Abstract
Chronic inflammation caused by gut dysbiosis is associated with the pathophysiology of metabolic disease. Synbiotics are useful for ameliorating gut dysbiosis; however, it remains unclear what types of bacteria act as key markers for synbiotic-driven improvement of chronic inflammation. Here, we performed a post hoc analysis of a 24-week randomized controlled study using synbiotics to investigate the association between gut microbiota and inflammatory markers. We characterized the responders who showed lower interleukin-6 (IL-6) levels in response to synbiotic supplementation among 86 obese patients with type 2 diabetes mellitus. In our baseline analysis, the relative abundances of Bifidobacterium adolescentis and Alistipes onderdonkii correlated positively with IL-6, lipopolysaccharide binding protein (LBP), and high-sensitivity C-reactive protein (Hs-CRP) levels. The relative abundance of Eubacterium rectale correlated positively with LBP and Hs-CRP levels, and that of Bacteroides thetaiotaomicron correlated positively with LBP levels. Based on our responder analysis, patients with higher body mass indices (over 30 kg/m2 on average), low abundances of Bacteroides caccae and Parabacteroides merdae at baseline and 24 weeks, and minimal changes in the relative abundance of E. rectale and Shannon index from baseline showed decreased IL-6 levels compared with baseline. However, glycemic control in responders was unchanged. In conclusion, we identified four bacterial species (B. adolescentis, A. onderdonkii, E. rectale, and B. thetaiotaomicron) related to chronic inflammation and predictive markers (B. caccae, P. merdae, and severity of obesity) in responders to synbiotic supplementation among obese patients with type 2 diabetes.
Keywords: diabetes mellitus, chronic inflammation, gut microbiota, synbiotic, obesity
INTRODUCTION
Type 2 diabetes mellitus (T2DM) is a complex metabolic disease. Multiple factors, including genetics, lifestyle, diet, and aging, contribute to either the onset or progression of this disease, if not both [1]. Among these factors, diet is an important environmental factor affecting the gut microbiota, and diet-induced changes in microbial composition are involved in human physiology and disease processes [2]. We previously demonstrated that obese patients with T2DM had chronic inflammatory states accompanied by gut dysbiosis and bacterial translocation [3]. Therefore, the gut microbiota is a new therapeutic target for treating chronic inflammation in T2DM.
To date, probiotics, prebiotics, and synbiotics (the combination of one or more probiotics and prebiotics) have been reported to be useful for inhibiting bacterial translocation [4] and improving the intestinal environment in metabolic disease [5] and other diseases [6]. Therefore, we previously performed a 24-week randomized controlled study to investigate the effects of daily intake of a synbiotic comprising Lacticaseibacillus paracasei (previously known as Lactobacillus casei) strain Shirota YIT 9029, Bifidobacterium breve strain Yakult YIT 12272, and galacto-oligosaccharides on chronic inflammation and gut microbiota in 86 obese patients with T2DM [7]. This synbiotic did not reduce plasma interleukin-6 (IL-6) levels as the primary outcome, although numerous bacterial species that showed significant changes in response to synbiotic supplementation were identified by using 16S rRNA amplicon gene analysis [7].
The presence of responders and non-responders to diet and probiotics in relation to cholesterol metabolism and insulin sensitivity has already been reported in obese individuals [8] and T2DM [9]. However, little is known about the clinical characteristics of responders with improvement of chronic inflammation in response to synbiotic supplementation among obese patients with T2DM. Therefore, it is important to identify clinical or microbial biomarkers for predicting responders to synbiotic supplementation in order to pave the way for personalized nutrition.
Recently, the associations between specific bacterial species and inflammation have been reported in some diseases, such as inflammatory bowel diseases [10] and autoimmune diseases [11]. However, bacterial species relating to inflammatory markers, such as IL-6, high-sensitivity C-reactive protein (Hs-CRP), and lipopolysaccharide binding protein (LBP), have yet to be investigated in obese patients with T2DM. Based on this background information, we performed a post hoc analysis of a randomized controlled study using synbiotics to investigate gut microbiota related to chronic inflammation. We also sought to identify clinical and microbial markers among responders to synbiotic supplementation in obese patients with T2DM.
MATERIALS AND METHODS
Study participants
In this post hoc analysis, inclusion and exclusion criteria were applied as previously described [7]. Briefly, the main inclusion criteria were 1) age ≥30 but <80 years, 2) HbA1c (National Glycohemoglobin Standardization Program, NGSP) ≥6.0 but <9.0%, and 3) body mass index (BMI) ≥25.0 kg/m2. The exclusion criteria were 1) serious kidney disease (serum creatinine level ≥1.5 mg/dL and/or hemodialysis), 2) serious liver disease excluding fatty liver, and 3) inflammatory bowel disease. A total of 86 patients with T2DM who met the requirements of the above inclusion and exclusion criteria were recruited into the interventional study examined [7]. As we aimed to perform the analysis by having as many subjects as possible, the baseline data of these subjects (n=86) before synbiotic supplementation were used to investigate correlations between baseline inflammatory markers (Hs-CRP, LBP, and IL-6) and relative abundances of the gut microbiota obtained by 16S rRNA gene sequencing. Next, we selected the change in IL-6 as an index of responders to synbiotic supplementation because it was the primary outcome in the synbiotic interventional study [7], and we divided the synbiotic recipients (n=44) into responders (n=23) and non-responders (n=21) depending on whether or not they displayed a decrease in IL-6 from baseline to 24 weeks after synbiotic supplementation. As the synbiotic intervention, the following agents were administered orally: 3.0 g dry powder containing at least 3×108 living L. paracasei strain Shirota YIT 9029, 3×108 living B. breve strain Yakult YIT 12272, and 7.5 g galacto-oligosaccharides (GOS) per day (product name: Yakult Super Synbiotics LBG-P, Yakult Honsha Co., Ltd., Tokyo, Japan). Patients were instructed to take the synbiotic twice a day (2.0 g dry powder and 5.0 g GOS at breakfast and 1.0 g dry powder and 2.5 g GOS at dinner). The synbiotic recipients consumed the aforementioned doses every day for 24 weeks. The protocol of the post hoc analysis was approved by the Human Ethics Committee of Juntendo University (approval number: H21-0041). Written informed consent was not obtained from each participant, as this was a retrospective study. Additionally, participants were given the opportunity to opt out of the study (https://www.gcprec.juntendo.ac.jp/kenkyu/6/detail/3357).
Analysis of the gut microbiota and inflammatory markers
We used previously described methods to perform a gut microbiota analysis using 16S rRNA gene sequencing [7]. Sequences generated from the MiSeq platform were analyzed using the open-source software package Quantitative Insights Into Microbial Ecology 2 (QIIME2; 2020.2) [12], and the SILVA138 database (https://www.arb-silva.de/) was used to annotate taxonomic information. Alpha diversities represented as the number of observed operational taxonomic units (OTUs), the Shannon index, and phylogenetic diversity (PD) were estimated for 5,000 randomly selected sequences to account for differences in sampling effort between the samples. Lastly, biochemical assays for HbA1c, lipids, fecal organic acids, and inflammatory markers (Hs-CRP, LBP, and IL-6) were performed as described previously [7]. These data were reanalyzed for the present post hoc analysis.
Statistical analysis
All data are presented as the mean ± standard deviation (SD), and they were analyzed using StatFlex ver. 7 (Artech Co., Osaka, Japan). Comparisons of the groups of responders and non-responders were analyzed by Student’s t-test. The relationships between microbiota and inflammatory markers were investigated by Pearson’s correlation coefficient analysis. p<0.05 was considered statistically significant.
RESULTS
Clinical characteristics of the study participants at baseline
Table 1 shows the clinical characteristics of the study participants at baseline. The average age of the study participants was 58.5 ± 11.1 years, and 21 participants were women. The average BMI was 29.3 ± 4.1 kg/m2, and only nine patients were not taking any medications. The medications of the participants during the 24-week study period are presented in Supplementary Table 1. A DPP-4 inhibitor or SGLT2 inhibitor was added to the medications taken by each of the non-responders and responders, respectively.
Table 1. Clinical characteristics of the study participants at baseline.
| n | 86 |
| Sex (male/female) | 65/21 |
| Age (years) | 58.5 ± 11.1 |
| BMI (kg/m2) | 29.3 ± 4.1 |
| HbA1c (%) | 7.3 ± 0.8 |
| C-peptide (ng/mL) | 2.4 ± 1.3 |
| T-CHO (mg/dL) | 192.6 ± 41.0 |
| HDL-C (mg/dL) | 50.1 ± 10.0 |
| TG (mg/dL) | 183.8 ± 274.5 |
| eGFR (mL/min/1.73m2) | 77.7 ± 20.7 |
| Hs-CRP (ng/mL) | 2,157.3 ± 4,530.9 |
| IL-6 (pg/mL) | 2.5 ± 1.7 |
| LBP (μg/mL) | 6.0 ± 3.5 |
| Medication for diabetes | |
| No medication | 9 (10.5) |
| Insulin only or with oral therapy | 28 (32.6) |
| Oral therapy only | |
| SU | 8 (9.3) |
| Metformin | 55 (64.0) |
| Thiazolidine | 12 (14.0) |
| DPP-4 inhibitor | 42 (48.8) |
| Glinide | 6 (7.0) |
| SGLT2 inhibitor | 42 (48.8) |
| GLP-1 receptor agonist | 10 (11.6) |
Data are expressed as mean ± SD.
Numbers in parentheses are percentages (%).
BMI: Body mass index; T-CHO: Total cholesterol; HDL-C: high-density lipoprotein cholesterol; TG: triglycerides; eGFR: estimated glomerular filtration rate; Hs-CRP: high-sensitivity C-reactive protein; IL-6: interleukin-6; LBP: lipopolysaccharide-binding protein; SU: sulfonylurea; DPP-4: dipeptidyl peptidase-4; SGLT2: sodium-dependent glucose cotransporter-2; GLP-1: glucagon-like peptide-1.
Correlations between gut microbiota and inflammatory markers
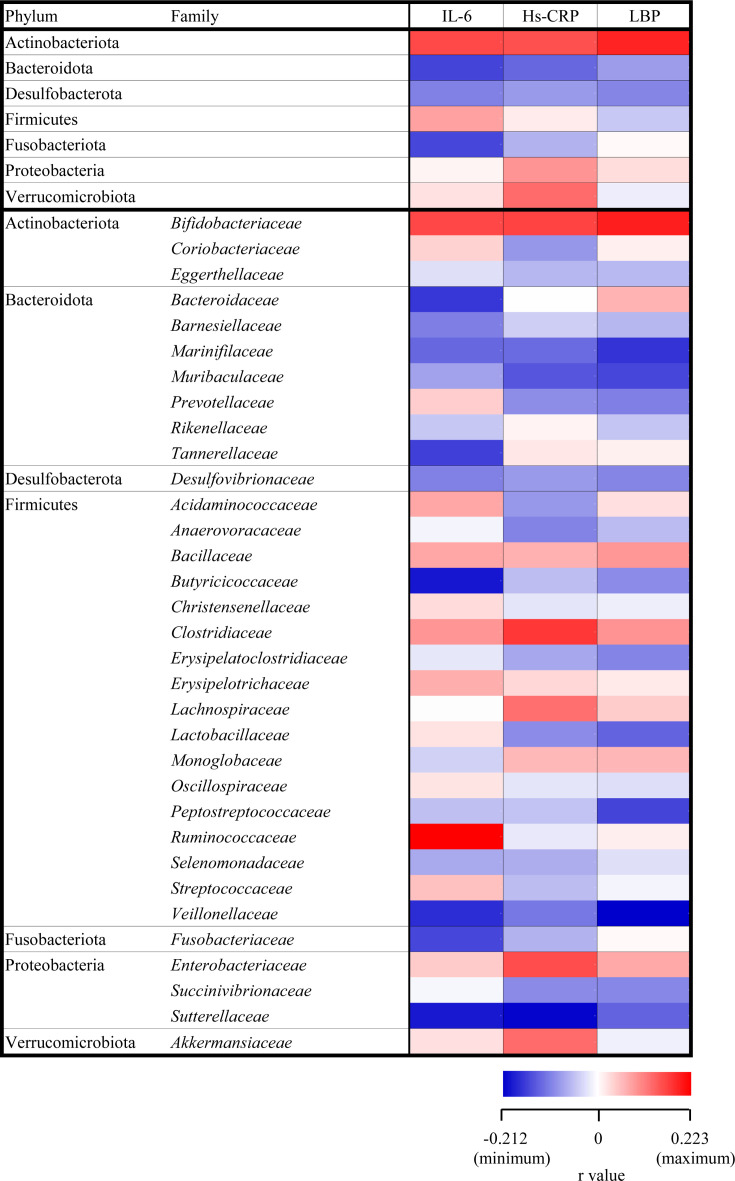
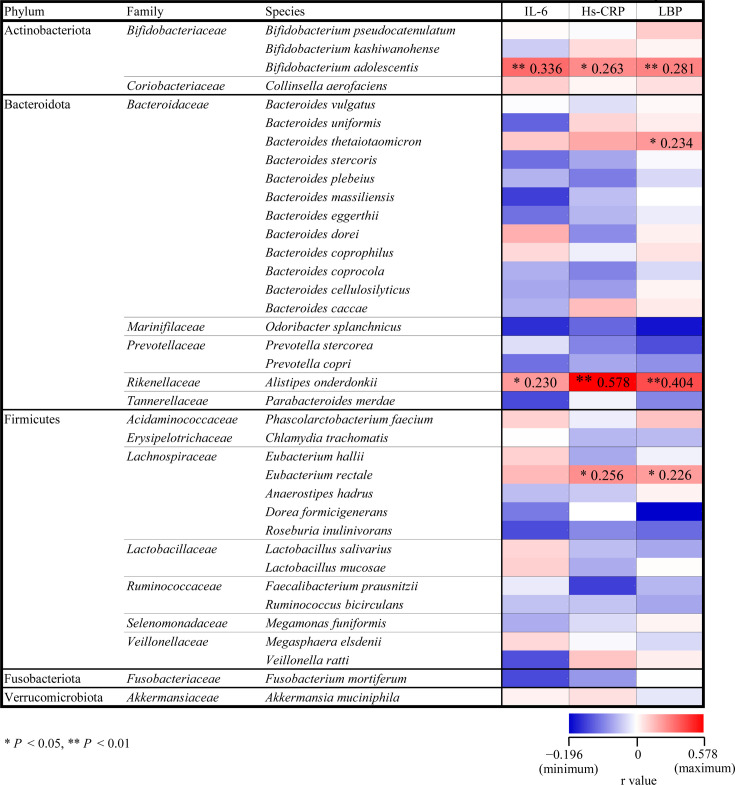
At baseline, the relative abundances of seven bacteria phyla and 33 bacterial families assigned based on the SILVA database showed no significant correlations with inflammatory markers (Fig. 1). However, among 37 bacterial species (Fig. 2), we found that the relative abundance of Bifidobacterium adolescentis was positively associated with IL-6 (r=0.336, p<0.01, Supplementary Fig. 1A), LBP (r=0.281, p<0.01, Supplementary Fig. 1B), and Hs-CRP levels (r=0.263, p<0.05, Supplementary Fig. 1C). Moreover, the relative abundance of Alistipes onderdonkii correlated positively with IL-6 (r=0.230, p<0.05, Supplementary Fig. 2A), LBP (r=0.404, p<0.01, Supplementary Fig. 2B), and Hs-CRP levels (r=0.578, p<0.01, Supplementary Fig. 2C). The relative abundance of Eubacterium rectale also correlated positively with LBP (r=0.226, p<0.05, Supplementary Fig. 3A) and Hs-CRP levels (r=0.256, p<0.05, Supplementary Fig. 3B), while the abundance of Bacteroides thetaiotaomicron correlated positively with LBP levels (r=0.234, p<0.05, Supplementary Fig. 3C).
Fig. 1.
Heat map of Pearson correlation coefficients between inflammatory markers and relative abundances of gut microbiota from the phylum to family levels. These relative abundances show no significant correlations with inflammatory markers.
Fig. 2.
Heat map of Pearson correlation coefficients between inflammatory markers and relative abundances of gut microbial species. Red and blue columns indicate positive and negative correlations, respectively. Correlation coefficients are visualized by color gradient (blue, minimum < r < maximum, red). Columns with asterisks shows significant correlations (*p<0.05; **p<0.01).
Clinical characteristics of the non-responders and responders
BMI and plasma IL-6 levels at baseline were significantly higher in patients that were responders compared with non-responders, and BMI at 24 weeks in responders was also higher than in non-responders (Table 2). Plasma IL-6 levels in responders showed a significant negative change from baseline compared with non-responders, and the change in plasma LBP levels in responders was significantly smaller than in non-responders. There were no significant differences between the two groups for the other parameters measured, including HbA1c.
Table 2. Clinical parameters before and after synbiotic supplementation in the non-responders and responders.
| Baseline | 24 weeks | change | ||
|---|---|---|---|---|
| n (male/female) | Non-responders | 21 (15/6) | ||
| Responders | 23 (16/7) | |||
| Age (years) | Non-responders | 59.8 ± 8.9 | ||
| Responders | 62.2 ± 13.1 | |||
| BMI (kg/m2) | Non-responders | 28.1 ± 2.2 | 27.8 ± 2.0 | −0.25 ± 0.79 |
| Responders | 30.8 ± 5.5* | 31.0 ± 5.6* | 0.16 ± 0.56 | |
| HbA1c (%) | Non-responders | 7.3 ± 0.7 | 7.3 ± 0.6 | 0.0 ± 0.6 |
| Responders | 7.4 ± 0.8 | 7.8 ± 1.3 | 0.4 ± 0.9 | |
| C-peptide (ng/mL) | Non-responders | 2.0 ± 1.1 | 1.9 ± 1.2 | −0.1 ± 0.6 |
| Responders | 2.6 ± 1.2 | 2.8 ± 1.6 | 0.3 ± 1.1 | |
| T-CHO (mg/dL) | Non-responders | 195.7 ± 31.8 | 190.3 ± 29.7 | −5.4 ± 23.5 |
| Responders | 182.3 ± 35.5 | 188.8 ± 34.2 | 6.4 ± 26.8 | |
| HDL-C (mg/dL) | Non-responders | 52.7 ± 10.2 | 50.8 ± 9.6 | −1.9 ± 6.4 |
| Responders | 49.0 ± 9.0 | 49.1 ± 9.6 | 0.1 ± 4.4 | |
| TG (mg/dL) | Non-responders | 146.2 ± 85.2 | 185.2 ± 150.4 | 39.0 ± 104.7 |
| Responders | 137.3 ± 57.5 | 178.3 ± 117.1 | 40.9 ± 87.1 | |
| eGFR (mL/min/1.73m2) | Non-responders | 75.1 ± 18.4 | 75.4 ± 23.7 | 0.3 ± 8.7 |
| Responders | 81.1 ± 26.4 | 83.6 ± 24.3 | 2.5 ± 12.3 | |
| Hs-CRP (ng/mL) | Non-responders | 1,780.8 ± 4,393.3 | 1,407.4 ± 1,755.8 | −373.4 ± 3,816.8 |
| Responders | 2,983.8 ± 6,728.4 | 1,772.0 ± 2,146.4 | −1,211.8 ± 6,736.1 | |
| IL-6 (pg/mL) | Non-responders | 1.8 ± 0.9 | 2.7 ± 1.0 | 0.9 ± 0.7 |
| Responders | 3.5 ± 2.5** | 2.3 ± 1.4 | −1.3 ± 1.8** | |
| LBP (μg/mL) | Non-responders | 5.4 ± 2.3 | 8.8 ± 2.9 | 3.4 ± 2.8 |
| Responders | 7.3 ± 5.3 | 7.9 ± 2.9 | 0.6 ± 4.8* |
Data are expressed as mean ± SD. Change is expressed as the value measured at 24 weeks minus the baseline value. *p<005, **p<0.01 vs. Non-responders.
BMI: Body mass index; T-CHO: Total cholesterol; HDL-C: high-density lipoprotein cholesterol; TG: triglycerides; eGFR: estimated glomerular filtration rate; Hs-CRP: high-sensitivity C-reactive protein; IL-6: interleukin-6; LBP: lipopolysaccharide-binding protein.
Relative abundances of bacterial phyla and families in the non-responders and responders
Supplementary Table 3 shows the relative abundances of different bacterial families in the responders and non-responders. At baseline, the relative abundance of Monoglobaceae in responders was significantly lower than that of non-responders, and it increased from baseline compared with that in non-responders. The changes in the relative abundances of Bifidobacteriaceae and Lachnospiraceae in responders were significantly smaller than those in non-responders. As shown by Supplementary Table 2, there were no significant differences in the relative abundances of bacterial phyla between the two groups.
Relative abundances of bacterial species in the non-responders and responders
At both baseline and 24 weeks, the relative abundances of Bacteroides caccae and Parabacteroides merdae in responders were significantly lower than in non-responders. At 24 weeks, the relative abundance of Collinsella aerofaciens in responders was significantly higher than in non-responders. In addition, the relative abundance of E. rectale in responders showed a significant positive change from baseline compared with non-responders (Table 3).
Table 3. Relative abundances at the species level before and after synbiotic supplementation in the non-responders and responders.
| Phylum | Family | Species | Relative abundance (%) | Change (%) | ||
|---|---|---|---|---|---|---|
| Baseline | 24 weeks | 24 weeks | ||||
| Actinobacteriota | Bifidobacteriaceae | Bifidobacterium pseudocatenulatum | Non-responders | 1.3 ± 1.9 | 6.5 ± 8.5 | 5.2 ± 7.4 |
| Responders | 2.8 ± 6.8 | 4.2 ± 4.3 | 1.3 ± 5.7 | |||
| Bifidobacterium kashiwanohense | Non-responders | 0.01 ± 0.03 | 0.01 ± 0.05 | 0.00 ± 0.02 | ||
| Responders | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | |||
| Bifidobacterium adolescentis | Non-responders | 2.3 ± 3.5 | 7.1 ± 9.2 | 4.8 ± 7.2 | ||
| Responders | 4.3 ± 6.4 | 7.6 ± 8.1 | 3.4 ± 7.6 | |||
| Coriobacteriaceae | Collinsella aerofaciens | Non-responders | 0.38 ± 0.40 | 0.46 ± 0.48 | 0.09 ± 0.29 | |
| Responders | 0.78 ± 0.93 | 0.97 ± 0.64* | 0.19 ± 0.76 | |||
| Bacteroidota | Bacteroidaceae | Bacteroides vulgatus | Non-responders | 0.11 ± 0.40 | 0.06 ± 020 | −0.05 ± 0.21 |
| Responders | 0.65 ± 1.53 | 0.44 ± 1.24 | −0.23 ± 0.49 | |||
| Bacteroides uniformis | Non-responders | 2.9 ± 2.8 | 1.9 ± 2.0 | −1.0 ± 2.4 | ||
| Responders | 2.5 ± 3.1 | 2.2 ± 3.4 | −0.4 ± 2.0 | |||
| Bacteroides thetaiotaomicron | Non-responders | 0.47 ± 0.52 | 0.49 ± 0.80 | 0.02 ± 0.67 | ||
| Responders | 0.58 ± 0.57 | 0.35 ± 0.48 | −0.22 ± 0.40 | |||
| Bacteroides stercoris | Non-responders | 2.0 ± 4.0 | 1.8 ± 3.6 | −0.3 ± 0.9 | ||
| Responders | 1.9 ± 3.2 | 1.9 ± 3.4 | −0.1 ± 2.6 | |||
| Bacteroides plebeius | Non-responders | 1.6 ± 3.8 | 2.2 ± 4.8 | 0.6 ± 2.6 | ||
| Responders | 3.0 ± 7.7 | 2.3 ± 5.1 | −0.9 ± 4.5 | |||
| Bacteroides massiliensis | Non-responders | 1.00 ± 2.80 | 0.40 ± 1.41 | −0.59 ± 1.47 | ||
| Responders | 0.66 ± 1.36 | 0.67 ± 1.57 | −0.03 ± 0.70 | |||
| Bacteroides eggerthii | Non-responders | 0.67 ± 1.53 | 0.44 ± 1.06 | −0.23 ± 0.61 | ||
| Responders | 0.09 ± 0.38 | 0.02 ± 0.06 | −0.07 ± 0.35 | |||
| Bacteroides dorei | Non-responders | 0.7 ± 1.9 | 0.9 ± 2.7 | 0.2 ± 0.8 | ||
| Responders | 1.9 ± 4.4 | 1.5 ± 3.3 | −0.4 ± 1.8 | |||
| Bacteroides coprophilus | Non-responders | 0.18 ± 0.58 | 0.09 ± 3.2 | −0.09 ± 0.36 | ||
| Responders | 0.82 ± 2.80 | 0.37 ± 1.59 | −0.49 ± 2.54 | |||
| Bacteroides coprocola | Non-responders | 0.58 ± 2.32 | 0.69 ± 3.01 | 0.11 ± 0.72 | ||
| Responders | 0.14 ± 0.51 | 0.03 ± 0.08 | −0.12 ± 0.49 | |||
| Bacteroides cellulosilyticus | Non-responders | 0.15 ± 0.32 | 0.08 ± 0.21 | −0.08 ± 0.28 | ||
| Responders | 0.08 ± 0.19 | 0.13 ± 0.41 | 0.06 ± 0.24 | |||
| Bacteroides caccae | Non-responders | 0.68 ± 0.81 | 0.78 ± 1.12 | 0.10 ± 0.59 | ||
| Responders | 0.24 ± 0.38* | 0.25 ± 0.44* | 0.01 ± 0.43 | |||
| Marinifilaceae | Odoribacter splanchnicus | Non-responders | 0.16 ± 0.14 | 0.13 ± 0.16 | −0.03 ± 0.12 | |
| Responders | 0.13 ± 0.16 | 0.11 ± 0.13 | −0.02 ± 0.17 | |||
| Prevotellaceae | Prevotella stercorea | Non-responders | 0.75 ± 2.30 | 0.20 ± 0.50 | −0.55 ± 1.94 | |
| Responders | 0.35 ± 1.34 | 0.35 ± 1.01 | 0.02 ± 1.10 | |||
| Prevotella copri | Non-responders | 0.6 ± 2.3 | 0.4 ± 1.3 | −0.2 ± 1.1 | ||
| Responders | 1.1 ± 2.9 | 1.0 ± 1.9 | 0.2 ± 2.1 | |||
| Rikenellaceae | Alistipes onderdonkii | Non-responders | 0.25 ± 0.52 | 0.12 ± 0.28 | −0.12 ± 0.45 | |
| Responders | 0.11 ± 0.26 | 0.15 ± 0.27 | 0.05 ± 0.21 | |||
| Tannerellaceae | Parabacteroides merdae | Non-responders | 1.41 ± 1.60 | 1.29 ± 1.22 | −0.11 ± 1.45 | |
| Responders | 0.55 ± 0.68* | 0.50 ± 0.61* | 0.03 ± 0.52 | |||
| Firmicutes | Acidaminococcaceae | Phascolarctobacterium faecium | Non-responders | 0.30 ± 0.48 | 0.18 ± 0.38 | −0.12 ± 0.38 |
| Responders | 0.29 ± 0.54 | 0.34 ± 0.67 | 0.03 ± 0.41 | |||
| Erysipelotrichaceae | Chlamydia trachomatis | Non-responders | 0.16 ± 0.72 | 0.22 ± 1.01 | 0.06 ± 0.29 | |
| Responders | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | |||
| Lachnospiraceae | Eubacterium hallii | Non-responders | 0.8 ± 1.2 | 0.5 ± 0.7 | −0.3 ± 0.7 | |
| Responders | 0.6 ± 1.0 | 0.4 ± 0.6 | −0.2 ± 0.6 | |||
| Eubacterium rectale | Non-responders | 3.4 ± 4.4 | 2.0 ± 3.3 | −1.4 ± 3.3 | ||
| Responders | 2.1 ± 3.8 | 2.5 ± 4.3 | 0.3 ± 1.6* | |||
| Anaerostipes hadrus | Non-responders | 1.7 ± 2.1 | 1.0 ± 1.3 | −0.7 ± 2.1 | ||
| Responders | 1.5 ± 2.0 | 1.9 ± 2.7 | 0.3 ± 1.3 | |||
| Dorea formicigenerans | Non-responders | 0.19 ± 0.21 | 0.20 ± 0.23 | 0.01 ± 0.23 | ||
| Responders | 0.13 ± 0.17 | 0.12 ± 0.11 | −0.01 ± 0.14 | |||
| Roseburia inulinivorans | Non-responders | 0.51 ± 0.84 | 0.21 ± 0.39 | −0.30 ± 0.57 | ||
| Responders | 0.19 ± 0.34 | 0.14 ± 0.23 | −0.05 ± 0.22 | |||
| Lactobacillaceae | Lactobacillus salivarius | Non-responders | 0.04 ± 0.15 | 0.08 ± 0.24 | 0.03 ± 0.11 | |
| (now Ligilactobacillus salivarius) | Responders | 0.30 ± 0.85 | 0.20 ± 0.80 | −0.12 ± 0.43 | ||
| Lactobacillus mucosae | Non-responders | 0.04 ± 0.18 | 0.13 ± 0.40 | 0.09 ± 0.25 | ||
| (now Limosilactobacillus mucosae) | Responders | 0.36 ± 0.76 | 0.45 ± 1.17 | 0.08 ± 1.00 | ||
| Ruminococcaceae | Faecalibacterium prausnitzii | Non-responders | 0.35 ± 0.59 | 0.25 ± 0.38 | −0.11 ± 0.52 | |
| Responders | 0.35 ± 0.55 | 0.31 ± 0.48 | −0.05 ± 0.53 | |||
| Ruminococcus bicirculans | Non-responders | 0.67 ± 1.27 | 0.94 ± 1.90 | 0.26 ± 1.00 | ||
| Responders | 0.42 ± 1.03 | 0.52 ± 0.92 | 0.08 ± 0.57 | |||
| Selenomonadaceae | Megamonas funiformis | Non-responders | 0.56 ± 1.19 | 0.56 ± 1.57 | 0.00 ± 1.27 | |
| Responders | 0.64 ± 2.20 | 0.18 ± 0.48 | −0.49 ± 1.96 | |||
| Veillonellaceae | Megasphaera elsdenii | Non-responders | 0.08 ± 0.20 | 0.15 ± 0.33 | 0.07 ± 0.23 | |
| Responders | 0.13 ± 0.44 | 0.18 ± 0.68 | 0.14 ± 0.54 | |||
| Veillonella ratti | Non-responders | 0.48 ± 1.01 | 0.74 ± 2.03 | 0.26 ± 1.51 | ||
| Responders | 0.06 ± 0.27 | 0.06 ± 0.30 | 0.01 ± 0.03 | |||
| Fusobacteriota | Fusobacteriaceae | Fusobacterium mortiferum | Non-responders | 0.93 ± 2.51 | 0.38 ± 1.51 | −0.55 ± 1.42 |
| Responders | 0.19 ± 0.88 | 0.01 ± 0.03 | −0.18 ± 0.90 | |||
| Verrucomicrobiota | Akkermansiaceae | Akkermansia muciniphila | Non-responders | 0.29 ± 0.88 | 0.03 ± 0.08 | −0.26 ± 0.86 |
| Responders | 0.13 ± 0.45 | 0.16 ± 0.52 | 0.03 ± 0.71 | |||
Change is expressed as the difference between the value measured at 24 weeks and the baseline value.
*p<0.05 vs. Non-responders.
Microbial diversity and fecal organic acids in the non-responders and responders
The Shannon index showed a significant positive change from baseline in responders compared with non-responders. However, there were no significant differences in the other indices between the two groups (Table 4). Furthermore, fecal organic acids levels showed no significant changes in comparisons between the two groups (Supplementary Table 4).
Table 4. Microbial diversity before and after synbiotic supplementation in the non-responders and responders.
| Measured values | Change | |||
|---|---|---|---|---|
| Baseline | 24 weeks | 24 weeks | ||
| Phylogenic diversity | Non-responders | 26.4 ± 5.3 | 25.3 ± 5.5 | −1.0 ± 2.7 |
| Responders | 26.8 ± 7.2 | 27.0 ± 6.5 | 0.2 ± 3.4 | |
| Observed OTU | Non-responders | 220.7 ± 61.4 | 209.7 ± 65.8 | −11.0 ± 45.7 |
| Responders | 225.8 ± 70.5 | 225.6 ± 61.6 | 0.9 ± 40.3 | |
| Shannon index | Non-responders | 6.12 ± 0.60 | 5.82 ± 0.62 | −0.30 ± 0.39 |
| Responders | 6.06 ± 0.75 | 6.10 ± 0.66 | 0.06 ± 0.57* | |
Change is expressed as the difference between the value measured at 24 weeks and the baseline value. *p<0.05 vs. Non-responders. OTU: observed operational taxonomic unit.
DISCUSSION
This is the first study to investigate associations between inflammatory markers and specific bacterial species and bacterial markers in responders to synbiotic supplementation among obese patients with T2DM. Based on analysis of baseline data, we found that the relative abundances of B. adolescentis, A. onderdonkii, E. rectale, and B. thetaiotaomicron correlated positively with plasma inflammatory markers.
Genus Bifidobacterium is a predominant bacterium in the human gut and contains more than 50 species, including several subspecies [13]. However, the details of the functional roles of these species in humans remain unclear. To date, some Bifidobacterium species have been reported to reduce intestinal endotoxin levels in mice [14] and have anti-inflammatory effects due to inhibition of lipopolysaccharide-induced nuclear factor-κB activation in vitro [15]. Furthermore, a previous report showed that B. adolescentis supplementation ameliorates visceral fat accumulation and insulin sensitivity in an experimental model of metabolic syndrome [16]. Since some beneficial effects of Bifidobacterium species on anti-inflammation have been reported, as described above, B. adolescentis may be involved in improving chronic inflammation by regulating intestinal barrier function and anti-inflammatory effects.
Genus Alistipes is a relatively new bacterial genus that comprises 13 species isolated primarily from human samples [17]. As shown by Table 3, the relative abundance of A. onderdonkii in feces is low compared with Bifidobacterium. Previous reports indicated that genus Alistipes is involved in liver fibrosis [17] and colorectal cancer in Il-10−/− mice [18]. In particular, A. onderdonkii tended to be reduced in non-alcoholic fatty liver disease (NAFLD) patients with advanced fibrosis [19]. Although there have been no reports regarding the roles of this species in T2DM, chronic inflammation activation is a characteristic of NAFLD [20], which is highly prevalent in T2DM [21]. Therefore, A. onderdonkii, which showed significant correlations with inflammatory markers, may play an important role in either the onset or progression of NAFLD in T2DM, if not both the onset and progression.
Genus Eubacterium, which is assigned to the phylum Firmicutes, is a part of the core human gut microbiota and is phylogenetically diverse [22]. E. rectale, one of the most extensively studied Eubacterium species, was first isolated from the feces of healthy Japanese-Hawaiian males and was identified as a major butyrate producer [23]. To date, this species has been reported to be negatively correlated with IL-6 and IL-1β levels, and the E. rectale relatives group has been reported to be correlated with IL-6 and IL-8 in the elderly [24], while the abundance of E. rectale and relatives has been reported to be correlated negatively with IL-1β, NLR family pyrin domain containing 3, and CXC motif chemokine ligand 2 in cognitively impaired elderly [25]. In addition, E. rectale was reported to be among the gut bacteria that were positively associated with lower postprandial glycemic response [26]. Based on these findings, E. rectale may be linked to the pathophysiology of T2DM via various mechanisms.
B. thetaiotaomicron exhibited a considerable increase in obese patients following a weight-loss intervention [27]. Experimentally, B. thetaiotaomicron administration protected mice against adiposity [27]. In addition, B. thetaiotaomicron modulates the intestinal mucus barrier by modifying goblet cells and mucin glycosylation in rats [28]. As a positive correlation was found between the relative abundance of B. thetaiotaomicron and the LBP level in the present study, this species is also considered to be involved in chronic inflammation by regulating mucus barrier function (i.e., goblet cell differentiation, expression of mucus-related genes, and the ratio of sialylated to sulfated mucins).
In our responder analysis, we found that diabetes patients with higher BMIs (over 30 kg/m2 on average) and IL-6 levels at baseline showed a significant reduction in IL-6 by the end of the study, suggesting that severe obesity is a clinical factor that predicts responders to synbiotics. Furthermore, as shown by the BMI data in Table 2, responders with higher BMIs (over 30 kg/m2 on average) had higher levels of IL-6. Therefore, the patients with higher BMIs might have had more severe chronic inflammation, suggesting that the patients with higher levels of chronic inflammation at baseline were more sensitive to synbiotic supplementation. The relative abundance of E. rectale in responders was stable compared with non-responders. Since E. rectale has some antidiabetic effects, as discussed above, the increase in feces in response to synbiotics may be a biological marker for responders. The relative abundances of two bacterial species, B. caccae and P. merdae, in responders were significantly lower at both baseline and 24 weeks; however, their abundances did not significantly change over time. Although the functional roles of P. merdae in chronic inflammation remain unclear, TonB-linked outer membrane protein (OmpW) produced by B. caccae is known to be associated with perinuclear anti-neutrophil cytoplasmic antibodies, and the anti-OmpW immunoglobulin A levels were found to be elevated in inflammatory bowel disease [29]. Furthermore, B. caccae is enriched in clinical gout [30]. Therefore, the low relative abundances of B. caccae and P. merdae may be microbial markers for responders to synbiotic supplementation.
The Shannon index accounts for both abundance and evenness of bacterial species, and a higher value is indicative of greater α-diversity, which is known to be lower in obese children [31]. Interestingly, in the present study, the index was stable in responders throughout the study compared with non-responders, suggesting that a stable α-diversity might play some possible roles for responders to synbiotic supplementation.
The present study has several limitations. First, we were unable to confirm a causal relationship between inflammatory markers and the gut microbiota because our study used a retrospective design. Second, the number of participants used in the responder analysis was small. Third, the differences in the daily diets between the responders and non-responders were not evaluated even though they may affect the gut microbiota. Therefore, a large-scale study with a dietary intake assessment is necessary in the future. Finally, species-level identification in this study was based on sequence information for the 16S rRNA gene V1-2 region obtained by MiSeq. Therefore, evaluation by quantitative PCR using species-specific primers is necessary in the future.
In conclusion, we identified the specific bacterial species related to inflammatory markers in obese patients with T2DM. Furthermore, the severity of obesity and presence of two bacterial species (B. caccae and P. merdae) may be predictive markers for responders to synbiotic supplementation.
FUNDING
None.
CONFLICT OF INTEREST
AK has received lecture fees from Sanofi, Novartis Pharmaceuticals, Daiichi Sankyo Inc., Mitsubishi Tanabe Pharma, Takeda Pharmaceutical Co., and MSD, as well as research funds from Mitsubishi Tanabe Pharma. HW has received lecture fees from Boehringer Ingelheim, Sanofi, Ono Pharmaceutical Co., Novo Nordisk Pharma, Novartis Pharmaceuticals, Eli Lilly, Sanwa Kagaku Kenkyusho, Daiichi Sankyo Inc., Takeda Pharmaceutical Co., MSD, Dainippon Sumitomo Pharma, and Kowa Co., as well as research funds from Boehringer Ingelheim, Pfizer, Mochida Pharmaceutical Co., Sanofi, Novo Nordisk Pharma, Novartis Pharmaceuticals, Sanwa Kagaku Kenkyusho, Terumo Corp., Eli Lilly, Mitsubishi Tanabe Pharma, Daiichi Sankyo Inc., Takeda Pharmaceutical Co., MSD, Shionogi Pharma, Dainippon Sumitomo Pharma, Kissei Pharma, and AstraZeneca. AM and YYo are employed by the Yakult Central Institute. The other authors declare no conflicts of interest.
Supplementary Material
REFERENCES
- 1.Kolb H, Martin S. 2017. Environmental/lifestyle factors in the pathogenesis and prevention of type 2 diabetes. BMC Med 15: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gentile CL, Weir TL. 2018. The gut microbiota at the intersection of diet and human health. Science 362: 776–780. [DOI] [PubMed] [Google Scholar]
- 3.Sato J, Kanazawa A, Ikeda F, Yoshihara T, Goto H, Abe H, Komiya K, Kawaguchi M, Shimizu T, Ogihara T, Tamura Y, Sakurai Y, Yamamoto R, Mita T, Fujitani Y, Fukuda H, Nomoto K, Takahashi T, Asahara T, Hirose T, Nagata S, Yamashiro Y, Watada H. 2014. Gut dysbiosis and detection of “live gut bacteria” in blood of Japanese patients with type 2 diabetes. Diabetes Care 37: 2343–2350. [DOI] [PubMed] [Google Scholar]
- 4.Sato J, Kanazawa A, Azuma K, Ikeda F, Goto H, Komiya K, Kanno R, Tamura Y, Asahara T, Takahashi T, Nomoto K, Yamashiro Y, Watada H. 2017. Probiotic reduces bacterial translocation in type 2 diabetes mellitus: a randomised controlled study. Sci Rep 7: 12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonai M, Shigehisa A, Kigawa I, Kurasaki K, Chonan O, Matsuki T, Yoshida Y, Aida M, Hamano K, Terauchi Y. 2017. Galacto-oligosaccharides ameliorate dysbiotic Bifidobacteriaceae decline in Japanese patients with type 2 diabetes. Benef Microbes 8: 705–716. [DOI] [PubMed] [Google Scholar]
- 6.Sugawara G, Nagino M, Nishio H, Ebata T, Takagi K, Asahara T, Nomoto K, Nimura Y. 2006. Perioperative synbiotic treatment to prevent postoperative infectious complications in biliary cancer surgery: a randomized controlled trial. Ann Surg 244: 706–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanazawa A, Aida M, Yoshida Y, Kaga H, Katahira T, Suzuki L, Tamaki S, Sato J, Goto H, Azuma K, Shimizu T, Takahashi T, Yamashiro Y, Watada H. 2021. Effects of synbiotic supplementation on chronic inflammation and the gut microbiota in obese patients with type 2 diabetes mellitus: a randomized controlled study. Nutrients 13: 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korpela K, Flint HJ, Johnstone AM, Lappi J, Poutanen K, Dewulf E, Delzenne N, de Vos WM, Salonen A. 2014. Gut microbiota signatures predict host and microbiota responses to dietary interventions in obese individuals. PLoS One 9: e90702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mobini R, Tremaroli V, Ståhlman M, Karlsson F, Levin M, Ljungberg M, Sohlin M, Bertéus Forslund H, Perkins R, Bäckhed F, Jansson PA. 2017. Metabolic effects of Lactobacillus reuteri DSM 17938 in people with type 2 diabetes: a randomized controlled trial. Diabetes Obes Metab 19: 579–589. [DOI] [PubMed] [Google Scholar]
- 10.Prosberg M, Bendtsen F, Vind I, Petersen AM, Gluud LL. 2016. The association between the gut microbiota and the inflammatory bowel disease activity: a systematic review and meta-analysis. Scand J Gastroenterol 51: 1407–1415. [DOI] [PubMed] [Google Scholar]
- 11.Alpizar-Rodriguez D, Lesker TR, Gronow A, Gilbert B, Raemy E, Lamacchia C, Gabay C, Finckh A, Strowig T. 2019. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann Rheum Dis 78: 590–593. [DOI] [PubMed] [Google Scholar]
- 12.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, 2nd, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37: 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hidalgo-Cantabrana C, Delgado S, Ruiz L, Ruas-Madiedo P, Sánchez B, Margolles A. 2017. Bifidobacteria and their health-promoting effects. Microbiol Spectr 5: 5. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths EA, Duffy LC, Schanbacher FL, Qiao H, Dryja D, Leavens A, Rossman J, Rich G, Dirienzo D, Ogra PL. 2004. In vivo effects of bifidobacteria and lactoferrin on gut endotoxin concentration and mucosal immunity in Balb/c mice. Dig Dis Sci 49: 579–589. [DOI] [PubMed] [Google Scholar]
- 15.Riedel CU, Foata F, Philippe D, Adolfsson O, Eikmanns BJ, Blum S. 2006. Anti-inflammatory effects of bifidobacteria by inhibition of LPS-induced NF-kappaB activation. World J Gastroenterol 12: 3729–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Wang R, Li XF, Wang RL. 2012. Bifidobacterium adolescentis supplementation ameliorates visceral fat accumulation and insulin sensitivity in an experimental model of the metabolic syndrome. Br J Nutr 107: 1429–1434. [DOI] [PubMed] [Google Scholar]
- 17.Parker BJ, Wearsch PA, Veloo ACM, Rodriguez-Palacios A. 2020. The genus Alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol 11: 906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moschen AR, Gerner RR, Wang J, Klepsch V, Adolph TE, Reider SJ, Hackl H, Pfister A, Schilling J, Moser PL, Kempster SL, Swidsinski A, Orth Höller D, Weiss G, Baines JF, Kaser A, Tilg H. 2016. Lipocalin 2 protects from inflammation and tumorigenesis associated with gut microbiota alterations. Cell Host Microbe 19: 455–469. [DOI] [PubMed] [Google Scholar]
- 19.Rau M, Rehman A, Dittrich M, Groen AK, Hermanns HM, Seyfried F, Beyersdorf N, Dandekar T, Rosenstiel P, Geier A. 2018. Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United European Gastroenterol J 6: 1496–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo Y, Lin H. 2021. Inflammation initiates a vicious cycle between obesity and nonalcoholic fatty liver disease. Immun Inflamm Dis 9: 59–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwok R, Choi KC, Wong GL, Zhang Y, Chan HL, Luk AO, Shu SS, Chan AW, Yeung MW, Chan JC, Kong AP, Wong VW. 2016. Screening diabetic patients for non-alcoholic fatty liver disease with controlled attenuation parameter and liver stiffness measurements: a prospective cohort study. Gut 65: 1359–1368. [DOI] [PubMed] [Google Scholar]
- 22.Mukherjee A, Lordan C, Ross RP, Cotter PD. 2020. Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes 12: 1802866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore WE, Holdeman LV. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl Microbiol 27: 961–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, Nikkïla J, Monti D, Satokari R, Franceschi C, Brigidi P, De Vos W. 2010. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One 5: e10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cattaneo A, Cattane N, Galluzzi S, Provasi S, Lopizzo N, Festari C, Ferrari C, Guerra UP, Paghera B, Muscio C, Bianchetti A, Volta GD, Turla M, Cotelli MS, Gennuso M, Prelle A, Zanetti O, Lussignoli G, Mirabile D, Bellandi D, Gentile S, Belotti G, Villani D, Harach T, Bolmont T, Padovani A, Boccardi M, Frisoni GB, INDIA-FBP Group2017. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol Aging 49: 60–68. [DOI] [PubMed] [Google Scholar]
- 26.Martínez I, Lattimer JM, Hubach KL, Case JA, Yang J, Weber CG, Louk JA, Rose DJ, Kyureghian G, Peterson DA, Haub MD, Walter J. 2013. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J 7: 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, Shi J, Zhao S, Liu W, Wang X, Xia H, Liu Z, Cui B, Liang P, Xi L, Jin J, Ying X, Wang X, Zhao X, Li W, Jia H, Lan Z, Li F, Wang R, Sun Y, Yang M, Shen Y, Jie Z, Li J, Chen X, Zhong H, Xie H, Zhang Y, Gu W, Deng X, Shen B, Xu X, Yang H, Xu G, Bi Y, Lai S, Wang J, Qi L, Madsen L, Wang J, Ning G, Kristiansen K, Wang W. 2017. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med 23: 859–868. [DOI] [PubMed] [Google Scholar]
- 28.Wrzosek L, Miquel S, Noordine ML, Bouet S, Joncquel Chevalier-Curt M, Robert V, Philippe C, Bridonneau C, Cherbuy C, Robbe-Masselot C, Langella P, Thomas M. 2013. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol 11: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei B, Dalwadi H, Gordon LK, Landers C, Bruckner D, Targan SR, Braun J. 2001. Molecular cloning of a Bacteroides caccae TonB-linked outer membrane protein identified by an inflammatory bowel disease marker antibody. Infect Immun 69: 6044–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Z, Zhang J, Wang Z, Ang KY, Huang S, Hou Q, Su X, Qiao J, Zheng Y, Wang L, Koh E, Danliang H, Xu J, Lee YK, Zhang H. 2016. Intestinal microbiota distinguish gout patients from healthy humans. Sci Rep 6: 20602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Sun H, Jiang F, Shen Y, Li X, Hu X, Shen X, Wei P. 2020. Alteration of the gut microbiota associated with childhood obesity by 16S rRNA gene sequencing. PeerJ 8: e8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.