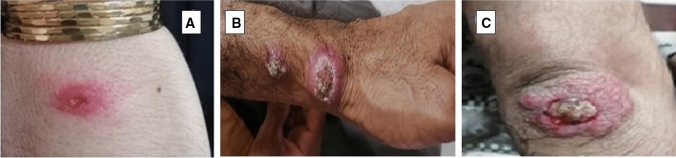
Most of the lesions are clinically wet form (Fig. 3) which is in consistent with L. major infection. The only isolation of L. tropica was from a dry lesion of 10-year-old girl, without any travel history (the photo was not available due to no authorization of her parents). The majority of patients (60%) had just 1 lesion, with the number of lesions ranging from 1–10. A 27-year-old male with 10 lesions was found to have the most lesions (Fig. 2B). In general, the most frequent location of the infection was on the leg (40%), followed by the hands-arms with 31.6%. Most patients were referred to the health center laboratory in less than 2 months after the onset of the skin lesions. The age distribution of the patients varied from 1 to 60 years, most of them were male, 33 out of 60. It seems that all the patients had got the infected bites in the study area, as they have no history of travel to other endemic provinces; only 28 cases (46.7%) have traveled to Agha-Ali Abas Shrine, as a religious place, which is located 80 km far from Aran o Bidgol.
Fig. 3.
Lesions of cutaneous leishmaniasis in the patients forwarded to Laboratory health Centers of Aran o Bidgol between the years 2018 and 2020 (Original images)
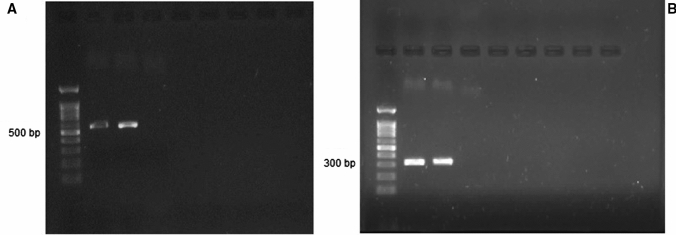
LRV2 Detection in Leishmania Isolates Based on the RdRp-Semi-nested PCR and Phylogenetic Characteristics
The first and second rounds of RdRp-semi-nested PCR of LRV2-positive sample revealed approximately 526 and 315 bp products, respectively. One out of 13 clinical cultures was LRV2 positive (Fig. 4) based on the amplification products. This isolation was done from L. major parasite which isolated from a lesion on the hand of 54-year-old women (Fig. 3A). Sequencing of the only LRV2 isolates of the current study carried out by Sanger sequencing method and sequences were deposited to GenBank (accession Number: MZ182275). Using BLAST, a range of homology (94.8–100%) was seen between the sequence of the current isolate and GenBank available sequences. The highest homology (100%) belonged to some L. major LRV2 isolates of Golestan and Isfahan province.
Fig. 4.
Gel electrophoresis of viral dsRNA, A: first round of RdRp-semi-nested PCR (– 526 bp): lane 1: molecular marker (100 bp, Cinnagen company), line 2: positive control of LRV2 (LRV2/IR/2014/HM-2), lines 3: LRV2-positive sample (current study), line 4: negative control. B: second round of RdRp-semi-nested PCR (– 315 bp): lane 1: molecular marker (100 bp, Cinnagen company), lane 2: positive control (LRV2/IR/2014/HM-2), lane 3: LRV2-positive sample (current study), line 5: negative control
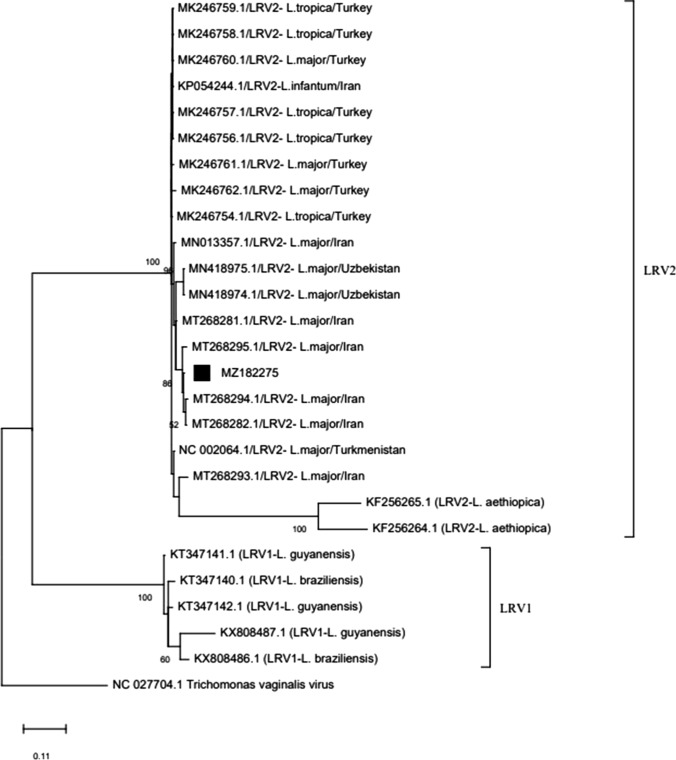
A maximum likelihood tree based on the sequence of the current LRV2 isolate and sequences in GenBank is built (Fig. 5). The current isolate was located among the LRV2 isolates from Iran, Turkey, Ethiopia, Uzbekistan, and Turkmenistan and, as expected, in a separate clade from LRV1 belongs to the new world.
Fig. 5.
Neighbor-joining tree inferred from sequences of current Leishmania RNA virus 2 isolates (marked with) and LRV1 (from new world) and LRV2 (from old world) sequences in GenBank
Discussion
Leishmaniasis is a poverty-related disease with 2 main clinical forms: visceral and cutaneous. The clinical presentation of CL depends on some factors associated with the virulence of the parasite, individual immune response and the site of lesions [28]. Among the various parasite-related factors, the endosymbiotic LRV started attracting attention recently as a virulence factor, with enhancing the disease severity or by treatment failure [11, 16, 17]. The current study was performed to investigate the presence of LRV in the isolated Leishmania spp from the skin lesions of patients in a ZCL focus in Isfahan province, Aran o Bidgol, that the increasing trend of CL cases was worrying in the 3 past years. Iran is among the endemic countries for CL in the world, and based on a met-analysis done in 2021, the highest prevalence of CL was reported for Isfahan (66%) followed by Golestan (64%) and Fars province (63%) [7].
In general, 60 parasitologically positive samples were collected from the CL-suspected patients. Most of the cases (98.3%) were due to L. major based on HaeIII restriction profile and based on a few sequences. It was not far from our expectation, since this area was known as a ZCL focus [7, 29]. However, L. tropica appears to be circulating in the study area, because the only patient with L. tropica lesion has not reported any travel history. Males (55%) outnumbered females, which is supported by other studies conducted in Iran, or overseas [30–32]. This may be in terms of their more exposure to the sand fly bites because of the type and time of their occupations and their less coverage, which makes males more prone to insect bites in general. Even though all age groups are affected by CL, but the disease was more prevalent in the age group 21–30 years (20%), because they spent more time outside as active workforce [7, 31, 32], and there was no patient under 1 year old. Corroborating with some other researches, which could be in terms of their complete coverage and staying mostly indoors, resulting in lesser contact with vectors, as well as the long latency of disease [4, 31]. The proboscis of Phlebotominae sandflies are short and they can feed only on the exposed part of the body [33], as a results, most of the lesions appears on the most open parts of the human skin, confirming the results here (71.6% on extremities). Besides, Leishmania parasites manipulate the sand fly vectors by the production of Promastigote Secretory Gel (PSG), leading to blocked sand fly; subsequently, this phenomena forces them to bite repeatedly or on multiple hosts to achieve a complete blood meal [34]. Infected bite frequency caused multi-lesion; in this study, 40% of the patients had more than 1 lesion. The maximum number of lesion [10] was observed in a man who stayed in his workplace. When we asked him about his background, he said he acquired the nocturnal bites while working in a partially finished building. He has no knowledge of the disease, and he was unaware of the higher risk of infection in such places. In endemic regions, public health education might be very beneficial. Most individuals in the study region, however, seemed to be acquainted with CL symptoms, since the majority of patients (86%) had seen a doctor in less than 2 months. We surveyed 59 samples of L. major and 1 L. tropica isolated from infected patients. But, LRV2 detection was done on 13 samples due to the cultivation failure; just 1 isolate of L. major was LRV2 positive, which is the first report of LRV2 detection in Aran o Bidgol and 5th in Iran [21, 22, 35, 36].
This positive sample was achieved from a lesion on woman’s hand; she had 2 more lesions, one on her hand and the other on her eyelid. The direct follow-up was not possible in terms of the COVID-19 restriction travel and the fear of the patient for a person visit.
Unlike the new world, there is not much data on LRV in the Old World. The history of LRV2 in the old world was backed in 1995, by the first isolation of LRV2 from L. major [15], and followed by some other researchers in Iran, Turkey and Uzbekistan [21–24, 35]. LRV’s global prevalence was estimated at 26.2% [37] and the highest prevalence of LRV2 was reported recently in Golestan province, Iran (69.4%), attributed to the “climate conditions, ecologic and epidemiologic aspects, and possibly population movement to/from Turkmenistan for trade”. As a limitation, parasite cultivation was not successful in most cases and it was failed due to some reasons: (1) contamination, (2) patients’ self-medication which sometimes made sampling impossible, and (3) delay in receiving the samples. Therefore, all genetic analyses were done based on the RdRp sequence obtained from the single LRV isolate in this study. BLAST analysis showed 94.8–100% homology between the RdRp sequence here and the ones which were submitted to the GenBank. In general, high genetic diversity in the virus whole genome and in RdRp sequence has been reported by other researchers [14, 22]. The highest similarity was observed between our isolate and LRV2 isolates belong to Isfahan province (MT629924.1 and MT629923.1) and some isolates from Golestan province (MT268290.1 and MT268288.1). Moreover, the lowest similarity (94.8%) also was found between the current isolate and one isolate from Isfahan (MT629920.1). It is assumed that LRV2 originated in Iran from Turkmenistan [22], the LRV2 isolate here showed relatively high similarity (97.75%) to Turkmenistan isolate (U32108.1).
Phylogenetic analysis of RdRp sequence of current isolate and those available in GenBank revealed 2 distinct clades, including LRV1 and LRV2, which seems quite reasonable due to the high genetic differences between LRV1 and LRV2 (< 40%) [14], resulting from separate geographical distributions and different evolutionary path. In the clade harboring LRV2 sequences, the current isolate stands near the other isolates from the old world (Iran, Turkey, Turkmenistan, Uzbekistan, and Ethiopia) [15, 20, 21, 23, 24].
The association between Leishmania pathogenicity and the presence of LRV is an important issue that has been more addressed in the new world. Many projects are undergoing in the old world, as well in Iran, to find a connection between LRV and Leishmania pathogenesis. Although the results of this study indicate the presence of LRV in this ZCL foci, but it is not possible to link the presence of LRV and the severity of disease in this study. Comprehensive investigations on more clinical samples are needed to provide a better view of the possible association of LRV2 and Leishmania pathogenesis. Future studies could provide helpful information for applied research in different fields of leishmaniasis such as treatment, control, and vaccine candidates.
Conclusion
In conclusion, CL is still an important public health problem in Aran o Bidgol. This study is detected L. major as the most frequent agent causing CL and 1 L. tropica suggesting this species is circulating in the study area. Besides that, we described the first report on the presence of LRV2 in Aran o Bidgol and the 5th in Iran. RdRp-semi-nested-PCR seems a sensitive method for LRV2 detection in clinical samples.
Acknowledgements
The project was financially supported by the Research Department of the School of Medicine, Shahid Beheshti University of Medical Sciences (Grant no.: 20101). The article has been extracted from the thesis written by Mrs. Fatemeh Zare Hoseinabadi (Registration no: 220). We would like to thank the administrators, authorities and personnel of Kashan Health center for their kind cooperation during the sample collection and all staff in the department of Medical Parasitology and Mycology, School of Public Health, Tehran University of Medical Sciences. We are also grateful of Dr. Farokhi for their kind assistance. Most importantly, the authors would like to thank all individuals for giving their permission to collect samples and for their participation in this study.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization. Fact sheets on leishmaniasis. Available at: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis
- 2.Ghatee MA, Taylor WR, Karamian M. The geographical distribution of cutaneous leishmaniasis causative agents in Iran and its neighboring countries a review. Front Public Health. 2020;8:11. doi: 10.3389/fpubh.2020.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohebali M. Visceral leishmaniasis in Iran: review of the epidemiological and clinical features. Iran J Parasitol. 2013;8(3):348–58. [PMC free article] [PubMed] [Google Scholar]
- 4.Norouzinezhad F, Ghaffari F, Norouzinejad A, Kaveh F, Gouya MM. Cutaneous leishmaniasis in Iran: results from an epidemiological study in urban and rural provinces. Asian Pac J Trop Biomed. 2016;6(7):614–619. doi: 10.1016/j.apjtb.2016.05.005. [DOI] [Google Scholar]
- 5.Yaghoobi-Ershadi M. Phlebotomine sand flies (Diptera: Psychodidae) in Iran and their role on Leishmania transmission. J Arthropod Borne Dis. 2012;6(1):1–17. [PMC free article] [PubMed] [Google Scholar]
- 6.Hajjaran H, Saberi R, Borjian A, Fakhar M, Hosseini SA, Ghodrati S, Mohebali M. The geographical distribution of human cutaneous and visceral leishmania species identified by molecular methods in Iran: a systematic review with meta-analysis. Front Public Health. 2021;9:835. doi: 10.3389/fpubh.2021.661674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabzevari S, Teshnizi SH, Shokri A, Bahrami F, Kouhestani F. Cutaneous leishmaniasis in Iran: a systematic review and meta-analysis. Microb Pathog. 2021;152:104721. doi: 10.1016/j.micpath.2020.104721. [DOI] [PubMed] [Google Scholar]
- 8.Shiee MR, Mohebali M, Doroodgar A, Teimouri A, Afzali H, Shirzadi MR. A molecular and parasitological survey on cutaneous leishmaniasis patients from historical city of Kashan in Isfahan province, center of Iran. Asian Pac J Trop Dis. 2012;2(6):421–5. doi: 10.1016/S2222-1808(12)60093-0. [DOI] [Google Scholar]
- 9.Sunter J, Gull K. Shape, form, function and leishmania pathogenicity: from textbook descriptions to biological understanding. Open Biol. 2017;7(9):170165. doi: 10.1098/rsob.170165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grybchuk D, Kostygov AY, Macedo DH, d'Avila-Levy CM, Yurchenko V. RNA viruses in trypanosomatid parasites: a historical overview. Mem Inst Oswaldo Cruz. 2018;113(4):e170487. doi: 10.1590/0074-02760170487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Carvalho RVH, Lima-Junior DS, da Silva MVG, Dilucca M, Rodrigues TS, Horta CV, et al. Leishmania RNA virus exacerbates leishmaniasis by subverting innate immunity Via TLR3-mediated NLRP3 inflammasome inhibition. Nat Commun. 2019;10(1):5273. doi: 10.1038/s41467-019-13356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bourreau E, Ginouves M, Prevot G, Hartley MA, Gangneux JP, Robert-Gangneux F, et al. Presence of leishmania RNA virus 1 in leishmania guyanensis increases the risk of first-line treatment failure and symptomatic relapse. J Infect Dis. 2016;213(1):105–11. doi: 10.1093/infdis/jiv355. [DOI] [PubMed] [Google Scholar]
- 13.Hartley MA, Ronet C, Zangger H, Beverley SM, Fasel N. Leishmania RNA virus: when the host pays the toll. Front Cell Infect Microbiol. 2012;2:99. doi: 10.3389/fcimb.2012.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheffter SM, Ro YT, Chung IK, Patterson JL. The complete sequence of Leishmania RNA virus LRV2-1, a virus of an old world parasite strain. Virolology. 1995;212(1):84–90. doi: 10.1006/viro.1995.1456. [DOI] [PubMed] [Google Scholar]
- 15.Widmer G, Dooley S. Phylogenetic analysis of leishmania RNA virus and leishmania suggests ancient virus-parasite association. Nucleic Acids Res. 1995;23(12):2300–4. doi: 10.1093/nar/23.12.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adaui V, Lye LF, Akopyants NS, Zimic M, Llanos-Cuentas A, Garcia L, et al. Association of the endobiont double-stranded RNA virus LRV1 with treatment failure for human leishmaniasis caused by Leishmania braziliensis in Peru and Bolivia. J Infect Dis. 2016;213(1):112–21. doi: 10.1093/infdis/jiv354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ives A, Ronet C, Prevel F, Ruzzante G, Fuertes-Marraco S, Schutz F, et al. Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Sci. 2011;331(6018):775–8. doi: 10.1126/science.1199326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kariyawasam R, Lau R, Valencia BM, Llanos-Cuentas A, Boggild AK. Leishmania RNA virus 1 (LRV-1) in Leishmania (Viannia) braziliensis isolates from Peru: a description of demographic and clinical correlates. Am J Trop Med Hyg. 2020;102(2):280–285. doi: 10.4269/ajtmh.19-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kariyawasam R, Mukkala AN, Lau R, Valencia BM, Llanos-Cuentas A, Boggild AK. Virulence factor RNA transcript expression in the Leishmania Viannia subgenus: influence of species, isolate source, and Leishmania RNA virus-1. Trop Med Health. 2019;47:25. doi: 10.1186/s41182-019-0153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zangger H, Hailu A, Desponds C, Lye LF, Akopyants NS, Dobson DE, et al. Leishmania aethiopica field isolates bearing an endosymbiontic dsRNA virus induce pro-inflammatory cytokine response. PLoS Negl Trop Dis. 2014;8(4):e2836. doi: 10.1371/journal.pntd.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hajjaran H, Mahdi M, Mohebali M, Samimi-Rad K, Ataei-Pirkooh A, Kazemi-Rad E, et al. Detection and molecular identification of Leishmania RNA virus (LRV) in Iranian Leishmania species. Arch Virol. 2016;161(12):3385–90. doi: 10.1007/s00705-016-3044-z. [DOI] [PubMed] [Google Scholar]
- 22.Saberi R, Fakhar M, Hajjaran H, Ataei-Pirkooh A, Mohebali M, Taghipour N, et al. Presence and diversity of Leishmania RNA virus in an old zoonotic cutaneous leishmaniasis focus, northeastern Iran: haplotype and phylogenetic based approach. Int J Infect Dis. 2020;101:6–13. doi: 10.1016/j.ijid.2020.08.033. [DOI] [PubMed] [Google Scholar]
- 23.Nalcaci M, Karakus M, Yilmaz B, Demir S, Ozbilgin A, Ozbel Y, et al. Detection of Leishmania RNA virus 2 in Leishmania species from Turkey. Trans R Soc Trop Med Hyg. 2019;113(7):410–7. doi: 10.1093/trstmh/trz023. [DOI] [PubMed] [Google Scholar]
- 24.Kleschenko Y, Grybchuk D, Matveeva NS, Macedo DH, Ponirovsky EN, Lukashev AN, et al. Molecular characterization of Leishmania RNA virus 2 in Leishmania major from Uzbekistan. Genes (Basel) 2019;10(10):830. doi: 10.3390/genes10100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasti S, Delavari M, Arani TS, Mousavi SG. Epidemiological and clinical study on the cutaneous leishmaniasis in Aran and Bidgol, center of Iran. Int Arch Health Sci. 2018;5:72–5. doi: 10.4103/iahs.iahs_26_18. [DOI] [Google Scholar]
- 26.Schonian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HD, Presber W, et al. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis. 2003;47(1):349–58. doi: 10.1016/s0732-8893(03)00093-2. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mokni M. Cutaneous leishmaniasis. Ann Dermatol Venereol. 2019;146(3):232–46. doi: 10.1016/j.annder.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Hajjaran H, Mohebali M, Mamishi S, Vasigheh F, Oshaghi MA, Naddaf SR, et al. Molecular identification and polymorphism determination of cutaneous and visceral leishmaniasis agents isolated from human and animal hosts in Iran. Biomed Res Int. 2013;2013:789326. doi: 10.1155/2013/789326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saberi R, Moin-Vaziri V, Hajjaran H, Niyyati M, Taghipour N, Kheirandish F, et al. Identification of Leishmania species using N-acetylglucosamine-1-phosphate transferase gene in a zoonotic cutaneous leishmaniasis focus of Iran. J Vector Borne Dis. 2018;55(1):14–9. doi: 10.4103/0972-9062.234621. [DOI] [PubMed] [Google Scholar]
- 31.Behravan M, Moin-Vaziri V, Haghighi A, Rahbarian N, Taghipour N, Abadi A, et al. Molecular identification of Leishmania species in a re-emerged focus of cutaneous leishmaniasis in Varamin district. Iran. J Arthropod Borne Dis. 2017;11(1):124–31. [PMC free article] [PubMed] [Google Scholar]
- 32.Bisetegn H, Zeleke AJ, Gadisa E, Shumie G, Damte D, Fenta T, et al. Clinical, parasitological and molecular profiles of cutaneous leishmaniasis and its associated factors among clinically suspected patients attending Borumeda hospital, north-east Ethiopia. PLoS Negl Trop Dis. 2020;14(8):e0008507. doi: 10.1371/journal.pntd.0008507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bates PA. Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. Int J Parasitol. 2007;37(10):1097–106. doi: 10.1016/j.ijpara.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Courtenay O, Peters NC, Rogers ME, Bern C. Combining epidemiology with basic biology of sand flies, parasites, and hosts to iNform leishmaniasis transmission dynamics and control. PLoS Pathog. 2017;13(10):e1006571. doi: 10.1371/journal.ppat.1006571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abtahi M, Eslami G, Cavallero S, Vakili M, Hosseini SS, Ahmadian S, et al. Relationship of Leishmania RNA virus (LRV) and treatment failure in clinical isolates of Leishmania major. BMC Res Notes. 2020;13(1):126. doi: 10.1186/s13104-020-04973-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farrokhi-Karibozorg M, Ghayour-Najafabadi Z, Hejazi SH, Ataei-Pirkooh A, Mohebali M, Teimouri P, et al. Molecular identification of Leishmania RNA virus in cutaneous leishmaniasis patients and rodent reservoirs in Isfahan province. Iran. Infect Genet Evol. 2022;98:105222. doi: 10.1016/j.meegid.2022.105222. [DOI] [PubMed] [Google Scholar]
- 37.Saberi R, Fakhar M, Mohebali M, Anvari D, Gholami S. Global status of synchronizing Leishmania RNA virus in Leishmania parasites: a systematic review with meta-analysis. Transbound Emerg Dis. 2019;66(6):2244–51. doi: 10.1111/tbed.13316. [DOI] [PubMed] [Google Scholar]