Abstract
Increasing temperatures impact plant biochemistry, but the effects can be highly variable. Both external and internal factors modulate how plants respond to rising temperatures. One such factor is the time of day or season the temperature increase occurs. This timing significantly affects plant responses to higher temperatures altering the signaling networks and affecting tolerance levels. Increasing overlaps between circadian signaling and high temperature responses have been identified that could explain this sensitivity to the timing of heat stress. ELF3, a circadian clock component, functions as a thermosensor. ELF3 regulates thermoresponsive hypocotyl elongation in part through its cellular localization. The temperature sensitivity of ELF3 depends on the length of a polyglutamine region, explaining how plant temperature responses vary between species. However, the intersection between the circadian system and increased temperature stress responses is pervasive and extends beyond this overlap in thermosensing. Here, we review the network responses to increased temperatures, heat stress, and the impacts on the mechanisms of gene expression from transcription to translation, highlighting the intersections between the elevated temperature and heat stress response pathways and circadian signaling, focusing on the role of ELF3 as a thermosensor.
Keywords: circadian clock, eukaryotic gene expression, gene regulatory networks, plant signal transduction, temperature sensing, temperature stress
Introduction
Extreme temperatures are a consistent challenge for crop growth. As global temperatures increase, research studying how plants adapt to these new climates has likewise grown with the hope of developing crop varieties that can withstand higher temperatures and deliver food security in the face of changing climates.
Exposure to elevated temperatures and heat stress are not synonymous. Heat stress occurs when the temperature reaches a point that damages the cellular components affecting functions such as metabolism, signaling, structure, and transport [1–4]. Molecular changes occur before plants reach this state of heat stress as the plant adapts to the higher temperatures. It would benefit this research area if a distinction was made between plant responses to increased temperature and heat stress. However, this is not easy, no constant threshold temperature exists, and exceeding the optimal growing range does not always invoke cellular damage or death.
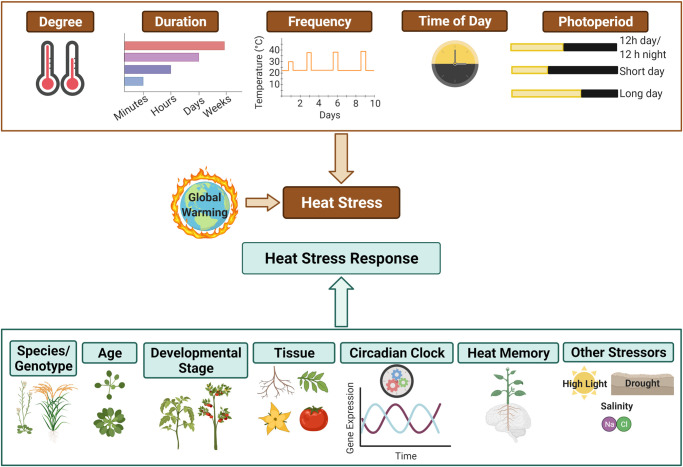
Both extrinsic factors related to the temperature event and intrinsic factors reflecting the state of the plant together determine if a plant can withstand a given temperature event (Figure 1). The extrinsic parameters depend on the heat intensity, frequency of heat events, duration, time of day, and time of year [5–12]. Most information on heat stress responses is based on experiments using transient, high temperatures, but field-grown plants tend to experience long-term, moderate heat stress. Prolonged heat stress and heat shock result in different physiological, metabolic, and transcriptomic responses [13]. Even mild heat stress can be damaging over a long period [14]. Moreover, because temperature and air moisture content are inherently linked, the combined effects of elevated temperatures and changes in vapor pressure are critical to identifying how elevated temperatures and heat stress affect actual yields in field conditions [15,16].
Figure 1. Plant heat stress response is a complex interaction between features of heat stress and internal factors in plants.
Heat stress is a combination of heat intensity, duration, and frequency. The time of day and photoperiod when the increased temperature occurs are important factors that impact the severity of heat stress. Internal factors also influence how plants respond to elevated temperatures. These include the genotype, age, developmental stage, tissue, stage of the internal clock, and prior exposure to heat or other stressors. The figure was created with BioRender.com.
Intrinsic, plant-centric parameters that affect heat responses include the plant genotype [17], developmental stage [18–22], prior exposure to heat stress [23–28], the time relative to the plants’ internal circadian clock [5,6,8], and the presence of other stresses (Figure 1) [29–33]. Even the same genotype can have significantly varied tolerance levels and responses to the same temperature increase depending on when the stress occurs [5–7,9,34,35]. The combinations of these extrinsic and intrinsic parameters result in a wide gradient of plant responses. Approaches that incorporate these variables have enabled new insights into heat stress tolerance. We review how time of day and circadian networks overlap with elevated temperature and heat stress responses through multiple levels of gene expression and the role of ELF3, a circadian clock component, in temperature sensing.
Interactions between the circadian clock and elevated temperatures
Increased temperatures affect most aspects of plant biochemistry including mRNA stability [36,37], splicing [38–42], translation [43–45], post-translational modifications [46–51], and protein degradation [52–56]. These pathways are themselves circadian-regulated (Figure 2) [52,53,56–63]. Thus, there is significant cross-regulation between the response to elevated temperatures, heat stress and circadian clock networks and their downstream targets. Such interactions between heat stress response and circadian-regulatory networks are supported by the fact that thermotolerance is gated by the time of the day when the elevated temperature occurs. In controlled environments, plants are more sensitive to elevated temperatures at dawn than at dusk [5,9,64]. About 68% of heat-responsive genes are gated by the time of day [5]. This large overlap suggests that, in part, temporal sensitivity of thermotolerance is due to interactions between the circadian-regulatory and heat stress response networks.
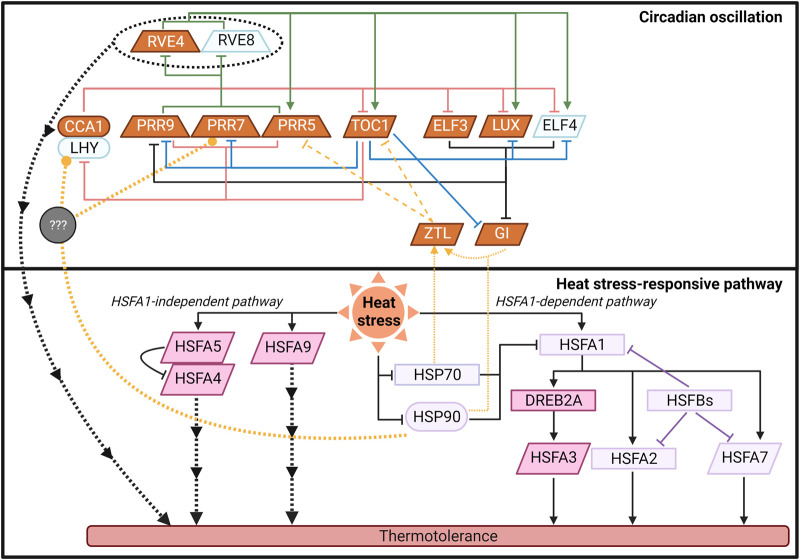
Figure 2. Crosstalk between the circadian clock and heat stress-responsive pathways.
The diagram of circadian oscillation (top) was adapted from [147]. The colors of the clock components represent their expression level in response to elevated temperatures from Grinevich et al. [5], and Blair et al. [8]. Increased expression is shown in orange and decreased expression in light blue. The heat stress-responsive pathway (bottom) was adapted from [2]. The diagram includes only HSFs with known variation in basal expression between times of day in Grinevich et al. [5]. Colors indicate whether HSFs are targeted by the clock proteins based on the ChIP-Seq data [70–76]. Direct targets of circadian components are shown in dark pink, and light purple means they are not the direct targets. For both sections, the shapes of the text box represent their expression peaks under normal temperature. Ovals, trapezoids, and parallelogram mean high expression in the morning, afternoon, and evening, respectively. Rectangles mean no difference in expression between dawn and dusk. Solid lines indicate that the regulation occurs via transcriptional activation or repression. Dashed lines indicate regulation via post-translational modification. Thin dotted lines represent the regulation through a chaperone function. Thick dotted lines indicate that the regulatory mechanisms are unknown. The figure was created with BioRender.com.
Conversely, elevated temperatures also affect the expression of circadian clock components (Figure 2) [8,65]. This complex coordination between the plant circadian clock and elevated temperature responses indicates that considering the effects of both factors is essential to acquiring a predictive understanding of plant responses to increased temperatures.
Time of day and circadian regulation of heat stress response networks
Daily expression of HSFs
The transcriptional response to elevated temperatures centers around the roles and regulation of Heat Shock Factors (HSFs) and Heat Shock Proteins (HSPs). In response to heat stress, type-A HSFs (HSFA1s) trimerize and bind Heat Shock Elements (HSEs) in the promoters of their target genes. This results in the induction of their targets, such as the transcription factor DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2A (DREB2A) and co-activator MULTIPROTEIN BRIDGING FACTOR 1C (MBF1C) [2,66,67].
Upon activation, HSFA1s trigger a gene expression cascade that results in new proteins that change the cellular composition improving tolerance. Critically, the network the response to elevated temperature traverses depends entirely on the molecules available at the time of the stress. This molecular landscape of the specific gene expression activity, chromatin state, mRNA abundance, protein composition, and metabolite levels varies significantly depending on the time of the increased temperature event (Figure 1). For example, in Arabidopsis and rice (Oryza sativa L.), the mRNA levels of HSFs, HSPs, and other regulators of heat stress vary significantly depending on the time of day [5,68] (Figure 2). In non-heat stress conditions in Arabidopsis, 17 out of 21 HSF genes show rhythmic expressions under diel conditions [5,69]. But the patterns of expression vary. For example, HSFA1a is highly expressed in the morning while HSFA1b, 1d, and 1e have a peak expression in the evening. A few HSF genes and some of the HSF targets, such as DREB2A, are the direct targets of the circadian clock components proteins (Figure 2) [70–76]. For example, HSFA3 is a CCA1 target [5,8], and its expression is altered in cca1/lhy double mutant under elevated temperature [8]. The HSFs and other heat-response regulators that show robust rhythmic expression could be responding to diel changes in light and temperature or indirectly regulated by the clock components via unknown mechanisms.
Time of day and circadian regulation of heat shock responses
The circadian clock plays a significant role in heat-responsive gene expression. In response to heat shock, cca1/lhy double mutants and prr7/9 double mutants have fewer differentially expressed genes and decreased expression-change magnitude. The reduced transcriptional response in these circadian mutants highlights the importance of clock components in the transcriptional response under elevated temperatures [8].
HSFA1s are primary regulators of heat shock responses. Most HSFA1 target genes are up-regulated in response to heat shock at dawn [5]. HSFA1b is induced by increased temperatures at dusk but not at dawn, suggesting that HSFA1b induction alone is insufficient to regulate most HSFA1 targets [5]. This disconnect between the expression of these regulators and their targets indicates that multiple network strategies for heat stress responses exist and vary depending on the time of day. Critically, these variations in responses can be used to understand how connections between regulators and targets are wired and may explain the specific roles of genes otherwise thought to be redundant. For example, HSFA2 is a target of HSFA1s, is a key regulator of heat acclimation and acquired thermotolerance [26,77], and is up-regulated under elevated temperatures at both dawn and dusk (Figure 2). Although HSFA2 responds to increased temperatures at both times of day, its targets HSP21, HSP22, HSP18.2, and ASCORBATE PEROXIDASE2 (APX2) are significantly induced under increased temperatures near dawn yet show substantially lower expression levels when plants are exposed to increased temperatures in the evening [5]. This may indicate required interactions between HSFA2 and other morning responsive HSFA1 targets in the induction of these genes. Another memory protein, HSFA32 [28], also accumulates to higher levels in response to elevated temperatures in the morning than in the evening [5]. The effects of these temporal differences in response on heat stress memory have not been evaluated.
Even though HSFA1 is a master regulator of the heat shock response, ∼40% of heat-responsive genes are HSFA1 independent [9]. The circadian clock proteins RVE4 and RVE8 control the expression of heat-responsive genes in an HSF-independent manner (Figure 2) [9]. As RVE4 and RVE8 are highly expressed in the afternoon, this pathway is possibly responsible for regulating thermotolerance during the daytime.
HSFA4, HSFA5, and HSFA9 proteins also function independently of HSFA1, but the downstream targets that confer thermotolerance are less understood (Figure 2) [2]. In Lily (Lilium Longiflorum), HSFA4 regulates ROS metabolism to enhance basal thermotolerance [78]. ROS metabolism is also a primary target of the circadian clock [79–81]. Exploring how time alters these transcriptional responses can provide new insights into how the heat stress response networks are wired.
The observed interaction between the circadian clock and heat shock networks persists downstream of transcription. At the global translational level, the time of day gates about one-third of circadian-regulated heat-responsive translatome [65].
Intersection between heat stress response and circadian networks in gene expression
Transcriptional activation
Both A and B classes of HSFs have an affinity for components of the basal transcription machinery [82]. This association with the core transcriptional components provides a mechanism for how increased HSF occupancy at promoters increases RNA Polymerase II (RNAP II) binding and induces transcription of their target genes.
RNAP II activity is regulated by post-translational modifications of its largest subunit's carboxy-terminal domain (CTD) [83]. Phosphorylation of RNAPII CTD by CYCLIN DEPENDENT KINASE C;2 (CDKC;2) is required for a proper circadian period in Arabidopsis (Figure 3) [84]. Increased temperatures change these RNAP II CTD modifications to alter transcription. In barley anthers, high temperatures result in hyperphosphorylation of the CTD's Serine 5 (Ser5) residue, which alters anther-specific gene expression, leading to an arrest of anther development [85]. In Arabidopsis, RNAP II CTD Ser2 and Ser5 phosphorylation increase during heat shock recovery, coinciding with increased thermotolerant-related gene expression. Mutation of CTD phosphatase-like 1 (CPL1, also known as RCF2/FIERY2) which dephosphorylates the CTD Ser5, reduces the expression of most HSFs, and decreases thermotolerance in Arabidopsis [86–88]. This suggests that proper regulation of RNAPII CTD phosphorylation is critical for heat-responsive gene expression. However, CPL1 also dephosphorylates the transcription factor NAC109, which induces the expression of several HSFs [89]. CPL1 and its close homolog CPL2 are also part of the plant growth, cell morphogenesis, and other stress response networks, including biotic stress response [86,87,90]. CPL2 is circadian-regulated and is induced in response to elevated temperatures in the morning but not in the evening [5,91]. In mammalian systems, the core transcriptional machinery, including phosphorylation of Ser5, is circadian-regulated. However, our understanding of the circadian regulation of core transcriptional machinery remains limited in plants. The intersection between the circadian and heat-responsive regulations of the RNAP II activity may explain how some variation in elevated temperature responses is clock-dependent (Figure 3) [92].
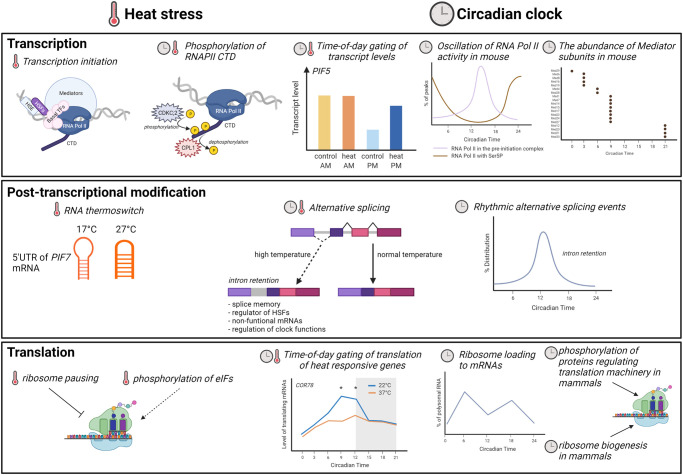
Figure 3. Overlap between heat stress response and circadian-regulatory pathways in gene expression.
Elevated temperature responses, heat stress and circadian-regulatory pathways affect multiple regulatory mechanisms controlling gene expression. The expression of PIF5 under heat shock at dawn vs dusk is from Grinevich et al. [5]. The oscillation of RNA Pol II activity and protein abundance of Mediator subunits in mouse are adapted from Koike et al., and Wang et al., respectively [97,98]. The rhythmic alternative splicing events are based on Yang et al. [109]. Ribosome loading to mRNAs and the gating of COLD-REGULATED 78 (COR78) translating mRNA are from Missra et al. and Bonnot and Nagel, respectively [65,148]. The figure was created with BioRender.com.
HSFA1s’ specificity for target genes may also occur through the recruitment of co-activators. HSFA1s recruit the transcriptional co-activator complex, Mediator, to the promoters of the heat-inducible genes [93]. However, not all HSFA1 target genes were equally affected in plants with disrupted Mediator complex. For example, loss of the structural hub subunits of Mediator, med14, or med17 resulted in significantly reduced induction of HSFA1 transcription factor targets, such as DREB2A and other HSFs, but had little effect on the heat-induced expression of HSPs [93]. This suggests that Mediator can fine-tune the network responses, even for the targets of the same master regulator, perhaps explaining some of the time of day variation in the induction of HSFA1 targets. The specific downstream network of HSFA1 could be tuned to provide different network responses based on the particular HSFA1 activation mechanism [93] or tailored based on tissue-specific [94] or environmentally induced [95,96] changes in Mediator subunit composition. Loss of specific mediator subunits reveals distinct roles in regulating which genes are expressed under heat stress in Arabidopsis, supporting the idea that different mediator compositions could modulate unique targets [96]. In particular, loss of MED16 resulted in misregulation of many heat-responsive genes, consistent with the role of this subunit in heat responses in S. cerevisiae [96]. In plants, most experimental evidence of potential mediator subunit composition differences is based on changes in mRNA levels. However, in mammalian systems, variation in the Mediator subunit composition has been shown at the protein level, including variation based on the time of the day [97]. In addition, the recruitment of basal transcription factors and activation of RNAP II vary across the time of day in mammals (Figure 3) [98]. Yet the complete transcriptional landscape over the time of the day in plants has not been examined. As the magnitude of transcript levels of heat-responsive genes is influenced by the time plants are exposed to elevated temperatures (Figure 3) [5,8], studying the fluctuation of transcription machinery during the day would provide a better insight into the time-of-day gating of basal thermotolerance in plants.
Post-transcriptional modification
The secondary structure of single-stranded mRNA is important for polyadenylation, splicing, translation, and turnover [99]. Bacteria have RNA thermometers, RNAs with temperature-sensing sequences, to control translation efficiency in response to temperature changes [100]. Unfolded RNA thermometers increase translation efficiency under heat stress [100]. In Arabidopsis, the expression of PHYTOCHROME INTERACTING FACTOR7 (PIF7) and HSFA2 are proposed to be regulated via an RNA thermoswitch [101]. PIF7 is a bHLH transcription factor regulating thermomorphogenesis [102], suggesting that RNA structure could be another mechanism to temporally regulate thermoresponsive growth. In rice, heat shock unfolds mRNA, promoting mRNA degradation [36]. Some evidence for circadian regulation of RNA stability exists in Arabidopsis [60]. For example, CCA1 is a target for m6A RNA methylation in response to blue light, which accelerates its degradation [103]. However, most plant studies that examine either elevated temperature responses, circadian regulation, or both measure only steady-state RNA levels without distinguishing between transcriptional activation and mRNA degradation. Therefore, this area needs further investigation to determine if it contributes to the temporal variation in increased temperature responses.
Alternative splicing (AS) is a way to generate transcript variants from a single gene. These variants can affect mRNA stability or result in different protein isoforms. Heat stress induces AS in Arabidopsis, wheat [38], tomato pollen [39], grape [40], and moss [41]. One hypothesis for this increase in AS is that heat stress increases splicing errors [104]. Heat acclimation, priming plants with a non-lethal temperature that enhances tolerance to severe heat stress [25], also affects AS [105]. For example, in Arabidopsis, primed plants have fewer intron retention (IR) events after severe heat stress than non-primed plants [105]. This result indicates that heat acclimation creates splicing memory, maintaining correct splicing after severe heat stress [105].
AS also regulates the expression of HSFs and HSPs [42]. Several HSFs and HSPs undergo AS under heat stress [39,41,106]. In Arabidopsis, heat stress induces AS on HSFA2, generating a splice variant HSFA2-III which self-regulates HSFA2 transcription [42]. In lily (Lilium spp.), the protein encoded from the heat-inducible splice variant LlHSFA3B-III interacts with the HSFA3 ortholog, LlHSFA3A-I, reducing the accumulation of LlHSFA3A-I, resulting in an attenuation of the heat stress responses [107]. AS is also regulated by the time of day and the circadian clock [108–110]. The core circadian clock genes themselves have temperature-dependent AS, suggesting one mechanism by which the clock and heat stress responses can interact [111–114]. However, variation in splicing after elevated temperatures has not been examined at different times of the day.
Translation
Under heat stress, the time of day gates about one-third of the circadian-regulated heat-responsive translatome in Arabidopsis (Figure 3) [65], but the molecular mechanisms underlying this phenomenon are still elusive. In Neurospora crassa, circadian control of Eukaryotic Elongation Factor2 (eEF2) [115] and the phosphorylation of Eukaryotic Initiation Factor 2a (eIF2ɑ) [116] result in circadian-regulated translation. In wheat, the phosphorylation state of eIF4A and eIF4B is altered under heat shock while other translation factors, eIF4F, eIFiso4F, eIF2α, and eIF2β remain the same [117]. However, the variation in this phosphorylation in response to elevated temperatures has not been examined at different times of the day. Mammalian studies also show that the circadian clock regulates the translation machinery via ribosome biogenesis and phosphorylation of proteins in initiation and elongation steps (Figure 3, review in [118]). Although translation in plants is circadian regulated [119,120], the specific mechanisms driving clock regulation of translation in plants are unknown [121].
Regulation of the circadian clock under heat stress
As these multiple levels of interaction between the circadian and elevated temperature response networks affect heat stress responses, they also affect circadian regulation. Elevated temperatures alters the expression of circadian genes (Figure 2) [8,65]. CCA1, LHY, PRR7, and PRR9 mRNA levels change between 22°C and 37°C. HSPs cooperate with circadian components and impact both heat stress responses and circadian rhythms. HSP90 interacts with ZTL in protein degradation under heat stress [122]. HSP90 and HSP70 work together with GI in ZTL protein maturation [123]. Moreover, HSP90 plays a role in circadian oscillation, interacting with the morning loop [124]. The hsp90.2.3 mutant has a longer period in temperature cycles (22/16°C) and a phase advance in the late morning. CCA1, LHY, and PRR7, which are part of the morning loop, are required for the period lengthening by HSP90. However, how HSP90 mediates the expression of CCA1, LHY, and PRR7 is still unclear [124]. HSFB2b binds the PRR7 promoter and represses PRR7 expression in the morning, and temperature compensation, a fundamental characteristic of circadian rhythms, requires HSFB2b [69].
Although the levels of several core clock components are altered between 22°C and 37°C, the daily expression pattern remains unchanged [8,65], suggesting that the circadian clock is functional at 37°C in Arabidopsis. The ability to retain the same period despite the influences of increased temperature is a feature of the circadian clock known as temperature compensation (reviewed in [125]). However, temperature compensation has limits, and outside of these ranges, the disrupted clock could compound the impacts of elevated temperatures and heat stress. Many questions remain about temperature compensation limits: Do limits vary across developmental stages or between genotypes? Does acclimation increase the limits within which the plant can be temperature compensated? What intrinsic and extrinsic factors affect the temperature compensation range? These questions about temperature compensation limits are beginning to be examined in Arabidopsis, and much less is known about these limits in other plant species or in natural conditions.
The role of the circadian component, ELF3, in temperature sensing
Before activating cellular responses to combat the effects of heat stress, plants must first sense a temperature increase. Multiple temperature sensing mechanisms have been described in plants. These include messenger RNA (mRNA) stability, protein degradation, and histone modifications and have been reviewed recently [126–128]. New research indicates connections between the circadian clock and thermosensing in Arabidopsis through the roles of ELF3 [129–131].
ELF3 is part of the evening complex (EC) with LUX and ELF4. The EC is so named because these components are highly expressed in the evening. As a member of the EC, ELF3 regulates hypocotyl elongation through transcriptional and post-transcriptional control of the basic helix–loop–helix transcription factors, PIF4 and PIF5 [132]. PIF4 and PIF5 positively contribute to light- and temperature-dependent growth, indicating that they integrate light and temperature cues into cellular pathways [133]. In Arabidopsis, increased temperature induces hypocotyl elongation. This thermoresponsive hypocotyl elongation is lost in elf3-1 mutant plants and can be restored by complementing with exogenous ELF3. Thus, ELF3 is critical for connecting increasing temperature to the physiological response of hypocotyl elongation [129]. ELF3's role in sensing ambient temperature changes extends beyond hypocotyl elongation. ELF3 also functions as a thermosensor in the temperature entertainment of the circadian clock [134].
ELF3 is a central component of temperature-responsive hypocotyl elongation
ELF3 regulates thermoresponsive hypocotyl elongation transcriptionally and posttranscriptionally. The EC restricts hypocotyl elongation through transcriptional control by repressing the expression of PIF4 and PIF5 [132]. Elevated temperatures reduce ELF3 occupancy at the PIF4 promoter, resulting in PIF4-mediated thermomorphogenesis [135]. In fact, the entire EC complex appears to dissociate from the DNA at higher temperatures [76,136]. However, high ELF4 levels can stabilize EC-DNA binding and overcome this temperature-induced dissociation of the EC from the target DNA [136].
In addition to the transcriptional control of PIF4 expression through the EC, ELF3 also regulates PIF4 activity [135,137,138]. ELF3 binds the bHLH domain of PIF4 directly, preventing PIF4 from activating its transcriptional targets [139]. Thus, ELF3 regulates thermoresponsive hypocotyl elongation through control of PIF4 at multiple levels.
ELF3 is a temperature sensor
Using thermoresponsive growth to test ELF3's temperature-sensing role, Jung et al. showed that the prion-like domain (PrD) provides part of this temperature-sensing function (Figure 4A) [129]. The PrD varies between species and contains a polyglutamine (PolyQ) tract. Increasing the length of the PolyQ tract increases the thermoresponsiveness in Arabidopsis. Chimeric Arabidopsis ELF3 versions with PrDs from plants from warmer climates reduce thermoresponsiveness [129]. Nevertheless, plants overexpressing ELF3 without a PrD are still thermally responsive, unlike the elf3-1 mutant, indicating that other features of ELF3 also function in temperature sensing [129].
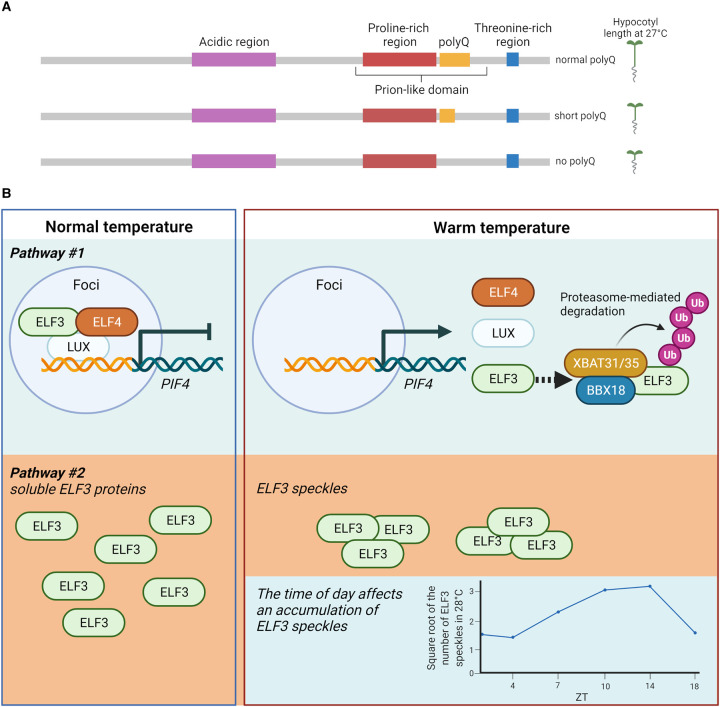
Figure 4. ELF3 functions as a thermosensor in plants.
(A) ELF3 protein contains 695 amino acids. It can be divided into four regions based on the composition of amino acids: an acidic region (206–320), a proline-rich region (440–540), a threonine-rich region (636–652), and glutamine repeats or polyQ (544–585) [149]. The prion-like domain (PrD) has recently been identified as a thermosensor, and it covers amino acids 430–609 [129]. The length of polyQ inside the PrD determines the degree of thermoresponsiveness. (B) The current model of ELF3 in response to warm temperatures in Arabidopsis is based on [129–131,143,144]. Under normal temperature, ELF3 interacts with ELF4 and LUX to form an evening complex in sub-nuclear areas called foci. The EC in the foci then binds to target genes such as PIF4 to repress their expression. Under warm temperatures, the EC disperses and leaves the foci [131]. The E3 ubiquitin ligase XBAT31/35 binds ELF3 with help from BBX18, targeting ELF3 for proteasome degradation [143,144]. Another model proposes a mechanism to reduce active ELF3 under warm temperatures via the PrD [129]. ELF3 proteins are in a soluble form under normal temperature. As temperatures increase, ELF3 proteins aggregate into speckles. The formation of ELF3 speckles varies throughout the day, as the speckles are more detectable in the afternoon [130]. The figure was created with BioRender.com.
Jung et al. observed that the PrD affects ELF3 localization in cells [129]. At lower temperatures, ELF3 is soluble and diffused in the cells. However, at higher temperatures, the ELF3 proteins form speckles in root cells and heterologous yeast systems [129]. The formation of these concentrated regions of ELF3 is reversible. When returned to low temperatures, the protein diffuses again. Replacing the Arabidopsis PrD with the equivalent region from Brachypodium distachyon, a warm temperature grass that lacks a detectable PrD reduces speckle formation at higher temperatures. This speckle formation is suggested to facilitate remembering warm daytime temperatures during nighttime hypocotyl growth [130]. The nighttime hypocotyl expansion depends on the nighttime temperature and the prior daytime temperature [130]. Warm daytime temperatures increase the nuclear level of PIF4 during the day, and the active PIF4 from daytime temperature affects hypocotyl growth at night (Figure 4B) [130,140]. This hysteretic PIF4 pattern requires ELF3 [130]. ELF3 speckle formation correlates with PIF4 promoter activity; increased ELF3 localization into speckles increases PIF4 transcription [130]. Warm daytime temperatures reduce speckle formation in hypocotyl cells during the morning and increase speckle formation during the afternoon suggesting connections between the circadian regulation of these components and their temperature sensing and memory functions [130].
Another study observed a different ELF3 localization response under warm temperatures and proposed an alternative mechanism connecting the temperature-sensing ELF3 and downstream physiological changes [131]. In the nucleus, ELF3 and other EC components localize in foci. This localization is important for suppressing EC-target gene expression [141,142]. Ronald et al. observed that warm temperatures disrupt the localization of ELF3 to these nuclear foci in hypocotyl and root cells, and ELF4 is not required for this process (Figure 4B) [131]. At warm temperatures (29°C), ELF3 is no longer localized to the foci and is targeted by E3 ubiquitin ligases, XBAT31 and XBAT35 (Figure 4B). Ubiquitinated ELF3 is degraded by the 26S proteasome removing the repressive effects of ELF3 on PIF4 and allowing hypocotyl elongation [143,144]. The interaction between XBAT31/35 requires B-box domain protein BBX18, which possibly acts as a scaffold protein enhancing the XBAT31/35-ELF3 interaction [143,144]. In this model, heat stress releases ELF3 from the nuclear foci, increasing PIF4 promoter activity. Ronald et al. describe potential experimental differences that could account for the apparent opposite effect of warm temperatures on ELF3 accumulation or diffusion, requiring future studies to distinguish between these mechanisms [129,131].
Intriguingly, XBAT31/35 are direct CCA1 targets with peak expression in the evening, coinciding with ELF3 expression [72,91]. However, BBX18 expression peaks in the morning [91]. BBX18 is a target of PRR5 [72,91] and interacts with PRR 5, 7, and 9 [73,91,145]. XBAT31 is down-regulated in response to heat stress in the morning, when BBX18 accumulates in response to heat stress [5]. XBAT31 is unaffected by heat stress at night while BBX18 remains at low levels and is not induced in the evening [5]. Identifying when the proposed interactions between XBAT31/35 and BBX18 occur under heat stress will require finer temporal resolution, examining the protein levels of XBAT31/35 and BBX18, and the activity of XBAT31/35. The lack of overlap in the transcriptional responses combined with changes in protein stability could provide a gating mechanism for thermosensing.
These studies demonstrate a role for ELF3 in sensing ambient changes in temperature (27-35°C) in Arabidopsis. It will be interesting to observe if the ELF3 PrD structure and subcellular localization influence the canonical heat-responsive gene expression network or if ELF3 plays a role in heat-stress response pathways at higher temperatures. PolyQ domains are not unique to plant circadian components; the animal Clock gene contains a PolyQ region [146], suggesting a fundamental relationship between circadian regulation and temperature sensing.
Conclusions
The combination of extrinsic and intrinsic factors complicates the study of elevated temperature responses in plants.
Plants show time of day variation in their susceptibility to heat stress. The connections between heat stress response and circadian clock networks extend throughout the regulatory cascade. Therefore, considering the effects of the time of day when studying elevated temperature and heat stress responses is critical.
ELF3 functions as a thermosensor regulating thermoresponsive growth. Increasing ambient temperatures affect ELF3 distribution in the cell. Two mechanisms have been proposed: (1) Increased temperatures alter ELF3 conformation leading to protein aggregation, or (2) warmer temperatures destabilize the EC routing ELF3 to proteasome degradation.
Perspectives
New research demonstrates complex connections between increased temperatures, heat stress responses, and plant circadian systems that can enlighten our understanding of how changing climates will impact plants and improve the selection of genotypes with enhanced yields in future climates.
From the function of ELF3 as a thermosensor to the overlapping regulation of heat stress and the circadian systems on every step of gene expression, the connections between circadian regulation and heat stress responses are pervasive in plants.
Examining the interactions between elevated temperature response, heat stress, and circadian clock pathways may identify the mechanisms for emergent properties that coordinate the timing of temperature responses. Initial studies were in Arabidopsis under controlled environments. Therefore, research in field conditions is needed to ascertain how these factors interact to improve plant yields even in a changing climate.
Acknowledgements
K.L. was supported by the Development and Promotion of Science and Technology Talents Project (DPST), Thailand. Additional support was provided by USDA National Institute of Food and Agriculture Project 1002035. We would like to thank the reviewers for their suggestions and insights.
Abbreviations
- CTD
carboxy-terminal domain
- DREB2A
DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2A
- EC
evening complex
- HSFs
heat shock factors
- HSPs
heat shock proteins
- PIF7
PHYTOCHROME INTERACTING FACTOR7
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contributions
Kanjana Laosuntisuk: Original draft preparation; Kanjana Laosuntisuk and Colleen J. Doherty: Writing, Reviewing, and Editing.
References
- 1.Bokszczanin, K.L. (2013) Solanaceae Pollen Thermotolerance Initial Training Network (SPOT-ITN) Consortium, Fragkostefanakis S. Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front. Plant Sci. 4, 315 10.3389/fpls.2013.00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohama, N., Sato, H., Shinozaki, K. and Yamaguchi-Shinozaki, K. (2017) Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22, 53–65 10.1016/j.tplants.2016.08.015 [DOI] [PubMed] [Google Scholar]
- 3.Nijabat, A., Bolton, A., Mahmood-ur-Rehman, M., Shah, A.I., Hussain, R., Naveed, N.H.et al. (2020) Cell membrane stability and relative cell injury in response to heat stress during early and late seedling stages of diverse carrot (Daucus carota L.) germplasm. HortScience 55, 1446–1452 10.21273/HORTSCI15058-20 [DOI] [Google Scholar]
- 4.Higashi, Y. and Saito, K. (2019) Lipidomic studies of membrane glycerolipids in plant leaves under heat stress. Prog. Lipid Res. 75, 100990 10.1016/j.plipres.2019.100990 [DOI] [PubMed] [Google Scholar]
- 5.Grinevich, D.O., Desai, J.S., Stroup, K.P., Duan, J., Slabaugh, E. and Doherty, C.J. (2019) Novel transcriptional responses to heat revealed by turning up the heat at night. Plant Mol Biol. 101, 1–19 10.1007/s11103-019-00873-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rienth, M., Torregrosa, L., Luchaire, N., Chatbanyong, R., Lecourieux, D., Kelly, M.T.et al. (2014) Day and night heat stress trigger different transcriptomic responses in green and ripening grapevine (vitis vinifera) fruit. BMC Plant Biol. 14, 108 10.1186/1471-2229-14-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balla, K., Karsai, I., Bónis, P., Kiss, T., Berki, Z., Horváth, Á.et al. (2019) Heat stress responses in a large set of winter wheat cultivars (Triticum aestivum L.) depend on the timing and duration of stress. PLoS ONE 14, e0222639 10.1371/journal.pone.0222639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blair, E.J., Bonnot, T., Hummel, M., Hay, E., Marzolino, J.M., Quijada, I.A.et al. (2019) Contribution of time of day and the circadian clock to the heat stress responsive transcriptome in Arabidopsis. Sci. Rep. 9, 4814 10.1038/s41598-019-41234-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, B., Gao, Z., Liu, X., Sun, D. and Tang, W. (2019) Transcriptional profiling reveals a time-of-day-specific role of REVEILLE 4/8 in regulating the first wave of heat shock–induced gene expression in Arabidopsis. Plant Cell 31, 2353–2369 10.1105/tpc.19.00519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janni, M., Gullì, M., Maestri, E., Marmiroli, M., Valliyodan, B., Nguyen, H.T.et al. (2020) Molecular and genetic bases of heat stress responses in crop plants and breeding for increased resilience and productivity. J. Exp. Bot. 71, 3780–3802 10.1093/jxb/eraa034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lobell, D.B., Hammer, G.L., McLean, G., Messina, C., Roberts, M.J. and Schlenker, W. (2013) The critical role of extreme heat for maize production in the United States. Nat. Clim. Chang. 3, 497–501 10.1038/nclimate1832 [DOI] [Google Scholar]
- 12.Lobell, D.B., Schlenker, W. and Costa-Roberts, J. (2011) Climate trends and global crop production since 1980. Science 333, 616–620 10.1126/science.1204531 [DOI] [PubMed] [Google Scholar]
- 13.Wang, L., Ma, K.-B., Lu, Z.-G., Ren, S.-X., Jiang, H.-R., Cui, J.-W.et al. (2020) Differential physiological, transcriptomic and metabolomic responses of arabidopsis leaves under prolonged warming and heat shock. BMC Plant Biol. 20, 86 10.1186/s12870-020-2292-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittler, R., Finka, A. and Goloubinoff, P. (2012) How do plants feel the heat? Trends Biochem. Sci. 37, 118–125 10.1016/j.tibs.2011.11.007 [DOI] [PubMed] [Google Scholar]
- 15.Galic, V., Franic, M., Jambrovic, A., Ledencan, T., Brkic, A., Zdunic, Z.et al. (2019) Genetic correlations between photosynthetic and yield performance in maize are different under Two heat scenarios during flowering. Front. Plant Sci. 10, 566 10.3389/fpls.2019.00566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, P., Li, D., Liao, W., Rigden, A. and Wang, W. (2019) Contrasting evaporative responses of ecosystems to heatwaves traced to the opposing roles of vapor pressure deficit and surface resistance. Water Resour. Res. 55, 4550–4563 10.1029/2019WR024771 [DOI] [Google Scholar]
- 17.Leon-Garcia, I.V. and Lasso, E. (2019) High heat tolerance in plants from the Andean highlands: Implications for paramos in a warmer world. PLoS ONE 14, e0224218 10.1371/journal.pone.0224218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahsan, N. and Komatsu, S. (2009) Comparative analyses of the proteomes of leaves and flowers at various stages of development reveal organ-specific functional differentiation of proteins in soybean. Proteomics 9, 4889–4907 10.1002/pmic.200900308 [DOI] [PubMed] [Google Scholar]
- 19.Siebers, M.H., Yendrek, C.R., Drag, D., Locke, A.M., Rios Acosta, L., Leakey, A.D.B.et al. (2015) Heat waves imposed during early pod development in soybean (Glycine max) cause significant yield loss despite a rapid recovery from oxidative stress. Glob. Chang. Biol. 21, 3114–3125 10.1111/gcb.12935 [DOI] [PubMed] [Google Scholar]
- 20.Siebers, M.H., Slattery, R.A., Yendrek, C.R., Locke, A.M., Drag, D., Ainsworth, E.A.et al. (2017) Simulated heat waves during maize reproductive stages alter reproductive growth but have no lasting effect when applied during vegetative stages. Agric. Ecosyst. Environ. 240, 162–170 10.1016/j.agee.2016.11.008 [DOI] [Google Scholar]
- 21.Cheabu, S., Moung-Ngam, P., Arikit, S., Vanavichit, A. and Malumpong, C. (2018) Effects of heat stress at vegetative and reproductive stages on spikelet fertility. Rice Sci. 25, 218–226 10.1016/j.rsci.2018.06.005 [DOI] [Google Scholar]
- 22.Ran, X., Chen, X., Shi, L., Ashraf, M., Yan, F., Chen, Y.et al. (2020) Transcriptomic insights into the roles of HSP70-16 in sepal's responses to developmental and mild heat stress signals. Environ. Exp. Bot. 179, 104225 10.1016/j.envexpbot.2020.104225 [DOI] [Google Scholar]
- 23.Friedrich, T., Oberkofler, V., Trindade, I., Altmann, S., Brzezinka, K., Lämke, J.et al. (2021) Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in arabidopsis. Nat. Commun. 12, 3426 10.1038/s41467-021-23786-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lämke, J., Brzezinka, K., Altmann, S. and Bäurle, I. (2016) A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J. 35, 162–175 10.15252/embj.201592593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charng, Y.-Y., Liu, H.-C., Liu, N.-Y., Hsu, F.-C. and Ko, S.-S. (2006) Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol. 140, 1297–1305 10.1104/pp.105.074898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charng, Y.-Y., Liu, H.-C., Liu, N.-Y., Chi, W.-T., Wang, C.-N., Chang, S.-H.et al. (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143, 251–262 10.1104/pp.106.091322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, M.-Y., Chai, K.-H., Ko, S.-S., Kuang, L.-Y., Lur, H.-S. and Charng, Y.-Y. (2014) A positive feedback loop between HEAT SHOCK PROTEIN101 and HEAT STRESS-ASSOCIATED 32-KD PROTEIN modulates long-Term acquired thermotolerance illustrating diverse heat stress responses in rice varieties. Plant Physiol. 164, 2045–2053 10.1104/pp.113.229609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu, T.-Y., Juan, Y.-T., Hsu, Y.-H., Wu, S.-H., Liao, H.-T., Fung, R.W.M.et al. (2013) Interplay between heat shock proteins HSP101 and HSA32 prolongs heat acclimation memory posttranscriptionally in Arabidopsis. Plant Physiol. 161, 2075–2084 10.1104/pp.112.212589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sewelam, N., Brilhaus, D., Bräutigam, A., Alseekh, S., Fernie, A.R. and Maurino, V.G. (2020) Molecular plant responses to combined abiotic stresses put a spotlight on unknown and abundant genes. J. Exp. Bot. 71, 5098–5112 10.1093/jxb/eraa250 [DOI] [PubMed] [Google Scholar]
- 30.Prasch, C.M. and Sonnewald, U. (2013) Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol. 162, 1849–1866 10.1104/pp.113.221044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hussain, H.A., Men, S., Hussain, S., Chen, Y., Ali, S., Zhang, S.et al. (2019) Interactive effects of drought and heat stresses on morpho-physiological attributes, yield, nutrient uptake and oxidative status in maize hybrids. Sci. Rep. 9, 3890. 10.1038/s41598-019-40362-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Handayani, T. and Watanabe, K. (2020) The combination of drought and heat stress has a greater effect on potato plants than single stresses. Plant Soil Environ. 66, 175–182 10.17221/126/2020-PSE [DOI] [Google Scholar]
- 33.Zandalinas, S.I., Mittler, R., Balfagón, D., Arbona, V. and Gómez-Cadenas, A. (2018) Plant adaptations to the combination of drought and high temperatures. Physiol. Plantarum. 162, 2–12 10.1111/ppl.12540 [DOI] [PubMed] [Google Scholar]
- 34.Kipp, E. (2008) Heat stress effects on growth and development in three ecotypes of varying latitude of Arabidopsis. Appl. Ecol. Environ. Res. 6, 1–14 10.15666/aeer/0604_001014 [DOI] [Google Scholar]
- 35.Wos, G. and Willi, Y. (2015) Temperature-stress resistance and tolerance along a latitudinal cline in North American Arabidopsis lyrata. PLoS ONE 10, e0131808 10.1371/journal.pone.0131808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su, Z., Tang, Y., Ritchey, L.E., Tack, D.C., Zhu, M., Bevilacqua, P.C.et al. (2018) Genome-wide RNA structurome reprogramming by acute heat shock globally regulates mRNA abundance. Proc. Natl Acad. Sci. U.S.A. 115, 12170–5 10.1073/pnas.1807988115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeRidder, B.P., Shybut, M.E., Dyle, M.C., Kremling, K.A.G. and Shapiro, M.B. (2012) Changes at the 3′-untranslated region stabilize rubisco Activase transcript levels during heat stress in Arabidopsis. Planta 236, 463–476 10.1007/s00425-012-1623-0 [DOI] [PubMed] [Google Scholar]
- 38.Liu, Z., Qin, J., Tian, X., Xu, S., Wang, Y., Li, H.et al. (2018) Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum L.). Plant Biotechnol. J. 16, 714–726 10.1111/pbi.12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keller, M., Hu, Y., Mesihovic, A., Fragkostefanakis, S., Schleiff, E. and Simm, S. (2017) Alternative splicing in tomato pollen in response to heat stress. DNA Res. 24, 205–217 10.1093/dnares/dsx006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang, J., Liu, X., Liu, C., Liu, G., Li, S. and Wang, L. (2017) Integrating omics and alternative splicing reveals insights into grape response to high temperature. Plant Physiol. 173, 1502–1518 10.1104/pp.16.01305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang, C.-Y., Lin, W.-D. and Tu, S.-L. (2014) Genome-wide analysis of heat-Sensitive alternative splicing in physcomitrella patens. Plant Physiol. 165, 826–840 10.1104/pp.113.230540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, J., Sun, N., Liu, M., Liu, J., Du, B., Wang, X.et al. (2013) An autoregulatory loop controlling arabidopsis HsfA2 expression: role of heat shock-Induced alternative splicing. Plant Physiol. 162, 512–521 10.1104/pp.112.205864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ueda, K., Matsuura, H., Yamaguchi, M., Demura, T. and Kato, K. (2012) Genome-wide analyses of changes in translation state caused by elevated temperature in Oryza sativa. Plant Cell Physiol. 53, 1481–1491 10.1093/pcp/pcs092 [DOI] [PubMed] [Google Scholar]
- 44.Yángüez, E., Castro-Sanz, A.B., Fernández-Bautista, N., Oliveros, J.C. and Mar Castellano, M. (2013) Analysis of genome-wide changes in the translatome of Arabidopsis seedlings subjected to heat stress. PLoS ONE 8, e71425 10.1371/journal.pone.0071425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merret, R., Descombin, J., Juan, Y.-T., Favory, J.-J., Carpentier, M.-C., Chaparro, C.et al. (2013) XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Rep. 5, 1279–1293 10.1016/j.celrep.2013.11.019 [DOI] [PubMed] [Google Scholar]
- 46.Rytz, T.C., Miller, M.J., McLoughlin, F., Augustine, R.C., Marshall, R.S., Juan, Y.-T.et al. (2018) SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO ligase SIZ1 during heat stress. Plant Cell 30, 1077–1099 10.1105/tpc.17.00993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller, M.J., Scalf, M., Rytz, T.C., Hubler, S.L., Smith, L.M. and Vierstra, R.D. (2013) Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis. Mol. Cell. Proteomics 12, 449–463 10.1074/mcp.M112.025056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, G.-T., Jiang, J.-F., Liu, X.-N., Jiang, J.-Z., Sun, L., Duan, W.et al. (2019) New insights into the heat responses of grape leaves via combined phosphoproteomic and acetylproteomic analyses. Hortic. Res. 6, 100 10.1038/s41438-019-0183-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu, X., Wu, L., Zhao, F., Zhang, D., Li, N., Zhu, G.et al. (2015) Phosphoproteomic analysis of the response of maize leaves to drought, heat and their combination stress. Front. Plant Sci. 6, 298 10.3389/fpls.2015.00298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen, X., Zhang, W., Zhang, B., Zhou, J., Wang, Y., Yang, Q.et al. (2011) Phosphoproteins regulated by heat stress in rice leaves. Proteome Sci. 9, 37 10.1186/1477-5956-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao, Q., Chen, W., Bian, J., Xie, H., Li, Y., Xu, C.et al. (2018) Proteomics and phosphoproteomics of heat stress-responsive mechanisms in spinach. Front. Plant Sci. 9, 800 10.3389/fpls.2018.00800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choudhary, M.K., Nomura, Y. and Wang, L. (2015) Quantitative circadian phosphoproteomic analysis of Arabidopsis reveals extensive clock control of key components in physiological, metabolic, and signaling pathways. Mol. Cell. Proteomics 14, 2243–2260 10.1074/mcp.M114.047183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen, L.L., van den Burg, H.A. and van Ooijen, G. (2017) Sumoylation contributes to timekeeping and temperature compensation of the plant circadian clock. J. Biol. Rhythms 32, 560–569 10.1177/0748730417737633 [DOI] [PubMed] [Google Scholar]
- 54.Morimoto, K., Ohama, N., Kidokoro, S., Mizoi, J., Takahashi, F., Todaka, D.et al. (2017) BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis. Proc. Natl Acad. Sci. U.S.A. 114, E8528–E8536 10.1073/pnas.1704189114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li, L.-M., Lü, S.-Y. and Li, R.-J. (2017) The Arabidopsis endoplasmic reticulum associated degradation pathways are involved in the regulation of heat stress response. Biochem. Biophys. Res. Commun. 487, 362–367 10.1016/j.bbrc.2017.04.066 [DOI] [PubMed] [Google Scholar]
- 56.Krahmer, J., Hindle, M., Perby, L.K., Mogensen, H.K., Nielsen, T.H., Halliday, K.J.et al. (2021) The circadian clock gene circuit controls protein and phosphoprotein rhythms in Arabidopsis thaliana. Mol. Cell. Proteomics 21, 100172 10.1016/j.mcpro.2021.100172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H.S., Han, B., Zhu, T.et al. (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113 10.1126/science.290.5499.2110 [DOI] [PubMed] [Google Scholar]
- 58.Lidder, P., Gutiérrez, R.A., Salomé, P.A., McClung, C.R. and Green, P.J. (2005) Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol. 138, 2374–2385 10.1104/pp.105.060368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michael, T.P., Mockler, T.C., Breton, G., McEntee, C., Byer, A., Trout, J.D.et al. (2008) Network discovery pipeline elucidates conserved time-of-day–specific cis-regulatory modules. PLoS Genet. 4, e14 10.1371/journal.pgen.0040014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanchez, S.E., Petrillo, E., Beckwith, E.J., Zhang, X., Rugnone, M.L., Hernando, C.E.et al. (2010) A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468, 112–116 10.1038/nature09470 [DOI] [PubMed] [Google Scholar]
- 61.Filichkin, S.A., Breton, G., Priest, H.D., Dharmawardhana, P., Jaiswal, P., Fox, S.E.et al. (2011) Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS ONE 6, e16907 10.1371/journal.pone.0016907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ito, S., Song, Y.H. and Imaizumi, T. (2012) LOV domain-containing F-box proteins: light-dependent protein degradation modules in arabidopsis. Mol. Plant 5, 573–582 10.1093/mp/sss013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanchez, S.E. and Kay, S.A. (2016) The plant circadian clock: from a simple timekeeper to a complex developmental manager. Cold Spring Harb. Perspect. Biol. 8, a027748 10.1101/cshperspect.a027748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dickinson, P.J., Kumar, M., Martinho, C., Yoo, S.J., Lan, H., Artavanis, G.et al. (2018) Chloroplast signaling gates thermotolerance in Arabidopsis. Cell Rep. 22, 1657–1665 10.1016/j.celrep.2018.01.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bonnot, T. and Nagel, D.H. (2021) Time of the day prioritizes the pool of translating mRNAs in response to heat stress. Plant Cell 33, 2164–2182 10.1093/plcell/koab113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Al-Whaibi, M.H. (2011) Plant heat-shock proteins: a mini review. J. King Saud Univ. Sci. 23, 139–150 10.1016/j.jksus.2010.06.022 [DOI] [Google Scholar]
- 67.Guo, M., Liu, J.-H., Ma, X., Luo, D.-X., Gong, Z.-H. and Lu, M.-H. (2016) The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 7, 114 10.3389/fpls.2016.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mody, T., Bonnot, T. and Nagel, D.H. (2020) Interaction between the circadian clock and regulators of heat stress responses in plants. Genes 11, 156 10.3390/genes11020156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kolmos, E., Chow, B.Y., Pruneda-Paz, J.L. and Kay, S.A. (2014) HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc. Natl Acad. Sci. U.S.A. 111, 16172–7 10.1073/pnas.1418483111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu, T., Carlsson, J., Takeuchi, T., Newton, L. and Farre, E.M. (2013) Direct regulation of abiotic responses by the A rabidopsis circadian clock component PRR 7. Plant J. 76, 101–114 10.1111/tpj.12276 [DOI] [PubMed] [Google Scholar]
- 71.Liu, T.L., Newton, L., Liu, M.-J., Shiu, S.-H. and Farré, E.M. (2016) A G-box-like motif Is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol. 170, 528–539 10.1104/pp.15.01562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nagel, D.H., Doherty, C.J., Pruneda-Paz, J.L., Schmitz, R.J., Ecker, J.R. and Kay, S.A. (2015) Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl Acad. Sci. U.S.A. 112, E4802–E4810 10.1073/pnas.1513609112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakamichi, N., Kiba, T., Kamioka, M., Suzuki, T., Yamashino, T., Higashiyama, T.et al. (2012) Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl Acad. Sci. U.S.A. 109, 17123–8 10.1073/pnas.1205156109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adams, S., Grundy, J., Veflingstad, S.R., Dyer, N.P., Hannah, M.A., Ott, S.et al. (2018) Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol. 220, 893–907 10.1111/nph.15415 [DOI] [PubMed] [Google Scholar]
- 75.Huang, W., Pérez-García, P., Pokhilko, A., Millar, A.J., Antoshechkin, I., Riechmann, J.L.et al. (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336, 75–79 10.1126/science.1219075 [DOI] [PubMed] [Google Scholar]
- 76.Ezer, D., Jung, J.-H., Lan, H., Biswas, S., Gregoire, L., Box, M.S.et al. (2017) The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 3, 17087 10.1038/nplants.2017.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeh, C.-H., Kaplinsky, N.J., Hu, C. and Charng, Y.-Y. (2012) Some like it hot, some like it warm: phenotyping to explore thermotolerance diversity. Plant Sci. 195, 10–23 10.1016/j.plantsci.2012.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang, C., Zhou, Y., Yang, X., Zhang, B., Xu, F., Wang, Y.et al. (2022) The heat stress transcription factor LlHsfA4 enhanced basic thermotolerance through regulating ROS metabolism in lilies (Lilium Longiflorum). Int. J. Mol. Sci. 23, 572 10.3390/ijms23010572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lai, A.G., Doherty, C.J., Mueller-Roeber, B., Kay, S.A., Schippers, J.H.M. and Dijkwel, P.P. (2012) CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl Acad. Sci. U.S.A. 109, 17129–17134 10.1073/pnas.1209148109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiménez, A., Sevilla, F. and Martí, M.C. (2021) Reactive oxygen species homeostasis and circadian rhythms in plants. J. Exp. Bot. 72, 5825–5840 10.1093/jxb/erab318 [DOI] [PubMed] [Google Scholar]
- 81.Román, Á., Li, X., Deng, D., Davey, J.W., James, S., Graham, I.A.et al. (2021) Superoxide is promoted by sucrose and affects amplitude of circadian rhythms in the evening. Proc. Natl Acad. Sci. U.S.A. 118, e2020646118 10.1073/pnas.2020646118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Czarnecka-Verner, E., Pan, S., Salem, T. and Gurley, W.B. (2004) Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol. Biol. 56, 57–75 10.1007/s11103-004-2307-3 [DOI] [PubMed] [Google Scholar]
- 83.Hajheidari, M., Koncz, C. and Eick, D. (2013) Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends Plant Sci. 18, 633–643 10.1016/j.tplants.2013.07.001 [DOI] [PubMed] [Google Scholar]
- 84.Uehara, T.N., Nonoyama, T., Taki, K., Kuwata, K., Sato, A., Fujimoto, K.J.et al. (2022) Phosphorylation of RNA polymerase II by CDKC;2 maintains the arabidopsis circadian clock period. Plant Cell Physiol. 63, 450–462 10.1093/pcp/pcac011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abiko, M., Akibayashi, K., Sakata, T., Kimura, M., Kihara, M., Itoh, K.et al. (2005) High-temperature induction of male sterility during barley (Hordeum vulgare L.) anther development is mediated by transcriptional inhibition. Sex Plant Reprod. 18, 91–100 10.1007/s00497-005-0004-2 [DOI] [Google Scholar]
- 86.Koiwa, H., Barb, A.W., Xiong, L., Li, F., McCully, M.G., Lee, B.-H.et al. (2002) C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate arabidopsis thaliana abiotic stress signaling, growth, and development. Proc. Natl Acad. Sci. U.S.A. 99, 10893–8 10.1073/pnas.112276199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thatcher, L.F., Foley, R., Casarotto, H.J., Gao, L.-L., Kamphuis, L.G., Melser, S.et al. (2018) The Arabidopsis RNA polymerase II carboxyl terminal domain (CTD) phosphatase-Like1 (CPL1) is a biotic stress susceptibility gene. Sci. Rep. 8, 13454 10.1038/s41598-018-31837-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koiwa, H., Hausmann, S., Bang, W.Y., Ueda, A., Kondo, N., Hiraguri, A.et al. (2004) Arabidopsis C-terminal domain phosphatase-like 1 and 2 are essential Ser-5-specific C-terminal domain phosphatases. Proc. Natl Acad. Sci. U.S.A. 101, 14539–14544 10.1073/pnas.0403174101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guan, Q., Yue, X., Zeng, H. and Zhu, J. (2014) The protein phosphatase RCF2 and Its interacting partner NAC019 Are critical for heat stress–Responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 26, 438–453 10.1105/tpc.113.118927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang, B., Yang, G., Chen, Y., Zhao, Y., Gao, P., Liu, B.et al. (2016) C-terminal domain (CTD) phosphatase links Rho GTPase signaling to Pol II CTD phosphorylation in Arabidopsis and yeast. Proc. Natl Acad. Sci. U.S.A. 113, E8197–E8206 10.1073/pnas.1605871113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Covington, M.F., Maloof, J.N., Straume, M., Kay, S.A. and Harmer, S.L. (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9, R130 10.1186/gb-2008-9-8-r130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tognacca, R.S., Guillermina Kubaczka, M., Servi, L., Rodríguez, F.S., Godoy Herz, M.A. and Petrillo, E. (2020) Light in the transcription landscape: chromatin, RNA polymerase II and splicing throughout Arabidopsis thaliana’s life cycle. Transcription 11, 117–133 10.1080/21541264.2020.1796473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ohama, N., Moo, T.L. and Chua, N.-H. (2021) Differential requirement of MED14/17 recruitment for activation of heat inducible genes. New Phytol. 229, 3360–3376 10.1111/nph.17119 [DOI] [PubMed] [Google Scholar]
- 94.Pasrija, R. and Thakur, J.K. (2013) Tissue specific expression profile of mediator genes in Arabidopsis. Plant Signal. Behav. 8, e23983 10.4161/psb.23983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pasrija, R. and Thakur, J.K. (2012) Analysis of differential expression of mediator subunit genes in Arabidopsis. Plant Signal. Behav. 7, 1676–1686 10.4161/psb.22438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Crawford, T., Karamat, F., Lehotai, N., Rentoft, M., Blomberg, J., Strand, Å.et al. (2020) Specific functions for mediator complex subunits from different modules in the transcriptional response of Arabidopsis thaliana to abiotic stress. Sci. Rep. 10, 5073 10.1038/s41598-020-61758-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang, Y., Song, L., Liu, M., Ge, R., Zhou, Q., Liu, W.et al. (2018) A proteomics landscape of circadian clock in mouse liver. Nat. Commun. 9, 1553 10.1038/s41467-018-03898-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koike, N., Yoo, S.-H., Huang, H.-C., Kumar, V., Lee, C., Kim, T.-K.et al. (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 10.1126/science.1226339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bevilacqua, P.C., Ritchey, L.E., Su, Z. and Assmann, S.M. (2016) Genome-wide analysis of RNA secondary structure. Annu. Rev. Genet. 50, 235–266 10.1146/annurev-genet-120215-035034 [DOI] [PubMed] [Google Scholar]
- 100.Kortmann, J. and Narberhaus, F. (2012) Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 10, 255–265 10.1038/nrmicro2730 [DOI] [PubMed] [Google Scholar]
- 101.Chung, B.Y.W., Balcerowicz, M., Di Antonio, M., Jaeger, K.E., Geng, F., Franaszek, K.et al. (2020) An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat. Plants 6, 522–532 10.1038/s41477-020-0633-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fiorucci, A.-S., Galvão, V.C., Ince, Y.Ç, Boccaccini, A., Goyal, A., Allenbach Petrolati, L.et al. (2020) PHYTOCHROME INTERACTING FACTOR 7 is important for early responses to elevated temperature in Arabidopsis seedlings. New Phytol. 226, 50–58 10.1111/nph.16316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang, X., Jiang, B., Gu, L., Chen, Y., Mora, M., Zhu, M.et al. (2021) A photoregulatory mechanism of the circadian clock in Arabidopsis. Nat. Plants 7, 1397–1408 10.1038/s41477-021-01002-z [DOI] [PubMed] [Google Scholar]
- 104.Ling, Y., Alshareef, S., Butt, H., Lozano-Juste, J., Li, L., Galal, A.A.et al. (2017) Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant J. 89, 291–309 10.1111/tpj.13383 [DOI] [PubMed] [Google Scholar]
- 105.Ling, Y., Serrano, N., Gao, G., Atia, M., Mokhtar, M., Woo, Y.H.et al. (2018) Thermopriming triggers splicing memory in Arabidopsis. J. Exp. Bot. 69, 2659–2675 10.1093/jxb/ery062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kannan, S., Halter, G., Renner, T. and Waters, E.R. (2018) Patterns of alternative splicing vary between species during heat stress. AoB Plants 10, ly013 10.1093/aobpla/ply013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu, Z., Liang, J., Wang, C., Ding, L., Zhao, X., Cao, X.et al. (2019) Alternative splicing provides a mechanism to regulate LlHSFA3 function in response to heat stress in lily. Plant Physiol. 181, 1651–1667 10.1104/pp.19.00839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang, X., Wu, F., Xie, Q., Wang, H., Wang, Y., Yue, Y.et al. (2012) SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24, 3278–3295 10.1105/tpc.112.100081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang, Y., Li, Y., Sancar, A. and Oztas, O. (2020) The circadian clock shapes the Arabidopsis transcriptome by regulating alternative splicing and alternative polyadenylation. J. Biol. Chem. 295, 7608–7619 10.1074/jbc.RA120.013513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Romanowski, A., Schlaen, R.G., Perez-Santangelo, S., Mancini, E. and Yanovsky, M.J. (2020) Global transcriptome analysis reveals circadian control of splicing events in Arabidopsis thaliana. Plant J. 103, 889–902 10.1111/tpj.14776 [DOI] [PubMed] [Google Scholar]
- 111.James, A.B., Syed, N.H., Bordage, S., Marshall, J., Nimmo, G.A., Jenkins, G.I.et al. (2012) Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24, 961–981 10.1105/tpc.111.093948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Park, M.-J., Seo, P.J. and Park, C.-M. (2012) CCA1 alternative splicing as a way of linking the circadian clock to temperature response in Arabidopsis. Plant Signal. Behav. 7, 1194–1196 10.4161/psb.21300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kwon, Y.-J., Park, M.-J., Kim, S.-G., Baldwin, I.T. and Park, C.-M. (2014) Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biol. 14, 136 10.1186/1471-2229-14-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dantas, L.L.B., Calixto, C.P.G., Dourado, M.M., Carneiro, M.S., Brown, J.W.S. and Hotta, C.T. (2019) Alternative splicing of circadian clock genes correlates with temperature in field-grown sugarcane. Front. Plant Sci. 10, 1614 10.3389/fpls.2019.01614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Caster, S.Z., Castillo, K., Sachs, M.S. and Bell-Pedersen, D. (2016) Circadian clock regulation of mRNA translation through eukaryotic elongation factor eEF-2. Proc. Natl Acad. Sci. U.S.A. 113, 9605–9610 10.1073/pnas.1525268113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Karki, S., Castillo, K., Ding, Z., Kerr, O., Lamb, T.M., Wu, C.et al. (2020) Circadian clock control of eIF2α phosphorylation is necessary for rhythmic translation initiation. Proc. Natl Acad. Sci. U.S.A. 117, 10935–10945 10.1073/pnas.1918459117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gallie, D.R., Le, H., Caldwell, C., Tanguay, R.L., Hoang, N.X. and Browning, K.S. (1997) The phosphorylation state of translation initiation factors is regulated developmentally and following heat shock in wheat. J. Biol. Chem. 272, 1046–1053 10.1074/jbc.272.2.1046 [DOI] [PubMed] [Google Scholar]
- 118.Parnell, A.A., De Nobrega, A.K. and Lyons, L.C. (2021) Translating around the clock: Multi-level regulation of post-transcriptional processes by the circadian clock. Cell Signal. 80, 109904 10.1016/j.cellsig.2020.109904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Urquidi Camacho, R.A., Lokdarshi, A. and von Arnim, A.G. (2020) Translational gene regulation in plants: a green new deal. Wiley Interdiscip. Rev. RNA 11, e1597 10.1002/wrna.1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Enganti, R., Cho, S.K., Toperzer, J.D., Urquidi-Camacho, R.A., Cakir, O.S., Ray, A.P.et al. (2017) Phosphorylation of ribosomal protein RPS6 integrates light signals and circadian clock signals. Front. Plant Sci. 8, 2210 10.3389/fpls.2017.02210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McLoughlin, F., Basha, E., Fowler, M.E., Kim, M., Bordowitz, J., Katiyar-Agarwal, S.et al. (2016) Class I and II small heat shock proteins together with HSP101 protect protein translation factors during heat stress. Plant Physiol. 172, 1221–1236 10.1104/pp.16.00536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gil, K.-E., Kim, W.-Y., Lee, H.-J., Faisal, M., Saquib, Q., Alatar, A.A.et al. (2017) ZEITLUPE contributes to a thermoresponsive protein quality control system in Arabidopsis. Plant Cell 29, 2882–2894 10.1105/tpc.17.00612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cha, J.-Y., Kim, J., Kim, T.-S., Zeng, Q., Wang, L., Lee, S.Y.et al. (2017) GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat. Commun. 8, 3 10.1038/s41467-016-0014-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Davis, A.M., Ronald, J., Ma, Z., Wilkinson, A.J., Philippou, K., Shindo, T.et al. (2018) HSP90 contributes to entrainment of the Arabidopsis circadian clock via the morning loop. Genetics 210, 1383–1390 10.1534/genetics.118.301586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gil, K.-E. and Park, C.-M. (2019) Thermal adaptation and plasticity of the plant circadian clock. New Phytol. 221, 1215–1229 10.1111/nph.15518 [DOI] [PubMed] [Google Scholar]
- 126.Delker, C., van Zanten, M. and Quint, M. (2017) Thermosensing enlightened. Trends Plant Sci. 22, 185–187 10.1016/j.tplants.2017.01.007 [DOI] [PubMed] [Google Scholar]
- 127.Vu, L.D., Gevaert, K. and De Smet, I. (2019) Feeling the heat: searching for plant thermosensors. Trends Plant Sci. 24, 210–219 10.1016/j.tplants.2018.11.004 [DOI] [PubMed] [Google Scholar]
- 128.Lamers, J., van der Meer, T. and Testerink, C. (2020) How plants sense and respond to stressful environments. Plant Physiol. 182, 1624–1635 10.1104/pp.19.01464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jung, J.-H., Barbosa, A.D., Hutin, S., Kumita, J.R., Gao, M., Derwort, D.et al. (2020) A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585, 256–260 10.1038/s41586-020-2644-7 [DOI] [PubMed] [Google Scholar]
- 130.Murcia, G., Nieto, C., Sellaro, R., Prat, S. and Casal, J.J. (2022) Hysteresis in PHYTOCHROME-INTERACTING FACTOR 4 and EARLY-FLOWERING 3 dynamics dominates warm daytime memory in Arabidopsis. The Plant Cell 34, 2188–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ronald, J., Wilkinson, A.J. and Davis, S.J. (2021) EARLY FLOWERING3 sub-nuclear localization responds to changes in ambient temperature. Plant Physiol. 187, 2352–2355 10.1093/plphys/kiab423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nusinow, D.A., Helfer, A., Hamilton, E.E., King, J.J., Imaizumi, T., Schultz, T.F.et al. (2011) The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402 10.1038/nature10182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhao, H. and Bao, Y. (2021) PIF4: integrator of light and temperature cues in plant growth. Plant Sci. 313, 111086 10.1016/j.plantsci.2021.111086 [DOI] [PubMed] [Google Scholar]
- 134.Zhu, Z., Quint, M. and Anwer, M.U. (2022) Arabidopsis EARLY FLOWERING 3 controls temperature responsiveness of the circadian clock independently of the evening complex. J. Exp. Bot. 73, 1049–1061 10.1093/jxb/erab473 [DOI] [PubMed] [Google Scholar]
- 135.Box, M.S., Huang, B.E., Domijan, M., Jaeger, K.E., Khattak, A.K., Yoo, S.J.et al. (2015) ELF3 controls thermoresponsive growth in Arabidopsis. Curr. Biol. 25, 194–199 10.1016/j.cub.2014.10.076 [DOI] [PubMed] [Google Scholar]
- 136.Silva, C.S., Nayak, A., Lai, X., Hutin, S., Hugouvieux, V., Jung, J.-H.et al. (2020) Molecular mechanisms of evening complex activity in Arabidopsis. Proc. Natl Acad. Sci. U.S.A. 117, 6901–6909 10.1073/pnas.1920972117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Raschke, A., Ibañez, C., Ullrich, K.K., Anwer, M.U., Becker, S., Glöckner, A.et al. (2015) Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4-dependent auxin response genes. BMC Plant Biol. 15, 197 10.1186/s12870-015-0566-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mizuno, T., Nomoto, Y., Oka, H., Kitayama, M., Takeuchi, A., Tsubouchi, M.et al. (2014) Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol. 55, 958–976 10.1093/pcp/pcu030 [DOI] [PubMed] [Google Scholar]
- 139.Nieto, C., López-Salmerón, V., Davière, J.-M. and Prat, S. (2015) ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr. Biol. 25, 187–193 10.1016/j.cub.2014.10.070 [DOI] [PubMed] [Google Scholar]
- 140.Legris, M., Nieto, C., Sellaro, R., Prat, S. and Casal, J.J. (2017) Perception and signalling of light and temperature cues in plants. Plant J. 90, 683–697 10.1111/tpj.13467 [DOI] [PubMed] [Google Scholar]
- 141.Herrero, E., Kolmos, E., Bujdoso, N., Yuan, Y., Wang, M., Berns, M.C.et al. (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24, 428–443 10.1105/tpc.111.093807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Anwer, M.U., Boikoglou, E., Herrero, E., Hallstein, M., Davis, A.M., Velikkakam James, G.et al. (2014) Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock. eLife 3, e02206 10.7554/eLife.02206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang, L.-L., Li, W., Tian, Y.-Y., Davis, S.J. and Liu, J.-X. (2021) The E3 ligase XBAT35 mediates thermoresponsive hypocotyl growth by targeting ELF3 for degradation in Arabidopsis. J. Integr. Plant Biol. 63, 1097–1103 10.1111/jipb.13107 [DOI] [PubMed] [Google Scholar]
- 144.Zhang, L.L., Shao, Y.J., Ding, L., Wang, M.J., Davis, S.J. and Liu, J.X. (2021) XBAT31 regulates thermoresponsive hypocotyl growth through mediating degradation of the thermosensor ELF3 in Arabidopsis. Sci. Adv. 7, eabf4427 10.1126/sciadv.abf4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yuan, L., Yu, Y., Liu, M., Song, Y., Li, H., Sun, J.et al. (2021) BBX19 fine-tunes the circadian rhythm by interacting with PSEUDO-RESPONSE REGULATOR proteins to facilitate their repressive effect on morning-phased clock genes. Plant Cell 33, 2602–2617 10.1093/plcell/koab133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Allada, R., White, N.E., So, W.V., Hall, J.C. and Rosbash, M. (1998) A mutant Drosophila homolog of mammalian clock disrupts circadian rhythms and transcription of period and timeless. Cell 93, 791–804 10.1016/S0092-8674(00)81440-3 [DOI] [PubMed] [Google Scholar]
- 147.Hsu, P.Y. and Harmer, S.L. (2014) Wheels within wheels: the plant circadian system. Trends Plant Sci. 19, 240–249 10.1016/j.tplants.2013.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Missra, A., Ernest, B., Lohoff, T., Jia, Q., Satterlee, J., Ke, K.et al. (2015) The circadian clock modulates global daily cycles of mRNA ribosome loading. Plant Cell 27, 2582–2599 10.1105/tpc.15.00546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hicks, K.A., Albertson, T.M. and Wagner, D.R. (2001) EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13, 1281–1292 10.1105/TPC.010070 [DOI] [PMC free article] [PubMed] [Google Scholar]