Abstract
Two major emerging bands (a 350-bp band and a 650-bp band) within the RISA (ribosomal intergenic spacer analysis) profile of a soil bacterial community spiked with Hg(II) were selected for further identification of the populations involved in the response of the community to the added metal. The bands were cut out from polyacrylamide gels, cloned, characterized by restriction analysis, and sequenced for phylogenetic affiliation of dominant clones. The sequences were the intergenic spacer between the rrs and rrl genes and the first 130 nucleotides of the rrl gene. Comparison of sequences derived from the 350-bp band to The GenBank database permitted us to identify the bacteria as being mostly close relatives to low G+C firmicutes (Clostridium-like genera), while the 650-bp band permitted us to identify the bacteria as being mostly close relatives to β-proteobacteria (Ralstonia-like genera). Oligonucleotide probes specific for the identified dominant bacteria were designed and hybridized with the RISA profiles derived from the control and spiked communities. These studies confirmed the contribution of these populations to the community response to the metal. Hybridization of the RISA profiles from subcommunities (bacterial pools associated with different soil microenvironments) also permitted to characterize the distribution and the dynamics of these populations at a microscale level following mercury spiking.
Recent major advances in the field of molecular biology have made it possible to develop community DNA fingerprinting methods with which to monitor complex bacterial communities in their natural environment without the need for isolation and cultivation steps. These methods involve in situ extraction of DNA from the microbial community, the PCR amplification of sequences of interest, and analysis of part of the genetic information. The sequences most commonly used as markers of communities are the genes of the ribosomal operon. The diversity of the amplified sequences is then resolved by differential electrophoretic migration on agarose or polyacrylamide gels, based on their size (terminal-restriction fragment length polymorphism, ribosomal intergenic spacer analysis [RISA]) or sequence (denaturing gel gradient electrophoresis, thermal gel gradient electrophoresis). These so-called fingerprinting methods provide band profiles that are representative of the genetic structure of the community as a whole or of a section of it, as defined by selected primers (for a review, see reference 26). These methods are valuable tools for characterizing complex bacterial communities and detecting shifts following environmental perturbations and are less time-consuming and labor-intensive than strategies such as small-subunit rRNA gene clone library construction (for a review, see reference 38). Individual bands can also be excised from a fingerprint profile, cloned and sequenced, or challenged with a range of probes providing precise information on the phylogenetic groups constituting a community (25, 17).
We previously looked at the modifications that occurred in a soil bacterial community and in various subcommunities (bacterial populations associated with different microenvironments) in response to a short exposure to inorganic mercury using RISA fingerprinting (36). This method is based on the length polymorphism of intergenic spacer (IGS) sequences between the small (16S) and the large (23S) subunit rRNA genes amplified with universal eubacterial primers directly on the DNA extracted from the community. RISA has also been used to contrast bacterial community structures associated with different soil microenvironments (37), in soils with different vegetation covers (6), or in the rhizosphere treated with different antibiotics (40).
The present study was done to determine the phylogenetic affiliations of bacterial populations responding to mercury spiking, as indicated by changes in the RISA band profiles of the soil community (36). Two major emerging bands (a 350-bp band and a 650-bp band) were excised from the gel derived from the RISA profile of the whole community, cloned, and further characterized by constructing and analyzing IGS clone libraries. Neither band was organism specific, since distinct and distantly related organisms had IGSs of different sequences but with similar sizes. Consequently, representative samples of clones (70 to 100) obtained from each RISA band were screened by amplified ribosomal DNA restriction analysis to determine pattern abundance profiles. Dominant clones in each RISA band were sequenced for phylogenetic affiliation and designing of oligonucleotide probes specific for the dominant responsive bacteria. Hybridization to the RISA profiles derived from the bacterial community before and after mercury spiking demonstrated the enrichment of these populations during adaptation of the community. Hybridization to the RISA profiles derived from the subcommunities associated with microenvironments highlighted the distribution and the dynamics of these responding populations at a microscale level.
MATERIALS AND METHODS
Soil origin and microcosm setup.
A soil containing a background of 72.3 ng of Hg · g−1 (dry weight) was collected from a cultivated silt-loam soil at La Côte Saint André (LCSAc, France). Soil sampling, storage, and microcosm setup were as described by Ranjard et al. (34). Briefly, microcosms containing 10 g (dry weight) of soil received 1 ml of a mercuric chloride (HgCl2) solution to obtain a final concentration of 10 μg of Hg(II) per g [e.g., 50 μM Hg(II)] and were incubated at 22°C for 30 days. Soil washing (34) and physical fractionation (35) were used to separate bacteria located outside aggregates and thus easily washed from the surface of aggregates (outer fraction) and to further separate the stable aggregates according to their size (size fractions). Details of the methods used to obtain various microenvironments, as well as the properties of the microenvironments have been published (36, 37).
RISA fingerprinting of bacterial communities.
Bacterial DNA was extracted, purified, and quantified from soil samples (35). The IGS region between the small (16S) and the large (23S) subunits of ribosomal sequences was amplified by PCR using primers S-D-Bact-1522-b-S-20 (5′-TGCGGCTGGATCCCCTCCTT-3′) and L-D-Bact-132-a-A-18 (5′-CCGGGTTTCCCCATTCGG-3′). PCR, and electrophoresis conditions were as previously described (36).
Purification of RISA products.
Two major emerging bands, one of 350 bp (RISA band 1) and another of 650 bp (RISA band 2) were detected in the RISA profiles of the Hg(II)-spiked community (36). They were cut out of the gel and purified by electroelution with a Mini-Protean II (Bio-Rad, Ivry sur Seine, France) according to the manufacturer's instructions. The DNA was recovered in 20 μl of ultrapure water, and the concentrations were evaluated by comparing the fluorescent-band intensities on 2% (wt/vol) agarose gels to the fluorescent-band intensities of known concentrations of the standard Smart ladder (Eurogentec, Seraing, Belgium).
Clone library construction.
Clone libraries from PCR products (the excised and purified DNA corresponding to RISA band 1 and RISA band 2) were constructed with the SureClone Ligation Kit (Pharmacia, Orsay, France) according to the manufacturer's instructions. PCR products were ligated to the vector pUC18 (Promega, Charbonnières, France). Ligation and transformation into Escherichia coli DH5α competent cells (Life Biotechnologies, Cergy Pontoise, France) were carried out according to the manufacturer's protocol. Cells were grown in Luria-Bertani medium at 37°C for 24 h. A total of 115 white clones were sampled for RISA band 1 and 200 for RISA band 2.
Analysis of IGS clone libraries.
The plasmid inserts were obtained from each clone by amplification with primers M13r and M13f, which are specifically designed to complement the polylinker of the vector pUC18 (Promega). The amplicons were run in 1.5% (wt/vol) agarose gel to determine the insert size. IGS diversity was investigated by digesting the amplified inserts from 5 μl of the PCR product with the restriction enzymes AluI, TaqI, and HaeIII (Boehringer Mannheim, Meylan, France) according to the manufacturer's instructions. The resulting fragments were separated by gel electrophoresis in 3.5% (wt/vol) Metaphor agarose (FMC Bioproducts, Le Perray en Yvelines, France). Individual clones were grouped into restriction groups or phylotypes based on a 100% identity threshold of the restriction patterns for the three enzymes used. The diversity of the phylotypes in the two RISA bands was compared by the Shannon diversity index (46).
Determination of nucleotide sequences and phylogenetic analysis.
Sequencing was performed by Act Gene-Euro Sequence Gene Services (Grenoble, France) on an ABI 377 sequencer FS (Perkin-Elmer, Norwalk, Conn.) using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq DNA Polymerase FS (Perkin-Elmer). The M13r and M13f primers were used to sequence both strands of the insert. Sequences from clones were aligned using CLUSTAL W (47) and were compared with databases available at the National Center for Biotechnology Information (NCBI) using the BLAST program (16).
Hybridization.
Two 20-nucleotide probes were designed based on the alignments of the sequenced RISA bands. Alignment of the sequences obtained from RISA band 1 allowed to design a probe (5′-ACGCTCGGTAGCTCTTTGAC-3′) located in the IGS zone that is specific for the dominant phylotypes 1, 2, 3, and 5. Alignment of the sequenced RISA band 2 allowed to design a probe (5′-GCATGCCTGATATACAAACG-3′) located in the IGS zone between the two tRNAs that is specific for the dominant phylotypes 1 and 2. These probes were hybridized with RISA profiles of the spiked and control soil communities and subcommunities. Profiles were separated on 5% (wt/vol) polyacrylamide gels as described above, and the DNA was electrotransferred to nylon membranes (Genescreen Plus, Life Science Products, Boston, Mass.) using a TransBlot SD apparatus (Bio-Rad) according to the method of Muyzer et al. (27). Oligonucleotides were end labeled using T4 polynucleotide kinase (Boehringer Mannheim) as specified by the manufacturer, and hybridizations were performed at 57°C for the RISA 1 probe and at 53°C for the RISA 2 probe, according to the method of Sambrook et al. (44).
IGS length database.
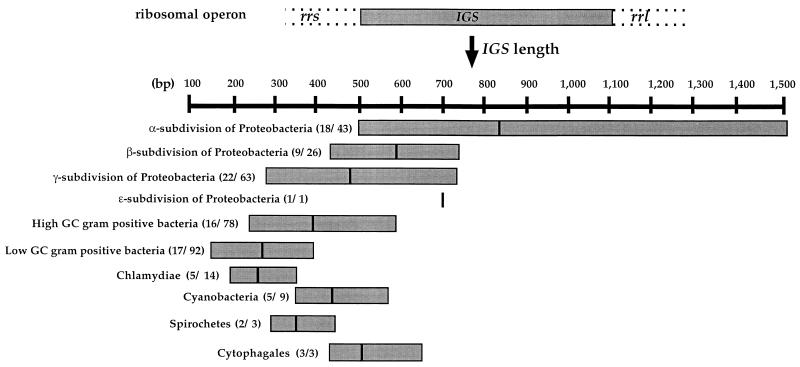
An overview of the size of the IGS within the main bacterial groups was obtained by comparing published data (5, 8, 15, 28, 45) and data from GenBank (NCBI) available as of September 2000. These data were from 99 bacterial genera and 332 species of bacteria (phylum α-, β-, γ-, and ɛ-subdivisions of the genera Proteobacteria, low G+C firmicutes, high G+C firmicutes, Cyanobacteria, Chlamydiae, Cytophagales, and Spirochetes). All multiple sequences were recorded for a given species or strain, whether or not their lengths were different.
Nucleotide sequence accession numbers.
A total of 19 sequences obtained from RISA 1 and RISA 2 bands have been deposited in GenBank under the accession numbers listed in Table 1.
TABLE 1.
Frequencies and phylogenetic affiliations of clones derived from the 350-bp band (RISA band 1) and the 650-bp band (RISA band 2)
| Phylotype | No. (%) of clones | Accession no. of RISA band sequences | Phylogenetic affiliationa | Closest relativea | % similarity | Accession no. of closest relative |
|---|---|---|---|---|---|---|
| RISA band 1 | ||||||
| 1 | 41 (54) | AF268436, AF268437 | Gram-positive low G+C | Clostridium tyrobutiricum | 85 | L08062 |
| AF268438, AF268439 | ||||||
| 2 | 4 (5.2) | AF268434, AF268435 | Gram-positive low G+C | Clostridium tyrobutiricum | 85 | L08062 |
| 3 | 2 (2.6) | AF268440 | Gram-positive low G+C | Clostridium tyrobutiricum | 85 | L08062 |
| 5 | 1 (1.3) | AF268441 | Gram-positive low G+C | Clostridium tyrobutiricum | 85 | L08062 |
| 14 | 1 (1.3) | AF268442 | Gram-positive high G+C | Streptosporangium pseudovulgare | 85 | AF116234 |
| 17 | 1 (1.3) | AF268443 | Gram-positive high G+C | Streptomyces lividans | 89 | U39477 |
| 25 | 1 (1.3) | AF268444 | Gram-positive high G+C | Streptosporangium pseudovulgare | 85 | AF116234 |
| RISA band 2 | ||||||
| 1 | 15 (15) | AF268426 | Beta-proteo | Ralstonia pickettii | 91 | AF012421 |
| 2 | 13 (13) | AF268427 | Beta-proteo | Ralstonia pickettii | 90 | AF012421 |
| 3 | 7 (7) | AF268428 | Gamma-proteo | Coxiella burnetii | 87 | X79704 |
| 4 | 5 (5) | AF268429 | Gamma-proteo | Xanthomonas axonopodis | 89 | AF123089 |
| 5 | 4 (4) | AF268430 | Beta-proteo | Ralstonia pickettii | 85 | AF012421 |
| 6 | 3 (3) | AF268431 | Beta-proteo | Ralstonia solanacearum | 93 | AF012419 |
| 8 | 2 (2) | AF268432 | Gamma-proteo | Pseudomonas stutzeri | 85 | X87289 |
| 9 | 2 (2) | AF268433 | Beta-proteo | Ralstonia pickettii | 90 | AF012421 |
Unambiguously aligned regions were used for the sequence identification by BLAST, i.e., the partial flanked sequence of 130 bp of the 23S rDNA. Beta- and gamma-proteo, β- and γ-proteobacteria, respectively.
RESULTS AND DISCUSSION
Changes in the RISA profiles of the whole community.
Fingerprinting of bacterial community by electrophoretic separation of amplified IGS sequences between the rrs and rrl genes (RISA) allows the quick characterization of a community within various environmental contexts (6, 11, 40). Using this approach, changes in the structure of a bacterial community in soil spiked for 30 days with HgCl2 were noted, but there was no change in a control community (36). These changes were likely to be the consequence of an Hg(II) addition due to the negligible concentration of chloride added in soil and the recognized toxicity of mercury for living cells. Shifts within RISA profiles were due to the emergence of four new bands and the loss of four preexisting bands and to variations in the relative intensity of eleven bands (36). The most noticeable changes were the increase in the intensity of the RISA band 1 at about 350 bp and the appearence of the new RISA band 2 at about 650 bp. These two bands were chosen for restriction analysis and sequencing to examine both the diversity associated with them and the community adaptation through the phylogenetic affiliation of responding bacterial populations.
Sequence variations in a one-size RISA band and phylogenetic affiliation.
Of the 115 and 200 clones sampled from RISA band 1 and RISA band 2, 76 and 99, respectively, had an insert of the expected size. Restriction analysis with AluI, TaqI, and HaeIII resulted in 30 phylotypes for RISA band 1 and 54 phylotypes for RISA band 2. A dominant restriction pattern accounted for 54% of the RISA band 1 inserts analyzed. Restriction groups 2, 3, and 4 were represented by 4, 2, and 2 clones, respectively. The remaining patterns were each represented by a single clone. For RISA band 2, 2 major phylotypes were observed that accounted for 15 and 13% of all of the 99 clones; 7 phylotypes accounted for 2 to 7%, while the remaining 46 phylotypes were each represented by a single clone. Thus, RISA band 1 had a prominent rRNA dominance structure of the first phylotype (54%), while RISA band 2 showed no phylotype frequency greater than 15%. The Shannon diversity index was calculated for the two RISA bands; it was 0.66 for RISA band 1 and 0.87 for band 2. Such diversity in a single-size RISA band was expected, since several authors who have studied variations between and within genera or species (14, 15, 18, 28) have reported spacer length heterogeneity. Similarly, published data (Fig. 1) stresses the extent of IGS length and the large overlap between eubacterial phyla.
FIG. 1.
Length distribution of 428 IGSs between the rrs and rrl genes among the groups of eubacterial domains represented by approximately 99 genera and 332 species. The data were compiled from the literature and the GenBank database. The numbers in parentheses indicate the numbers of genera and species from each phylum. The vertical lines within the boxes indicate the median-size IGS for that phylum. The following genera were used to build the database: α-proteobacteria, Hyphomicrobium, Blastobacter, Rhodobacter, Rhodopseudomonas, Bartonella, Nitrobacter, Azospirillum, Agrobacterium, Rhizobium, Bradyrhizobium, Candidatus, Zymomonas, Gluconobacter, Acetobacter, Ochrobactrum, Brucella, Caulobacter, and Ehrlichia; β-proteobacteria, Acidithiobacillus, Thiobacillus, Ralstonia, Nitrosospira, Nitrosomonas, Burkholderia, Xylophilus, Nitrosolobus, Neisseria, and Microvirgula; γ-proteobacteria, Yersinia, Photorhabdus, Azotobacter, Haemophilus, Enterobacter, Citrobacter, Xanthomonas, Pseudomonas, Acinetobacter, Vibrio, Aeromonas, Klebsiella, Escherichia, Salmonella, Pasteurella, Actinobacillus, Thiobacillus, Dichelobacter, Piscirickettsia, Xylella, Erwinia, and Pectobacterium; ɛ-proteobacteria, Campylobacter; high-G+C-content gram-positive bacteria, Streptomyces, Rhodococcus, Frankia, Arthrobacter, Brevibacterium, Microbispora, Bifidobacterium, Corynebacterium, Staphylococcus, Mycobacterium, Renibacterium, Tropheryma, Thermonospora, Spirillospora, Excellospora, Actinocorallia, and Actinomadura; low-G+C-content gram-positive bacteria, Bacillus, Lactobacillus, Clostridium, Leuconostoc, Streptococcus, Acholeplasma, Anaeroplasma, Listeria, Enterococcus, Mycoplasma, Ureaplasma, Phytoplasma, Lactococcus, Pectinatus, Zymophilus, and Planococcus; chlamydiae, Chlamydia, Chlamydophila, Simkania, Parachlamydia, and Waddlia; cyanobacteria, Microcystis, Spirulina, Trichodesmium, Arthrospira, and Anacystis; spirochetes, Leptonema and Treponema; and cytophagales, Prevotella, Rhodothermus, and Flavobacterium.
Certain representative phylotypes were fully sequenced. Such was the case for four representatives of phylotype 1, two representatives of phylotype 2, and 1 representative each of five minor phylotypes (i.e., phylotypes 3, 5, 14, 17, and 25) for RISA band 1. Similarly, there was one representative for each of the RISA band 2 restriction groups 1, 2, 3, 4, 5, 6, 8, and 9. Alignment of the sequences confirmed the positions of the primers, the IGS, and the 130-bp 23S rDNA sequences. Sequences varied in length from 337 to 359 bp for RISA band 1 and from 634 to 664 bp for RISA band 2. Entire sequences (IGS plus the adjacent sequences) were used for the BLAST search and similarities were found within the partial 23S sequence for RISA band 1 and in both the partial 23S sequence and the IGS (tRNA isoleucine and tRNA alanine) for RISA band 2. The finding of these tRNAs is not surprising since they are present in the E. coli rRNA operons in many other eubacterial genera. Whereas the 23S sequence used to define similarity was short and corresponds to the large-subunit rRNA gene, for which fewer data are available than for the small subunit, this region was informative enough for a rough identification and was reported as being as informative as the 3′ end of the small subunit 16S gene (11). The names and accession numbers of cultured organisms that most closely matched each of the clones in the 23S rRNA gene sequence (calculated by BLAST as the percent similarity), as well as their tentative phylogenetic placements, are given in Table 1. Based on the BLAST analysis of the 23S sequences, similarities were found between RISA band 1 and sequences from low- and high-G+C genera, and sequences from RISA band 2 were found to be similar to β- and γ-proteobacteria, indicating that distantly related genera may have spacers of similar size, as shown in Fig. 1. Alignment of the sequenced inserts showed no differences between the IGS sequences of the clones within a phylotype, but various differences (ranging from 5 to 17) between the phylotypes. In the case of RISA band 1, the sequences obtained from the four representatives of phylotype 1, the two representatives of the phylotype 2, and the representatives of phylotypes 3 and 5 were all identical for the first 130 bp of the 23S gene but differed by from 2 to 16 nucleotides between phylotypes within the IGS sequences. The BLAST search indicated that the Clostridium genus was the most closely related to these sequences. The representatives of the remaining phylotypes were closely related to the Streptomyces (phylotype 17) or Streptosporangium (phylotypes 14 and 25) genera. The sequences of representatives RISA band 2 showed that phylotypes 1, 2, 5, 6, and 9 were close relatives to the genus Ralstonia, whereas the representatives of phylotypes 3, 4, and 8 were close relatives to the genera Coxiella, Xanthomonas, and Pseudomonas, respectively. BLAST comparisons based on the 23S partial sequences were found to be in agreement with the comparison based on the tRNA sequences (data not shown) at the genus level. The great similarity of the IGS regions of phylotypes 1 and 2 of RISA band 1 and phylotypes 1 and 2 of RISA band 2 suggests that these phylotypes belong to different strains of a single species of Clostridium (band 1) or Ralstonia (band 2), respectively.
Hybridization and the role of the identified populations in the adaptation of the whole community to mercury stress.
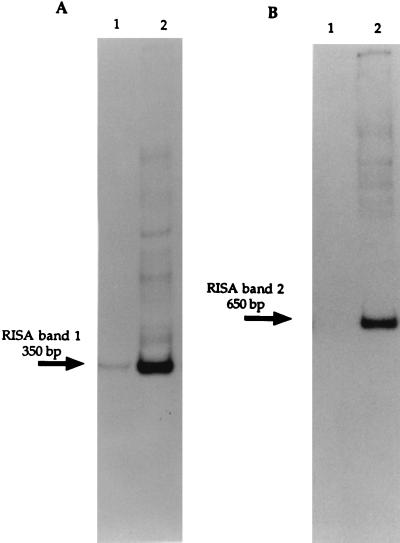
The 23S partial sequences indicated the phylogenetic affiliations of the bacterial population. The IGS, because of its variability, permits an analysis with a finer resolution and is particularly powerful for designing very specific probes (5). Two probes, RISA probe 1 and RISA probe 2, were designed from the IGS of the dominant clone sequences derived from RISA band 1 and RISA band 2. RISA probe 1 was designed to be specific for the IGS sequences similar to Clostridium (phylotypes 1, 2, 3, and 5 of RISA band 1), and RISA probe 2 was intended to be specific for the IGS sequences similar to Ralstonia (phylotypes 1 and 2 of RISA band 2). This specificity was confirmed by hybridization with the various Clostridium-like phylotypes and the five Ralstonia-like phylotypes (data not shown). The RISA profile hybridization with the oligonucleotide RISA probes 1 and 2 yielded a strong positive signal on the profiles of the spiked community at the expected positions of 350 and 650 bp, whereas little or no signal was found with the control community profiles (Fig. 2).
FIG. 2.
Autoradiogram of a Southern blot of RISA profile from soil DNA, hybridized at 58°C with RISA probe 1 (A) and at 53°C with RISA probe 2 (B). Lanes 1, RISA profile of DNA extracted from soil before adding Hg(II); lanes 2, RISA profile of DNA extracted from soil 30 days after adding Hg(II).
These results suggest that Clostridium-like populations and Ralstonia-like populations are enriched during adaptation of the community to Hg(II) (Fig. 2). Whether or not these populations were selected due to their resistance to mercury or were “opportunist” bacteria which benefited from the deleterious effect of mercury on sensitive cells can hardly be answered based on this study alone. However, it has been shown that mercury as an environmental factor induces an increase in mercury-resistant (Hg) phenotypes within the aerobic heterotrophic community (1, 21, 34). Similarly, mer determinants which specifically confer high resistance to mercury are frequently detected in Hg bacteria (32) and are selected within the whole bacterial community in polluted sites or after exposure of pristine environments to mercury in laboratory experiments (2, 3, 29).
In a previous study (34), an analysis of the culturable heterotrophic bacteria showed that mercury causes enrichment of gram-negative cells. The present study using RISA to examine the entire community of culturable and unculturable cells shows that some gram-positive bacterial populations can be stimulated by Hg(II) stress. The resistance of gram-positive bacteria (i.e., Bacillus, Streptomyces, Mycobacterium, Arthrobacter, and Corynebacterium spp.) to mercury has often been reported among isolates originating from sediment, from fresh water or marine environments, and from soils (32, 48), but the individual role of these genera in the whole community (both culturable and nonculturable cells) has never been investigated. Similarly, the role of gram-positive anaerobes in adaptation to mercury has been little studied, since studies usually involve bacterial selection on mercury plates and evaluate the contribution of aerobic genera (30, 31). However, the resistance of these organisms to mercury has been reported (33, 42). The role of Clostridium spp. in community adaptation to stress could also be linked to the high copy number of the ribosomal operon, since bacteria with multiple rRNA operons are favored (more opportunist) under fluctuating growth conditions (22). Clostridium perfringens has 9 rrn operons, and C. acetobutyricum and C. paradoxum have 14 and as many as 15 operons, respectively (12, 13, 39). Whether or not these various copies have similar length IGSs and sequences is not known but, if they do, the apparent enrichment in Clostridium-like populations may be an overestimate due to the contribution of several IGS sequences per cell to the increased intensity of the RISA band. However, further investigations are needed in order to ascertain the role of anaerobes in the community adaptation, since the responding populations were identified from short sequences of the rrl gene whose similarity to Clostridium-like genera was only 85%. There is also a paucity of data on the rrl gene, since less-diverse genera have been sequenced for that region of the rrn operon. The enrichment in anaerobes could then be examined using a culturable method.
Isolates of Ralstonia have been reported to be resistant to both mercury and several heavy metals (10, 24). These isolates have been detected in an industrial anthropogenic biotope heavily polluted with heavy metals (23) and can represent up to 40% of the culturable community in such hostile environments (9). Whether or not Ralstonia is frequently involved in the adaptation of the bacterial community to mercury has not been reported. However, it has to be noticed that strains previously assigned to various species of the Alcaligenes genus have been recently repositioned in the genus Ralstonia (7, 49). Many studies reported on the identification of Hg strains as Alcaligenes (19, 20, 30, 41, 48), but most of them were done prior to the description of the genus Ralstonia and/or when the identification relied on the use of phenotypic characters which may not discriminate between these two close relatives. One can thus assume that the contribution of the genus Ralstonia in adaptation to mercury may have been underestimated.
In this study, we did not look for the enrichment of clones belonging to the γ-proteobacteria by hybridization, but such enrichment could have occurred since heterotrophic bacteria such as Pseudomonas, Xanthomonas, or Enterobacter spp. from mercury-contaminated environments are often selected on mercury plates (4, 21, 32, 43).
Microscale distribution of responsive bacteria.
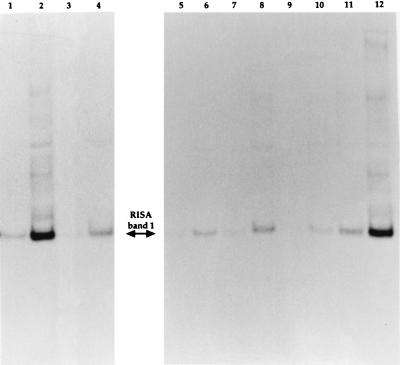
It was shown previously (36) that mercury spiking causes major changes in the RISA profiles of subcommunities associated with the outer and dispersible clay fractions but that there are only minor shifts in those of the macroaggregate and microaggregate fractions. The heterogeneous mercury impact at a microscale level was explained in part by differences in the bioaccessibility and bioavailability of metal to bacteria according to their location. We concluded that all microenvironments did not contribute similarly to the overall community response. In the present study, it was found that the two selected RISA bands could be easily detected on the profiles of the outer and dispersible clay fractions after contamination with Hg(II). Whether or not the major phylotypes of the whole soil community are also involved in the response of the subcommunities was investigated by performing hybridizations with the designed oligonucleotide probes on RISA profiles derived from each microenvironment before and after mercury contamination. Strong positive signals of the expected size were obtained with the subcommunities associated with the outer and dispersible clay fractions, but little or no signal was obtained for other subcommunities after hybridizations with RISA probe 1 (Fig. 3) and RISA probe 2 (data not shown). Little or no signal was detected for the outer and dispersible clay fraction before Hg(II) contamination. These results show the nonuniform distribution of these populations as well as the preferential enrichment in the outer and dispersible clay fractions. Therefore, populations of subcommunities associated with the outer and dispersible clay fractions are more involved in the whole soil community response and adaptation, as previously suggested by multivariate analysis of RISA profiles (36).
FIG. 3.
Autoradiograms of a Southern blot of RISA profiles from soil microenvironments before and after adding Hg(II) hybridized at 58°C with RISA probe 1. Lanes [before and after addition of Hg(II), respectively]: 1 and 2, outer fraction; 3 and 4, 250 to 2,000 μM; 5 and 6, 50 to 250 μM; 7 and 8, 20 to 50 μM; 9 and 10, 2 to 20 μM; 11 and 12, dispersible clay fraction.
Conclusion.
As previously shown (40), our study confirms that RISA is a valuable tool for monitoring the structure and dynamics of complex bacterial communities under stress and that excising, cloning, and sequencing shifted RISA bands can be used to identify the populations involved in the community adaptation. The variations within a single RISA band indicate that this method can be used to examine the diversity in soil communities and to focus on diversity within them, perhaps within even a single species, because of the large variability of the IGS. Responding bacterial populations were found to belong to distantly related genera and to be located in similar microenvironments. Isolating these populations and understanding the way they manage their adaptation to mercury and compete together will be our next challenge.
ACKNOWLEDGMENTS
We thank F. Gourbière for statistical analysis and M. Mergeay and P. Normand for comments on the manuscript.
REFERENCES
- 1.Barkay T, Olson B H. Phenotypic and genotypic adaptation of aerobic heterotrophic sediment bacterial communities to mercury stress. Appl Environ Microbiol. 1986;52:403–406. doi: 10.1128/aem.52.2.403-406.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barkay T, Liebert C, Gillman M. Hybridization of DNA probes with whole-community genome for detection of genes that encode microbial responses to pollutants: mer genes and Hg2+ resistance. Appl Environ Microbiol. 1989;55:1574–1577. doi: 10.1128/aem.55.6.1574-1577.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barkay T, Turner R R, Vandenbrook A, Liebert C. The relationships of Hg(II) volatilization from a freshwater pond to the abundance of mer genes in the gene pool of the indigenous microbial community. Microb Ecol. 1991;21:151–161. doi: 10.1007/BF02539150. [DOI] [PubMed] [Google Scholar]
- 4.Barbieri P, Bestetti G, Reniero D, Galli E. Mercury resistance in aromatic compound degrading Pseudomonas strains. FEMS Microbiol Ecol. 1996;62:375–384. [Google Scholar]
- 5.Barry T, Colleran G, Glennon M, Dunican L K, Gannon F. The 16S/23S ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1991;1:51–56. doi: 10.1101/gr.1.1.51. [DOI] [PubMed] [Google Scholar]
- 6.Borneman J, Triplett E. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coenye T, Falsen E, Vancanneyt M, Hoste B, Govan J R W, Kersters K, Vandamme P. Classification of Alcaligenes faecalis-like isolates from the environment and human clinical samples as Ralstonia gilardii sp. nov. Int J Syst Bacteriol. 1999;49:405–413. doi: 10.1099/00207713-49-2-405. [DOI] [PubMed] [Google Scholar]
- 8.Daffonchio D, Borin S, Frova G, Manachini P L, Sorlini C. PCR fingerprinting of whole genomes: the spacers between the 16S and 23S rRNA genes and of intergenic tRNA gene regions reveal a different intraspecific genomic variability of Bacillus cereus and Bacillus licheniformis. Int J Syst Bacteriol. 1998;48:107–116. doi: 10.1099/00207713-48-1-107. [DOI] [PubMed] [Google Scholar]
- 9.Diels L, Mergeay M. DNA probe-mediated detection of resistant bacteria from soils highly polluted by heavy metals. Appl Environ Microbiol. 1990;56:1485–1491. doi: 10.1128/aem.56.5.1485-1491.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diels L, Faelen M, Mergeay M, Nies D. Mercury transposons from plasmids governing multiple resistance to heavy metals in Alcaligenes eutrophus CH34. Arch Int Physiol Biochim. 1985;93:B27–B28. [Google Scholar]
- 11.Fischer M M, Triplett E. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl Environ Microbiol. 1999;65:4630–4636. doi: 10.1128/aem.65.10.4630-4636.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fogel G B, Collins C R, Li J, Brunk C F. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb Ecol. 1999;38:93–113. doi: 10.1007/s002489900162. [DOI] [PubMed] [Google Scholar]
- 13.Garnier T, Canard B, Cole S T. Cloning, mapping, and molecular characterization of the rRNA operons of Clostridium perfringens. J Bacteriol. 1991;173:5431–5438. doi: 10.1128/jb.173.17.5431-5438.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill S, Belles-Isles J, Brown G, Gagné S, Lemieux C, Mercier J-P. Identification of variability of ribosomal DNA spacer from Pseudomonas soil isolates. Can J Microbiol. 1994;40:541–547. doi: 10.1139/m94-087. [DOI] [PubMed] [Google Scholar]
- 15.Gürtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S–23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff S. Sequence analysis by electronic mail server. Trends Biochem Sci. 1993;18:267–268. doi: 10.1016/0968-0004(93)90179-q. [DOI] [PubMed] [Google Scholar]
- 17.Heuer H, Hartung K, Wielang G, Kramer I, Smalla K. Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl Environ Microbiol. 1999;65:1045–1049. doi: 10.1128/aem.65.3.1045-1049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen M A, Webster J A, Strauss N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jobling M G, Peters S E, Ritchie D A. Restriction patterns and polypeptide homology among plasmid borne mercury resistance determinants. Plasmid. 1988;20:106–112. doi: 10.1016/0147-619x(88)90013-3. [DOI] [PubMed] [Google Scholar]
- 20.Kelly W J, Reanney D C. Mercury resistance among soil bacteria: ecology and transferability of genes encoding resistance. Soil Biol Biochem. 1984;16:1–8. [Google Scholar]
- 21.Khesin R B, Karasyova E V. Mercury-resistant plasmids in bacteria from a mercury and antimony deposit area. Mol Gen Genet. 1984;197:280–285. doi: 10.1007/BF00330974. [DOI] [PubMed] [Google Scholar]
- 22.Klappenbach J A, Dunbar J M, Schmidt T M. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mergeay M, Nies D, Schlegel H G, Gerits J, Charles P, van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985;162:328–344. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mergeay M. Bacteria adapted to industrial biotopes: metal-resistant Ralstonia. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 403–414. [Google Scholar]
- 25.Muyzer G, Teske A, Wirsen C O, Jannash H W. Phylogenetic relationships of Thiomicrospora species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–171. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 26.Muyzer G. Structure, function and dynamics of microbial communities: the molecular biological approach. In: Carvalho G R, editor. Advances in molecular ecology. Amsterdam, The Netherlands: IOS Press; 1998. pp. 87–118. [Google Scholar]
- 27.Muyzer G, Brinhoff T, Nübel U, Santegoeds C, Schafër H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Vol. 3. 1998. pp. 1–27. 4.4. Kluwer Academic Publishers, Dordrecht, The Netherlands. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1–27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, vol. 3.4.4. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 28.Nagpal M L, Fox K F, Fox A. Utility of 16S–23S rRNA spacer region methodology: how similar are interspace regions within a genome and between strains for closely related organisms? J Microbiol Methods. 1998;33:211–219. [Google Scholar]
- 29.Nakamura K, Silver S. Molecular analysis of mercury-resistant Bacillus isolates from sediment of Minamata Bay, Japan. Appl Environ Microbiol. 1994;60:4596–4599. doi: 10.1128/aem.60.12.4596-4599.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson B H, Lester J N, Cayless S M, Ford S. Distribution of mercury resistance determinants in bacterial communities of river sediments. Water Res. 1989;23:1209–1217. [Google Scholar]
- 31.Osborn A M, Bruce K D, Strike P, Ritchie D A. Polymerase chain reaction-restriction fragment length polymorphism analysis shows divergence among mer determinants from gram-negative soil bacteria indistinguishable by DNA-DNA hybridization. Appl Environ Microbiol. 1993;59:4024–4030. doi: 10.1128/aem.59.12.4024-4030.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osborn A M, Bruce K D, Strike P, Ritchie D A. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol Rev. 1997;19:239–262. doi: 10.1111/j.1574-6976.1997.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 33.Pan-Hou H S K, Nobusama H. Role of hydrogen sulfide in mercury resistance determined by plasmid of Clostridium cochlearum T-2. Arch Microbiol. 1981;129:49–52. doi: 10.1007/BF00417179. [DOI] [PubMed] [Google Scholar]
- 34.Ranjard L, Richaume A, Jocteur Monrozier L, Nazaret S. Response of soil bacteria to Hg(II) in relation to soil characteristics and cell location. FEMS Microbiol Ecol. 1997;24:321–331. [Google Scholar]
- 35.Ranjard L, Poly F, Combrisson J, Richaume A, Nazaret S. A single procedure to recover DNA from the surface or inside aggregates and in various size fractions of soil suitable for PCR-based assays of bacterial communities. Eur J Soil Biol. 1998;34:89–97. [Google Scholar]
- 36.Ranjard L, Nazaret S, Gourbière F, Thioulouse J, Linet P, Richaume A. A soil microscale study to reveal the heterogeneity of Hg(II) impact on indigenous bacteria by quantification of adapted phenotypes and analysis of community DNA fingerprints. FEMS Microbiol Ecol. 2000;31:107–115. doi: 10.1111/j.1574-6941.2000.tb00676.x. [DOI] [PubMed] [Google Scholar]
- 37.Ranjard L, Poly F, Combrisson J, Richaume A, Gourbière F, Thioulouse J, Nazaret S. Heterogeneous cell density and genetic structure of bacterial pools associated with various soil microenvironments as determined by enumeration and DNA fingerprinting approach (RISA) Microb Ecol. 2000;39:263–272. [PubMed] [Google Scholar]
- 38.Ranjard L, Poly F, Nazaret S. Monitoring complex bacterial communities using culture-independent molecular techniques: application to soil environment. Res Microbiol. 2000;151:1–11. doi: 10.1016/s0923-2508(00)00136-4. [DOI] [PubMed] [Google Scholar]
- 39.Rainey F A, Ward-Rainey N L, Jansen P H, Hippe H, Stackebrandt E. Clostridium paradoxum DSM 7308T contains multiple 16S rRNA genes with heterogeneous intervening sequences. Microbiology. 1996;142:2087–2095. doi: 10.1099/13500872-142-8-2087. [DOI] [PubMed] [Google Scholar]
- 40.Robleto E A, Borneman J, Triplett E. Effects of bacterial antibiotic production on rhizosphere microbial communities from a culture-independent perspective. Appl Environ Microbiol. 1998;64:5020–5022. doi: 10.1128/aem.64.12.5020-5022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rochelle P A, Wetherbee M K, Olson B H. DNA sequences encoding narrow- and broad-spectrum mercury resistance. Appl Environ Microbiol. 1991;57:1581–1589. doi: 10.1128/aem.57.6.1581-1589.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudrik J T, Bawdon R E, Guss S P. Determination of mercury organomercurial resistance in obligate anaerobic bacteria. Can J Microbiol. 1985;31:276–281. doi: 10.1139/m85-051. [DOI] [PubMed] [Google Scholar]
- 43.Sabaté J, Villaneuva A, Prieto M J. Isolation and characterization of a mercury-resistant broad host-range plasmid from Pseudomonas cepacia. FEMS Microbiol Lett. 1994;119:345–350. [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Scheinert P, Krausse R, Ullman U, Soller R, Krupp G. Molecular differentiation of bacteria by PCR amplification of the 16S–23S rRNA spacer. J Microbiol Methods. 1996;26:103–117. [Google Scholar]
- 46.Shannon C E, Weaver W. The mathematical theory of communication. Urbana, Ill: University of Illinois Press; 1949. [Google Scholar]
- 47.Thompson J D, Higgins D G, Gibson T J. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trajanovska S, Britz M L, Bhave M. Detection of heavy metal ion resistance genes in Gram-positive and Gram-negative bacteria isolated from a lead-contaminated site. Biodegradation. 1997;8:113–124. doi: 10.1023/a:1008212614677. [DOI] [PubMed] [Google Scholar]
- 49.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1986) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]