Abstract
Objective
Cysteine conjugate beta-lyase 2 (CCBL2), also known as kynurenine aminotransferase 3 (KAT3) or glutamine transaminase L (GTL), plays an essential role in transamination and cytochrome P450. Its correlation with some other cancers has been explored, but breast cancer (BC) not yet.
Methods
The mRNA and protein expression of CCBL2 in BC cell lines and patient samples were detected by RT-qPCR and immunohistochemistry (IHC). BC patients’ clinical information and RNA-Seq expression were acquired via The Cancer Genome Atlas (TCGA) database. Patients were categorized into high/low CCBL2 expression groups based on the optimal cutoff value (8.973) determined by receiver operating characteristic (ROC) curve. We investigated CCBL2 and clinicopathological characteristics’ relationship using Chi-square tests, estimated diagnostic capacity using ROC curves and drew survival curves using Kaplan–Meier estimate. We compared survival differences using Cox regression and externally validated using Gene Expression Omnibus (GEO) database. We evaluated enriched signaling pathways using gene set enrichment analysis (GSEA), explored CCBL2 and relevant genes’ relationship using tumor immunoassay resource (TIMER) databases and used the human protein atlas (HPA) for pan-cancer analysis and IHC.
Results
CCBL2 was overexpressed in normal human cell lines and tissues. CCBL2 expression was lower in BC tissues (n = 1104) than in normal tissues (n = 114), validated by GEO database. Several clinicopathologic features were related to CCBL2, especially estrogen receptor (ER), progesterone receptor (PR) and clinical stages. The low expression group exhibited poor survival. CCBL2’s area under curve (AUC) analysis showed finite diagnostic capacity. Multivariate cox-regression analysis indicated CCBL2 independently predicted BC survival. GSEA showed enriched pathways: early estrogen response, MYC and so on. CCBL2 positively correlated with estrogen, progesterone and androgen receptors. CCBL2 was downregulated in most cancers and was associated with their survival, including renal and ovarian cancers.
Conclusions
Low CCBL2 expression is a promising poor BC survival independent prognostic marker.
Introduction
Breast cancer (BC) is the most diagnosed cancer in women, accounting for 11.7% of all types of cancers worldwide and the highest morbidity rate in women [1–3]. This commonly diagnosed malignant tumor is also the leading cause of cancer deaths worldwide, just after lung cancer [4]. Approximate 2.1 million people were diagnosed with BC in 2018 [5]. As a heterogeneous disease, various biomarker-based diagnostic and prognostic approaches have emerged in recent years. ER, PR and human epidermal growth factor receptor-2 (HER2) have served as both diagnostic and prognostic biomarkers of BC [6]. Nowadays, with advances in sequencing technology, DNA methylation, miRNAs, autoantibodies, lipidomics and proteomics as well as identification of multiparameter gene signatures have facilitated the early diagnosis and prognosis of breast carcinoma [7–10]. These latest studies have sparked our interest in mining genes as biomarkers associated with BC.
CCBL2 has been found in mouse, rat, and human whose mRNA is widely expressed in several organs such as the liver, kidney, heart, and neuroendocrine tissues. However, the highest expression of CCBL2 is found in the kidney [11, 12]. CCBL2 can effectively catalyze the transamination of glutamine, methionine, histidine, phenylalanine, cysteine, asparagine, and kynurenine (KYN) to kynurenic acid (KYNA) as well as the pathway of drug metabolism by cytochrome P450 [11, 13]. All of these functions are involved in the important processes in human amino acid metabolism. According to the HUGO Gene Nomenclature Committee (HGNC), CCBL2 is identical to kynurenine aminotransferase 3 (KAT3) and glutamine transaminase L (GTL) genes [12, 14]. In mammalian cells, the essential amino acid tryptophan is degraded mainly through the kynurenine pathway. Kynurenine aminotransferases (KATs) catalyze the synthesis of KYNA, which is a metabolite of tryptophan and an endogenous antagonist of N-methyl-D-aspartate and alpha 7-nicotinic acetylcholine receptors [15–17]. And KYNA is a recognized neuroprotective and anticonvulsant agent involved in synaptic transmission and in the pathophysiology of various neurological disorders(11). Abnormal expression levels of CCBL2 are involved in the pathophysiological process of kidney injury, hospital-acquired VTE, depression and neurological disorders [12, 15, 18–22]. Recently, GTK (Glutamine Transaminase K, which is identical to KAT1 and CCBL1) has been reported to play an important role in pancreatic tumorigenesis through the glutamine pathway and cysteine conjugate beta-lyase (CCBL) had close relation with kidney cancer. Glutamine, as one of the catalytic substrates of CCBL2, plays biosynthetic roles in cells, as it is used in the biosynthesis of amino acids, proteins, lipids, and nucleotides which are essential to cell division, especially in cancer cells, also known as glutamine addiction, reported to be concerned with the process of pancreatic cancer. Furthermore, cysteine conjugate beta-lyase (CCBL) was found to be closely associated with the development of kidney cancer. Studies have shown that variants in the CCBL2 gene were significantly associated with the risk of chronic kidney disease due to a defect in reductive metabolism that leads to the formation of a cysteine conjugate, which is then converted to an active metabolite [20, 22, 23]. The findings piqued our curiosity in the role of the CCBL family member, CCBL2, in breast tissues, as well as the unknown association between CCBL2 and BC, which is now the most frequently diagnosed cancer worldwide. Moreover, we assessed whether CCBL2 could serve as a prognostic marker of survival in patients with BC.
Therefore, to initially ascertain whether CCBL2 expression levels affect BC prognosis, we studied the correlation between CCBL2 expression in BC tissues and clinicopathological characteristics, as well as with the survival status of patients with BC through analysis of The Cancer Genome Atlas-Breast Invasive Carcinoma (TCGA-BRCA) level 3 data. Additionally, the results were validated using Gene Expression Omnibus (GEO) datasets. At both mRNA and protein levels, the expression of CCBL2 was verified with real-time quantitative polymerase chain reaction (RT-qPCR) and immunohistochemistry (IHC) staining in human protein atlas (HPA) dataset, respectively. Furthermore, a pan-cancer analysis of CCBL2 was performed to explore the correlation between CCBL2 and various cancers.
Methods
Breast cell lines
During this study, two types of human BC cells, MCF-7 and MDA-MB-231, and human normal breast epithelial cell lines MCF-10A were used. MCF-7 cell lines, regarded as type of luminal, were cultured in DMEM (Gibco, USA). MDA-MB -231, regarded as type of basal, were grown in RPMI 1640 (Gibco, USA). Both cells were supplemented with 10% fetal calf serum (Gibco, USA) and 1% Penicillin-Streptomycin Solution (Beyotime China). MCF-10A cell lines were cultured in DMEM-F12 (Gibco, USA) with 5% equine serum, 1% Penicillin-Streptomycin Solution, 20ng/ml epidermal growth factor, 0.5ug/ml hydrocortisone, 0.1ug/ml cholera toxin and 10ug/ml insulin. The cell lines mentioned above were all obtained from the American Type Culture Collection (ATCC, USA) and cultured in a humid atmosphere of 5% CO2 at 37°C.
Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
The total RNA was isolated using the TRIzol reagent (Invitrogen, USA) and reverse transcription was implemented using the HiScript Ⅲ RT SuperMix for qPCR with gDNA wiper (Vazyme Biotech) to synthesize cDNA following the manufacturer’s instructions. The RT-qPCR was performed by ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech) and run by the Mastercycler Ep Realplex (Eppendorf, Hamburg, Germany). The relative gene expression fold change was normalized using beta-tubulin 2A (TUBB2A) as an internal control and compared with MCF-10A. The primers sequences used in this study were as follows: CCBL2, F: 5ʹ-ATC CTT GTG ACA GTA GGA GCA-3ʹ, R: 5ʹ-GGG CTC ATA GCA GTC ATA GAA AG-3ʹ; TUBB2A, F: 5ʹ-TTG GGA GGT CAT CAG CGA TGA G-3ʹ, R: 5ʹ-AGG CTC CAG ATC CAC CAG GAT G-3ʹ. The independent experiments were performed at least three times.
Gene Set Enrichment Analyze (GSEA)
GSEA software3.0. (http://software.broadinstitute.org/gsea/downloads.jsp) was applied for Gene Set Enrichment Analysis (GSEA). The normalized enrichment score (NES) was obtained by alignment analysis 1000 times with FDR (false discovery rates)<0.25 and NOM p<0.05.
Gene correlation analysis
Tumor Immunoassay Resource (TIMER) databases (https://cistrome.shinyapps.io/timer/) (data based on TGGA) were employed to explore the association between CCBL2 and estrogen receptor 1 (ESR1), estrogen receptor 2 (ESR2), progesterone receptor (PGR), androgen receptor (AR), cytochrome P450 2B6 (CYP2B6), ribosomal protein S6 kinase B1(RPS6KB1) and MYC genes. Correlation analysis drew the expression scatterplots between a pair of genes in breast cancer, together with the Spearman’s rho value and estimated statistical significance.
Pan-cancer analysis of CCBL2
The expression differences between normal and tumor tissues were analyzed by TIMER databases (data based on TCGA). Analysis of survival probability of CCBL2 in pan-cancer was completed systemically in HPA databases (https://www.proteinatlas.org/). Relevant immunohistochemistry (IHC) staining images were also obtained from HPA.
Data mining
Breast cancer patients’ data of the clinical information as well as the level 3 RNA-Seq expression were acquired via TCGA database (https://cancergenome.nih.gov/), involving normal tissues (n = 114) and BC tissues (n = 1104). The analysis process applied the RNA-Seq by Expectation Maximization (RSEM) expression values. GEO datasets (GSE42568, GSE71053) [24, 25] were obtained via the GEO database (https://www.ncbi.nlm.nih.gov/geo/).
Statistical analysis
Using the ggplot2 package in R, the differential expression of discrete variables was processed into visible boxplots with Wilcoxon and Kruskal-Wallis test. Based on the optimal cutoff value (8.973) determined by the ROC curve, patients were categorized into high and low CCBL2 expression groups. With the implement of Chi-square test as well as Fisher exact test in R program (version 3.5.2), the analysis of relationship between the expression of CCBL2 and clinicopathological characteristics was performed. ROC curves of the subjects were plotted to estimate their diagnostic ability by applying the ROC package. Adopting the survival package of R, we used the Kaplan-Meier curves to compare respective differences in overall survival (OS) and relapse-free survival (RFS) between high and low groups. Kaplan-Meier Plotter (https://kmplot.com/analysis/) was used to further explore the relationship between the prognosis of patients with endocrine therapy and CCBL2 expression. Log-rank test was used for p values calculation. The clinicopathological characteristics were selected by the univariate and multivariate cox regression analysis. Independent experiment of RT-qPCR was done three times and measured data expressed as mean ± standard deviation. The result of RT-qPCR was plotted by GraphPad Prism 8. P value<0.05 was the significance threshold.
Results
CCBL2 expression in BC cell lines and tissues
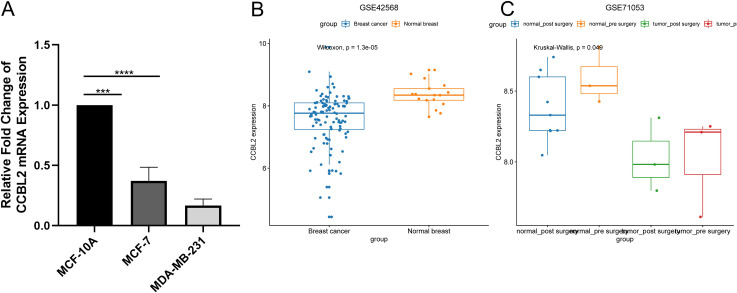
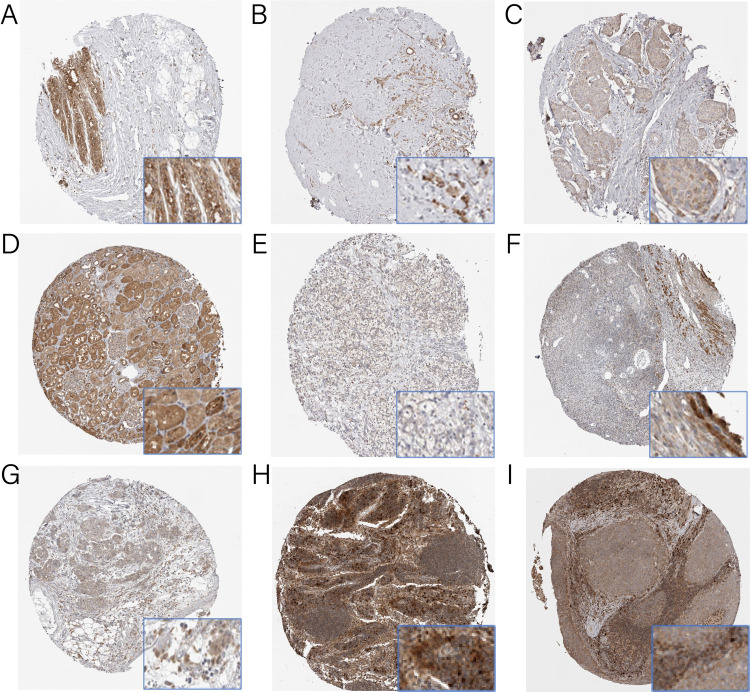
CCBL2 was overexpressed in the human normal breast epithelial cell line MCF-10A while downregulated in BC cell lines (Fig 1A) and tissues (Fig 2A–2C). In particular, MCF-7 cells showed higher CCBL2 expression than MDA-MB-231 cells. Out of a total of 24 samples of BC, 22 of them exhibited moderate IHC staining for CCBL2, other two of them showed weak IHC staining. However, normal tissues exhibited strong IHC staining for CCBL2 (Fig 2A–2C). This result indicated that CCBL2 expression varied at the protein level. In addition, it was validated in microarrays GSE42568 (p = 1.3e-05) and GSE71053 (p = 0.0490) that CCBL2 expression of BC was lower in tumor tissue than in normal breast tissues (Fig 1B and 1C).
Fig 1. Overexpressed CCBL2 in normal breast.
MCF-7 and MDA-MB-231 mRNA expression was detected lower than MCF-10A and MCF-7 was higher than MDA-MB-231 (A) (*p<0.05, **p<0.01). Microarrays related to CCBL2 expression in normal breast and breast cancer were shown (B and C).
Fig 2. Immunohistochemistry staining for CCBL2.
The expression of CCBL2 in breast cancer (B and C), renal cancer (E), ovarian cancer (G) and head and neck cancer (I) cells were decreased than in respective normal tissues (A, D, F and H). In breast cancer, infiltrating lobular carcinoma (B) expressed more CCBL2 than infiltrating ductal carcinoma (C).
Patient characteristics
Based on the TCGA-BRCA level 3 data, Table 1 showed the clinical characteristics of tumor samples, such as molecular subtype, histological type, menopause status, radiation therapy, margin status, neoadjuvant treatment, targeted molecular therapy, ER, PR, HER-2, TNM stage, clinical stage, vital status, lymph node status and sample type.
Table 1. Clinical characteristics of TCGA-BRCA level 3 cohort.
| Characteristics | Numbers of cases (%) |
|---|---|
| CCBL2 | |
| High | 746(67.57) |
| Low | 358(32.43) |
| Age | |
| <60 | 589(53.45) |
| > = 60 | 513(46.55) |
| Gender | |
| Female | 1090(98.73) |
| Male | 12(1.09) |
| NA | 2(0.18) |
| Histological type | |
| Infiltrating Ductal Carcinoma | 790(71.56) |
| Infiltrating Lobular Carcinoma | 204(18.48) |
| Other | 107(9.69) |
| NA | 3(0.27) |
| Molecular subtype | |
| Basal | 142(12.86) |
| HER-2 | 67(6.07) |
| Lum A | 422(38.22) |
| Lum B | 194(17.57) |
| Normal | 24(2.17) |
| NA | 255(23.1) |
| ER | |
| Indeterminate | 2(0.18) |
| Negative | 239(21.65) |
| Positive | 813(73.64) |
| NA | 50(4.53) |
| PR | |
| Indeterminate | 4(0.36) |
| Negative | 345(31.25) |
| Positive | 704(63.77) |
| NA | 51(4.62) |
| HER-2 | |
| Equivocal | 180(16.3) |
| Indeterminate | 12(1.09) |
| Negative | 565(51.18) |
| Positive | 164(14.86) |
| NA | 183(16.58) |
| T classification | |
| T1 | 281(25.45) |
| T2 | 640(57.97) |
| T3 | 138(12.5) |
| T4 | 40(3.62) |
| TX | 3(0.27) |
| NA | 2(0.18) |
| N classification | |
| N0 | 516(46.74) |
| N1 | 367(33.24) |
| N2 | 120(10.87) |
| N3 | 79(7.16) |
| NX | 20(1.81) |
| NA | 2(0.18) |
| M classification | |
| M0 | 917(83.06) |
| M1 | 22(1.99) |
| MX | 163(14.76) |
| NA | 2(0.18) |
| Stage | |
| I | 182(16.49) |
| II | 626(56.7) |
| III | 252(22.83) |
| IV | 20(1.81) |
| X | 14(1.27) |
| NA | 10(0.91) |
| Lymph node status | |
| No | 28(2.54) |
| Yes | 697(63.13) |
| NA | 379(34.33) |
| Vital status | |
| Deceased | 155(14.04) |
| Living | 947(85.78) |
| NA | 2(0.18) |
| Sample type | |
| Metastatic | 7(0.63) |
| Primary tumor | 1097(99.37) |
| Menopause status | |
| Inde | 34(3.08) |
| Peri | 40(3.62) |
| Post | 706(63.95) |
| Pre | 231(20.92) |
| NA | 93(8.42) |
| Margin status | |
| Close | 31(2.81) |
| Negative | 922(83.51) |
| Positive | 79(7.16) |
| NA | 72(6.52) |
| Radiation therapy | |
| NO | 445(40.31) |
| YES | 557(50.45) |
| NA | 102(9.24) |
| Neoadjuvant treatment | |
| No | 1088(98.55) |
| Yes | 13(1.18) |
| NA | 3(0.27) |
| Targeted molecular therapy | |
| NO | 46(4.17) |
| YES | 533(48.28) |
| NA | 525(47.55) |
| Os | |
| Alive | 933(85.83) |
| Dead | 154(14.17) |
| Rfs | |
| Relapse-free | 816(89.47) |
| Relapse | 96(10.53) |
Abbreviation: ER: estrogen receptor; PR: progesterone receptor; HER-2: human epidermal growth factor-2; T: tumor; M: metastasis; N: node; OS: overall survival; RFS: relapse-free survival; NA: not available.
CCBL2 expression in BC
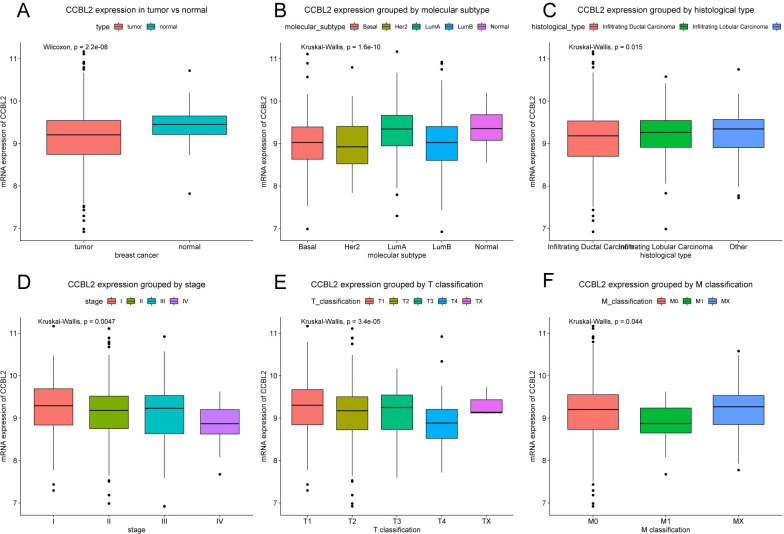
Compared with that in normal tissues (n = 114), the expression of CCBL2 was lower in BC tissues (n = 1104; p = 2.2e-08). Additionally, drawn in boxplots, CCBL2 expression varied with the molecular subtype (p = 1.6e-10), histological type (p = 0.0150), clinical stage (p = 0.0047), T classification (p = 3.4e-05), M classification (p = 0.0440) (Fig 3), while no statistical difference in patient age, menopause status, sample type, N classification, sample type etc. (data not shown).
Fig 3. Differences in CCBL2 expression shown in boxplots.
The subgroups included type (A), molecular subtype (B), histological type (C), clinical stage (D), T classification (E), and M classification (F). (p < 0.05).
The correlation between clinicopathological characteristics and CCBL2 expression in BC
Based on the optimal cutoff value (8.973) determined by the ROC curve, patients were categorized into high and low CCBL2 expression groups. Results of Chi-square or Fisher exact test demonstrated that several clinicopathologic features, including histological type (p = 0.0010), molecular subtype (p = 0.0005), ER (p = 0.0005), PR (p = 0.0005), HER2 (p = 0.0085), T classification (p = 0.0130), M classification (p = 0.0210), vital status (p = 0.0025), stage (p = 0.0320), OS (p = 0.0025) and RFS (p = 0.0375), were correlated with CCBL2 expression (Table 2).
Table 2. Correlations of CCBL2 expression in BC tissues with clinicopathologic features.
| Clinical characteristics | Variable | No. of cases | CCBL2 expression | χ2 | P value | |
|---|---|---|---|---|---|---|
| High n (%) | Low n (%) | |||||
| Age | <60 | 589 | 404 (68.59) | 185 (31.41) | 0.7858 | 0.4108 |
| > = 60 | 513 | 339 (66.08) | 174 (33.92) | |||
| Gender | Female | 1090 | 736 (67.52) | 354 (32.48) | 0.4564 | 0.5437 |
| Male | 12 | 7 (58.33) | 5 (41.67) | |||
| Histological type | Infiltrating Ductal Carcinoma | 790 | 505 (63.92) | 285 (36.08) | 16.1545 | 0.001 |
| Infiltrating Lobular Carcinoma | 204 | 156 (76.47) | 48 (23.53) | |||
| Other | 107 | 82 (76.64) | 25 (23.36) | |||
| Molecular subtype | Basal | 142 | 82 (57.75) | 60 (42.25) | 49.4216 | 0.0005 |
| Her2 | 67 | 35 (52.24) | 32 (47.76) | |||
| LumA | 422 | 332 (78.67) | 90 (21.33) | |||
| LumB | 194 | 110 (56.70) | 84 (43.30) | |||
| Normal | 24 | 19 (79.17) | 5 (20.83) | |||
| ER | Indeterminate | 2 | 1 (50.00) | 1 (50.00) | 19.3441 | 0.0005 |
| Negative | 239 | 134 (56.07) | 105 (43.93) | |||
| Positive | 813 | 578 (71.09) | 235 (28.91) | |||
| PR | Indeterminate | 4 | 1 (25.00) | 3 (75.00) | 20.1547 | 0.0005 |
| Negative | 345 | 205 (59.42) | 140 (40.58) | |||
| Positive | 704 | 507 (72.02) | 197 (27.98) | |||
| HER2 | Equivocal | 180 | 129 (71.67) | 51 (28.33) | 11.7483 | 0.0085 |
| Indeterminate | 12 | 9 (75.00) | 3 (25.00) | |||
| Negative | 565 | 382 (67.61) | 183 (32.39) | |||
| Positive | 164 | 91 (55.49) | 73 (44.51) | |||
| T classification | T1 | 281 | 207 (73.67) | 74 (26.33) | 12.7439 | 0.013 b |
| T2 | 640 | 422 (65.94) | 218 (34.06) | |||
| T3 | 138 | 91 (65.94) | 47 (34.06) | |||
| T4 | 40 | 20 (50.00) | 20 (50.00) | |||
| TX | 3 | 3 (100) | 0 (0) | |||
| N classification | N0 | 516 | 350 (67.83) | 166 (32.17) | 3.4757 | 0.5072 |
| N1 | 367 | 255 (69.48) | 112 (30.52) | |||
| N2 | 120 | 73 (60.83) | 47 (39.17) | |||
| N3 | 79 | 51 (64.56) | 28 (35.44) | |||
| NX | 20 | 14 (70.00) | 6 (30.00) | |||
| M classification | M0 | 917 | 613 (66.85) | 304 (33.15) | 7.8213 | 0.021 |
| M1 | 22 | 10 (45.45) | 12 (54.55) | |||
| MX | 163 | 120 (73.62) | 43 (26.38) | |||
| Stage | I | 182 | 134 (73.63) | 48 (26.37) | 10.766 | 0.032 |
| II | 626 | 421 (67.25) | 205 (32.75) | |||
| III | 252 | 164 (65.08) | 88 (34.92) | |||
| IV | 20 | 8 (40.00) | 12 (60.00) | |||
| X | 14 | 10 (71.43) | 4 (28.57) | |||
| Lymph node status | No | 28 | 18 (64.29) | 10 (35.71) | 0.5126 | 0.5277 |
| Yes | 697 | 492 (70.59) | 205 (29.41) | |||
| Vital status | Deceased | 155 | 88 (56.77) | 67 (43.23) | 9.3118 | 0.0025 |
| Living | 947 | 655 (69.17) | 292 (30.83) | |||
| Sample type | Metastatic | 7 | 4 (57.14) | 3 (42.86) | 0.3367 | 0.6982 |
| Primary Tumor | 1097 | 740 (67.46) | 357 (32.54) | |||
| Menopause status | Inde | 34 | 24 (70.59) | 10 (29.41) | 3.1873 | 0.3683 |
| Peri | 40 | 29 (72.50) | 11 (27.50) | |||
| Post | 706 | 462 (65.44) | 244 (34.56) | |||
| Pre | 231 | 164 (71.00) | 67 (29.00) | |||
| Margin status | Close | 31 | 23 (74.19) | 8 (25.81) | 0.9229 | 0.6322 |
| Negative | 922 | 617 (66.92) | 305 (33.08) | |||
| Positive | 79 | 55 (69.62) | 24 (30.38) | |||
| Radiation therapy | NO | 445 | 290 (65.17) | 155 (34.83) | 1.9235 | 0.1859 |
| YES | 557 | 386 (69.30) | 171 (30.70) | |||
| Neoadjuvant treatment | NO | 1088 | 735 (67.56) | 353 (32.44) | 0.2119 | 0.7686 |
| YES | 13 | 8 (61.54) | 5 (38.46) | |||
| Targeted molecular therapy | NO | 46 | 32 (69.57) | 14 (30.43) | 0.0035 | 1 |
| YES | 533 | 373 (69.98) | 160 (30.02) | |||
| OS | Alive | 933 | 646 (69.24) | 287 (30.76) | 9.7778 | 0.0025 |
| Dead | 154 | 87 (56.49) | 67 (43.51) | |||
| RFS | Relapse-free | 816 | 566 (69.36) | 250 (30.64) | 4.8181 | 0.0375 |
| Relapse | 96 | 56 (58.33) | 40 (41.67) | |||
Abbreviations: Bold values of P < 0.05 indicate statistically significant correlations.
bFisher’s exact test.
Note: High n (%) and low n (%) added up to 100% in each subgroup. For example, high BBCL2 expression n (%) of “Age<60” = 404/589 = 68.59%; low CCBL2 expression n (%) of “Age<60” = 185/589 = 31.41%
Diagnostic capacity of CCBL2 expression
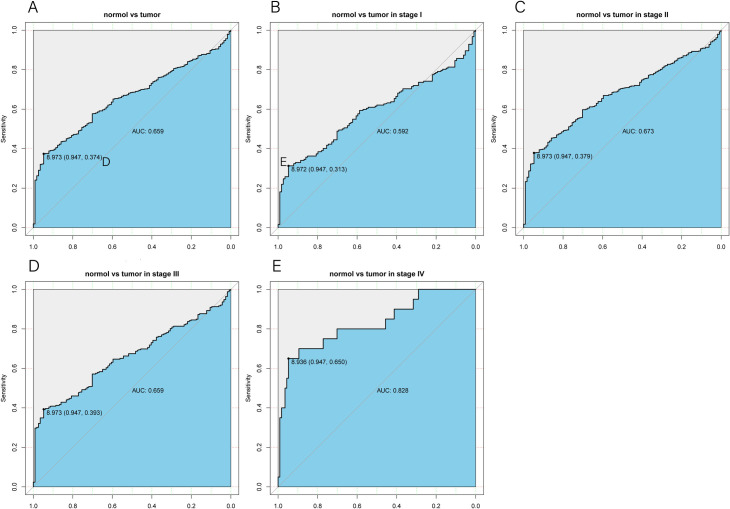
The ROC curve was plotted to assess the diagnostic capacity of CCBL2 and the area under the curve (AUC) showed a value of 0.659, implying a finite diagnostic capacity. In the subgroup analysis of different stages, CCBL2 showed a relatively valuable diagnostic capacity in patients with stage IV BC (AUG: 0.828) (AUC values of BC stages I, II, and III were 0.592, 0.673, and 0.659 respectively) (Fig 4).
Fig 4. The ROC curve of CCBL2 in breast carcinoma cohort.
Normal and tumor samples (A). The AUC (0.659) indicated a limited diagnostic capability. Subgroup analyses: Stage Ⅰ (B) (AUC: 0.592), Stage Ⅱ (C) (AUG:0.673), Stage Ⅲ (D) (AUG:0.659), Stage Ⅳ (E) (AUG:0.828). Abbreviations: AUC, area under the curve; ROC, receiver operating characteristic.
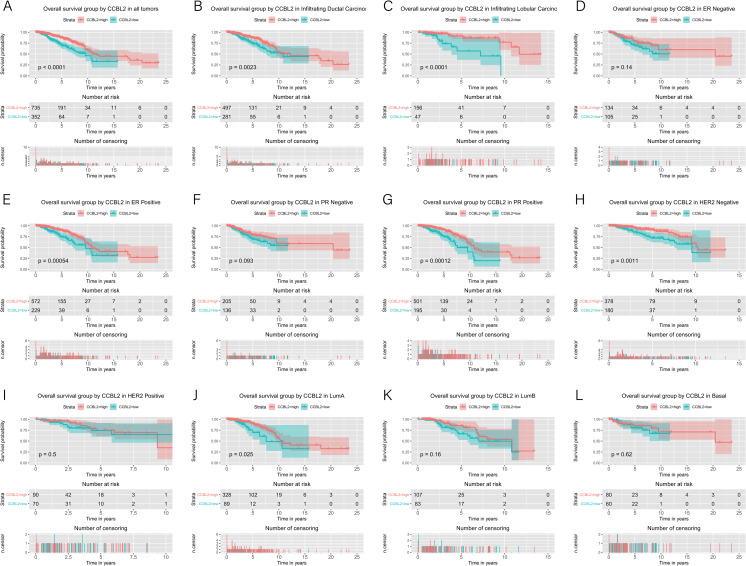
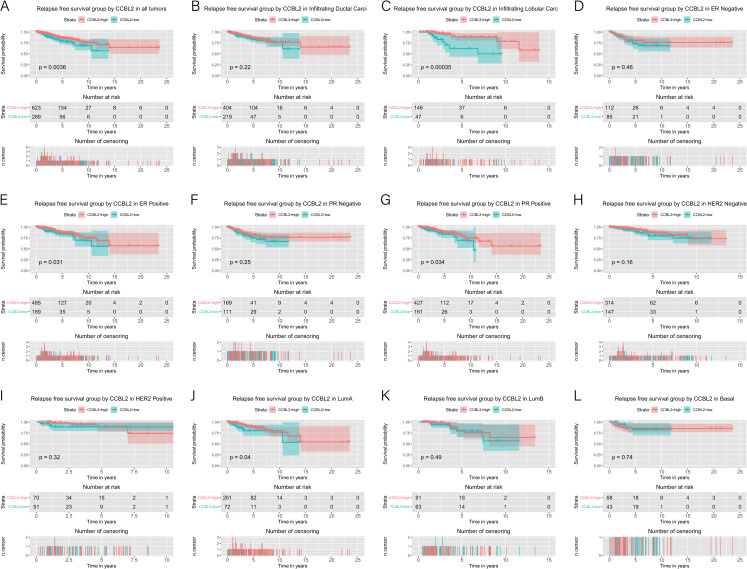
Correlation between CCBL2 expression and survival of patients with BC
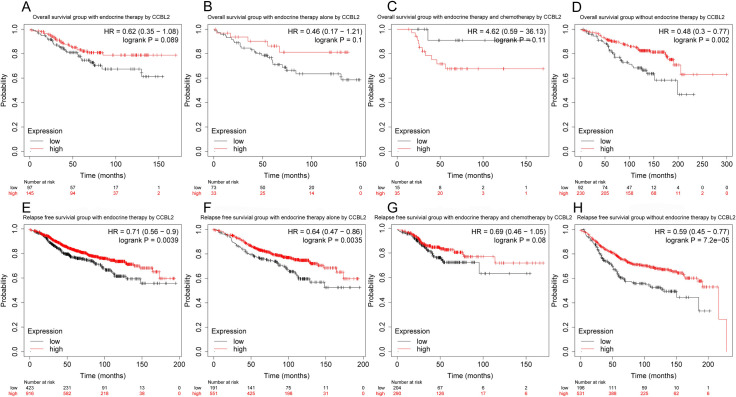
The correlation between CCBL2 expression and survival of patients with BC was determined using Kaplan–Meier curves. The log-rank tests indicated that low CCBL2 expression was associated with a low overall survival (OS) rate (p<0.0001) (Fig 5) as well as a low relapse-free survival (RFS) rate (p = 0.0036) (Fig 6). Subgroup analysis revealed that low CCBL2 expression was correlated with low OS in patients with ER-positive BC (p = 0.0005), PR-positive BC (p = 0.0001), HER-2-negative BC (p = 0.0011), infiltrating ductal carcinoma (p = 0.0023), infiltrating lobular carcinoma (p<0.0001), and luminal A (p = 0.0250) (Fig 5). The analysis also revealed that low CCBL2 expression was associated with low RFS in patients with ER-positive BC (p = 0.0310), PR-positive BC (p = 0.0340), luminal A BC (p = 0.0400), and infiltrating ductal carcinoma (p = 0.0004) (Fig 6). Additionally, Kaplan–Meier analysis was conducted based on whether patients with ER-positive BC had received endocrine therapy. The log-rank tests indicated that high CCBL2 expression was associated with a high RFS rate in patients receiving endocrine therapy (with or without chemotherapy) (p = 0.0039) and in a subgroup of patients receiving endocrine therapy alone (without chemotherapy) (p = 0.0035). Furthermore, high CCBL2 expression was associated with better OS (p = 0.0020) and RFS (p = 7.2e−05) rate in patients with ER-positive BC without endocrine therapy (Fig 7).
Fig 5. Kaplan–Meier curves of overall survival in breast cancer according to CCBL2 expression in breast cancer tissues.
Overall survival analysis and subgroup analyses of histological type (B and C), ER (D and E), PR (F and G), HER-2 (H and I) and molecular subtype (J, K and L). High CCBL2 expression had relations with the high overall survival. (p < 0.0001) (A).
Fig 6. Kaplan–Meier curves of relapse free survival in breast cancer according to CCBL2 expression in breast cancer tissues.
Relapse free survival analysis and subgroup analyses of histological type (B and C), ER (D and E), PR (F and G), HER-2 (H and I) and molecular subtype (J, K and L). High CCBL2 expression had relations with the high relapse free survival (p = 0.0036) (A).
Fig 7. Kaplan–Meier curves of overall survival and relapse free survival in ER-positive breast cancer according to CCBL2 expression in breast cancer tissues.
Subgroups analysis included the overall survival and relapse free survival of patients with endocrine therapy (A and E), with endocrine therapy alone (B and F), with endocrine therapy and chemotherapy (C and G), and without endocrine therapy (D and H). P value<0.05 was the significance threshold.
Independent prognostic value of low CCBL2 expression in BC
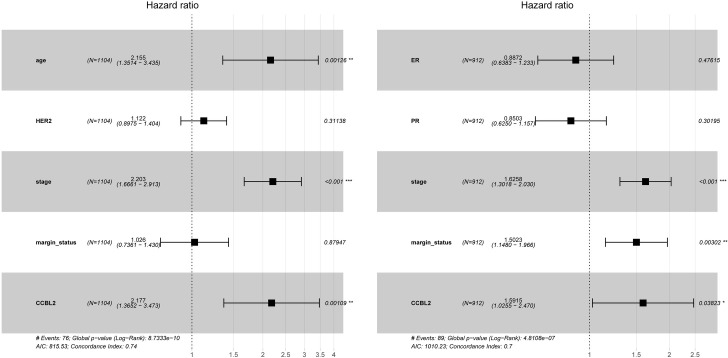
Univariate and multivariate analyses were performed to demonstrate the prognostic value of clinicopathological characteristics, which were subsequently used in the evaluation of the impacts of CCBL2 on the survival of patients with BC. Age, clinical stage, HER-2, margin status and CCBL2 expression were linked with poor OS according to the results of the univariate analysis (Table 3). Likewise, ER, PR, margin status, clinical stage, and CCBL2 expression were linked with an unfavorable RFS (Table 4). Subsequently, multivariate analysis was performed, the results of which were shown in the forest plot (Fig 8). Low CCBL2 expression served as an independent prognostic biomarker for low OS (p = 0.0011; HR: 2.18, 95% CI: 1.37–3.47) and low RFS (p = 0.0382; HR: 1.59, 95% CI: 1.03–2.47) (Tables 3 and 4).
Table 3. Univariate and multivariate analyses of overall survival in breast cancer patients.
| Parameters | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | P value | HR | 95%CI | P value | |
| Age | 1.91 | 1.39–2.63 | 0 | 2.16 | 1.36–3.45 | 0.0013 |
| Histological type | 0.93 | 0.74–1.17 | 0.543 | |||
| Molecular subtype | 1.01 | 0.88–1.16 | 0.901 | |||
| ER | 0.85 | 0.71–1.02 | 0.074 | |||
| PR | 0.87 | 0.73–1.03 | 0.096 | |||
| HER-2 | 1.29 | 1.05–1.57 | 0.013 | 1.12 | 0.90–1.40 | 0.297 |
| Menopause status | 1.16 | 0.94–1.43 | 0.165 | |||
| Stage | 1.64 | 1.40–1.91 | 0 | 2.20 | 1.67–2.91 | 0.0000 |
| Margin status | 1.42 | 1.11–1.81 | 0.005 | 1.03 | 0.74–1.43 | 0.8795 |
| Lymph node status | 1.10 | 0.93–1.30 | 0.274 | |||
| CCBL2 | 2.12 | 1.53–2.93 | 0 | 2.18 | 1.37–3.47 | 0.0011 |
Abbreviations: HR Hazard Ratio, CI confidence interval, bold values of P < 0.05 indicate statistically significant correlations
Table 4. Univariate and multivariate analyses of relapse free survival in breast cancer patients.
| Parameters | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | P value | HR | 95%CI | P value | |
| Age | 1.45 | 0.97–2.16 | 0.072 | |||
| Histological type | 0.86 | 0.65–1.14 | 0.290 | |||
| Molecular subtype | 0.99 | 0.82–1.2 | 0.945 | |||
| ER | 0.78 | 0.63–0.97 | 0.026 | 0.89 | 0.64–1.23 | 0.4762 |
| PR | 0.78 | 0.64–0.96 | 0.019 | 0.85 | 0.63–1.16 | 0.3020 |
| HER-2 | 0.93 | 0.7–1.22 | 0.596 | |||
| Menopause status | 0.95 | 0.74–1.22 | 0.713 | |||
| Stage | 1.71 | 1.4–2.08 | 0 | 1.63 | 1.30–2.03 | 0.0000 |
| Lymph node status | 0.86 | 0.7–1.06 | 0.159 | |||
| Margin status | 1.59 | 1.23–2.06 | 0 | 1.50 | 1.15–1.97 | 0.0030 |
| CCBL2 | 1.82 | 1.21–2.73 | 0.004 | 1.59 | 1.03–2.47 | 0.0382 |
Abbreviations: HR Hazard Ratio, CI confidence interval, bold values of P < 0.05 indicate statistically significant correlations
Fig 8. Multivariate analysis was shown in the forest plot.
Gene set enrichment analysis (GSEA) of CCBL2
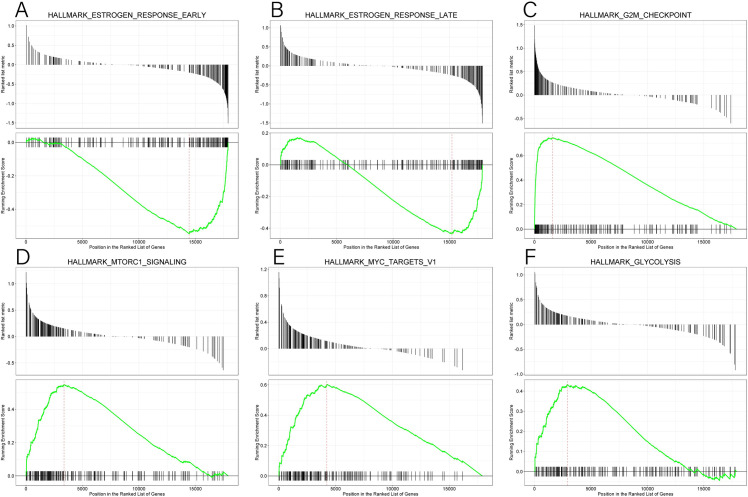
GSEA was performed between the low and high CCBL2 expression datasets, which was significantly different in h.all.v6.2.symbols.gmt of the MsigDB database (FDR<0.25, NOM p<0.05) (Table 5). On the basis of the normalized enrichment score (NES), the most significantly enriched pathways included estrogen response early and estrogen response late, indicating that the estrogen response was downregulated when CCBL2 expression was low. In addition, the correlated pathway, androgen response, was also declined (Table 5). The oppositely regulated and enriched pathways included the G2M checkpoint, MYC, mTorc1 signaling, and glycolysis (Fig 9).
Table 5. Gene sets which were significantly enriched.
| Description of gene set | NES | NOM (p value) | FDR (q value) |
|---|---|---|---|
| HALLMARK_ESTROGEN_RESPONSE_EARLY | -2.047 | 0.001 | 0.005 |
| HALLMARK_ESTROGEN_RESPONSE_LATE | -1.632 | 0.001 | 0.005 |
| HALLMARK_ANDROGEN_RESPONSE | -1.464 | 0.015 | 0.016 |
| HALLMARK_G2M_CHECKPOINT | 3.164 | 0.004 | 0.005 |
| HALLMARK_MYC_TARGETS_V1 | 2.558 | 0.005 | 0.005 |
| HALLMARK_MTORC1_SIGNALING | 2.337 | 0.005 | 0.005 |
| HALLMARK_GLYCOLYSIS | 1.844 | 0.005 | 0.005 |
Abbreviation: NES, normalized enrichment score. FDR, false discovery rate. NOM, nominal p value.
Fig 9. GSEA results of CCBL2 in breast cancer.
GSEA results showed the different enrichment of estrogen response early (A), estrogen response late (B), G2M checkpoint (C), mTorc1 signaling (D), MYC targets V1 (E) and glycolysis (F) in CCBL2 related to breast cancer. Abbreviation: GSEA, Gene Set Enrichment Analysis.
Correlation of CCBL2 with other genes in BC
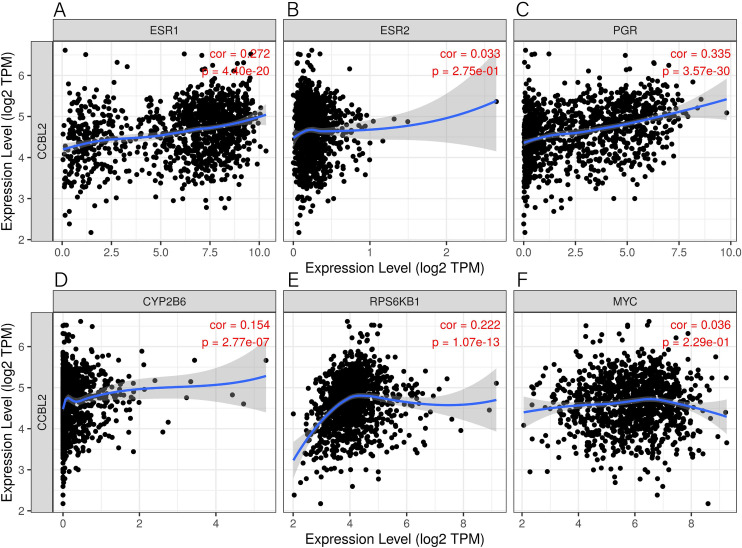
By the use of TIMER database, we showed that the expression of CCBL2 was significantly associated with that of ESR1 (r = 0.2718, p = 4.40e-20), PGR (r = 0.3346, p = 3.57e-30), AR (r = 0.3412, p = 4.38e-31) (S1 Fig), CYP2B6 (r = 0.1542, p = 2.77e-07) and RPS6KB1 (r = 0.2215, p = 1.07e-13), while no significant association with that of ESR2 (r = 0.0330, p = 2.75e-01) and MYC (r = 0.0363, p = 2.29e-01) (Fig 10).
Fig 10. Correlation analysis between CCBL2 and relevant genes.
Results included ESR1 (A), ESR2 (B), PGR (C), CYP2B6 (D), RPS6KB1 (E), and MYC (F) through TIMER databases. Abbreviation: TIMER, Tumor Immunoassay Resource.
Pan-cancer analysis of CCBL2 expression
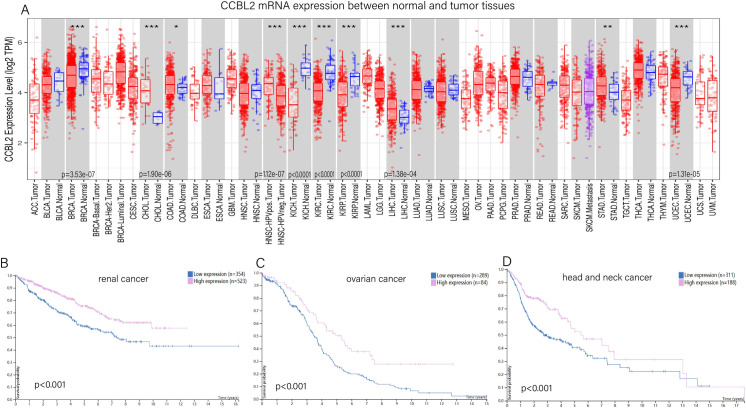
The differential expression of CCBL2 between normal and tumor tissues was analyzed using the TIMER database. According to the results, the expression of CCBL2 was significantly higher in normal tissues compared with that in tumor tissues in not only BC but also in renal cancer (p<0.0001), ovarian cancer (p<0.0001), and uterine corpus endometrial carcinoma (p = 1.31e-05). In cholangiocarcinoma (p = 1.90e-06) and liver hepatocellular carcinoma (p = 1.38e-04), the expression of CCBL2 was lower in normal tissues compared with tumor tissues. Considering the results of both TIMER and HPA databases, monogenic pan-cancer analysis of CCBL2 expression (data from HPA) was performed, and the results indicated that apart from BC, low expression of CCBL2 was associated with poor prognosis of renal, ovarian and head and neck cancers (p<0.0010). There was no significant relation between CCBL2 and prognosis of patients with uterine corpus endometrial carcinoma (p = 0.1600) and liver cancer (cholangiocarcinoma and liver hepatocellular carcinoma) (p = 0.2600) (Fig 11).
Fig 11. Pan-cancer analysis of CCBL2.
Differential expression of CCBL2 between normal and tumor tissues(A). Low CCBL2 expression had relations with the low survival probability of renal cancer (B) and ovarian cancer (C) and head and neck cancer (D) (p < 0.0010). Data were obtained from Human Protein Atlas Dataset available from proteinatlas.org and TIMER databases.
Low CCBL2 expression in patient-derived tissue samples of breast, renal, ovarian and head and neck cancers
The results of IHC staining were downloaded from HPA, and IHC staining was employed to verify the protein expression of CCBL2 in breast, renal, ovarian and head and neck cancers. Compared with the strong IHC staining in normal tissues, there were totally 24 samples of breast cancer. 22 tissue samples exhibited moderate IHC staining and 2 tissue samples exhibited weak staining. Out of 24 renal cancer samples, 3 of them were detected moderate staining, and 21 of them were detected weak staining. For ovarian cancer, 5 tissue samples presented strong staining, 16 showed moderate staining and 5 showed weak staining. A total of 8 tissue samples of head and neck cancer were analyzed, out of which 5 showed moderate staining, and 3 showed weak staining. The first three types of cancer tissues showed evidently weaker staining than their respective normal tissues (para-tumor tissues); however, no change in the staining intensity was observed in tissue samples of head and neck cancer. In particular, in BC, infiltrating lobular carcinoma presented stronger staining than infiltrating ductal carcinoma (Fig 2).
Discussion
Based on the data acquired from the TCGA database, CCBL2 showed lower expression in tumor tissues compared with normal tissues, and this result was validated in GEO datasets. In addition, RT-qPCR and IHC staining demonstrated the enhanced expression of CCBL2 in the human normal breast epithelial cell line MCF-10A and its diminished expression in BC cell lines as well as BC tissues. Low CCBL2 expression was correlated with an unfavorable survival, and we could come to a conclusion that CCBL2 was a prognostic biomarker in BC. To the best of our knowledge, this is the first study to elucidate the correlation between CCBL2 expression and BC survival based on TCGA data analysis.
CCBL2, a gene located on chromosome 1p22.2 [11], encodes an aminotransferase that transaminates kynurenine to form kynurenic acid, which is a metabolite of tryptophan. According to previous studies, CCBL2 facilitated the clearance of nephrotoxic substances [26]. The expression of CCBL2 was also decreased in patients with hyperoxaluria [27]. Moreover, CCBL2 expression was positively correlated with the occurrence of hospital-acquired VTE [19]. As important paralogs, evidence has shown the correlation between CCBL1 (identical to KAT1 and GTK) and pancreatic, prostate, and bladder cancers [28]. Furthermore, CCBL2 (identical to KAT2) plays an important role in several neurological diseases such as Huntington’s disease, Alzheimer’s disease and depression [15, 18, 29]. However, limited information is available regarding the expression of CCBL2 in tumors, especially BC. In our study, we demonstrated that CCBL2 expression was lower in tumor tissues than in normal tissues based on both the TCGA database and microarray datasets GSE42568 and GSE71053.
Low expression of CCBL2 was correlated with several clinicopathologic characteristics, including histological type, ER, PR, HER2, molecular subtype, T classification, M classification, vital status, and stage. Several research groups have reported that the abovementioned clinicopathologic features could guide the diagnosis, treatment and prognosis of BC, which promoted us to further explore the correlation between CCBL2 and BC [9, 30]. As confirmed above, CCBL2 had significantly strong relation with ER (p = 0.0005) and PR (p = 0.0005) status, accounting for the lower expression of CCBL2 in tumor cells of basal-like/Her-2-enriched BC and higher expression in luminal A (ER/PR-positive) BC cells. Therefore, lower OS and RFS were linked with lower CCBL2 expression because of the low survival rate and poor prognosis of basal-like/Her-2 enriched BC. The prognosis of infiltrating ductal carcinoma was found to be worse than that of infiltrating lobular carcinoma [31]. And the analysis results showed that the expression of CCBL2 was lower in infiltrating ductal carcinoma than in infiltrating lobular carcinoma, which was consistent with the IHC staining results that lower CCBL2 expression was linked to worse BC survival. A recent study showed that CCBL1 (identical to GTK) was involved in glutamine utilization through the GLS1 and glutaminase II pathways to generate glutamate [23], while the role of CCBL2 (identical to GTL) was unclarified, although glutamine was one of its metabolic substrates [15]. For this reason, when CCBL2 expression is low, it can be estimated that glutamine is relatively abundant. Glutamine plays an important role in the biosynthesis of amino acids, proteins, lipids, and nucleotides, which are essential to cell division, especially in cancer cells, also known as glutamine addiction. Therefore, the proliferating BC cells consumed glutamine at a very high rate [32, 33]. Thus, among all T stages, T4 had the lowest CCBL2 levels and relatively the highest glutamine levels, with the fastest tumor cell growth and angiogenesis. In the case of the M stage, low CCBL2 levels tended to be related to distant metastasis [23, 34]. Many cancer cells, especially those driven by the Myc gene (involving BC, as confirmed by GSEA results), were metabolically reprogrammed to consume more glutamine. When the expression of CCBL2 was low, the pathway of Myc in BC cells was upregulated. Altered glutamine metabolism in Myc-driven cancer, BC, resulted in glutamine addiction, which caused worse survival rate [35].
Our study shows that low expression of CCBL2 is associated with low OS in BC, especially in ER-positive tumors, PR positive tumors, HER-2 negative tumors, luminal A tumors, and invasive ductal and lobular carcinomas. Estrogen plays an important role in BC progression. Through GSEA, we found several relevant pathways, including estrogen response, enriched in CCBL2. Estrogen response pathway was downregulated when CCBL2 exhibited low expression, indicating that CCBL2 is positively correlated with this pathway. Oshi et al. found that the ESR1-associated early estrogen response was upregulated in ER-positive BC, indicating a better OS [36]. Therefore, that the low expression of CCBL2 was correlated with the worse OS of ER-positive BC, which was consistent with the results of our survival analysis. However, the pathway of estrogen response (early) involves 200 relevant genes [36] and the underlying molecular mechanism remains unclear. Additionally, CCBL2 could favorably predict the response to endocrine therapy in patients with ER-positive BC. Conventionally, RFS is used to evaluate the therapeutic effect of adjuvant therapy in carcinomas. The results of the Kaplan-Meier analysis indicated that with higher CCBL2 expression, patients who received endocrine therapy showed better RFS rates than lower CCBL2 groups. More specifically, patients receiving endocrine therapy alone (without chemotherapy) with higher CCBL2 expression presented a significant better RFS rate, whereas patients receiving both endocrine therapy and chemotherapy showed no increase in OS rate and insignificant increase in RFS rate. Therefore, CCBL2 possesses a significant prognostic value for ER-positive BC patients with or after endocrine therapy, particularly in the subgroup receiving only endocrine therapy, but little prognostic value in the subgroup receiving both endocrine therapy and chemotherapy. In patients with ER-positive BC without endocrine therapy, high CCBL2 expression indicated a favorable OS and RFS. In other words, CCBL2 also exhibited a valuable prognostic capacity in BC patients without or before endocrine therapy. Nowadays, the administration of endocrine therapy is mainly based on ER status (ER-positive tumors). However, patient compliance is poor due to the requirement for long-term medicine and associated side effects, with 20–50% of patients failing to finish the treatment cycle [37, 38]. Additionally, drug resistance has become a growing problem. Hence, novel biomarkers, such as CCBL2, are needed to evaluate the benefit of endocrine therapy, which would allow for a better prognosis. This will not only improve patient compliance but also assist in the selection of patients who are likely to benefit from neoadjuvant endocrine therapy.
Similarly, the mTORC1-signaling pathway is also correlated with ER-positive BC, which was downregulated when the CCBL2 expression was high. The regulatory targets of rapamycin (mTOR) are involved in protein translation, metabolism, cell growth, and proliferation [39]. As an enzyme complex, MTORC1, mainly binds with S6 kinases (S6Ks) to mediate its function. Studies have demonstrated that S6K1 (one of the S6 kinases) and some other relevant kinases contributed to the activation of ERα through its phosphorylation [40, 41]. It was further shown that S6K1 and ERα formed a positive feed-forward loop. The phosphorylation of ERα by S6K1 facilitated the process, promoting the transcription of RPS6KB1, which in turn regulated he proliferation of BC cells [42, 43]. To conclude, lower expression of CCBL2 in ER-positive or luminal A BC is related to the higher transcription of RPS6KB1 and subsequent BC cell proliferation. For the reasons mentioned above, the consensus can be reached that lower CCBL2 expression is associated with worse OS in ER-positive or luminal A BC. Moreover, the Myc pathway is negatively correlated with CCBL2 according to the GSEA. In triple-negative breast cancer (TNBC), MYC was found to regulate polyamine metabolism and a plasma polyamine signature was related to the development and progression of TNBC [44]. Therefore, for TNBC, the lower expression of CCBL2 was related to worse OS and RSF, consistent with the Kaplan–Meier survival analysis. However, to date, not much is known about the correlation between CCBL2 and Myc, thereby warranting further experimental validation. Other results of GSEA stated that G2M-checkpoint and glycolysis pathways were negatively correlated with CCBL2 expression, both of which have close correlation with the proliferation of most types of malignant tumors [45]. This is obviously consistent with the results of our analysis.
In addition to ER and PR, AR is also a hormone receptor that is expressed on the surface of mammary cells, regarded as a novel biomarker arousing heated discussion. Our study showed that the expression of CCBL2 was positively correlated with that of the AR gene and that the pathway of androgen response was upregulated when CCBL2 expression was high. According to a prospective clinical study by Anand et al., higher AR expression was correlated with earlier stage (p < 0.03), lower axillary burden (p < 0.04), higher ER (p = 0.002) and PR (p = 0.001) expression [46]. More specifically, Lin et al. showed that AR expression was positively correlated with a better prognosis of patients with HER2-positive breast carcinomas [47]. In TNBC, several studies have confirmed that the luminal androgen receptor (LAR) subtype, defined as AR-positive subtype, was associated with the highest OS compared with other subtypes [48–50]. We can conclude that CCBL2 could be a potential prognostic biomarker based on the confirmed relationship between higher AR and better BC prognosis, as well as the findings of the study mentioned above.
Furthermore, we found some proofs to confirm our conclusions with the assistance of website of http://guotosky.vip:13838/GPSA/. In this website, 3048 gene knock out RNAseq datasets were performed GSEA with four source of gene sets, including TCGA, Genotype-Tissue Expression (GTEx), and Cancer Cell Line Encyclopedia(CCLE). The result (S1 Table) showed the negative fold change values of ESR1 and AR with knock-down CCBL2 gene. It could be assumed that when CCBL2 gene was knocked down, ESR1 and AR genes also declined.
Using multivariate analysis, it was confirmed that low CCBL2 expression might serve as an independent prognostic marker, which was correlated with the unfavorable OS and RFS of BC using multivariate analysis. Then, by analyzing the AUC value, we found that CCBL2 possessed a moderate diagnostic efficacy between tumor and normal tissues, especially in stage IV. Therefore, CCBL2 can be regarded as a novel biomarker in the field of diagnosis and prognosis in BC.
The pan-cancer analysis revealed differential expression of CCBL2 between normal human tissues and various types of cancers. In addition to BC, some other cancers exhibited significantly different CCBL2 expression levels. Combined with the HPA results, renal, ovarian and head and neck cancers had significantly different CCBL2-related survival rates. According to the findings of RT-qPCR and IHC, CCBL2 may be a favorable prognostic biomarker in most cancer types. Further experimental verification was underway. Further in silico and in vitro research is needed to explore the correlation between CCBL2 and pan-cancers.
In this study, we initially discussed the value of CCBL2 expression as an independent prognostic marker for BC. Due to the limited samples, further explorations still need to be carried out based on the large samples data to verify our consequence. Additionally, the results of this study also promote the subsequent work, involving the further cell function test through gene overexpression or knock-down.
Conclusion
Compared with normal tissues, the expression of CCBL2 was lower in BC. The results revealed that low CCBL2 expression could act as an independent prognostic marker associated with survival of BC.
Supporting information
Abbreviation: AR, Androgen Receptor; TIMER, Tumor Immunoassay Resource.
(TIF)
Abbreviations: LogFC Log2 fold-change, AveExpr average log2-expression, P value< 0.05 indicate statistically significant correlations.
(DOCX)
(XLSX)
(ZIP)
(ZIP)
(ZIP)
Acknowledgments
The authors would like to thank for the public database for its convenience.
Throughout the writing of this article, I have received a great deal of support and assistance. I would first like to thank my supervisor, Dr. Keren Wang, whose expertise was invaluable in formulating the research questions and methodology. Your insightful feedback pushed me to sharpen my thinking and brought my work to a higher level. I was particularly grateful to my tutors, Dr. Zhaoying Yang, for his valuable guidance throughout my studies. You provided me with the tools that I needed to choose the right direction and successfully complete my article. I would also like to acknowledge my teammate Dr. Wenjing Song, for her wonderful collaboration and patient support. In addition, I would like to thank my parents for their wise counsel and sympathetic ear. You are always there for me.
List of abbreviations
- AR
Androgen receptor
- AUC
Area under the curve
- BC
Breast cancer
- CCBL2
Cysteine conjugate beta-lyase 2
- ER
Estrogen receptor
- GEO
Gene Expression Omnibus
- GSEA
Gene Set Enrichment Analysis
- GTK
Glutamine Transaminase K
- GTL
Glutamine Transaminase L
- HER-2
human epidermal growth factor-2
- HPA
The human protein atlas
- HR
Hazard ratio
- IHC
Immunohistochemistry
- KAT3
Kynurenine aminotransferase 3
- OS
Overall survival
- PR
progesterone receptor
- RFS
Relapse-free survival
- ROC
Receiver operating characteristic
- RT-qPCR
Real-time quantitative polymerase chain reaction
- TCGA
The Cancer Genome Atlas
- TIMER
Tumor Immunoassay Resource
Data Availability
All files are available from the TGGA database and GEO database (accession numbers GSE42568, GSE71053.) The relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Fahad Ullah M. Breast Cancer: Current Perspectives on the Disease Status. Advances in experimental medicine and biology. 2019;1152:51–64. doi: 10.1007/978-3-030-20301-6_4 [DOI] [PubMed] [Google Scholar]
- 2.Anastasiadi Z, Lianos GD, Ignatiadou E, Harissis HV, Mitsis M. Breast cancer in young women: an overview. Updates Surg. 2017;69(3):313–7. doi: 10.1007/s13304-017-0424-1 [DOI] [PubMed] [Google Scholar]
- 3.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021. [DOI] [PubMed] [Google Scholar]
- 4.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64(1):52–62. doi: 10.3322/caac.21203 [DOI] [PubMed] [Google Scholar]
- 5.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018. 68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 6.Weaver O, Leung JWT. Biomarkers and Imaging of Breast Cancer. AJR Am J Roentgenol. 2018;210(2):271–8. doi: 10.2214/AJR.17.18708 [DOI] [PubMed] [Google Scholar]
- 7.Loke SY, Lee ASG. The future of blood-based biomarkers for the early detection of breast cancer. European Journal of Cancer. 2018;92:54–68. doi: 10.1016/j.ejca.2017.12.025 [DOI] [PubMed] [Google Scholar]
- 8.Li G, Hu J, Hu G. Biomarker Studies in Early Detection and Prognosis of Breast Cancer. Advances in experimental medicine and biology. 2017;1026:27–39. doi: 10.1007/978-981-10-6020-5_2 [DOI] [PubMed] [Google Scholar]
- 9.A MB, R VS, J ME, A AO. Breast cancer biomarkers: risk assessment, diagnosis, prognosis, prediction of treatment efficacy and toxicity, and recurrence. Current pharmaceutical design. 2014;20(30):4879–98. doi: 10.2174/1381612819666131125145517 [DOI] [PubMed] [Google Scholar]
- 10.Kyle RA, Yee GC, Somerfield MR, Flynn PJ, Halabi S, Jagannath S, et al. American Society of Clinical Oncology 2007 Clinical Practice Guideline Update on the Role of Bisphosphonates in Multiple Myeloma. Journal of Clinical Oncology. 2007;25(17):2464–72. doi: 10.1200/JCO.2007.12.1269 [DOI] [PubMed] [Google Scholar]
- 11.Yu P, Li Z, Zhang L, Tagle DA, Cai T. Characterization of kynurenine aminotransferase III, a novel member of a phylogenetically conserved KAT family. Gene. 2006;365:111–8. doi: 10.1016/j.gene.2005.09.034 [DOI] [PubMed] [Google Scholar]
- 12.Yang C, Zhang L, Han Q, Liao C, Lan J, Ding H, et al. Kynurenine aminotransferase 3/glutamine transaminase L/cysteine conjugate beta-lyase 2 is a major glutamine transaminase in the mouse kidney. Biochemistry and biophysics reports. 2016;8:234–41. doi: 10.1016/j.bbrep.2016.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han Q, Robinson H, Cai T, Tagle DA, Li J. Biochemical and structural properties of mouse kynurenine aminotransferase III. Molecular and cellular biology. 2009;29(3):784–93. doi: 10.1128/MCB.01272-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinto JT, Krasnikov BF, Alcutt S, Jones ME, Dorai T, Villar MT, et al. Kynurenine Aminotransferase III and Glutamine Transaminase L Are Identical Enzymes that have Cysteine S-Conjugate β-Lyase Activity and Can Transaminate l-Selenomethionine*. Journal of Biological Chemistry. 2014;289(45):30950–61. doi: 10.1074/jbc.M114.591461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han Q, Cai T, Tagle DA, Li J. Structure, expression, and function of kynurenine aminotransferases in human and rodent brains. Cellular and Molecular Life Sciences. 2010;67(3):353–68. doi: 10.1007/s00018-009-0166-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stone TW. Kynurenic acid blocks nicotinic synaptic transmission to hippocampal interneurons in young rats. European Journal of Neuroscience. 2007;25(9):2656–65. doi: 10.1111/j.1460-9568.2007.05540.x [DOI] [PubMed] [Google Scholar]
- 17.Birch PJ, Grossman CJ, Hayes AG. Kynurenic acid antagonises responses to NMDA via an action at the strychnine-insensitive glycine receptor. European Journal of Pharmacology. 1988;154(1):85–7. doi: 10.1016/0014-2999(88)90367-6 [DOI] [PubMed] [Google Scholar]
- 18.Gao Y, Mu J, Xu T, Linghu T, Zhao H, Tian J, et al. Metabolomic analysis of the hippocampus in a rat model of chronic mild unpredictable stress-induced depression based on a pathway crosstalk and network module approach. Journal of pharmaceutical and biomedical analysis. 2021;193:113755-. doi: 10.1016/j.jpba.2020.113755 [DOI] [PubMed] [Google Scholar]
- 19.Voils SA, Shahin MH, Garrett TJ, Frye RF. Metabolomic association between venous thromboembolism in critically ill trauma patients and kynurenine pathway of tryptophan metabolism. Thrombosis research. 2018;165:6–13. doi: 10.1016/j.thromres.2018.03.003 [DOI] [PubMed] [Google Scholar]
- 20.Lock EA, Keane P, Rowe PH, Foster JR, Antoine D, Morris CM. Trichloroethylene-induced formic aciduria in the male C-57 B1/6 mouse. Toxicology. 2017;378:76–85. doi: 10.1016/j.tox.2017.01.004 [DOI] [PubMed] [Google Scholar]
- 21.Yaqoob N, Evans A, Foster JR, Lock EA. Trichloroethylene and trichloroethanol-induced formic aciduria and renal injury in male F-344 rats following 12 weeks exposure. Toxicology. 2014;323:70–7. doi: 10.1016/j.tox.2014.06.004 [DOI] [PubMed] [Google Scholar]
- 22.Cristofori P, Sauer AV, Trevisan A. Three common pathways of nephrotoxicity induced by halogenated alkenes. Cell biology and toxicology. 2015;31(1):1–13. doi: 10.1007/s10565-015-9293-x [DOI] [PubMed] [Google Scholar]
- 23.Udupa S, Nguyen S, Hoang G, Nguyen T, Quinones A, Pham K, et al. Upregulation of the Glutaminase II Pathway Contributes to Glutamate Production upon Glutaminase 1 Inhibition in Pancreatic Cancer. Proteomics. 2019;19(21–22). doi: 10.1002/pmic.201800451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke C, Madden SF, Doolan P, Aherne ST, Joyce H, O’Driscoll L, et al. Correlating transcriptional networks to breast cancer survival: a large-scale coexpression analysis. Carcinogenesis. 2013;34(10):2300–8. doi: 10.1093/carcin/bgt208 [DOI] [PubMed] [Google Scholar]
- 25.Pedersen IS, Thomassen M, Tan Q, Kruse T, Thorlacius-Ussing O, Garne JP, et al. Differential effect of surgical manipulation on gene expression in normal breast tissue and breast tumor tissue. Molecular medicine (Cambridge, Mass). 2018;24(1):57. doi: 10.1186/s10020-018-0058-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper AJL, Krasnikov BF, Niatsetskaya ZV, Pinto JT, Callery PS, Villar MT, et al. Cysteine S-conjugate beta-lyases: important roles in the metabolism of naturally occurring sulfur and selenium-containing compounds, xenobiotics and anticancer agents. Amino Acids. 2011;41(1):7–27. doi: 10.1007/s00726-010-0552-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han Q, Yang C, Lu J, Zhang Y, Li J. Metabolism of Oxalate in Humans: A Potential Role Kynurenine Aminotransferase/Glutamine Transaminase/Cysteine Conjugate Beta-lyase Plays in Hyperoxaluria. Current medicinal chemistry. 2019;26(26):4944–63. doi: 10.2174/0929867326666190325095223 [DOI] [PubMed] [Google Scholar]
- 28.Cooper AJL, Dorai T, Dorai B, Krasnikov BF, Li J, Hallen A, et al. Role of Glutamine Transaminases in Nitrogen, Sulfur, Selenium, and 1-Carbon Metabolism. In: Rajendram R, Preedy VR, Patel VB, editors. Glutamine in Clinical Nutrition. New York, NY: Springer New York; 2015. p. 37–54. [Google Scholar]
- 29.Passera E, Campanini B, Rossi F, Casazza V, Rizzi M, Pellicciari R, et al. Human kynurenine aminotransferase II—reactivity with substrates and inhibitors. Febs Journal. 2011;278(11):1882–900. doi: 10.1111/j.1742-4658.2011.08106.x [DOI] [PubMed] [Google Scholar]
- 30.Harbeck N, Gnant M. Breast cancer. The Lancet. 2017;389(10074):1134–50. [DOI] [PubMed] [Google Scholar]
- 31.Zhao H. The prognosis of invasive ductal carcinoma, lobular carcinoma and mixed ductal and lobular carcinoma according to molecular subtypes of the breast. Breast cancer (Tokyo, Japan). 2021;28(1):187–95. doi: 10.1007/s12282-020-01146-4 [DOI] [PubMed] [Google Scholar]
- 32.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell metabolism. 2012;15(1):110–21. doi: 10.1016/j.cmet.2011.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, Cui H. Targeting Glutamine Induces Apoptosis: A Cancer Therapy Approach. International journal of molecular sciences. 2015;16(9):22830–55. doi: 10.3390/ijms160922830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polet F, Feron O. Endothelial cell metabolism and tumour angiogenesis: glucose and glutamine as essential fuels and lactate as the driving force. J Intern Med. 2013;273(2):156–65. doi: 10.1111/joim.12016 [DOI] [PubMed] [Google Scholar]
- 35.Bernfeld E, Foster DA. Glutamine as an Essential Amino Acid for KRas-Driven Cancer Cells. Trends Endocrinol Metab. 2019;30(6):357–68. doi: 10.1016/j.tem.2019.03.003 [DOI] [PubMed] [Google Scholar]
- 36.Oshi M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Endo I, et al. Degree of Early Estrogen Response Predict Survival after Endocrine Therapy in Primary and Metastatic ER-Positive Breast Cancer. Cancers. 2020;12(12). doi: 10.3390/cancers12123557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walsh EM, Smith KL, Stearns V. Management of hormone receptor-positive, HER2-negative early breast cancer. Seminars in oncology. 2020;47(4):187–200. doi: 10.1053/j.seminoncol.2020.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huiart L, Ferdynus C, Giorgi R. A meta-regression analysis of the available data on adherence to adjuvant hormonal therapy in breast cancer: summarizing the data for clinicians. Breast cancer research and treatment. 2013;138(1):325–8. doi: 10.1007/s10549-013-2422-4 [DOI] [PubMed] [Google Scholar]
- 39.Hare SH, Harvey AJ. mTOR function and therapeutic targeting in breast cancer. 2017. [PMC free article] [PubMed] [Google Scholar]
- 40.Sridharan S, Basu A. Distinct Roles of mTOR Targets S6K1 and S6K2 in Breast Cancer. International journal of molecular sciences. 2020;21(4). doi: 10.3390/ijms21041199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamnik RL, Digilova A, Davis DC, Brodt ZN, Murphy CJ, Holz MK. S6 kinase 1 regulates estrogen receptor alpha in control of breast cancer cell proliferation. The Journal of biological chemistry. 2009;284(10):6361–9. doi: 10.1074/jbc.M807532200 [DOI] [PubMed] [Google Scholar]
- 42.Maruani DM, Spiegel TN, Harris EN, Shachter AS, Unger HA, Herrero-González S, et al. Estrogenic regulation of S6K1 expression creates a positive regulatory loop in control of breast cancer cell proliferation. Oncogene. 2012;31(49):5073–80. doi: 10.1038/onc.2011.657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng H, Zhang XT, Wang ML, Zheng HY, Liu LJ, Wang ZY. ER-α36-mediated rapid estrogen signaling positively regulates ER-positive breast cancer stem/progenitor cells. PloS one. 2014;9(2):e88034. doi: 10.1371/journal.pone.0088034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fahrmann JF, Vykoukal J, Fleury A, Tripathi S, Dennison JB, Murage E, et al. Association Between Plasma Diacetylspermine and Tumor Spermine Synthase With Outcome in Triple-Negative Breast Cancer. J Natl Cancer Inst. 2020;112(6):607–16. doi: 10.1093/jnci/djz182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35(8):427–33. doi: 10.1016/j.tibs.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anand A, Singh KR, Kumar S, Husain N, Kushwaha JK, Sonkar AA. Androgen Receptor Expression in an Indian Breast Cancer Cohort with Relation to Molecular Subtypes and Response to Neoadjuvant Chemotherapy—a Prospective Clinical Study. Breast care (Basel, Switzerland). 2017;12(3):160–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin Fde M, Pincerato KM, Bacchi CE, Baracat EC, Carvalho FM. Coordinated expression of oestrogen and androgen receptors in HER2-positive breast carcinomas: impact on proliferative activity. Journal of clinical pathology. 2012;65(1):64–8. doi: 10.1136/jclinpath-2011-200318 [DOI] [PubMed] [Google Scholar]
- 48.Masuda H, Baggerly KA, Wang Y, Zhang Y, Gonzalez-Angulo AM, Meric-Bernstam F, et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19(19):5533–40. doi: 10.1158/1078-0432.CCR-13-0799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thike AA, Yong-Zheng Chong L, Cheok PY, Li HH, Wai-Cheong Yip G, Huat Bay B, et al. Loss of androgen receptor expression predicts early recurrence in triple-negative and basal-like breast cancer. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2014;27(3):352–60. doi: 10.1038/modpathol.2013.145 [DOI] [PubMed] [Google Scholar]
- 50.Loibl S, Müller BM, von Minckwitz G, Schwabe M, Roller M, Darb-Esfahani S, et al. Androgen receptor expression in primary breast cancer and its predictive and prognostic value in patients treated with neoadjuvant chemotherapy. Breast cancer research and treatment. 2011;130(2):477–87. doi: 10.1007/s10549-011-1715-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Abbreviation: AR, Androgen Receptor; TIMER, Tumor Immunoassay Resource.
(TIF)
Abbreviations: LogFC Log2 fold-change, AveExpr average log2-expression, P value< 0.05 indicate statistically significant correlations.
(DOCX)
(XLSX)
(ZIP)
(ZIP)
(ZIP)
Data Availability Statement
All files are available from the TGGA database and GEO database (accession numbers GSE42568, GSE71053.) The relevant data are within the paper and its Supporting Information files.