Abstract
Over the last decade, developments in molecular profiling have radically altered the diagnosis, classification, and management of numerous cancer types, with primary brain tumors being no exception. Although historically brain tumors have been classified based on their morphological characteristics, recent advances have allowed refinement of tumor classification based on molecular alterations. This shift toward molecular classification of primary brain tumors is reflected in the 2021 5th edition of the WHO classification of central nervous system tumors (WHO 2021). In this review, we will discuss the most recent updates to the classification of adult‐type diffuse gliomas, a group of highly infiltrative and largely incurable CNS malignancies. It is our hope continued that refinement of molecular criteria will improve diagnosis, prognostication, and eventually treatment of these devastating tumors.
Keywords: astrocytoma, glioblastoma, glioma, IDH, oligodendroglioma
The 2021 5th edition of the WHO classification of central nervous system tumors (WHO 2021) further solidifies the centrality of molecular markers in the classification of adult type diffuse gliomas, generating 3 primary diagnoses: IDH‐mutant, 1p/19q codeleted oligodendroglioma; IDH‐mutant, non‐codeleted astrocytoma; and IDH‐wildtype glioblastoma. These changes represent a step towards more uniform disease entities, thereby enabling rational clinical decision making and advancing the field toward a future of distinct and targetable, molecularly‐defined disease entities.
1. INTRODUCTION
Adult‐type diffuse gliomas are the most common malignant tumors of the central nervous system [1]. Survival varies greatly depending on the subtype of glioma present, with low‐grade gliomas having 5‐year survival rates as high as 80%, while high‐grade gliomas have 5‐year survival rates under 5% [1]. Regardless of grade and prognosis, gliomas are highly infiltrative and resistant to therapy, rendering them largely incurable. Historically, classification and prognostication for diffuse gliomas has been based on morphologic features of the tumor in question. These histopathological categories (e.g., astrocytoma, oligodendroglioma, and glioblastoma) were conceptually based on presumed cell of origin. However, in recent years these tumors have been subjected to significant molecular characterization and it has become clear that molecular markers yield more uniform disease entities and better predict clinical behavior [2, 3, 4]. Beyond this, it is expected that as therapeutic toolkits expand, molecular—rather than histopathologic—classification will allow for better therapeutic selection and improved clinical outcomes.
WHO 2021 expands upon the trend started in 2016, using key molecular biomarkers to define neoplastic entities and greatly reducing the dependency on morphologic features for tumor classification. Terminology around tumor grading has also been simplified, with molecular features dictating classification and joint histopathologic and molecular analysis determining grade. The classification of diffuse gliomas under the 2021 update is dependent largely on isocitrate dehydrogenase (IDH1/2) mutation status and 1p/19q codeletion status, resulting in 3 primary disease groups: IDH‐mutant, 1p/19q codeleted oligodendroglioma; IDH‐mutant, non‐codeleted astrocytoma; and IDH‐wildtype glioblastoma. This revision further demarcates IDH‐mutant from IDH‐wildtype disease, a necessity given the wide gap in survival between IDH‐mutant and IDH‐wildtype tumors, even those bearing the same histopathologic classification [5]. For the remainder of this review the term oligodendroglioma will refer to IDH‐mutant, 1p/19q codeleted diffuse glioma, astrocytoma will refer exclusively to IDH‐mutant, non‐codeleted diffuse glioma, and glioblastoma will refer to IDH‐wildtype diffuse glioma.
The increasing dependence of classification and prognostication on molecular features in turn increases the importance of laboratory assessment of biomarkers. Many relevant biomarkers—such as mutations involving IDH1/2 and ATRX—can be assessed with immunohistochemistry (IHC), while others—including 1p/19q codeletion—require focused molecular approaches like fluorescence in situ hybridization (FISH). Although sufficient in many cases, singleton assays have crucial limitations, discussed further below, and have been rendered increasingly inefficient in the face of the molecular profiling demands implicit in modern tumor classification schemes. This quandary has been largely addressed by the widespread implementation of multiplex and high‐dimensional assays based on platforms such as next‐generation sequencing (NGS) and global DNA methylome profiling. The expanding use of multiplex assays stands to both improve clinical decision making and deepen our understanding of these complex diseases.
2. ASTROCYTOMA, IDH‐MUTANT
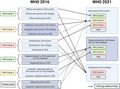
Astrocytoma, IDH‐mutant is now the preferred designation for all adult‐type gliomas that are IDH1‐ or IDH2‐mutant with absence of 1p/19q codeletion. These diffusely infiltrating gliomas frequently harbor inactivating mutations in TP53 and ATRX and can be defined as either CNS WHO grade 2, grade 3, or grade 4. Designations such as diffuse astrocytoma, IDH‐mutant; anaplastic astrocytoma, IDH‐mutant; and glioblastoma, IDH‐mutant are no longer preferred, having been replaced with astrocytoma, IDH‐mutant grade 2, grade 3, and grade 4, respectively (Figure 1). The association of multiple possible WHO grade designations with a specific diagnostic entity represents a departure from earlier CNS tumor classifications, which linked all named diagnoses with specific WHO grades, whether 1, 2, 3, or 4. Finally, disease entities such as diffuse astrocytoma, IDH‐wild type, and anaplastic astrocytoma, IDH‐wildtype are now encompassed by other entities not bearing the astrocytoma designation, most notably glioblastoma, IDH‐wildtype (see below).
FIGURE 1.
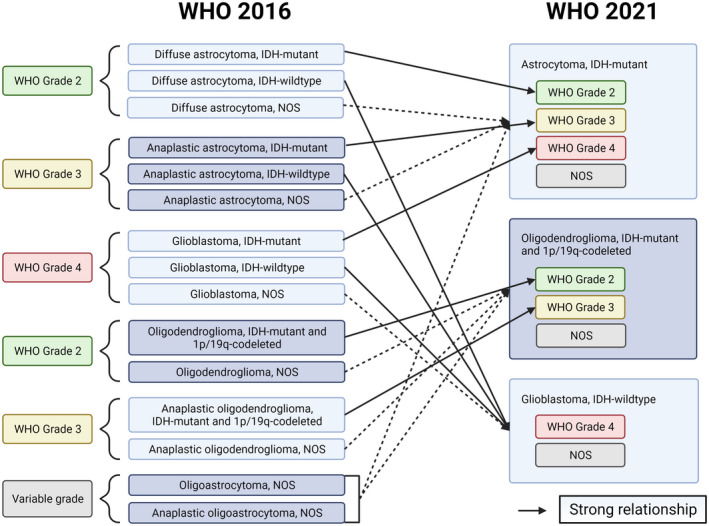
Schematic showing how the disease entities from WHO 2016 is now defined in WHO 2021. Solid lines denote strong correlations between the two classifications, while dotted lines denote how a WHO 2016 disease entity would likely, but not definitively, be defined
The histologic appearance of astrocytoma can vary greatly, from well‐differentiated, and minimally mitotic (CNS WHO grade 2) to overtly anaplastic, hypercellular, and proliferative (CNS WHO grades 3–4). Broadly, astrocytomas are highly infiltrative, with hypercellular regions intermixed with and entrapping normal brain. Nuclear morphology is key in identifying neoplastic cells, which commonly exhibit angular nuclei with uneven chromatin and hyperchromasia [6]. While the WHO does not define a firm threshold for proliferative activity, grade 3 designation is dependent on an elevated mitotic rate, while WHO grade 2 tumors are comparatively inert (Figure 2). Generally, grade 3 tumors have greater cellular crowding and nuclear atypia; abnormal mitoses and multinucleated tumor cells may also be present (Figure 3). Grade 4 IDH‐mutant astrocytomas possess, by definition, microvascular proliferation, necrosis, and/or homozygous deletion of cyclin‐dependent kinase inhibitor 2A/B (CDKN2A/B) (see below) [7]. Large foci of ischemic and pseudopalisading necrosis, typically associated with IDH‐wild type glioblastoma, are not infrequent [8].
FIGURE 2.
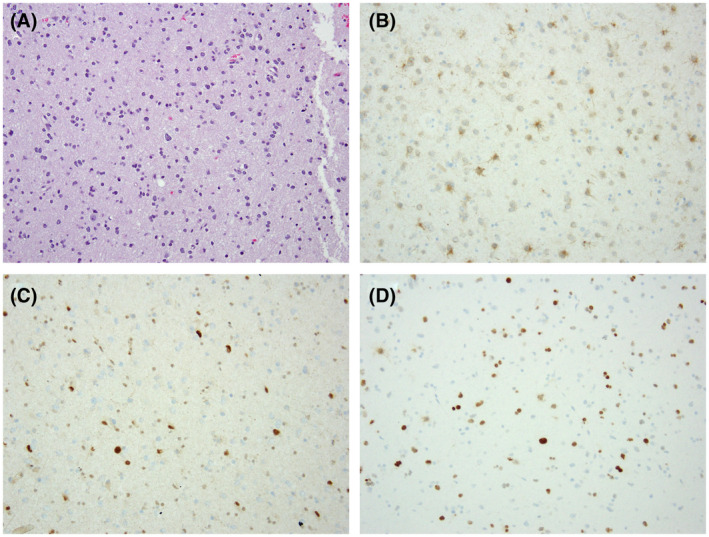
Astrocytoma, IDH‐mutant, WHO grade 2. H & E‐stained sections reveal an infiltrating glial neoplasm composed of pleomorphic tumor cells (A). Microvascular proliferation, necrosis, and mitotic activity are inapparent. Immunohistochemical stains reveal that the tumor cells express IDH1 R132H (B), while exhibiting loss of nuclear ATRX expression (C; note retained expression in background normal cellular constituents). p53 immunostaining is positive in tumor cells (D). (200X magnification)
FIGURE 3.
Astrocytoma, IDH‐mutant, WHO grade 3. H & E‐stained sections reveal a cellular astrocytic neoplasm (A). Immunohistochemical stains are positive for IDH1 R132H (B) and negative for ATRX (C; loss of nuclear expression). MIB1 (Ki‐67) immunostaining demonstrates an elevated proliferation index (D), consistent with WHO grade 3 classification. (200X magnification)
WHO 2021 fully formalizes IDH mutational status as a defining molecular feature of adult‐type astrocytomas and oligodendrogliomas. Glioma‐associated IDH1/2 mutations are exclusively missense and involve active site arginine residues, R132 for IDH1 and the analogous R172 for IDH2 [9, 10]. Under normal conditions, isocitrate dehydrogenase is responsible for the conversion of isocitrate to alpha‐ketoglutarate. However, the R132 and R172 mutations induce a neomorphic enzymatic activity, wherein alpha‐ketoglutarate is converted to D‐2‐hydroxyglutarate (2‐HG) [11]. Buildup of 2‐HG leads to significant epigenetic changes to the cell, altering methylation patterns of both histones and DNA. Increased DNA methylation and gene silencing genome‐wide induced by elevated 2‐HG is often referred to as the CpG island hypermethylator phenotype (G‐CIMP) [12, 13], and is thought to promote tumorigenesis through repression of normal cellular differentiation [14, 15]. Moreover, G‐CIMP tumors have been shown to broadly dysregulate genome topology, resulting in aberrant enhancer function and gene regulation. Beyond its epigenetic impact, IDH mutation also appears to have significant impacts on oncogenic HIF signaling [16, 17].
Concurrent with IDH1 and IDH2 mutations, astrocytomas frequently possess mutations in TP53 and ATRX [18, 19]. The aptly named “guardian of the genome,” TP53 encodes a central regulator of cell cycle arrest and apoptosis in response to cellular stress [20]. A canonical tumor suppressor, TP53, is the most commonly mutated gene in cancer, and its central role in multiple primary tumors of the CNS has been known for some time [21, 22]. By contrast, the impact of ATRX deficiency on tumor biology is only beginning to be explored, despite its recent association with multiple tumor types within and outside of the CNS. ATRX cooperates with death domain‐associated protein (Daxx) to form a key chromatin remodeling complex, which is responsible for the deposition of H3.3 in telomeric regions, pericentric heterochromatin, and other regions of repeat DNA [23, 24]. Loss of ATRX leads to genome instability, DNA damage, and global epigenetic dysregulation [25, 26]. Cells deficient in ATRX undergo p53‐dependent cell death, a phenomenon that at least partially explains the frequent co‐occurrence of ATRX and TP53 mutations in cancer [27, 28]. For reasons that remain obscure, patients with ATRX‐deficient, IDH‐mutant, non‐codeleted astrocytoma have a somewhat better prognosis than counterparts with ATRX‐intact, IDH‐mutant, non‐codeleted astrocytoma [29, 30]. Beyond their abnormal genomic and epigenomic phenotypes, ATRX‐deficient tumors display lengthening of telomeres via a telomerase‐independent process known as alternative lengthening of telomeres (ALT) [31]. Interestingly, IDH‐mutant and 1p/19q codeleted oligodendrogliomas frequently possess activating mutations to telomerase reverse transcriptase (TERT), which are mutually exclusive with ATRX mutations. In this way, virtually all IDH‐mutant tumors, both codeleted and non‐codeleted, exhibit some form of pathological telomere maintenance.
Laboratory assessment of molecular alterations relevant to the diagnosis of IDH‐mutant astrocytoma is generally feasible by routine IHC and FISH. Immunohistochemical interrogation of IDH1 R132H, p53, and ATRX is generally advisable. The IDH1 R132H mutation accounts for ~90% of all glioma‐associated IDH mutations, enabling highly sensitive immunohistochemical screening of IDH mutational status [10]. That being said, given the low but significant number of so‐called “non‐canonical” mutations in IDH1/2 characterizing both astrocytomas and oligodendrogliomas, it is recommended that indeterminate or negative staining for IDH1 R123H be followed up by confirmatory gene sequencing, particularly in the setting of ATRX deficiency or other compelling histopathological findings [9, 32]. Such follow‐up testing may not be necessary in patients over the age of 55 whose tumors exhibit WHO grade 4 histopathological features (e.g., microvascular proliferation and necrosis), as non‐canonical mutations in this clinical context have been found to be quite rare [33, 34].
Immunostaining for p53 mutation leverages the fact that missense mutations in TP53 increase the encoded protein's half‐life, leading to observable nuclear accumulation. Samples should be considered “positive” (consistent with underlying TP53 mutation) if >10% of their nuclei stain strongly for p53 [35, 36]. However, as non‐missense variants can account for 10% of TP53 mutations, p53 immunostaining results should be evaluated in the context of the larger molecular workup (e.g., ATRX and 1p/19q codeletion status), as well as tumor histomorphology. ATRX mutations frequently take the form of truncating alterations and an absence of protein expression, leading to loss of nuclear immunoreactivity [37]. ATRX expression in endothelial cells and neurons serves as an internal positive control and must be present for effective IHC interpretation [38]. Although mutations in ATRX and IDH1/2 frequently co‐occur, ATRX expression may also be lost in multiple IDH‐wildtype glioma variants. As such, absence of nuclear ATRX immunolabeling does not obviate the need for IDH1/2 mutational assessment. That being said, the combination of IDH1/2 mutation and ATRX deficiency in a diffuse glioma is sufficient for the diagnosis of IDH‐mutant astrocytoma and eliminates the need for 1p/19q codeletion testing to formally exclude oligodendroglioma.
Beyond the diagnostic biomarkers discussed above, several additional molecular features have been identified with significant prognostic implications for IDH‐mutant astrocytomas. For instance, focal amplification events involving CDK4 and MYC, along with mutations in PIK3R1, have each been associated with significant decreases in overall survival [39, 40, 41]. Most significantly, homozygous deletion of CDKN2A/B, both key cell cycle regulators and tumor suppressors, has been repeatedly associated with decreased progression free and overall survival [7, 40, 42, 43]. Accordingly, identification of homozygous deletion of CDKN2A/B is sufficient to classify IDH‐mutant glioma as WHO grade 4, independent of histologic characteristics, necessitating its formal assessment in the context of a complete workup.
Finally, recent work has confirmed that rare infratentorial variants of IDH‐mutant astrocytoma exhibit molecular and clinical distinctions from their supratentorial counterparts. Specifically, the rate of non‐canonical IDH mutations in infratentorial astrocytomas is almost 80% (relative to <20% in supratentorial astrocytomas), the rate of ATRX loss is under 50% (relative to 94%), and the rate of O(6)‐methylguanine‐DNA methyltransferase (MGMT) promoter methylation is just under 60% (relative to 96%). Patients with infratentorial IDH‐mutant astrocytomas also appear to have significantly reduced overall survival relative to those with supratentorial tumors. While this relatively unfavorable outcome measure may partially reflect the infratentorial localization of these tumors and their proximity to vital brainstem structures, the molecular distinctions described above speak to more fundamental biological properties driving tumor intrinsic malignancy factors. Further study is needed to explore these possibilities.
3. OLIGODENDROGLIOMA, IDH‐MUTANT, AND 1P/19Q CODELETED
Oligodendroglioma is defined on a molecular level by co‐occurrence of IDH mutation and complete deletion of the 1p and 19q chromosomal arms. Histologically, oligodendrogliomas consist of closely packed tumor cells with round‐to‐oval, monotonous nuclei (Figure 4). The presence of perinuclear clearing is frequently described as giving the cells a “fried egg” appearance. Dense, branching capillary networks within the tumors lead to a “chicken wire” appearance, and microcalcifications are frequently present [44]. Much like IDH‐mutant astrocytomas in WHO 2021, IDH‐mutant and 1p/19q‐codeleted oligodendrogliomas are now associated with multiple possible CNS WHO grades, either grade 2 or grade 3, reflecting significant differences in overall survival [45]. Histopathologically, Grade 3 tumors display some number of the following features: increased cellularity, marked atypia, greater mitotic activity, microvascular proliferation, and necrosis with or without palisading (Figure 5). Although ≥2.5 mitoses/mm2 has been identified as a cutoff of prognostic significance, there is no set proliferative standard differentiating grade 2 and grade 3 oligodendrogliomas [46]. Accordingly, Ki‐67 immunostaining and clinical features (e.g., rapid symptomatic progression) can be informative in borderline cases. Homozygous CDKN2A and/or CDKN2B deletion is present in a relatively small proportion of oligodendrogliomas; however, its association with poor prognosis independent of histologic features has prompted its designation as a molecular marker for grade 3 oligodendroglioma [42].
FIGURE 4.
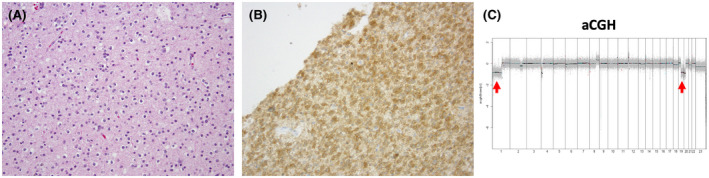
Oligodendroglioma, IDH‐mutant and 1p/19q codeleted, WHO grade 2. H & E‐stained sections reveal a diffusely infiltrating glial neoplasm characterized by round‐to‐oval nuclear monomorphism and perinuclear halos (A). Immunostaining is positive for IDH1 R132H (B). (200X magnification). Array‐based comparative genomic hybridization (aCGH) demonstrates whole‐arm loss of chromosomes 1p and 19q (arrows) consistent with 1p/19q codeletion (C)
FIGURE 5.
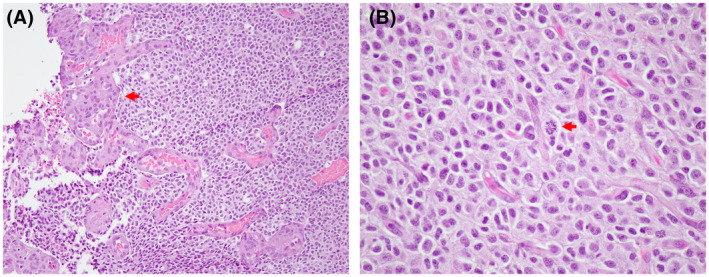
Oligodendroglioma, IDH‐mutant, and 1p/19q codeleted, WHO grade 3. H & E‐stained sections reveal a densely cellular oligodendroglial neoplasm with obvious glomeruloid microvascular proliferation (A; arrow; 200X magnification). Mitotic activity is readily apparent (B; arrow; 400X magnification)
As indicated above, 1p/19q codeletion distinguishes oligodendroglioma from astrocytoma on a foundational molecular level. This unique molecular alterations is the product of an unbalanced translocation between chromosomes 1 and 19, with subsequent loss of the product containing whole‐arm loss of 1p and 19q [47, 48]. Partial deletion of either or both arms does not constitute diagnostic 1p/19q codeletion, and can in fact occur in other relevant CNS tumors, most notably IDH‐wildtype glioblastoma [49]. Although it is broadly hypothesized that 1p/19q codeletion unmasks loss‐of‐function mutations in the retained chromosomal copies, with some genetic evidence substantiating this conjecture (CIC and FUBP1 mutations, see below), the full biological consequences of this complex structural abnormality remain unclear.
Several other molecular abnormalities occur frequently in oligodendrogliomas, including mutations in the TERT promoter, capicua transcriptional repressor (CIC), and far upstream element‐binding protein 1 (FUBP1) [50, 51, 52]. TERT encodes the catalytic subunit of telomerase, an enzymatic complex responsible for maintaining telomere length. Point mutations in the TERT promoter region characterize many solid tumor variants and are present in virtually all oligodendrogliomas. Functionally, TERT promoter mutations unmask binding motifs for ETS transcription factors, leading to TERT overexpression, telomere elongation, and cellular immortality [53, 54]. As indicated above, the mutual exclusivity of TERT and ATRX mutations in IDH‐mutant adult gliomas underscores the importance of abnormal telomere maintenance to the process of gliomagenesis. CIC encodes a transcriptional repressor of genes integral to growth and metabolism that are induced by receptor tyrosine kinase (RTK) signaling [55, 56]. Mutations to CIC mitigate its repression of RTK‐induced gene expression, leading to pro‐survival and proliferative transcriptional profiles. FUBP1 encodes a key regulator of tumor suppressor and oncogene alternative splicing, the loss of which leads to widespread alterations in RNA splicing [57, 58]. FUBP1 is also crucial for establishing a setpoint of MYC expression, with FUBP1 loss leading to setpoint dysregulation and increased MYC transcript levels [59, 60]. Mutations in CIC and FUBP1 occur in oligodendrogliomas at rates as high as 60% and 30%, respectively, and mutations in either or both are associated with shorter survival and faster time to recurrence [61]. Intriguingly, the genomic localization of CIC and FUBP1, on chromosomes 19q and 1p respectively, is consistent with the notion that 1p/19q codeletion partially disables key tumor suppressors resident on the 1p and 19q chromosomal arms.
Molecular assessments of IDH mutational status for oligodendrogliomas do not differ from approaches used for astrocytomas and are performed before a definitive diagnosis can be made. Evaluation of 1p/19q codeletion is somewhat more complicated, as effective approaches should identify coordinate absence of 2 chromosomal arms, rather than a single missense mutation or even single gene loss. FISH is the most commonly used technical approach and typically employs paired probes, one for the lost arm and one for the retained arm. The most common commercial probes localize to 1p36 and 1q25 on chromosome 1 and 19p13 and 19q13 on chromosome 19 [62, 63]. Although this method is cost effective and sensitive, the direct assessment of localized genomic regions as proxies for whole chromosome arms raises the possibility of false positive results. Accordingly, any 1p/19q codeletion testing methodology that does not explicitly evaluate whole‐arm chromosomal deletion should be interpreted with caution, particularly in unusual clinicopathological contexts. Alternative methods to directly assess whole‐arm 1p/19q codeletion include cytogenomic array, next‐generation sequencing (with appropriately tiled capture probe sets), and global DNA methylation profiling. This latter technology will likely enjoy more widespread usage in the years to come, due to its notable capabilities in designating both known and novel subgroups of CNS neoplasms [64].
Finally, the historical designation of oligoastrocytoma, referring to tumors exhibiting histopathological features of both oligodendroglioma and astrocytoma, is again absent in WHO 2021, as in the previous 2016 update to the 4th edition. It has been repeatedly demonstrated that virtually all of these tumors have molecular features of either oligodendroglioma or astrocytoma, but not both [18, 19, 32, 65]. Nevertheless, there have been several case reports describing IDH‐mutant gliomas exhibiting both mixed histopathologic and molecular features [66, 67]. Within these tumors, regions appearing morphologically oligodendroglial displayed 1p/19q codeletion, while regions appearing astrocytic harbored TP53 and ATRX mutations. The presence of truly mixed IDH‐mutant gliomas is rare enough that the creation of a specific subclass encompassing them was not deemed necessary. However, these tumors do offer insight into the natural history of IDH‐mutant disease. The presence of molecularly distinct astrocytic and oligodendroglial lineages emerging from the same IDH‐mutant parental subclone speaks to an evolutionary sequence of events where IDH mutation is acquired first following by either 1p/19q codeletion and TERT mutation or TP53 and ATRX mutations. These tumors also indicate that both oligodendrogliomas and astrocytomas can arise from a common cell of origin, demonstrating that despite their distinct naming, they share much histogenetically.
4. GLIOBLASTOMA, IDH‐WILDTYPE
WHO 2021 reserves the term glioblastoma specifically for IDH‐wildtype tumors, with IDH‐mutant glioblastoma having been effectively renamed astrocytoma, WHO grade 4. Mutations to histone variant 3 (H3) are also common in IDH‐wildtype diffuse glioma, particularly in pediatric and young adult populations, but these tumor variants are designated separately (see below). IDH‐wildtype glioblastomas are generally high grade and rapidly proliferating, with very poor prognosis [68]. Historically these tumors have been identified based on the histologic presence of florid microvascular proliferation and/or necrosis with or without pseudopalisading (Figure 6). Glioblastomas also tend to be poorly differentiated with brisk mitotic activity. Microvascular proliferation and/or necrosis are both sufficient to establish a diagnosis of glioblastoma in an IDH‐wildtype, H3‐wildtype diffuse glioma. However, WHO 2021 also delineates multiple defining molecular features for IDH‐wildtype glioblastoma, namely TERT promoter mutation, epidermal growth factor receptor (EGFR) amplification, and combined chromosome 7 gain/chromosome 10 loss (+7/−10) [7], with the latter most frequently involving the whole chromosomes 7 and 10 and less frequently restricted to only the 7q and 10q arms. These alterations effectively serve as “rule in” criteria for IDH‐wildtype glioblastoma, as diffuse gliomas harboring any one of them, even in the context of grade 2 or 3 histopathology, are characterized by poor clinical performance [41, 69, 70, 71, 72]. TERT promoter mutations in glioblastoma are thought to promote tumor growth in the same manner as in IDH‐mutant tumors (see above), while EGFR amplification leads to increased pro‐survival and pro‐cell division signaling downstream of the encoded receptor tyrosine kinase. By contrast, much less is known about the functional significance of +7/−10, although gains in chromosome 7 may facilitate oncogenic transformation in part by way of increased expression of secreted platelet‐derived growth factor α (PDGFA) [73]. Of note, recent work has questioned the robustness of isolated TERT mutation as a diagnostic criterion for IDH‐wildtype glioblastoma, particularly in the context of WHO grade 2 histopathology [74]. Additional studies should clarify the optimal application of this diagnostic feature moving forward.
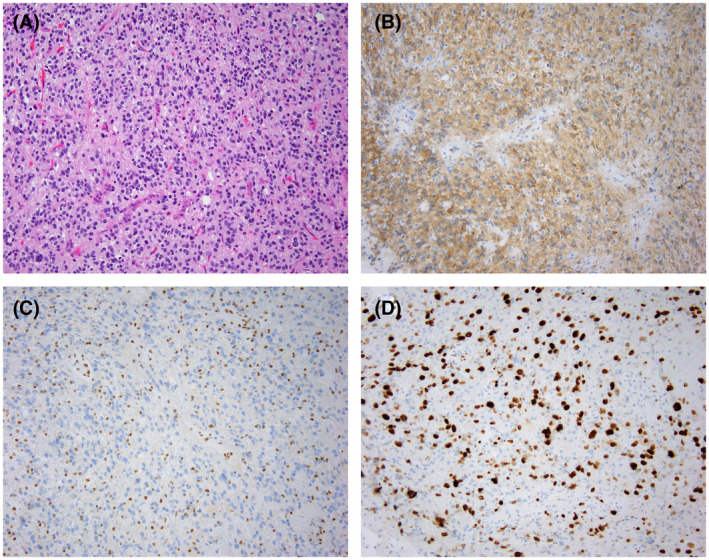
FIGURE 6.
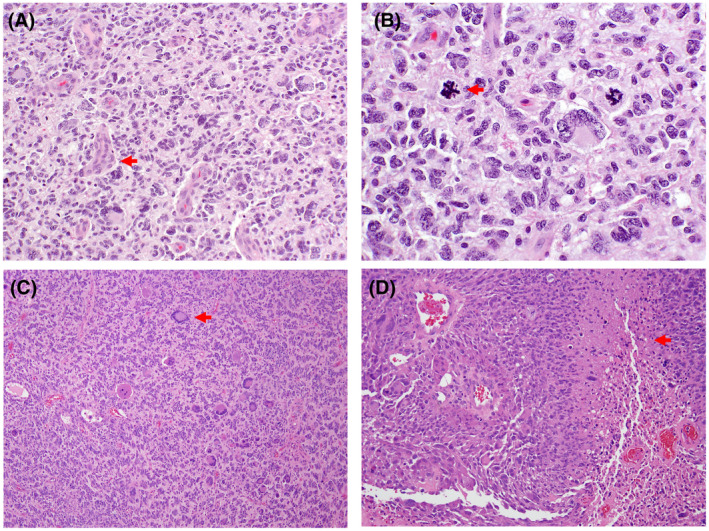
Glioblastoma, IDH‐wild type, WHO grade 4. H & E‐stained sections reveal a cellular, pleomorphic, glial neoplasm with glomeruloid microvascular proliferation (A; arrow; 200X magnification). Atypical mitotic figures (B; arrow; 400X magnification) are prominent. H & E‐stained sections of another case demonstrating notable nuclear pleomorphism and giant cell features (C; arrow; 100X magnification), along with pseudopalisading necrosis (D; arrow; 200X magnification)
Promoter methylation of the MGMT gene has been shown to play a significant role in the glioblastoma prognosis and therapeutic response [75]. MGMT encodes an enzyme responsible for removal of alkyl groups from the O6 position on guanine, which reduces the efficacy of alkylating agents. Promoter methylation impairs MGMT transcription, resulting in decreased enzymatic activity and mitigated therapeutic resistance. Assessment of MGMT promoter methylation yields the only validated predictive biomarker routinely employed in the management of IDH‐wildtype glioblastoma. Additional molecular alterations have been associated with prognostic relevance in glioblastoma, although results are less than unanimous. CDKN2A/B loss has been linked with reduced overall survival; however, some studies have found this to be true only in tumors without MGMT methylation [76, 77]. Recent work has also demonstrated variable associations between TERT mutation and unfavorable clinical course [77, 78]. These contrasting findings may reflect dependencies on specific clinical and/or molecular contexts that remain to be elucidated. In the coming years it is likely that more context‐dependent and independent prognostic factors will emerge, further enhancing our ability to predict outcomes for glioblastoma. IDH‐wildtype glioblastoma is characterized by marked molecular heterogeneity, on both intra‐ and intertumoral levels. Although IDH‐mutant astrocytoma and oligodendroglioma possess small numbers of defining molecular alterations, glioblastomas typically harbor many gene abnormalities spread across a number of classic oncogenic networks, including the p53 and retinoblastoma [79]. Amplification and/or mutation events involving receptor tyrosine kinase (RTK) genes not limited to EGFR are also common, activating a variety of downstream pro‐tumorigenic effectors. Alternative RTKs most commonly include platelet‐derived growth factor receptor α (PDGFRA), MET, and fibroblast growth factor receptor 3 (FGFR3). For the latter, fusion of the tyrosine kinase coding domain to the transforming acidic coiled‐coil 3 coding domain of the TACC3 gene represents the most frequent alteration, defining a small subset of IDH‐wildtype glioblastoma with distinctive histopathology [80, 81, 82]. The FGFR3‐TACC3 fusion protein possesses constitutive kinase activity and engages molecular pathways distinct from those typically downstream of canonical FGFR signaling. Finally, intratumoral variability in RTK composition is now a well‐documented phenomenon for IDH‐wildtype glioblastoma, with different populations of cells in a given tumor harboring distinct amplifications events in EGFR, PDGFRA, and/or MET [83, 84]. The implications of such cell‐by‐cell heterogeneity are profound, particularly with regards to the molecular assessment of clinical specimens, along with the development of truly viable targeted treatment approaches. Within a single tumor, multiple subclones can exist with unique genetic and transcriptional profiles, with unique clones recurring after therapy [85, 86, 87, 88]. Targeting multiple tumor cell clones with distinct therapeutic vulnerabilities and resistances represents a significant barrier to the treatment of glioblastoma and one which will need to be addressed as the field advances.
The considerable inter‐ and intratumoral heterogeneity described above emphasizes the inherent limitations of standard singleton approaches for the molecular workup of glioblastoma. As this fundamentally diverse diagnostic entity becomes further fragmented into molecularly specified subclasses—with each hopefully paired with more effective, targeted treatment approaches—the importance of evaluating many potential biomarkers simultaneously with high‐dimensional molecular assessment will become increasingly clear. The technological framework of this more complex diagnostic approach, encompassing next‐generation sequencing‐based genomics, and global DNA methylation profiling, is operative at many medical centers and should facilitate this transition in the coming years.
As a final note, it is important to remember that while the three basic tumor categories discussed above reflect our current conceptions of adult‐type diffuse glioma, other glioma variants bearing extensive histopathological similarity also arise in the adult population. These primarily consist of entities that normally affect children and adolescent age groups and include high‐grade histone 3‐mutant gliomas as well as low‐grade glial neoplasm with MYB or MYBL1 gene fusions or alterations involving constituents of the RAS/MAPK pathway [89, 90, 91, 92]. These tumors are discussed elsewhere in this issue.
4.1. Concluding remarks
WHO 2021 continues the trend of WHO 2016, further solidifying tumor classifications based on molecular alterations, thereby generating more uniform disease entities (Figure 7). This process has been enabled by the drastic increase in the number of relevant biomarkers identified in recent years and has considerably enhanced the prognostic stratification inherent to tumor classification schemes. The continued identification of potentially targetable molecular alterations—and classification of diseases based on these alterations—stands to inform clinical trial design, improve therapeutic selection, and increase the number of available therapies for adult‐type diffuse glioma.
FIGURE 7.
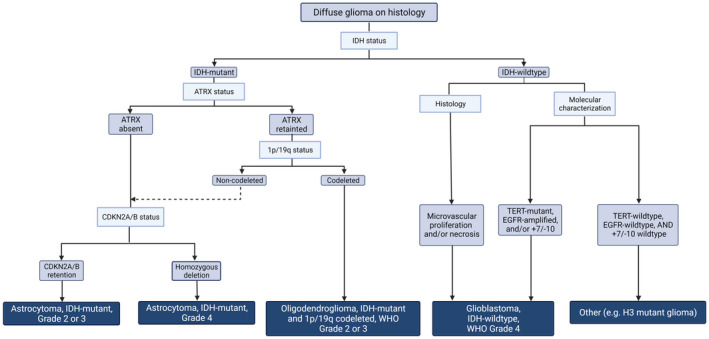
Flowchart showing the diagnostic progression of diffuse glioma in adults, based on the most relevant molecular markers. IDH mutation is used initially to divide diffuse gliomas into IDH‐mutant and IDH‐wildtype tumors. The IDH‐wildtype tumors are they subdivided into 1p/19q‐codeleted oligodendroglioma and 1p/19q‐non‐codeleted astrocytoma, based on both 1p/19q and ATRX status. CDKN2A then provides further prognostic value. IDH‐wildtype tumors will be classified as glioblastoma if they possess any one of five markers: (1) histologic evidence of necrosis, (2) histologic evidence of microvascular proliferation, (3) TERT promoter mutation, (4) EGFR amplification, and/or (5) whole gain of chromosome 7 and whole loss of chromosome 10. IDH‐wildtype tumors which do not possess any of these “rule‐in” criteria must be further examined for appropriate diagnosis
5. AUTHOR CONTRIBUTIONS
Benjamin T. Whitfield and Jason T. Huse: Wrote manuscript and provided final review.
Whitfield BT, Huse JT. Classification of adult‐type diffuse gliomas: Impact of the World Health Organization 2021 update. Brain Pathol. 2022;32:e13062. 10.1111/bpa.13062
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study
REFERENCES
- 1. Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro‐Oncol. 2019;21(Supplement_5):v1–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van den Bent MJ, Brandes AA, Taphoorn MJB, Kros JM, Kouwenhoven MCM, Delattre J‐Y, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long‐term follow‐up of EORTC Brain Tumor Group Study 26951. J Clin Oncol. 2013;31(3):344–50. [DOI] [PubMed] [Google Scholar]
- 3. Wick W, Hartmann C, Engel C, Stoffels M, Felsberg J, Stockhammer F, et al. NOA‐04 randomized phase iii trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009;27(35):5874–80. [DOI] [PubMed] [Google Scholar]
- 4. Cairncross G, Wang M, Shaw E, Jenkins R, Brachman D, Buckner J, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long‐term results of RTOG 9402. J Clin Oncol Off J Am Soc Clin Oncol. 2013;31(3):337–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hartmann C, Hentschel B, Simon M, Westphal M, Schackert G, Tonn JC, et al. Long‐term survival in primary glioblastoma with versus without isocitrate dehydrogenase mutations. Clin Cancer Res Off J Am Assoc Cancer Res. 2013;19(18):5146–57. [DOI] [PubMed] [Google Scholar]
- 6. Ichimura K, Narita Y, Hawkins CE. Diffusely infiltrating astrocytomas: pathology, molecular mechanisms and markers. Acta Neuropathol (Berl). 2015;129(6):789–808. [DOI] [PubMed] [Google Scholar]
- 7. Brat DJ, Aldape K, Colman H, Figrarella‐Branger D, Fuller GN, Giannini C, et al. cIMPACT‐NOW Update 5: Recommended Grading Criteria And Terminologies for IDH‐mutant astrocytomas. Acta Neuropathol (Berl). 2020;139(3):603–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15(19):6002–7. [DOI] [PubMed] [Google Scholar]
- 9. Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol (Berl). 2009;118(4):469–74. [DOI] [PubMed] [Google Scholar]
- 11. Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer‐associated IDH1 mutations produce 2‐hydroxyglutarate. Nature. 2009;462(7274):739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel‐Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rohle D, Popovici‐Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013. May 3;340(6132):626–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S‐H, et al. Oncometabolite 2‐hydroxyglutarate is a competitive inhibitor of α‐ketoglutarate‐dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma‐derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF‐1alpha. Science. 2009;324(5924):261–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3(7):709–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kannan K, Inagaki A, Silber J, Gorovets D, Zhang J, Kastenhuber ER, et al. Whole‐exome sequencing identifies ATRX mutation as a key molecular determinant in lower‐grade glioma. Oncotarget. 2012;3(10):1194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bykov VJN, Eriksson SE, Bianchi J, Wiman KG. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer. 2018;18(2):89–102. [DOI] [PubMed] [Google Scholar]
- 21. Louis DN. The p53 gene and protein in human brain tumors. J Neuropathol Exp Neurol. 1994;53(1):11–21. [DOI] [PubMed] [Google Scholar]
- 22. Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lewis PW, Elsaesser SJ, Noh K‐M, Stadler SC, Allis CD. Daxx is an H3.3‐specific histone chaperone and cooperates with ATRX in replication‐independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A. 2010;107(32):14075–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goldberg AD, Banaszynski LA, Noh K‐M, Lewis PW, Elsaesser SJ, Stadler S, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140(5):678–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Danussi C, Bose P, Parthasarathy PT, Silberman PC, Van Arnam JS, Vitucci M, et al. Atrx inactivation drives disease‐defining phenotypes in glioma cells of origin through global epigenomic remodeling. Nat Commun. 2018;9(1):1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y, Yang J, Wild AT, Wu WH, Shah R, Danussi C, et al. G‐quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX‐deficient malignant glioma. Nat Commun. 2019;10(1):943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Conte D, Huh M, Goodall E, Delorme M, Parks RJ, Picketts DJ. Loss of Atrx sensitizes cells to DNA damaging agents through p53‐mediated death pathways. PLoS One. 2012;7(12):e52167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bérubé NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ, et al. The chromatin‐remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest. 2005;115(2):258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wiestler B, Capper D, Holland‐Letz T, Korshunov A, von Deimling A, Pfister SM, et al. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol (Berl). 2013;126(3):443–51. [DOI] [PubMed] [Google Scholar]
- 30. Mur P, Mollejo M, Hernández‐Iglesias T, de Lope ÁR, Castresana JS, García JF, et al. Molecular classification defines 4 prognostically distinct glioma groups irrespective of diagnosis and grade. J Neuropathol Exp Neurol. 2015;74(3):241–9. [DOI] [PubMed] [Google Scholar]
- 31. Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333(6041):425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Comprehensive, integrative genomic analysis of diffuse lower‐grade gliomas. N Engl J Med. 2015;372(26):2481–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robinson C, Kleinschmidt‐DeMasters BK. IDH1‐mutation in diffuse gliomas in persons age 55 years and over. J Neuropathol Exp Neurol. 2017;76(2):151–4. [DOI] [PubMed] [Google Scholar]
- 34. DeWitt JC, Jordan JT, Frosch MP, Samore WR, Iafrate AJ, Louis DN, et al. Cost‐effectiveness of IDH testing in diffuse gliomas according to the 2016 WHO classification of tumors of the central nervous system recommendations. Neuro‐Oncol. 2017;19(12):1640–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takami H, Yoshida A, Fukushima S, Arita H, Matsushita Y, Nakamura T, et al. Revisiting TP 53 mutations and immunohistochemistry—a comparative study in 157 diffuse gliomas. Brain Pathol. 2014;25(3):256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gillet E, Alentorn A, Doukouré B, Mundwiller E, van Thuij H, Reijneveld JC, et al. TP53 and p53 statuses and their clinical impact in diffuse low grade gliomas. J Neurooncol. 2014;118(1):131–9. [DOI] [PubMed] [Google Scholar]
- 37. Reuss DE, Sahm F, Schrimpf D, Wiestler B, Capper D, Koelsche C, et al. ATRX and IDH1‐R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an “integrated” diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol (Berl). 2015;129(1):133–46. [DOI] [PubMed] [Google Scholar]
- 38. Tanboon J, Williams EA, Louis DN. The diagnostic use of immunohistochemical surrogates for signature molecular genetic alterations in gliomas. J Neuropathol Exp Neurol. 2016;75(1):4–18. [DOI] [PubMed] [Google Scholar]
- 39. Shirahata M, Ono T, Stichel D, Schrimpf D, Reuss DE, Sahm F, et al. Novel, improved grading system(s) for IDH‐mutant astrocytic gliomas. Acta Neuropathol (Berl). 2018;136(1):153–66. [DOI] [PubMed] [Google Scholar]
- 40. Yang RR, Shi Z‐F, Zhang Z‐Y, Chan A‐Y, Aibaidula A, Wang W‐W, et al. IDH mutant lower grade (WHO Grades II/III) astrocytomas can be stratified for risk by CDKN2A, CDK4 and PDGFRA copy number alterations. Brain Pathol. 2019;30(3):541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Aoki K, Nakamura H, Suzuki H, Matsuo K, Kataoka K, Shimamura T, et al. Prognostic relevance of genetic alterations in diffuse lower‐grade gliomas. Neuro‐Oncol. 2018;20(1):66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Appay R, Dehais C, Maurage C‐A, Alentorn A, Carpentier C, Colin C, et al. CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH‐mutant gliomas. Neuro‐Oncol. 2019;21(12):1519–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lu VM, O’Connor KP, Shah AH, Eichberg DG, Luther EM, Komotar RJ, et al. The prognostic significance of CDKN2A homozygous deletion in IDH‐mutant lower‐grade glioma and glioblastoma: a systematic review of the contemporary literature. J Neurooncol. 2020;148(2):221–9. [DOI] [PubMed] [Google Scholar]
- 44. Figarella‐Branger D, Mokhtari K, Dehais C, Jouvet A, Uro‐Coste E, Colin C, et al. Mitotic index, microvascular proliferation, and necrosis define 3 groups of 1p/19q codeleted anaplastic oligodendrogliomas associated with different genomic alterations. Neuro‐Oncol. 2014;16(9):1244–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cimino PJ, Zager M, McFerrin L, Wirsching H‐G, Bolouri H, Hentschel B, et al. Multidimensional scaling of diffuse gliomas: application to the 2016 World Health Organization classification system with prognostically relevant molecular subtype discovery. Acta Neuropathol Commun. 2017;5:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Giannini C, Scheithauer BW, Weaver AL, Burger PC, Kros JM, Mork S, et al. Oligodendrogliomas: reproducibility and prognostic value of histologic diagnosis and grading. J Neuropathol Exp Neurol. 2001;60(3):248–62. [DOI] [PubMed] [Google Scholar]
- 47. Griffin CA, Burger P, Morsberger L, Yonescu R, Swierczynski S, Weingart JD, et al. Identification of der(1;19)(q10;p10) in Five Oligodendrogliomas Suggests Mechanism of Concurrent 1p and 19q Loss. J Neuropathol Exp Neurol. 2006;65(10):988–94. [DOI] [PubMed] [Google Scholar]
- 48. Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66(20):9852–61. [DOI] [PubMed] [Google Scholar]
- 49. Vogazianou AP, Chan R, Bäcklund LM, Pearson DM, Liu LU, Langford CF, et al. Distinct patterns of 1p and 19q alterations identify subtypes of human gliomas that have different prognoses. Neuro‐Oncol. 2010;12(7):664–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self‐renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Arita H, Narita Y, Fukushima S, Tateishi K, Matsushita Y, Yoshida A, et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol (Berl). 2013;126(2):267–76. [DOI] [PubMed] [Google Scholar]
- 52. Bettegowda C, Agrawal N, Jiao Y, Sausen M, Wood LD, Hruban RH, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333(6048):1453–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bell RJA, Rube HT, Xavier‐Magalhães A, Costa BM, Mancini A, Song JS, et al. Understanding TERT promoter mutations: a common path to immortality. Mol Cancer Res MCR. 2016;14(4):315–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–61. [DOI] [PubMed] [Google Scholar]
- 55. Astigarraga S, Grossman R, Díaz‐Delfín J, Caelles C, Paroush Z, Jiménez G. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 2007;26(3):668–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ahmad ST, Rogers AD, Chen MJ, Dixit R, Adnani L, Frankiw LS, et al. Capicua regulates neural stem cell proliferation and lineage specification through control of Ets factors. Nat Commun. 2019;10(1):2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Elman JS, Ni TK, Mengwasser KE, Jin D, Wronski A, Elledge SJ, et al. Identification of FUBP1 as a long tail cancer driver and widespread regulator of tumor suppressor and oncogene alternative splicing. Cell Rep. 2019;28(13):3435–3449.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hwang I, Cao D, Na Y, Kim D‐Y, Zhang T, Yao J, et al. Far upstream element‐binding protein 1 regulates LSD1 alternative splicing to promote terminal differentiation of neural progenitors. Stem Cell Rep. 2018;10(4):1208–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhou W, Chung YJ, Parrilla Castellar ER, Zheng Y, Chung H‐J, Bandle R, et al. Far upstream element binding protein plays a crucial role in embryonic development, hematopoiesis, and stabilizing Myc expression levels. Am J Pathol. 2016;186(3):701–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zheng Y, Dubois W, Benham C, Batchelor E, Levens D. FUBP1 and FUBP2 enforce distinct epigenetic setpoints for MYC expression in primary single murine cells. Commun Biol. 2020;3(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chan AK‐Y, Pang JC‐S, Chung NY‐F, Li KK‐W, Poon WS, Chan DT‐M, et al. Loss of CIC and FUBP1 expressions are potential markers of shorter time to recurrence in oligodendroglial tumors. Mod Pathol Off J U S Can Acad Pathol Inc. 2014;27(3):332–42. [DOI] [PubMed] [Google Scholar]
- 62. Horbinski C, Miller CR, Perry A. Gone FISHing: clinical lessons learned in brain tumor molecular diagnostics over the last decade. Brain Pathol. 2010;21(1):57–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Woehrer A, Hainfellner JA. Molecular diagnostics: techniques and recommendations for 1p/19q assessment. CNS Oncol. 2015;4(5):295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation‐based classification of central nervous system tumours. Nature. 2018;555(7697):469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mueller W, Hartmann C, Hoffmann A, Lanksch W, Kiwit J, Tonn J, et al. Genetic signature of oligoastrocytomas correlates with tumor location and denotes distinct molecular subsets. Am J Pathol. 2002;161(1):313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huse JT, Diamond EL, Wang L, Rosenblum MK. Mixed glioma with molecular features of composite oligodendroglioma and astrocytoma: a true “oligoastrocytoma”? Acta Neuropathol (Berl). 2015;129(1):151–3. [DOI] [PubMed] [Google Scholar]
- 67. Wilcox P, Li CCY, Lee M, Shivalingam B, Brennan J, Suter CM, et al. Oligoastrocytomas: throwing the baby out with the bathwater? Acta Neuropathol (Berl). 2015;129(1):147–9. [DOI] [PubMed] [Google Scholar]
- 68. Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro‐Oncol. 2014;16(Suppl 4):iv1–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Aibaidula A, Chan AK‐Y, Shi Z, Li Y, Zhang R, Yang R, et al. Adult IDH wild‐type lower‐grade gliomas should be further stratified. Neuro‐Oncol. 2017;19(10):1327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Reuss DE, Kratz A, Sahm F, Capper D, Schrimpf D, Koelsche C, et al. Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol (Berl). 2015;130(3):407–17. [DOI] [PubMed] [Google Scholar]
- 71. Stichel D, Ebrahimi A, Reuss D, Schrimpf D, Ono T, Shirahata M, et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol (Berl). 2018;136(5):793–803. [DOI] [PubMed] [Google Scholar]
- 72. Wijnenga MMJ, Dubbink HJ, French PJ, Synhaeve NE, Dinjens WNM, Atmodimedjo PN, et al. Molecular and clinical heterogeneity of adult diffuse low‐grade IDH wild‐type gliomas: assessment of TERT promoter mutation and chromosome 7 and 10 copy number status allows superior prognostic stratification. Acta Neuropathol (Berl). 2017;134(6):957–9. [DOI] [PubMed] [Google Scholar]
- 73. Ozawa T, Riester M, Cheng Y‐K, Huse J, Squatrito M, Helmy K, et al. Most human non‐GCIMP glioblastoma subtypes evolve from a common proneural‐like precursor glioma. Cancer Cell. 2014;26(2):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Berzero G, Di Stefano AL, Ronchi S, Bielle F, Villa C, Guillerm E, et al. IDH‐wildtype lower‐grade diffuse gliomas: the importance of histological grade and molecular assessment for prognostic stratification. Neuro‐Oncol. 2021;23(6):955–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Esteller M, Garcia‐Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA‐repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–4. [DOI] [PubMed] [Google Scholar]
- 76. Funakoshi Y, Hata N, Takigawa K, Arita H, Kuga D, Hatae R, et al. Clinical significance of CDKN2A homozygous deletion in combination with methylated MGMT status for IDH‐wildtype glioblastoma. Cancer Med. 2021;10(10):3177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ma S, Rudra S, Campian JL, Dahiya S, Dunn GP, Johanns T, et al. Prognostic impact of CDKN2A/B deletion, TERT mutation, and EGFR amplification on histological and molecular IDH‐wildtype glioblastoma. Neuro‐Oncol Adv. 2020;2(1):vdaa126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Labussière M, Di Stefano AL, Gleize V, Boisselier B, Giry M, Mangesius S, et al. TERT promoter mutations in gliomas, genetic associations and clinico‐pathological correlations. Br J Cancer. 2014;111(10):2024–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Di Stefano AL, Fucci A, Frattini V, Labussiere M, Mokhtari K, Zoppoli P, et al. Detection, characterization and inhibition of FGFR‐TACC fusions in IDH wild type glioma. Clin Cancer Res Off J Am Assoc Cancer Res. 2015;21(14):3307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099):1231–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bielle F, Di Stefano A‐L, Meyronet D, Picca A, Villa C, Bernier M, et al. Diffuse gliomas with FGFR3‐TACC3 fusion have characteristic histopathological and molecular features. Brain Pathol Zurich Switz. 2018;28(5):674–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–7. [DOI] [PubMed] [Google Scholar]
- 84. Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109(8):3041–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single‐cell RNA‐seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sottoriva A, Spiteri I, Piccirillo SGM, Touloumis A, Collins VP, Marioni JC, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci. 2013;110(10):4009–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kim H, Zheng S, Amini SS, Virk SM, Mikkelsen T, Brat DJ, et al. Whole‐genome and multisector exome sequencing of primary and post‐treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25(3):316–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy‐driven evolution of recurrent glioma. Science. 2014;343(6167):189–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wefers AK, Stichel D, Schrimpf D, Coras R, Pages M, Tauziède‐Espariat A, et al. Isomorphic diffuse glioma is a morphologically and molecularly distinct tumour entity with recurrent gene fusions of MYBL1 or MYB and a benign disease course. Acta Neuropathol (Berl). 2020;139(1):193–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang L, Shao L, Li H, Yao K, Duan Z, Zhi C, et al. Histone H3.3 G34‐mutant diffuse gliomas in adults. Am J Surg Pathol. 2022;46(2):249–57. [DOI] [PubMed] [Google Scholar]
- 91. Meyronet D, Esteban‐Mader M, Bonnet C, Joly M‐O, Uro‐Coste E, Amiel‐Benouaich A, et al. Characteristics of H3 K27M‐mutant gliomas in adults. Neuro‐Oncol. 2017;19(8):1127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Riva G, Cima L, Villanova M, Ghimenton C, Sina S, Riccioni L, et al. Low‐grade neuroepithelial tumor: unusual presentation in an adult without history of seizures. Neuropathol Off J Jpn Soc Neuropathol. 2018;38(5):557–60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study


