Abstract
The nutritional requirements of Lactobacillus helveticus CRL 1062 were determined with a simplified chemically defined medium (SCDM) and compared with those of L. helveticus CRL 974 (ATCC 15009). Both strains were found to be prototrophic for alanine, glycine, asparagine, glutamine, and cysteine. In addition, CRL 1062 also showed prototrophy for lysine and serine. The microorganisms also required riboflavin, calcium pantothenate, pyridoxal, nicotinic acid, and uracil for growth in liquid SCDM. The growth rate and the synthesis of their cell membrane-bound serine proteinases, but not of their intracellular leucyl-aminopeptidases, were influenced by the peptide content of the medium. The highest proteinase levels were found during cell growth in basal SCDM, while the synthesis of this enzyme was inhibited in SCDM supplemented with Casitone, Casamino Acids, or β-casein. Low-molecular-mass peptides (<3,000 Da), extracted from Casitone, and the dipeptide leucylproline (final concentration, 5 mM) play important roles in the medium-dependent regulation of proteinase activity. The addition of the dipeptide leucylproline (5 mM) to SCDM reduced proteinase activity by 25%.
Lactobacillus helveticus is an industrially important starter used for the manufacture of hard cheeses, such as Grana and Provolone (6). This microorganism produces high levels of lactic acid in milk and can lower the pH of this medium in 24 h to values between 3.3 and 3.5 (25). L. helveticus has complex nutritional requirements for growth. Morishita et al. (28) determined the absolute requirements of amino acids, vitamins, and bases for the growth of L. helveticus ATCC 15009 in a glucose-salts defined medium. This strain is auxotrophic for 14 amino acids, 4 vitamins, and uracil. The amino acids l-serine, glycine, l-cysteine, and l-alanine, as well as folic acid, p-aminobenzoic acid, adenine, guanine, and xanthine, are not essential but stimulate cell growth on solid media. The concentrations of essential amino acids in milk are very limited; thus, sustained growth of L. helveticus in this medium, like that of other lactic acid bacteria (e.g., Lactococcus), depends on the production of a proteinase, peptidases, and specific peptide and amino acid transport systems (14, 15, 24). In addition to the vital role of the complex proteolytic system of lactic acid bacteria for bacterial growth in milk, it also contributes to the texture, development of flavor, and bitterness of cheeses (19, 33).
There has been an extensive effort to characterize the cell envelope-associated proteinases of lactic acid bacteria as well as the regulation of expression of these enzymes. The genetics and biochemistry of these extracellular proteinases in Lactococcus lactis have been intensively studied (14, 15). It was found that L. lactis strains AM1, E8, and Wg2 produced more proteinase in milk than in laboratory media (12). Cell envelope-associated proteinases of thermophilic lactobacilli have been isolated and characterized, mainly from Lactobacillus delbrueckii subsp. bulgaricus and L. helveticus (8, 9, 18, 21). The genes encoding proteinases from L. delbrueckii subsp. bulgaricus (8) and Lactobacillus paracasei subsp. paracasei (11) have been sequenced. However, no information is available about the regulation of proteinase production in lactobacilli.
L. helveticus CRL 1062 is a microorganism currently used as a starter culture in the manufacture of Argentinian hard cheeses; this organism produces a cell membrane-bound proteinase (9). We report here the minimal nutritional requirements for CRL 1062 and the effects of various nitrogen sources on the proteinase and aminopeptidase N activities of two L. helveticus strains (CRL 1062 and CRL 974 [ATCC 15009]).
MATERIALS AND METHODS
Microorganisms, media, and growth conditions.
L. helveticus CRL 974 (ATCC 15009) and CRL 1062 were obtained from Centro de Referencia para Lactobacilos, San Miguel de Tucuman, Argentina. Cultures were stored at −70°C in 10% sterile reconstituted skim milk containing 0.5% yeast extract and 10% glycerol; cultures were reactivated in MRS (3) broth at 40°C for 16 h.
In this study, the basal medium described by Morishita et al. (28) and a simplified chemically defined medium (SCDM) (Table 1) were used. SCDM without amino acids was called CDMWA (chemically defined medium without amino acids). Chemically defined media (pH 6.5) were prepared from concentrated individual stock solutions which were stored at −4°C after filtration, except for the cysteine solution, which was freshly prepared. Stock solutions were composed of 100-fold-concentrated solutions of each amino acid, base, and vitamin, 20% glucose; Tween 80; and salts. All amino acids, vitamins, purines, pyrimidines, and inorganic salts were of analytical grade (Sigma Chemical Co., St. Louis, Mo.). Media and stock solutions were sterilized by filtration through a cellulose acetate membrane (0.20-μm-pore size; Sartorius AG, Göttingen, Germany).
TABLE 1.
Composition of SCDM
| Constituent | Concn (g · liter−1) |
|---|---|
| Glucose | 10 |
| Sodium acetate | 6 |
| KH2PO4 | 3 |
| K2HPO4 | 3 |
| MgSO4 · 7H2O | 0.2 |
| Tween 80 | 1 |
| l-Alanine | 0.10 |
| l-Arginine | 0.10 |
| l-Asparagine | 0.20 |
| l-Aspartic acid | 0.20 |
| l-Cysteine | 0.20 |
| l-Glutamine | 0.20 |
| l-Glutamic acid | 0.20 |
| Glycine | 0.10 |
| l-Histidine | 0.10 |
| l-Isoleucine | 0.10 |
| l-Leucine | 0.10 |
| l-Lysine | 0.10 |
| l-Methionine | 0.10 |
| l-Phenylalanine | 0.10 |
| l-Proline | 0.10 |
| l-Serine | 0.10 |
| l-Threonine | 0.10 |
| l-Tryptophan | 0.10 |
| l-Tyrosine | 0.10 |
| l-Valine | 0.10 |
| Nicotinic acid | 0.001 |
| Calcium pantothenate | 0.001 |
| Pyridoxal | 0.002 |
| Riboflavin | 0.001 |
| Uracil | 0.01 |
Low-molecular-mass (<3,000 Da) peptides (LMMP) of Casitone (Difco Laboratories, Detroit, Mich.) were separated from high-molecular-mass peptides (HMMP) of Casitone by centrifugal filtration (3,000 × g) using filter units with a nominal molecular mass limit of 3,000 Da (Centricon-3 concentrators; Amicon, Beverly, Mass.).
When needed, SCDM and CDMWA were supplemented with 1% (wt/vol) Casitone, 1% (wt/vol) Casamino Acids (Difco), 0.1% (wt/vol) β-casein (Sigma), 1% (wt/vol) LMMP, 1% (wt/vol) HMMP, and the following di- or tripeptides: glycylmethionine, glycylproline, glycyltyrosine, leucylleucine, leucylproline, prolylalanine, prolylleucine, tyrosylglycine, leucylglycylglycine, and valylprolylleucine (final concentrations, 1 to 5 mM; Sigma).
Working cultures of L. helveticus were propagated in MRS broth at 40°C for 16 h. To eliminate carryover nutrients, the cells were harvested by centrifugation at 8,000 × g for 15 min, washed twice in sterile 50 mM sodium phosphate (pH 7.0), and resuspended in this buffer to the original volume. This cell suspension was used to inoculate the different media at an initial optical density (determined with Spectronic 2000; Bausch & Lomb, Rochester, N.Y.) at 560 nm (OD560) of 0.07. Bacterial growth was monitored by measuring the OD560, and these measurements were used for correlation with cell dry weight determinations. Cells were harvested by filtration (0.2-μm-pore-size filter), washed once with deionized water, and dried to a constant weight at 60°C under partial vacuum (200 mm Hg). A change of 1 unit of optical density was shown to be equivalent to 0.50 g of dry matter.
Cell suspensions and cell extracts.
Cells grown in the different media were harvested by centrifugation (10,000 × g, 10 min, 4°C) at the exponential growth phase (OD560 = 0.65), washed twice with 0.85% (wt/vol) saline supplemented with 10 mM CaCl2, and resuspended to a final OD560 of approximately 10 in 100 mM sodium phosphate (pH 7.0).
Cells extracts were prepared by adding glass beads (0.15- to 0.25-mm diameter; Sigma) to the bacterial cell suspensions and mixing these suspensions for 7 min at 4°C in a vortex mixer (Reax 2000; Heidolph, Schwabach, Germany) at maximum speed. Glass beads, cell debris, and unbroken cells were removed by centrifugation (10,000 × g, 10 min, 4°C).
Enzyme assays.
The proteinase (PrtH) activities of whole-cell suspensions in 50 mM sodium phosphate (pH 7.0) at 40°C were measured with the chromogenic substrate succinyl-alanyl-alanyl-prolyl-phenylalanine-p-nitroaniline (S-Ala; Sigma) as described by Exterkate (5). One unit of proteinase was defined as the amount required to liberate 1 nmol of nitroanilide per min; specific activity was expressed as that unit per milligram of protein.
Leucyl-aminopeptidase (PepN) activity was determined at 40°C with l-leucine-p-nitroaniline (20 mM; Sigma) as the substrate. Reaction mixtures contained 100 μl of substrate stock solution, 850 μl of 0.2 M Tris-HCl (pH 7.0), and 50 μl of enzyme extract. The reaction was stopped by the addition of 0.5 ml of 80% acetic acid. The samples were centrifuged (20,000 × g, 10 min, 4°C), and the release of nitroaniline was subsequently monitored spectrophotometrically at 410 nm. One unit of PepN activity was defined as the amount of enzyme that hydrolyzed 1 μmol of substrate per min.
Cell lysis was determined by monitoring the release of lactate dehydrogenase (LDH) by the method of Thomas (31).
Casein hydrolysis.
Washed cells, harvested from the different media, were suspended in 100 mM sodium phosphate (pH 7.0). The suspensions were allowed to utilize the residual intracellular amino acids for 30 min at 40°C. Casein degradation was carried out as described previously (10). Washed whole cells (OD560 = 10) were incubated with 5 mg of substrate per ml, dissolved in 100 mM sodium phosphate (pH 7.0) at a ratio of 1:1. As the substrate, α-, β-, or κ-casein (Sigma) was used. The resulting mixtures were incubated for 30 min or 1, 2, or 3 h at 40°C. The samples were then centrifuged (10,000 × g, 10 min, 4°C), and the supernatants were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (17). Either Coomassie brilliant blue R-250 or silver staining (Bio-Rad Laboratories, Richmond, Calif.) was used to visualize the proteins after SDS-PAGE.
Protein determination.
Protein concentration was determined by use of a protein assay according to the manufacturer's instructions (Bio-Rad).
RESULTS
Nutritional requirements of L. helveticus.
To identify the absolute nutritional requirements of L. helveticus CRL 1062, the single- or multiple-omission technique was applied to each component of the synthetic growth medium described by Morishita et al. (28). In these experiments, L. helveticus CRL 974 (ATCC 15009) was included as a control. Bacterial growth was not affected when ZnCl2, MnSO4 · 4H2O, and FeSO4 · 7H2O were omitted individually or together. However, when MgSO4 · 7H2O or potassium phosphate was removed, growth stopped rapidly, indicating an absolute requirement for these elements. The addition of ammonium citrate to the growth medium had no effect, whereas a slightly lower OD560 (about 6%) was found in the absence of sodium acetate. The omission of Tween 80 resulted in a reduction of growth by approximately 70%. Calcium pantothenate, riboflavin, nicotinic acid, and pyridoxal were essential for the growth of both L. helveticus strains. d-Biotin, cyanocobalamin, thiamine, p-aminobenzoic acid, folic acid, and niacinamide could be omitted simultaneously with only a small reduction (10%) of the final cell density. Adenine, guanine, deoxyguanosine, inosine, thymidine, hypoxanthine, and xanthine could be removed from the medium without a reduction of the maximum growth rates of both strains by more than 8%. However, the omission of uracil prevented growth completely. Other compounds assayed, such as CaCl2 and orotic acid, did not have any effect on cell growth.
On the basis of these nutritional requirements, SCDM was formulated (Table 1) and used in further experiments. SCDM contains, in addition to the 18 amino acids of the medium developed by Morishita et al. (28), 0.2 g each of glutamine and asparagine per liter. In this medium, L. helveticus CRL 1062 and CRL 974 showed similar growth rates (0.34 and 0.28 h−1, respectively), as in the initial medium. In SCDM, the stationary phase was reached after 10 and 14 h for L. helveticus CRL 1062 and CRL 974, respectively.
To characterize the amino acid requirements of L. helveticus CRL 1062 and CRL 974, the cells were grown in SCDM from which individual amino acids or an entire metabolic family of amino acids had been removed (Table 2). No growth was observed for any strains when arginine, glutamic acid, histidine, isoleucine, leucine, methionine, phenylalanine, proline, threonine, tryptophan, tyrosine, or valine was removed, suggesting that these amino acids were essential. L. helveticus CRL 974 showed a requirement for lysine and serine in addition to the amino acids necessary for the growth of L. helveticus CRL 1062. No growth was observed, even after 72 h, when aspartic acid and asparagine were omitted, indicating that aspartic acid was also essential. The single omission of aspartic acid, asparagine, glutamine, or cysteine did not affect the growth of either strain. However, these strains did not grow when only the essential amino acids were present. Furthermore, the removal of alanine or glycine resulted in a decreased growth rate and in a prolonged lag phase for L. helveticus CRL 1062 (Table 2). Also, an important decrease of the growth rate (70%) of CRL 1062 was observed when the serine-glycine-cysteine family was removed, an effect essentially attributed to serine (Table 2). The omission of each of the remaining families resulted in total growth inhibition, because of the absence of essential amino acids in these families.
TABLE 2.
Effect of omission of amino acids from SCDM on the growth rates of L. helveticus CRL 974 and CRL 1062
| Omitted amino acid(s) | Growth ratesa of:
|
|
|---|---|---|
| CRL 974 | CRL 1062 | |
| Ala | 94 | 58 |
| Arg | 0 | 0 |
| Asn | 100 | 95 |
| Asp | 98 | 97 |
| Cys | 100 | 100 |
| Gln | 100 | 95 |
| Glu | 0 | 0 |
| Gly | 100 | 60 |
| His | 0 | 0 |
| Ile | 0 | 0 |
| Leu | 0 | 0 |
| Lys | 0 | 91 |
| Met | 0 | 0 |
| Phe | 0 | 0 |
| Pro | 0 | 0 |
| Ser | 0 | 35 |
| Thr | 0 | 0 |
| Trp | 0 | 0 |
| Tyr | 0 | 0 |
| Val | 0 | 0 |
| Phe, Trp, Tyr | 0 | 0 |
| Ser, Gly, Cys | 0 | 30 |
| Gln, Glu, Pro, Arg | 0 | 0 |
| Thr, Lys, Asn, Met, Asp | 0 | 0 |
| Ile, Leu, Val | 0 | 0 |
Growth rates are expressed as a percentage of the maximum growth rate obtained in SCDM. Maximum growth rates were 0.28 and 0.34 h−1 for L. helveticus CRL 974 and CRL 1062, respectively. Values are the averages of three independent experiments.
Growth on different nitrogen sources.
The growth rates of L. helveticus CRL 1062 and CRL 974 were evaluated with different media (MRS broth, SCDM, and CDMWA) as well as with minimal media (SCDM and CDMWA) supplemented with various nitrogen sources: Casitone, Casamino Acids, β-casein, specific di- and tripeptides, and LMMP and HMMP fractions isolated from Casitone (Table 3). In all media, L. helveticus CRL 1062 grew faster than CRL 974, although the final cell densities were similar for both strains (Table 3). The highest specific growth rate was obtained in MRS broth. When Casitone or LMMP were added to SCDM, specific growth rates of both strains increased 1.3-fold. L. helveticus CRL 974 and CRL 1062 were able to grow in CDMWA supplemented with Casitone as the sole amino acid source, with specific growth rates similar to those found in SCDM plus Casitone. However, no growth was observed when the free amino acids of SCDM were replaced with β-casein or Casamino Acids (Table 3). No effect on the growth of either strain was observed when SCDM was supplemented with HMMP or di- and tripeptides (data not shown).
TABLE 3.
Effect of supplementation of SCDM with various nitrogen sources on the growth of L. helveticus CRL 974 and L. helveticus CRL 1062a
| Medium | CRL 974
|
CRL 1062
|
||
|---|---|---|---|---|
| Growth rate (h−1) | Final cell density (OD560)b | Growth rate (h−1) | Final cell density (OD560)b | |
| Basal SCDM | 0.28 ± 0.02 | 1.4 ± 0.08 | 0.34 ± 0.02 | 1.5 ± 0.07 |
| SCDM + 10-fold excess of amino acids | 0.29 ± 0.02 | 1.4 ± 0.07 | 0.33 ± 0.02 | 1.6 ± 0.07 |
| SCDM + 1% Casitone | 0.36 ± 0.03 | 2.2 ± 0.18 | 0.45 ± 0.03 | 2.2 ± 0.19 |
| SCDM + LMMP from Casitone | 0.35 ± 0.03 | 2.3 ± 0.16 | 0.46 ± 0.04 | 2.1 ± 0.18 |
| SCDM + 1% Casamino Acids | 0.30 ± 0.02 | 1.8 ± 0.11 | 0.35 ± 0.03 | 1.7 ± 0.13 |
| SCDM + 1% β-casein | 0.31 ± 0.02 | 1.6 ± 0.12 | 0.36 ± 0.03 | 1.7 ± 0.12 |
| CDMWA + 1% Casitone | 0.33 ± 0.03 | 1.8 ± 0.12 | 0.41 ± 0.04 | 1.8 ± 0.13 |
| CDMWA + 1% Casamino Acids | NG | NG | NG | NG |
| CDMWA + 1% β-casein | NG | NG | NG | NG |
| MRS broth | 0.48 ± 0.04 | 2.9 ± 0.21 | 0.58 ± 0.04 | 3.0 ± 0.28 |
Values are the means and standard deviations from three independent experiments. NG, no growth.
OD560 measurements were obtained after 24 h of incubation at 40°C.
Effect of nitrogen source on proteinase and aminopeptidase activities.
The proteinase (PrtH) and aminopeptidase (PepN) activity levels were measured after the growth of L. helveticus CRL 974 and CRL 1062 in SCDM supplemented with different peptide sources (Table 4). The highest specific PrtH activity was observed at exponential growth phase for both strains (data not shown). PrtH production was decreased in L. helveticus cells grown in peptide-rich media. Thus, the PrtH activities from CRL 1062 cells grown in MRS broth and in SCDM supplemented with Casitone were approximately 32- and 11-fold lower, respectively, than those from cells grown in basal SCDM. The decrease of PrtH production was less marked in cells grown in SCDM supplemented with β-casein (about a 1.7-fold reduction) than in cells grown in SCDM supplemented with Casitone. Casitone is a pancreatic digest of casein consisting of mostly small peptides and free amino acids, in a proportion of 4:1, respectively (22). In control experiments, it was established that Casitone did not have any direct inhibitory effect on the activity of the enzyme PrtH (unpublished data). PrtH activities were also assayed after the growth of L. helveticus CRL 1062 in SCDM supplemented with Casamino Acids and in SCDM supplemented with high (10-fold) concentrations of each of 20 amino acids. Casamino Acids is an acid hydrolysate of casein in which free amino acids and small peptides are present in a ratio of 82 to 18%, respectively (according to the manufacturer). The PrtH activity of L. helveticus CRL 1062 decreased twofold when cells were grown in SCDM with Casamino Acids compared to when cells were grown in basal SCDM. This reduction was smaller than that obtained with the equivalent concentration of the peptide-rich nitrogen source Casitone (Table 4). Furthermore, no decrease of PrtH activity was observed in CRL 1062 cells grown in SCDM containing high concentrations of free amino acids (10-fold).
TABLE 4.
PrtH and PepN activities of L. helveticus CRL 974 and CRL 1062 after growth in SCDM supplemented with different nitrogen sourcesa
| Medium | Sp act of the following enzyme in the indicated strain:
|
|||
|---|---|---|---|---|
| PrtHb
|
Sp PepNc
|
|||
| CRL 974 | CRL 1062 | CRL 974 | CRL 1062 | |
| Basal SCDM | 129 ± 7 | 192 ± 8 | 468 ± 25 | 122 ± 9 |
| SCDM + 10-fold excess of amino acids | 132 ± 7 | 195 ± 7 | 460 ± 23 | 112 ± 8 |
| SCDM + 1% Casitone | 45 ± 3 | 17 ± 0.8 | 433 ± 26 | 140 ± 7 |
| SCDM + 1% Casamino Acids | 115 ± 8 | 94 ± 4 | 485 ± 32 | 121 ± 9 |
| SCDM + 1% β-casein | 72 ± 6 | 114 ± 5 | 402 ± 25 | 132 ± 9 |
| CDMWA + 1% Casitone | 46 ± 3 | 18 ± 0.9 | 430 ± 26 | 140 ± 8 |
| MRS broth | 6 ± 0.3 | 6 ± 0.4 | 405 ± 27 | 138 ± 8 |
Values are the means and standard deviations from three independent experiments.
Specific activity is expressed as nanomoles minute−1 milligram of protein−1. LDH activity was less than 0.06% the total activity in each cell extract.
Specific activity is expressed as micromoles minute−1 milligram of protein−1.
To determine which peptides are involved in the regulation of PrtH activity, eight specific dipeptides, two tripeptides, and two peptide fractions from Casitone (LMMP and HMMP) were evaluated for their effect on the PrtH activity of CRL 1062 cells after their addition to the growth medium. When L. helveticus CRL 1062 was grown in SCDM supplemented with LMMP, the PrtH activity levels were similar to those obtained in SCDM supplemented with Casitone (Table 5 and Fig. 1). With the exception of leucylproline (5 mM), none of the di- and tripeptides analyzed (final concentrations, 1 to 5 mM) nor HMMP influenced PrtH activity (Table 5). An increase in the leucylproline concentration (up to 5 mM) resulted in a reduction of PrtH activity by 25% compared to the activity obtained after growth in basal SCDM (Table 5).
TABLE 5.
PrtH activity of L. helveticus CRL 1062 after growth in SCDM supplemented with different peptidesa
| Addition to basal SCDM |
PrtH sp act (nmol min−1 mg of protein−1)b |
|---|---|
| None | 192 ± 8 |
| 1% LMMP | 18 ± 1 |
| 1% HMMP | 191 ± 8 |
| 1 mM Leu-Gly-Glyc | 190 ± 7 |
| 1 mM Leu-Leud | 189 ± 8 |
| 5 mM Leu-Leud | 185 ± 9 |
| 1 mM Leu-Pro | 191 ± 7 |
| 5 mM Leu-Pro | 144 ± 5 |
Values are the means and standard deviations from three independent experiments.
LDH activity was less than 0.06% the total activity in each cell extract.
The PrtH activity of L. helveticus CRL 1062 after growth in SCDM with the tripeptide Val-Pro-Leu was similar.
The PrtH activity of L. helveticus CRL 1062 after growth in SCDM with the dipeptides Gly-Met, Gly-Pro, Gly-Tyr, Pro-Ala, Pro-Leu, and Tyr-Gly was similar.
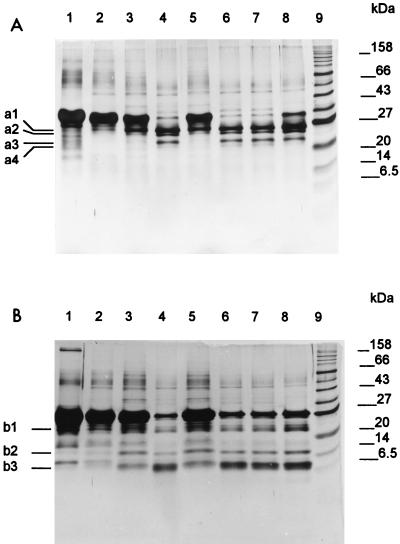
FIG. 1.
SDS-PAGE analysis of α-casein (A) and β-casein (B) hydrolysis by L. helveticus CRL 1062 after growth in MRS broth (lane 2), SCDM supplemented with 1% Casitone (lane 3), SCDM with HMMP from Casitone (lane 4), SCDM with LMMP from Casitone (lane 5), basal SCDM (lane 6), SCDM with 1 mM Leu-Pro (lane 7), and SCDM with 5 mM Leu-Pro (lane 8). Lane 1, starting substrate; lane 9, molecular mass markers.
The effect of the medium composition on the PrtH activity of L. helveticus CRL 974 was also studied. Cells of this strain showed a medium dependence of PrtH activity similar to that shown by cells of strain CRL 1062, but the effects were smaller (Table 4). The PrtH activity in cells of strain CRL 974 grown in MRS broth was 21-fold lower than that obtained in SCDM. In CRL 974 cells grown in SCDM supplemented with Casamino Acids, β-casein, or Casitone, the PrtH activities were about 1.2-, 2-, and 3-fold lower, respectively, than those in cells grown in basal SCDM (Table 4).
In contrast to PrtH activity, the highest levels of PepN activity were observed in L. helveticus CRL 974 (Table 4). For both strains, similar PepN activities were observed in cells grown in SCDM or in the peptide-rich MRS broth. Furthermore, PepN activity was not affected significantly by the addition of Casitone, Casamino Acids, or β-casein to SCDM.
Casein hydrolysis.
The ability of L. helveticus CRL 1062 to hydrolyze α-, β-, and κ-casein was tested after growth in MRS broth and SCDM supplemented with different peptide sources. The CRL 1062 proteinase hydrolyzed α- and β-casein predominantly and κ-casein at a much lower rate. Since LDH activity in whole-cell suspensions did not exceed 1% total cell LDH activity, it was concluded that the detected proteolytic activity was due to the action of a cell-wall-bound proteinase. The rates of α- and β-casein hydrolysis by cells grown in peptide-rich media, such as MRS broth or SCDM supplemented with 1% Casitone, were significantly lower than those observed for cells grown in basal SCDM (Fig. 1). With the exception of cells grown in SCDM plus leucylproline (5 mM), there were no apparent differences in the rates and patterns of hydrolysis of α- and β-casein by cells grown in basal SCDM and those grown in SCDM supplemented with di- and tripeptides (final concentrations, 1 to 5 mM) or with HMMP (Fig. 1). On the other hand, the proteolytic activity of L. helveticus CRL 1062 cells grown in SCDM was higher than that of cells grown in SCDM supplemented with LMMP (Fig. 1).
The α-casein hydrolysis patterns obtained with L. helveticus CRL 1062 after 3 h of incubation were characterized by four peptide products (a1 to a4) with approximate molecular masses of between 20 and 27 kDa (Fig. 1A). When the β-casein hydrolysate was studied by SDS-PAGE, three main peptides, b1, b2, and b3 (of about 24, 15, and 6.5 kDa, respectively), appeared after 3 h of incubation (Fig. 1B). With prolonged incubation times, all products were further hydrolyzed into smaller peptides, which were undetectable by SDS-PAGE.
DISCUSSION
This study was conducted to determine the nutritional requirements of L. helveticus CRL 1062 cells and to investigate the influence of nitrogen source nutrients, such as Casitone, Casamino Acids, and β-casein, on the proteinase and peptidase activities of this microorganism in a defined minimal medium.
Ledesma et al. (20) and Morishita et al. (28) have defined synthetic growth media for Lactobacillus which allow study of the minimal growth requirements of L. helveticus. The main differences between these media and the simple synthetic medium formulated in this work (SCDM) are in the vitamin and inorganic element compositions. The medium used in this work (SCDM) contains only 31 components and is less complex than previously described media. This medium supports sustained growth at a reasonably high rate for the L. helveticus strains tested. This result indicates that SCDM will be most suitable for physiological studies of these microorganisms and perhaps many other lactobacilli. In most synthetic media described in the literature, ferrous sulfate is present. We did not observe such a requirement for the growth of L. helveticus, in agreement with the results of Pandy et al. (29) for numerous lactic acid bacteria. The slightly lower specific growth rate found when sodium acetate was omitted from the minimal medium developed by Morishita et al. (28) could be attributed to the effect of acetate on the size of the cell (20) or to the low buffer capacity of the modified medium (26). Growth was not perturbed by a reduced ammonium ion content, indicating that the amino acids present in SCDM satisfy the nitrogen requirements for biomass synthesis. The vitamins nicotinic acid, calcium pantothenate, riboflavin, and pyridoxal were essential for growth. Nicotinic acid and calcium pantothenate are involved in coenzyme biosynthesis by Lactococcus (1). Riboflavin, a component of flavin coenzymes, appears to be essential for the growth of lactic acid bacteria (7). Pyridoxal is the prosthetic group of different enzymes and is involved in catalysis (4). Multiple omissions of all nonessential vitamins reduced the growth rate by 8%, in contrast to the results obtained for the amino acids.
L. helveticus CRL 1062 and CRL 974 require for growth more amino acids than other lactic acid bacteria, such as Lactobacillus plantarum (26, 30), Lactobacillus curvatus (26), Lactobacillus casei (28), and L. lactis (1, 13). L. helveticus is a homofermentative lactobacillus; therefore, strains of this species cannot use derivatives of the pentose pathway as amino acid precursors. The multiple-amino-acid auxotrophies of the two L. helveticus strains tested showed considerable similarities, although the cells of CRL 974 were more demanding. CRL 1062 and CRL 974 required 13 and 15 essential amino acids for growth in SCDM, respectively. Our data confirmed the amino acid requirements of CRL 974 described by Morishita et al. (28). However, in our experiments, serine was also shown to be essential for CRL 974 cells, while Morishita et al. (28) described it to be a nonessential but stimulatory amino acid for CRL 974 cell growth. We found that for L. helveticus strains, glutamine could be replaced by glutamate but not vice versa, indicating that glutamate is essential for growth and that glutamine is not necessary if glutamate is present.
The multiple auxotrophy of lactobacilli has been related to a single mutation in the RNA polymerase (27), associated with the presence of a fully inoperative citric acid cycle (2), or to single-step mutations accumulated in the amino acid biosynthetic pathways. The glutamate biosynthetic pathway is one of the most extensively affected pathways in lactic acid bacteria (28). This pathway is often blocked in the step leading to the synthesis of 2-oxoglutarate. Glutamate auxotrophy could not be reverted to prototrophy in L. plantarum, L. casei, L. helveticus, or Lactobacillus acidophilus (28). However, Morishita et al. (28) isolated mutants of L. helveticus and L. casei which became prototrophic for certain amino acids required for growth and suggested that in wild-type strains, the genes involved in their synthesis were present but not functional.
The growth rates of CRL 1062 and CRL 974 were affected by the nitrogen source present in the culture medium. They were higher in SCDM supplemented with Casitone, a pancreatic digest of casein containing mainly peptides, than in SCDM supplemented with β-casein or Casamino Acids.
The levels of PepN of L. helveticus CRL 974 and CRL 1062 were similar in SCDM with different peptide compositions. These results suggest that PepN does not have medium-dependent regulation. Relatively stable levels of PepN activity and its transcripts in cells of L. helveticus grown in MRS broth have also been described previously (32). For L. lactis, the regulation of intracellular peptidase activity has been described to be a strain-dependent phenomenon (23).
In contrast to the situation for PepN, medium dependence of PrtH synthesis was observed. The lowest PrtH activity was found in peptide-rich media (e.g., MRS broth). Proteinase synthesis in L. helveticus CRL 974 and CRL 1062 seems to be inhibited by casein degradation products other than amino acids. The PrtH activity of cells grown in basal SCDM was reduced by the addition of either Casitone, Casamino Acids, or LMMP to SCDM but not in SCDM supplemented with a 10-fold excess of amino acids. Inhibition of PrtH activity in SCDM supplemented with 1% β-casein was less extensive than that seen with the equivalent concentration of Casitone. These results suggest either that the production of inhibitory peptides from β-casein, after its hydrolysis by PrtH, is low or that the LMMP obtained from Casitone are more effective in inhibiting proteinase synthesis than are those liberated from β-casein.
The effect of nitrogen source on proteinase production in the strains tested suggests that the proteolytic system of L. helveticus is similar to that of L. lactis subsp. cremoris (16, 23), although the nature of the regulatory compound may be different. Like that in L. lactis, PrtH activity in L. helveticus was inhibited (25%) by leucylproline (5 mM), but inhibition of PrtH activity by this dipeptide was less effective than that reported for L. lactis (22, 23). Also, unlike that in L. lactis, PrtH production in L. helveticus seemed to be insensitive to the addition of prolylleucine (up to 5 mM) to SCDM. The mechanism of inhibition of PrtH synthesis by peptides and leucylproline is unknown. Additional work on the molecular charrization of the medium-dependent regulation of proteinase activity in L. helveticus is under way.
ACKNOWLEDGMENTS
This work was supported by grants from the Consejo Nacional de Investigaciones Científcas y Técnicas (CONICET), FONCYT, and the Consejo de Investigaciones de la Universidad Nacional de Tucumán (CIUNT).
REFERENCES
- 1.Cocaign-Bousquet M, Garrigues C, Novak L, Lindley N D, Loubiere P. Rational development of a simple synthetic medium for the sustained growth of Lactococcus lactis. J Appl Bacteriol. 1995;79:108–116. [Google Scholar]
- 2.Deguchi Y, Morishita T. Nutritional requirements in multiple auxotrophic lactic acid bacteria: genetic lesions affecting amino acid biosynthetic pathways in Lactococcus lactis, Enterococcus faecium, and Pediococcus acidilactici. Biosci Biotech Biochem. 1992;56:913–918. doi: 10.1271/bbb.56.913. [DOI] [PubMed] [Google Scholar]
- 3.De Man J C, Rogosa M, Sharpe M E. A medium for the cultivation of lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 4.Dempsey W B. Synthesis of pyridoxal phosphate. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 539–543. [Google Scholar]
- 5.Exterkate F A. Differences in short peptide-substrate cleavage by two cell-envelope-located serine proteinases of Lactococcus lactis subsp. cremoris are related to secondary binding specificity. Appl Microbiol Biotechnol. 1990;33:401–406. doi: 10.1007/BF00176654. [DOI] [PubMed] [Google Scholar]
- 6.Fortina M G, Nicastro G, Carminati D, Neviani E, Manachini P L. Lactobacillus helveticus heterogeneity in natural cheese starters: the diversity in phenotypic characteristics. J Appl Microbiol. 1998;84:72–80. doi: 10.1046/j.1365-2672.1997.00312.x. [DOI] [PubMed] [Google Scholar]
- 7.Foucaud C, Francois A, Richard J. Development of a chemically defined medium for the growth of Leuconostoc mesenteroides. Appl Environ Microbiol. 1997;63:301–304. doi: 10.1128/aem.63.1.301-304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert C, Atlan D, Blanc B, Portalier R, Germond J E, Lapierre L, Mollet B. A new cell surface proteinase: sequencing and analysis of the prtB gene from Lactobacillus delbrueckii subsp. bulgaricus. J Bacteriol. 1996;178:3059–3065. doi: 10.1128/jb.178.11.3059-3065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hébert E M, Raya R R, De Giori G S. Characterisation of a cell-envelope proteinase of Lactobacillus helveticus CRL 1062. Biotechnol Lett. 1999;21:831–834. [Google Scholar]
- 10.Hébert E M, Raya R R, Tailliez P, De Giori G S. Characterisation of natural isolates of Lactobacillus strains to be used as starter cultures in dairy fermentation. Int J Food Microbiol. 2000;59:19–27. doi: 10.1016/s0168-1605(00)00282-8. [DOI] [PubMed] [Google Scholar]
- 11.Holck A, Naes H. Cloning, sequencing and expression of the gene encoding the cell-envelope-associated proteinase from Lactobacillus paracasei subsp. paracasei NCDO151. J Gen Microbiol. 1992;138:1353–1364. doi: 10.1099/00221287-138-7-1353. [DOI] [PubMed] [Google Scholar]
- 12.Hugenholtz J, Exterkate F A, Konings W N. The proteolytic systems of Streptococcus cremoris: an immunological analysis. Appl Environ Microbiol. 1984;48:1105–1110. doi: 10.1128/aem.48.6.1105-1110.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen P R, Hammer K. Minimal requirements for exponential growth of Lactococcus lactis. Appl Environ Microbiol. 1993;59:4363–4366. doi: 10.1128/aem.59.12.4363-4366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunji E R S, Hagting A, De Vries C J, Juillard V, Haandrikman A J, Poolman B, Konings W N. Transport of β-casein-derived peptides by the oligopeptide transport system is a crucial step in the proteolytic pathway of Lactococcus lactis. J Biol Chem. 1995;270:1569–1574. doi: 10.1074/jbc.270.4.1569. [DOI] [PubMed] [Google Scholar]
- 15.Kunji E R S, Mierau I, Hagting A, Poolman B, Konings W N. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:187–221. doi: 10.1007/BF00395933. [DOI] [PubMed] [Google Scholar]
- 16.Laan H, Bolhuis H, Poolman B, Abee T, Konings W N. Regulation of proteinase synthesis in Lactococcus lactis. Acta Biotechnol. 1993;2:95–101. [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Laloi P, Atlan D, Blanc B, Gilbert C, Portalier R. Cell-wall-associated proteinase of Lactobacillus delbrueckii subsp. bulgaricus CNRZ 397: differential extraction, purification and properties of the enzyme. Appl Microbiol Biotechnol. 1991;36:196–204. doi: 10.1007/BF00164419. [DOI] [PubMed] [Google Scholar]
- 19.Law J, Haandrikman A. Proteolytic enzymes of lactic acid bacteria. Int Dairy J. 1997;7:1–11. [Google Scholar]
- 20.Ledesma O V, de Ruiz Holgado A A P, Oliver G, de Giori G S, Raibaud P, Galpin J V. A synthetic medium for comparative nutritional studies of lactobacilli. J Appl Bacteriol. 1977;42:123–133. doi: 10.1111/j.1365-2672.1977.tb00676.x. [DOI] [PubMed] [Google Scholar]
- 21.Martín-Hernández M C, Alting A C, Exterkate F A. Purification and characterization of the mature, membrane-associated cell-envelope proteinase of Lactobacillus helveticus L89. Appl Microbiol Biotechnol. 1994;40:828–834. [Google Scholar]
- 22.Marugg J D, Meijer W, van Kranenburg R, Laverman P, Bruinenberg P G, De Vos W M. Medium-dependent regulation of proteinase gene expression in Lactococcus lactis: control of transcription initiation by specific dipeptides. J Bacteriol. 1995;177:2982–2989. doi: 10.1128/jb.177.11.2982-2989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meijer W, Marugg J D, Hugenholtz J. Regulation of proteolytic enzyme activity in Lactococcus lactis. Appl Environ Microbiol. 1996;62:151–156. doi: 10.1128/aem.62.1.156-161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mierau I, Kunji E R S, Leenhouts K J, Hellendoorn M A, Haandrikman A J, Poolman B, Konings W N, Venema G, Kok J. Multiple-peptidase mutants of Lactococcus lactis are severely impaired in their ability to grow in milk. J Bacteriol. 1996;178:2794–2803. doi: 10.1128/jb.178.10.2794-2803.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morelli L, Vescovo M, Cocconcelli P S, Bottazzi V. Fast and slow milk-coagulating variants of Lactococcus helveticus HLM 1. Can J Microbiol. 1986;32:758–760. doi: 10.1139/m86-138. [DOI] [PubMed] [Google Scholar]
- 26.Møreto T, Hagen B F, Axelsson L. A new, completely defined medium for meat lactobacilli. J Appl Microbiol. 1998;85:715–722. [Google Scholar]
- 27.Morishita T, Fukada T, Shirota M, Yura T. Genetic basis of nutritional requirements in Lactobacillus casei. J Bacteriol. 1974;120:1078–1084. doi: 10.1128/jb.120.3.1078-1084.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morishita T, Deguchi Y, Yajima M, Sakurai T, Yura T. Multiple nutritional requirements of lactobacilli: genetic lesions affecting amino acid biosynthetic pathways. J Bacteriol. 1981;148:64–71. doi: 10.1128/jb.148.1.64-71.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pandy A, Bringel F, Meyer J M. Iron requirement and search for siderophores in lactic acid bacteria. Appl Microbiol Biotechnol. 1994;40:735–739. [Google Scholar]
- 30.Ruiz-Barba J L, Jiménez-Diaz R. Vitamin and amino acid requirements of Lactobacillus plantarum strains isolated from green olive fermentations. J Appl Bacteriol. 1994;76:350–355. doi: 10.1111/j.1365-2672.1994.tb01639.x. [DOI] [PubMed] [Google Scholar]
- 31.Thomas T D. Tagatose-1,6-diphosphate activation of lactate dehydrogenase from Streptococcus cremoris. Biochem Biophys Res Commun. 1975;63:1035–1042. doi: 10.1016/0006-291x(75)90673-7. [DOI] [PubMed] [Google Scholar]
- 32.Varmanen P, Vesanto E, Steele J L, Palva A. Characterization and expression of the pepN gene encoding a general aminopeptidase from Lactobacillus helveticus. FEMS Microbiol Lett. 1994;124:315–320. doi: 10.1111/j.1574-6968.1994.tb07302.x. [DOI] [PubMed] [Google Scholar]
- 33.Visser S. Proteolytic enzymes and their relation to cheese ripening and flavor: an overview. J Dairy Sci. 1991;76:329–350. [Google Scholar]