Abstract
Interactions between plant-associated microorganisms play important roles in suppressing plant diseases and enhancing plant growth and development. While competition between plant-associated bacteria and plant pathogens has long been thought to be an important means of suppressing plant diseases microbiologically, unequivocal evidence supporting such a mechanism has been lacking. We present evidence here that competition for plant-derived unsaturated long-chain fatty acids between the biological control bacterium Enterobacter cloacae and the seed-rotting oomycete, Pythium ultimum, results in disease suppression. Since fatty acids from seeds and roots are required to elicit germination responses of P. ultimum, we generated mutants of E. cloacae to evaluate the role of E. cloacae fatty acid metabolism on the suppression of Pythium sporangium germination and subsequent plant infection. Two mutants of E. cloacae EcCT-501R3, Ec31 (fadB) and EcL1 (fadL), were reduced in β-oxidation and fatty acid uptake, respectively. Both strains failed to metabolize linoleic acid, to inactivate the germination-stimulating activity of cottonseed exudate and linoleic acid, and to suppress Pythium seed rot in cotton seedling bioassays. Subclones containing fadBA or fadL complemented each of these phenotypes in Ec31 and EcL1, respectively. These data provide strong evidence for a competitive exclusion mechanism for the biological control of P. ultimum-incited seed infections by E. cloacae where E. cloacae prevents the germination of P. ultimum sporangia by the efficient metabolism of fatty acid components of seed exudate and thus prevents seed infections.
In recent years there has been considerable interest in exploiting soil microorganisms for the control of fungal and bacterial plant diseases. A substantial body of research has clearly shown that microorganisms introduced onto subterranean plant parts will colonize seeds and roots, where they provide promising levels of biological disease control of seed- and root-infecting fungal plant pathogens (17). However, the mechanisms of disease suppression are unclear, which has made the successful deployment of microorganisms unpredictable under different environmental conditions.
Current models for microbial control of soilborne plant diseases have focused on antibiotic biosynthesis, parasitism, induced systemic resistance, and microbial competition (57). Whereas there is compelling evidence to support those models involving antibiotic biosynthesis (5), parasitism (24), and induced systemic resistance (54), convincing evidence to support a microbial competition model has been difficult to obtain, in part because of our poor understanding of common critical resources for which introduced bacteria and plant pathogens can compete in rhizosphere and spermosphere habitats.
For competitive interactions to occur, both the introduced microbial strain and the plant pathogen must compete in time and/or space for some common resource. In rhizosphere and spermosphere habitats, limiting concentrations of common critical resources shared by both the pathogen and introduced microbial strains are unknown, and resources utilized by the pathogen during interactions with rhizosphere or spermosphere microorganisms have largely been ignored. We have chosen to study a system in which a common resource is known to be utilized by a plant pathogenic oomycete, Pythium ultimum, and the introduced bacterium, Enterobacter cloacae.
Our studies focus on P. ultimum because it is one of the most destructive plant pathogens, causing seed and root diseases of a wide variety of economically important plants (32). Despite their virulence, Pythium spp. are generally considered to be poor microbial competitors in the presence of other plant-associated bacteria (10, 52). To compensate for this, Pythium spp. rely on the production of survival propagules such as oospores and sporangia (49, 51), which germinate rapidly in response to fatty acids present in the exudates of germinating seeds (45). This allows Pythium species to infect seeds at a time when microbial activities, and thus competitive interactions, are low around the surface of the seed.
An approach for reducing this rapid response to exudates and the subsequent infection of plants by Pythium species has been to introduce bacteria on the surface of the seed at the time of planting. One of the more effective bacterial species studied for its Pythium suppressiveness is E. cloacae (32). A unique feature of some E. cloacae strains is that they can rapidly reduce the germination of survival propagules and subsequent seed colonization and infection by P. ultimum (53). This reduction in spore germination is expressed within 2 h after a seed is planted (36) and long before radicle emergence.
Although the mechanisms by which E. cloacae suppresses Pythium seed rot are unknown, we can eliminate mechanisms involving parasitism, antibiosis, or induced systemic resistance. For example, even though E. cloacae may attach to the hyphae of P. ultimum colonizing seed surfaces, no parasitism is known to occur (38). Unlike many well-studied species of Pseudomonas and Bacillus, E. cloacae does not produce antibiotics or other Pythium-inhibitory substances in the presence of seeds or seed exudates (53). Furthermore, suppression of Pythium occurs on the surface of the seed coat long before induced systemic resistance mechanisms are known to be expressed in plants (54). Together, these observations suggest a biological control process mediated by competition between E. cloacae and P. ultimum for specific molecules such as fatty acids released by germinating seeds, which results in the suppression of Pythium propagule germination and subsequent plant infection.
The current study was designed to test the hypothesis that competition for seed exudate fatty acids between E. cloacae and P. ultimum results in the suppression of Pythium seed rot. We focused on linoleic acid as a critical and potentially limiting resource since it is necessary for P. ultimum to break fungistasis and resume active vegetative growth and since it is also the most abundant stimulatory fatty acid found in cottonseed exudates (45). Although little is known about fatty acid metabolism in E. cloacae, much is known about this process in Escherichia coli (1), providing a useful model for understanding fatty acid metabolism in E. cloacae. Although there are a number of fatty acid degradation (fad) genes in E. coli, fadB and fadL genes are among the more important. The gene fadB encodes a protein which, together with the product of fadA, forms the complex responsible for five of the key β-oxidation enzymes (1), whereas fadL encodes an outer membrane protein involved in the binding and transport of fatty acids into the cell (2).
We report here the characterization of the β-oxidation genes fadB and fadL of E. cloacae EcCT-501R3. We provide both genetic and biochemical evidence to support a mechanism for the suppression of a plant disease in which the metabolism of seed exudate fatty acid stimulants of P. ultimum sporangium germination by E. cloacae reduces sporangium germination and subsequent seed infection. We hypothesize that competition for fatty acids in the spermosphere resulting in the suppression of Pythium propagule germination, seed colonization, and seed infection occurs when seed-associated populations of E. cloacae metabolize fatty acids (primarily linoleic acid) released from germinating seeds within this early 12-h window of seed germination.
MATERIALS AND METHODS
Microbial strains, plasmids, and culture conditions.
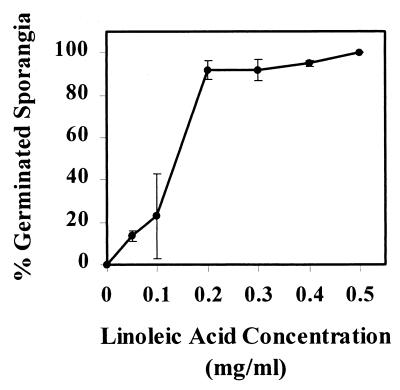
Bacterial strains and plasmids used in this study are described in Table 1. E. cloacae strains were routinely grown in Trypticase soy broth (TSB; Difco Laboratories, Detroit, Mich.) at 27°C, and E. coli strains were grown in Luria-Bertani (LB) medium (33) at 37°C. M9 medium without glucose (33) but supplemented with 25 μg of thiamine HCl/ml is referred to as M9. Sodium linoleate dissolved in Brij58 (Sigma, St. Louis, Mo.) was added to liquid M9 at a concentration of 0.35 mg/ml and to solid media at a concentration of 0.5 mg/ml (M9L). Concentrations of linoleic acid of between 0.2 and 0.5 mg/ml induced 80 to 100% germination of sporangia (Fig. 1).
TABLE 1.
Characteristics of bacterial strain and plasmids used in this study
| Designation | Relevant characteristics | Source or reference |
|---|---|---|
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169 (f80 lacZΔM15) hsdR16 recA1 endA1 gyrA96 thi-1 relA1, Nalr | Life Technologies |
| LS6164 (or D10) | fadR ΔfadL | 14 |
| LS6164Km2 | Tn5tac1 insertion in LS6164, Kmr | This study |
| 34 | ||
| Sm10λpir | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu, Kmr λpir | |
| Enterobacter cloacae | ||
| EcCT-501R3 | Wild type, spontaneous Rifr | 43 |
| Ec31 | fadB::Tn5phoA, Kmr | This study |
| EcL1 | ΔfadL::nptII | This study |
| Plasmids and phage | ||
| pWSK29 | Low-copy-number cloning vector, Ampr | 55 |
| pBCKS− | Cloning vector, Cmr | Stratagene |
| pUC19 | Cloning vector, Ampr | 58 |
| pBS II SK+ | Cloning vector, Ampr | Stratagene |
| PUTKm | Tn5-based delivery plasmid, Kmr Ampr | 20 |
| pN103 (or pACC) | Subclone containing E. coli fadL in pACYC177, Ampr | 2 |
| pRK2073 | A pRK2013 derivative with a TnZ insertion in Kmr gene: mobilizing | 11, 28 |
| pCPP2277 | Promoterless uidA adjacent to nptII lacking a transcriptional terminator in pBS II SK− | D. Bauer, Cornell |
| pKV311 | PCR clone of E. cloacae fadBA in pWSK29, Ampr | This study |
| pKVL1 | fadL on ∼30-kb fragment of EcCT-501R3 genomic DNA on pLAFR3 | This study |
| pKVL16 | PCR clone of E. cloacae fadL in pWSK29, Ampr | This study |
| λ::Tn5tac1 | Tn5 with outwardfacing Ptac | 4 |
FIG. 1.
Dose-response relationships between linoleic acid and P. ultimum sporangium germination. Sporangium germination was determined 3 h after exposure to linoleic acid. The error bars represent the standard deviations.
Glucose and succinate were added to a final concentration of 50 mM. Where appropriate, media were supplemented with kanamycin (Km; 50 μg/ml), ampicillin (Amp; 100 μg/ml), rifampycin, (Rif; 100 μg/ml), tetracycline (Tet, 20 μg/ml), chloramphenicol (Cm, 20 μg/ml), and penicillin G (Pen; 50 μg/ml). In media containing linoleic acid or in cottonseed exudate medium, 0.01% butylated hydroxytoluene (BHT; Sigma) was added to reduce fatty acid oxidation.
P. ultimum isolate P4 (53) was maintained on wheat leaves in 4 ml of sterile water at 21°C (48). Growth conditions and production of sporangia were as described previously (39, 53). For sporangium germination and seedling bioassay experiments, P. ultimum P4 was maintained on solid SM+L medium for 5 days at 24 C. This medium contained the following ingredients per liter: 180 mg of d-glucose, 1.3 mg of l-asparagine, 1 g of soy lecithin (as α-phosphatidylcholine; Sigma catalog number P-5638), 5.3 mg of (NH4)2SO4, 2.4 mg of MgSO4 · 7H2O, 1.1 mg of CaCl2, 3.1 mg of K2HPO4, 1.6 mg of KH2PO4, 1.7 μg of thiamine HCl, and 30 g of agar.
Preparation of cottonseed exudate.
A total of 48 g of cottonseeds (Gossypium hirsutum L. Acala SJ-2) was used for all seed exudate batch preparations. Seeds with no cracks or other visible deformations were surface disinfested for 5 min in ∼100 ml of 0.5% sodium hypochlorite containing 1 or 2 drops of Tween 20 (polyoxyethylene sorbitan monolaurate; Sigma) as a wetting agent. Seeds were filtered through a sterilized strainer and rinsed three times with sterile water. Surface disinfested seeds were added to 400 ml of sterile water (Ca. one seed/milliliter) and incubated at 27°C on a rotary shaker at 120 rpm. After 4 h, flask contents were filtered sequentially through two layers of Whatman no. 1 filter paper using 0.8-, 0.45-, and 0.2-μm (pore-size) filters; evaporated under vacuum at 50°C to a volume of 2 to 5 ml; lyophilized; and stored at −20°C. The specific activity of different exudate batches was determined by preparing a series of exudate dilutions in 10 mM potassium phosphate buffer (pH 6.0) and testing for the germination of P. ultimum sporangia. The minimum exudate concentration (generally 15 to 20 mg/ml) resulting in 90 to 100% sporangium germination was used in all subsequent experiments.
DNA manipulations, primers, DNA sequencing, and sequence data analysis.
Standard procedures were followed for DNA manipulations (46). Oligonucleotide synthesis and DNA sequencing were performed at the Cornell Biotechnology Resource Center. DNA sequence data were managed and analyzed using the DNAStar program (DNAStar, Madison, Wis.).
Mini-Tn5phoA mutagenesis of E. cloacae EcCT-501R3 and isolation of a fadB mutant.
E. cloacae EcCT-501R3 was mutagenized using mini-Tn5phoA as described by de Lorenzo et al. (7). Transconjugants of EcCT-501R3 were tested for growth on linoleic acid as a sole carbon source by replica plating onto solid M9L. Colonies showing little or no growth on M9L were selected and tested for growth on M9 containing 50 mM succinate to ensure that mini-Tn5phoA had not transposed into genes encoding proteins involved in the tricarboxylic or dicarboxylic acid cycles or in oxidative phosphorylation. Transconjugants unable to grow on M9L, but capable of growth on glucose and succinate, were subjected to sequence and complementation analyses to verify their identity as β-oxidation (specifically, fadBA) mutants.
Isolation of an E. cloacae fadL homolog and construction of an E. cloacae fadL mutant.
The presence of a fadL homolog in E. cloacae was confirmed by DNA gel blot analysis by using an internal EcoRV fragment of E. coli fadL as a probe. A pLAFR3 genomic library of E. cloacae EcCT-501R3 (30) was mobilized en masse into an E. coli fadL deletion strain, LS6164Km2, by triparental matings with an E. coli strain harboring pRK2073. E. coli LS6164Km2 was constructed by introducing a Km marker into E. coli LS6164 (FadL− FadR−) by infecting LS6164 with λ∷Tn5tac1 (4) using procedures described by de Bruijn and Lupski (6). Transconjugants of E. coli LS6164Km2 with the E. cloacae pLAFR3 library were selected on LB agar containing Km and Tet and transferred to Hybond N membranes (Amersham Life Sciences, Arlington Heights, Ill.) according to the manufacturer's instructions. Membranes were hybridized with a 2.8-kb EcoRV fragment of pN103 (2) labeled with 32P according to procedures of the Prime-It II random primer kit (Stratagene, La Jolla, Calif.).
One cosmid that hybridized to the E. coli fadL probe, pKVL1, was used to subclone a 12-kb HindIII fragment containing E. cloacae fadL into the low-copy-number vector pWSK29 to produce pKVL2. fadL was disrupted by deletion of an internal 850-kb fragment using unique EcoRV and KpnI sites followed by the insertion of a uidA nptII cassette. This construct was subsequently subcloned into pBCKS− to produce pKLV15. pKVL15 was then used for the homologous recombination of ΔfadL∷nptII into the EcCT-501R3 chromosome using the methods described by Roeder and Collmer (44).
Complementation of E. cloacae fadBA and fadL mutants.
The fadBA and fadL genes from E. cloacae EcCT-501R3 were PCR amplified using standard protocols (23). The primers introduced restriction sites for XbaI and XhoI to the 5′ and 3′ ends, respectively. Primer sequences were as follows: 5′-end fadBA primer, 5′-ATGATCTAGACAGGAGACTGACATGCTC3′; the 3′ fadBA primer, 5′-ATGACTCGAGTTACACTCTCTCAAACAC3′; the 5′ fadL primer, 5′-ATGATCTAGACTCAATGAGGTTATGGTC-3′, and the 3′ fadL primer, 5′-ATGACTCGAGTTAGAACGCGTAGTTGAA-3′. The PCR products were cloned into pWSK29, producing pKV311 and pKVL16 for fadBA and fadL, respectively. pKV311 and pKVL16 were electroporated into E. cloacae strains Ec31 and EcL1, respectively, and tested for their ability to rescue the growth of Ec31 and EcL1 on M9L (16).
Inactivation of cottonseed exudate and linoleic acid germination stimulating activity by E. cloacae strains.
Bacterial cultures were grown overnight in 25 ml of TSB, centrifuged at 5,000 rpm, rinsed, and resuspended in 0.01 M K2HPO4 (pH 6.0) buffer to 109 CFU/ml as determined by an optical density at 600 nm (OD600) of 0.6 for a 1:5 dilution. For growth determinations, 10 μl of the washed cultures were then added to 10 ml of cottonseed exudate (15 mg/ml in 0.01 M K2HPO4 [pH 6.0]). At 3, 6, 9, 12, and 24 h, the OD600 values were determined, and colony counts were obtained by plating aliquots from the cultures onto TSB plates.
For exudate inactivation assays, 0.5 ml of the bacterial suspensions (109 CFU/ml) were placed in 4.5 ml of cottonseed exudate (15 mg/ml in 0.01 M K2HPO4 [pH 6.0]), incubated in a rotary shaker at 27°C at 100 rpm, and 500-μl portions were removed at intervals, filtered, and frozen at −20°C under an argon atmosphere. These filtrates were assessed for germination-stimulating activity as described previously (53).
For linoleic acid inactivation assays, 2 ml of the bacterial suspensions (109 CFU/ml) was added to 18 ml of M9L, and the mixtures were incubated at 27°C on a rotary shaker at 100 rpm. At intervals, 700 μl was removed, filtered, and frozen at −20°C under an argon atmosphere. Filtrates were used in sporangium germination assays to determine the germination stimulating activity remaining in liquid M9L and to quantify by gas chromatography the linoleic acid remaining in these solutions (see below).
To determine whether linoleic acid could restore the germination stimulating activity of cottonseed exudate exposed to E. cloacae for 24 h, linoleic acid was added at a final concentration of 0.35 mg/ml, and the amended exudate solutions were examined for germination-stimulating activity.
Gas chromatographic analysis of linoleic acid in culture filtrates.
Linoleic acid was extracted from culture filtrates as described by Folch et al. (12) and esterified at 55°C overnight in 500 μl of methanol containing 2% H2SO4. Linoleic acid methyl esters were extracted with petroleum ether, dried under a stream of nitrogen, and dissolved in 50 μl of hexane. Then, 0.5-μl injections were made onto a HP5890 gas chromatograph equipped with a 25-m HP-255 GC gas chromatography column (0.2-mm diameter; 0.2-μm film thickness) and a flame ionization detector. The injector temperature was 220°C, and the detector temperature was 230°C. The gas chromatograph was programmed as follows. The initial oven temperature of 180°C was held for 1 min, followed by a 5°C min−1 temperature increase to 220°C, at which point the temperature remained constant for 7 min. The total run time was 17 min. Linoleic acid peak areas and retention times were integrated with a HP3393A integrator. Peak areas were quantified using an external linoleic acid standard. Data were statistically analyzed using pairwise t tests.
Seedling bioassays.
Soil containing inoculum of P. ultimum was prepared by mixing 1,600 cm3 of Arkport sandy loam soil, 500 ml of distilled deionized water, and 400 cm3 of sand that had been sieved to a particle size range of 0.5 to 1 mm. This mixture was amended with two macerated petri plate cultures of P. ultimum (5 days on SM+L medium; macerated with 100 ml of distilled deionized H2O) and planted repeatedly with 40 surface-disinfested cottonseeds until no seedlings emerged. This replant procedure was used to develop natural stable soil populations of P. ultimum. Prior to the bioassay setup, Pythium population levels were determined by plating soil dilutions on a Pythium-selective medium (35). Noninfested soil was amended with infested soil to adjust the Pythium inoculum density to ∼1.2 × 103 CFU/g of soil. Prior to its use in the bioassays, this soil was mixed with 2 volumes of sand. The resulting mixture had a water content of 3.4%.
Bacterial cultures were grown overnight in 30 ml of TSB, centrifuged at 5,000 rpm, rinsed with 0.02 M K2HPO4 (pH 7.0), and suspended in 6 ml of 0.02 M K2HPO4 to a cell density of ∼1011 CFU/ml.
Bioassays were set up essentially as described previously (53). Glass cylinders (2.2 cm high, 2.2 cm in diameter) were placed on sheets of moist blotter paper positioned on a Plexiglas plate. The paper was marked so that the center of the cylinders were 4 cm apart. Pythium-infested soil or noninoculated control soil (3.5 g) was placed in each glass cylinder. One surface-disinfested cottonseed was placed on top of each soil column, drenched with 200 μl of a bacterial cell suspension, covered with 3.5 g of soil, and moistened with 1 ml of K2HPO4 buffer. There were five cylinders per treatment (used to estimate seedling stands), and the experiments were replicated three times. The entire assembly was placed in a clear plastic box in a 24°C growth chamber with a 16-h photoperiod.
Cylinders were watered daily with distilled deionized water and, after 7 days, were rated for disease incidence and severity. Disease severity was rated on a scale of 0 to 3, where 0 was used to indicate no seedling emergence or an emerged seedling that was completely covered with mycelium; 1 indicated that the emerged seedlings failed to develop and had visible mycelial growth; 2 indicated seedlings that appeared healthy but had visible mycelial growth or seedlings that were less vigorous than those in the noninoculated control cylinders; and 3 represented seedlings that were healthy and had no visible signs of mycelial growth. The disease incidence was calculated as the percentage of remaining healthy seedlings (those receiving a rating of 2 or 3) out of the total seeds planted. Seedling stand data were statistically analyzed using pairwise t tests. Disease rating data were analyzed by the nonparametric Mann-Whitney test using pairwise mean comparisons with the aid of Minitab statistical software release 12.1.
Accession numbers.
The entire sequences of fadBA and fadL have been deposited in the GenBank database under accession numbers AF191029 and AF191030, respectively.
RESULTS
An E. cloacae fadBA mutant, Ec31, is unable to grow on linoleic acid as a sole carbon source.
To test our hypothesis about fatty acid competition between E. cloacae and P. ultimum, we built upon four important facts. (i) E. cloacae protects seeds from P. ultimum infection and can reduce the germination stimulating activity of seed exudates to P. ultimum sporangia (53). (ii) Sporangia of P. ultimum require fatty acids for germination (45) at concentrations of ≥0.2 mg/ml. (iii) Seed infection does not occur without the germination of sporangia (37), which occurs within 1.5 h of being exposed to germinating seeds (50). (iv) The first 12 h of seed germination represents the time frame during which competitive interactions must occur since removal of fatty-acid-containing exudates released within this time period greatly reduces or eliminates seed infections of plants subsequently exposed to P. ultimum (15, 18, 38, 40).
We therefore constructed a mutant of E. cloacae EcCT-501R3, Ec31, which failed to grow on linoleic acid but which retained the ability to grow on succinate. We reasoned that strains unable to take up or degrade fatty acids should not limit the availability of seed exudate fatty acids to P. ultimum for stimulation of sporangium germination and subsequent mycelial growth and would thus not reduce seed infection. Southern analysis revealed a single Tn5phoA insertion into the E. cloacae genome. Sequence analysis revealed that the disrupted gene encodes a deduced product of 729 amino acids with 92.7% amino acid identity with E. coli fadB. Immediately downstream of the fadB homolog was a second open reading frame with a deduced product of 365 amino acids and 94.6% identity with E. coli fadA. Similar to E. coli, the E. cloacae fadA and fadB genes are in a two-gene operon, with fadB transcribed first. A PCR clone of fadBA from E. cloacae, pKV311, restored the ability of Ec31 to grow on M9L (data not shown).
E. cloacae Ec31 cannot inactivate the germination-stimulating activity of linoleic acid and has a reduced ability to inactivate the germination-stimulating activity of cottonseed exudate.
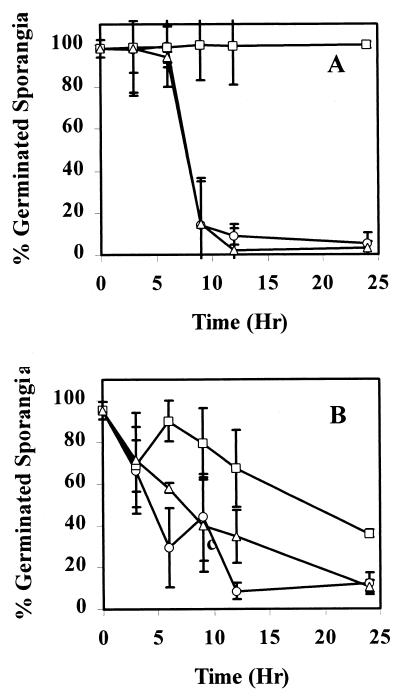
Ec31 was grown in M9L to determine if the fadB mutation affected the ability of Ec31 to inactivate the germination-stimulating activity of this growth medium. Even after 24 h of incubation in M9L, Ec31 failed to reduce the germination-stimulating activity of linoleic acid (culture filtrates induced 80 to 90% sporangium germination) (Fig. 2A). In addition to its inability to inactivate the germination-stimulating activity of linoleic acid, Ec31 also failed to inactivate the activity of other long-chain fatty acids, such as myristoleic, palmitoleic, oleic, and linolenic acids (data not shown). However, culture filtrates from EcCT-501R3 and Ec31(pKV311) reduced the germination-stimulating activity such that, within 9 h of growth, the remaining germination-stimulating activity induced only 15 to 20% sporangium germination.
FIG. 2.
Reduction in germination-stimulating activity of linoleic acid or cottonseed exudate due to growth of E. cloacae strains EcCT501R3 (○), Ec31 (□), and Ec31(pKV311) (▵). Overnight cultures of E. cloacae were adjusted to a cell density of 109 CFU/ml, Then, either 2 ml was placed in M9L medium (A) or 0.5 ml was placed in cottonseed exudate (15 mg/ml) (B). Cell aliquots of cultures at 3, 6, 9, 12, and 24 h were assessed for remaining germination-stimulating activity. The error bars represent the standard deviations. The error bars at some 24-h points are smaller than the marker.
Gas chromatographic analysis of culture filtrates confirmed that Ec31 failed to metabolize linoleic acid. After 12 h of incubation on this medium, 0.32 mg/ml remained in the Ec31 culture, whereas no linoleic acid could be detected in culture filtrates after 12 h of growth of EcCT-501R3 or Ec31(pKV311) (Table 2).
TABLE 2.
Linoleic acid levels in M9L after growth of E. cloacae strains
| E. cloacae strain | Remaining linoleic acid levels (mg/ml)a at:
|
||
|---|---|---|---|
| 0 h | 3 h | 12 h | |
| EcCt-501R3 | 0.35 | 0.14* | 0.00* |
| Ec31 | 0.35 | 0.35 | 0.32 |
| Ec31(pKV311) | 0.35 | 0.03* | 0.00* |
| EcL1 | 0.35 | 0.35 | 0.38 |
| EcL1 (pKVL16) | 0.35 | 0.08* | 0.00* |
Linoleic acid concentrations were determined by gas chromatography after periods of E. cloacae growth in M9 supplemented with 0.35 mg of sodium linoleate per ml. Numbers in each row followed by an asterisk differ significantly (P = 0.05) from the readings at 0 h according to t tests.
Given that the stimulatory activity of cottonseed exudate is due largely to the presence of fatty acids (45), we tested Ec31 further to determine whether the disruption of β-oxidation in E. cloacae had any effect on the bacterium's ability to inactivate the germination-stimulating activity of cottonseed exudate. E. cloacae strains EcCT-501R3, Ec31, and Ec31(pKV311) all grew similarly on cottonseed exudate (data not shown). However, Ec31 could not inactivate the germination-stimulating activity of cottonseed exudate as quickly and to the same level as that of EcCT-501R3 and Ec31(pKV311) (Fig. 2B). In one assay, EcCT-501R3 and Ec31(pKV311) reduced the germination stimulating activity to 5 to 10% within 3 h, while Ec31 reduced the germination stimulating activity to only 35 to 40% after 24 h of growth. When linoleic acid was added to EcCT-501R3 treated exudate to ensure that the inactivation of cottonseed exudate germination-stimulating activity by EcCT-501R3 was not due to the production of sporangium germination inhibitors, the germination-stimulating activity was restored to 100% compared to 1% in response to the EcCT-501R3-treated exudate (data not shown).
E. cloacae EcCT-501R3 has a fadL homolog necessary for linoleic acid metabolism by E. cloacae.
Because of the partial ability of Ec31 to inactivate cottonseed exudate germination-stimulating activity, we constructed an additional E. cloacae mutant with a disrupted fadL gene, which, in E. coli, encodes an outer membrane protein necessary for fatty acid uptake. Of approximately 1,500 cosmids screened, one clone, pKVL1, hybridized strongly to an E. coli fadL probe. Sequencing revealed fadL to be 1,329 bp, with a deduced product of 442 amino acids, and to have 78.0% identity with E. coli fadL. A ΔfadL::nptII construct was marker exchanged into the chromosome of EcCT-501R3, producing mutant EcL1. Marker exchange was confirmed by the Km resistant (Kmr), Cm-sensitive (Cms) phenotype of EcL1, by Southern analysis, and by the failure of EcL1 to grow on linoleic acid as a sole carbon source (data not shown). EcL1 was complemented with pKVL16, which contained the E. cloacae fadL open reading frame, and was restored in its ability to grow on linoleic acid as a sole carbon source (data not shown).
E. cloacae EcL1 cannot inactivate linoleic acid germination-stimulating activity and has a reduced ability to inactivate cottonseed exudate germination-stimulating activity.
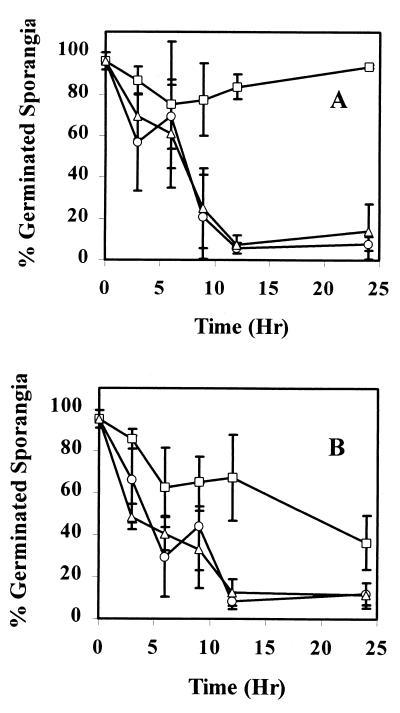
EcL1 possessed stimulant inactivation phenotypes similar to those of Ec31 (Fig. 3). EcL1 M9L culture filtrates induced 80 to 90% sporangium germination even after 24 h of incubation, whereas EcCT-501R3 and EcL1(pKVL16) removed the majority of the linoleic acid germination-stimulating activity within 9 h (15 to 20% sporangium germination) (Fig. 3A). As with Ec31, EcL1 also failed to inactivate the germination-stimulating activity of other selected unsaturated long-chain fatty acids (data not shown). Gas chromatographic analysis of culture filtrates confirmed that EcL1 failed to remove linoleic acid from the growth medium (Table 2). EcL1 was also impaired in its inactivation of cottonseed exudate germination-stimulating activity (Fig. 3B). After 12 h of growth on cottonseed exudate, a germination-stimulating activity of 65% remained in EcL1 culture filtrates, whereas a germination-stimulating activity of 10% remained in culture filtrates of EcCT-501R3 and EcL1(pKVL16).
FIG. 3.
Reduction in germination-stimulating activity of linoleic acid or cottonseed exudate due to growth of E. cloacae strains EcCT501R3 (○), EcL1 (□), and EcL1(pKVL16) (▵). Overnight cultures of E. cloacae were adjusted to a cell density of 109 cfu/ml. Then, either 2 ml was placed in M9L medium (A) or 0.5 ml was placed in cottonseed exudate (15 mg/ml) (B) Cell aliquots of cultures at 3, 6, 9, 12, and 24 h were assessed for remaining germination-stimulating activity. The error bars represent the standard deviations. The error bars at some 24-h points are smaller than the marker.
Neither Ec31 nor EcL1 can protect cotton seeds from P. ultimum seed infections.
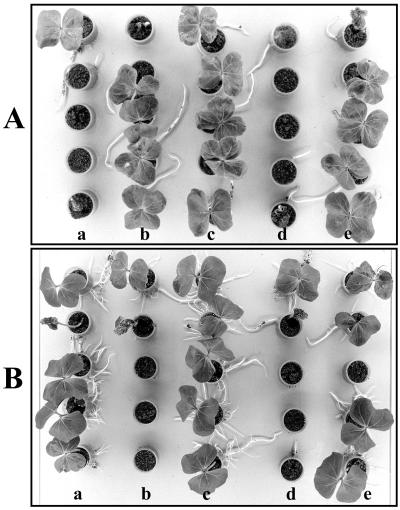
The effect of a fadBA or a fadL mutation on the biocontrol activity of E. cloacae EcCT-510R3 against P. ultimum-incited seed rot was tested in cotton seedling bioassays using a Pythium-infested soil. Both Ec31 and EcL1 failed to suppress Pythium seed rot (Table 3, Fig. 4). EcL1 or Ec31 populations of 6 × 109 per soil column gave rise to 13.3% or 6.7 seedling stands, respectively, and the seedling quality was poor (ratings of 0.8 and 0.4, respectively). Both pKV311 and pKVL16 restored the biocontrol ability of Ec31 or EcL1, respectively, giving rise to ≥80% seedling stands (Table 3, Fig. 4). Seedling stands arising from seeds treated with complemented mutant strains did not differ significantly from those treated with the wild-type strain. Moreover, seedling quality from seeds treated with EcL1(pKVL16) or Ec31(pKV311) did not differ significantly from seedling quality arising from seeds treated with EcCT-501R3.
TABLE 3.
Suppression of Pythium seed rot of cotton by wild-type and fatty acid mutant strains of E. cloacae
| E. cloacae strain | Seedling standa (%) | Seedling qualityb |
|---|---|---|
| Noninoculated Control | 86.7 | 2.5 |
| Nontreated (Pythium only) | 6.7* | 0.8* |
| EcCT-501R3 | 93.3 | 3.0 |
| Ec31 | 6.7* | 0.4* |
| Ec31(pKV311) | 86.7 | 2.6 |
| EcL1 | 13.3* | 0.8* |
| EcL1(pKVL16) | 80.0 | 2.4 |
Seedling stands determined 7 days after sowing. Percent stands are based on five individual seedlings per treatment. The results are the means of three replicate experiments. Values followed by an asterisk differ significantly from the noninoculated control according to simple t-tests.
Disease severity rated as on a scale of 0 to 3, where 0 = no seedling emergence or emerged seedling but completely covered with mycelium; 1 = the emerged seedlings failed to develop and had visible mycelial growth; 2 = seedlings appeared healthy but had visible mycelial growth or seedlings were less vigorous than those in the noninoculated control cylinders; and 3 = seedlings were healthy and had no visible signs of mycelial growth. Values followed by an asterisk differ significantly from the noninoculated control according to the Mann-Whitney test.
FIG. 4.
Cottonseed bioassays illustrating the ability of E. cloacae strains to suppress P. ultimum seed rot. The bioassays were incubated at 24°C with a 16-h photoperiod and were photographed 7 days later. (A) Cotton seeds were placed in P. ultimum-infested soil and drenched with KPO4 buffer (pH 7.0) (a), EcCT-501R3 (c) Ec31 (d), or Ec31(pKV311) (e). A noninoculated control is shown in column b. (B) Cottonseeds were placed in P. ultimum-infested soil and drenched with KPO4 buffer (pH7.0) (b), EcCT-501R3 (c) EcL1 (d), and EcL1(pKVL16) (e). A noninoculated control is shown in column a.
DISCUSSION
Our results provide strong evidence for fatty acid competition in the biological control of Pythium damping-off by E. cloacae. The dependency of P. ultimum developmental processes on fatty acids and other lipids (19, 25, 31, 56), coupled with the ability of E. cloacae to rapidly metabolize fatty acids, provides ideal conditions for competitive interactions between E. cloacae and P. ultimum. The strength of our conclusions comes from evidence from our current and past studies in which we have identified a common fatty acid resource and established its importance to the behavior of P. ultimum (45, 53). Furthermore, we have demonstrated here a mechanism by which E. cloacae can limit the availability of fatty acids to P. ultimum, thus establishing competition in favor of E. cloacae.
Conclusions have been made, based on other biological control studies with root-infecting pathogens, that microbial competition may be involved in the suppression of diseases (42). However, insufficient information is currently available to determine the limiting resources shared between the pathogens and introduced microbial inoculants. Studies with microbial siderophores have provided the only indirect evidence for iron competition in the biological control of some root-infecting pathogens (3, 29). However, the shared resources that may be potentially limiting to the interacting microbial populations have not been defined. Additionally, the iron requirements of the pathogens under study are not known and have only been inferred in a limited number of cases (8, 9, 27, 47). As a result, no solid evidence for iron competition exists, particularly from the perspective of the pathogen.
Our evidence for fatty acid competition between E. cloacae and P. ultimum comes not only from our understanding of the response of P. ultimum to fatty acids but from our characterization of fatty acid uptake and β-oxidation mutants (fadB and fadL) of E. cloacae EcCT-501R3. Based on our analysis of Ec31 and EcL1, the only differences between these strains and the wild type are dysfunctional fadB and fadL genes, respectively. Therefore, differences between the wild-type strain and the mutant strains in biological control efficacy, as well as reductions in germination-stimulating activity in the presence of seed exudates or linoleic acid, can be ascribed to these genes and their respective functions in β-oxidation.
The failure of Ec31 to grow on linoleic acid or reduce its germination-stimulating activity provides indirect evidence that the product of fadB is required for the β-oxidation of fatty acids. This β-oxidation impairment gave rise to several important phenotypes. First and most importantly, Ec31 failed to protect seeds from Pythium infections. Second, Ec31 failed to metabolize and thus inactivate the germination-stimulating activity of linoleic acid over a 24-h period. Third, Ec31 displayed a reduced ability to inactivate the germination-stimulating activity of cottonseed exudate compared to EcCT-501R3. Since the growth of Ec31 on seed exudate was similar to that of EcCT-501R3 over a 24-h period, this response cannot be explained simply by a general growth defect. Furthermore, since suppression of seed infections by P. ultimum must occur during the first 12 to 24 h after planting (15, 38, 40, 41), the inability of Ec31 to protect seeds from Pythium infection was due most likely to its reduced ability to remove fatty acids from the spermosphere during this time period.
Despite the dysfunctional fadB gene and the inability to reduce the germination-stimulating activity of linoleic acid and other unsaturated long-chain fatty acids, Ec31 retained some ability to inactivate the germination stimulating activity of cottonseed exudate. There are a number of possible explanations for this. First, although fatty acids comprise most of the germination-stimulating activity in cottonseed exudate, there are additional lipid exudate fractions that contain low levels of germination-stimulating activity (45). It is therefore possible that either Ec31 can still metabolize these weakly stimulatory molecules and thus slightly reduce cottonseed exudate germination-stimulating activity. Another possibility is that Ec31 can still take up and partially degrade fatty acids, albeit at a much slower rate than EcCT-501R3. Such a process has been described in β-oxidation mutants of E. coli (26).
Because Ec31 retained some capacity to inactivate a significant amount of cottonseed exudate germination-stimulating activity, we constructed a fadL mutant of E. cloacae, EcL1, since fadL is absolutely required for long-chain fatty acid uptake in E. coli. EcL1 shared similar phenotypes with Ec31. Like Ec31, EcL1 was reduced, but not completely impaired, in its ability to inactivate cottonseed exudate germination-stimulating activity. Its growth rate on cottonseed exudate was similar to that of EcCT-501R3 and Ec31 over a 24-h period. This indicates that compounds other than fatty acids in cottonseed exudate may also act as low-level stimulants for Pythium sporangium germination and that the ability of EcL1 and Ec31 to partially reduce the germination-stimulating activity of cottonseed exudate but not that of linoleic acid is due to the metabolism of molecules other than fatty acids with slight germination-stimulating activity. However, the inability to completely reduce the germination-stimulating activity of cottonseed exudate was sufficient to impair the capacity of both E. cloacae EcL1 and Ec31 to suppress Pythium seed infection.
Biological control of fungal pathogens by seed- and root-associated bacteria is undoubtedly a complex process, requiring a number of traits for a bacterium to be successful in suppressing diseases. Despite the importance of fatty acid competition in the suppression of Pythium seed infections by E. cloacae, it is unlikely that this trait alone determines the success or failure of E. cloacae as a seed protectant. Some of the other traits may be related to (i) complex surface phenomena that place bacteria in positions to suppress Pythium infections (21, 38), (ii) to the rapid growth rate around seeds (13), (iii) to the ability to produce antifungal volatiles (22), and (iv) to the ability to compete with other spermosphere organisms. A better understanding of each mechanism in this and other complex biological control systems will provide important insights into the nature of microbial interactions in soil habitats and ways in which such interactions may be exploited for the development of ecologically based plant disease control strategies.
ACKNOWLEDGMENTS
We thank Tom Ruttledge, Steve Winans, and Alan Collmer for helpful discussions; Jim Alfano and Raghida Bukhalid for expert technical advice; Joyce Loper for the pLAFR3 library of EcCT501R3; and Dan Roberts (USDA, ARS, Beltsville, Md.) and Michael Milgroom (Department of Plant Pathology, Cornell University, Ithaca, N.Y.) for helpful comments on the manuscript.
This work was supported, in part, by grants from the U.S. Department of Agriculture National Research Initiative, the U.S. Golf Association, The Cornell Biotechnology Program, and the New York State IPM Program.
REFERENCES
- 1.Black P N, DiRusso C C. Molecular and biochemical analyses of fatty acid transport, metabolism, and gene regulation in Escherichia coli. Biochem Biophys Act. 1994;1210:123–145. doi: 10.1016/0005-2760(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 2.Black P N, Kianian S F, DiRusso C C, Nunn W D. Long-chain fatty acid transport in Escherichia coli: cloning, mapping, and expression of the fadL gene. J Biol Chem. 1985;260:1780–1789. [PubMed] [Google Scholar]
- 3.Buyer J S, Kratzke M G, Sikora L J. Microbial siderophores and rhizosphere ecology. In: Manthey J A, Crowley D E, Luster D G, editors. Biochemistry of metal micronutrients in the rhizosphere. Boca Raton, Fla: CRC Press, Inc.; 1994. pp. 67–80. [Google Scholar]
- 4.Chow W Y, Berg D E. Tn5tac1, a derivative of transposon Tn5 that generates conditional mutations. Proc Natl Acad Sci USA. 1988;85:6468–6472. doi: 10.1073/pnas.85.17.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook R J, Thomashow L S, Weller D M, Fujimoto D, Mazzola M, Bangera G, Kim D. Molecular mechanisms of defense by rhizobacteria against root disease. Proc Natl Acad Sci USA. 1995;92:4197–4201. doi: 10.1073/pnas.92.10.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Bruijn F J, Lupski J R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids—a review. Gene. 1984;27:131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]
- 7.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dujff B J, Bakker P A H M, Schippers B. Suppression of Fusarium wilt of carnation by Pseudomonas putida WCS358 at different levels of disease incidence and iron availability. Biocont Sci Technol. 1994;4:279–288. [Google Scholar]
- 9.Dujff B J, Meijer J W, Bakker P A H M, Schippers B. Siderophore-mediated competition for iron and induced systemic resistance in the suppression of Fusarium wilt of carnation by fluorescent Pseudomonas spp. Netherlands J Plant Pathol. 1993;99:277–289. [Google Scholar]
- 10.Elad Y, Chet I. Possible role of competition for nutrients in biocontrol of Pythium damping-off by bacteria. Phytopathology. 1987;77:190–195. [Google Scholar]
- 11.Figurski D H, Helinski D R. Replication of an origin-containing derivative plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Folch J, Lees M, Sloane-Stanley G H. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 13.Fukui R, Schroth M N, Hendson M, Hancock J G, Firestone M K. Growth patterns and metabolic activity of pseudomonads in sugar beet spermospheres: relationship to pericarp colonization by Pythium ultimum. Phytopathology. 1994;84:1331–1338. [Google Scholar]
- 14.Ginsburgh C L, Black P N, Nunn W D. Transport of long-chain fatty acids in Escherichia coli: identification of a membrane protein associated with the fadL gene. J Biol Chem. 1984;259:8437–8443. [PubMed] [Google Scholar]
- 15.Hadar Y, Harman G E, Taylor A G, Norton J M. Effects of pre-germination of pea and cucumber seeds and of seed treatment with Enterobacter cloacae on rots caused by Pythium spp. Phytopathology. 1983;73:1322–1325. [Google Scholar]
- 16.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 17.Handelsman J, Stabb E V. Biocontrol of soilborne plant pathogens. Plant Cell. 1996;8:1855–1869. doi: 10.1105/tpc.8.10.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harman G E, Taylor A G. Improved seedling performance by integration of biological control agents at favorable pH levels with solid matrix priming. Phytopathology. 1988;78:520–525. [Google Scholar]
- 19.Hendrix J W. Physiology and biochemistry of growth and reproduction in Pythium. Proc Am Phytopathol Soc. 1974;1:207–210. [Google Scholar]
- 20.Herrero M, De Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hood M A, van Dijk K, Nelson E B. Factors affecting colonization of germinating seeds by Enterobacter cloacae. Microb Ecol. 1998;36:101–110. doi: 10.1007/s002489900097. [DOI] [PubMed] [Google Scholar]
- 22.Howell C R, Beier R C, Stipanovic R D. Production of ammonia by Enterobacter cloacae and its possible role in the biological control of Pythium preemergence damping-off by the bacterium. Phytopathology. 1988;78:1075–1078. [Google Scholar]
- 23.Innis M A, Gelfand D H, Sninsky J J, White T J. PCR protocols. San Diego, Calif: Academic Press, Inc.; 1990. [Google Scholar]
- 24.Jeffries P. Biology and ecology of mycoparasitism. Can J Bot. 1995;73:S1284–S1290. [Google Scholar]
- 25.Kerwin J L, Duddles N D. Reassessment of the role of phospholipids in sexual reproduction by sterol-auxotrophic fungi. J Bacteriol. 1989;171:3831–3839. doi: 10.1128/jb.171.7.3831-3839.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein K, Steinberg R, Fiethen B, Overath P. Fatty acid degradation in Escherichia coli: an inducible system for the uptake of fatty acids and further characterization of old mutants. Eur J Biochem. 1971;19:442–450. doi: 10.1111/j.1432-1033.1971.tb01334.x. [DOI] [PubMed] [Google Scholar]
- 27.Lemanceau P, Bakker P A H M, Jandekogel W, Alabouvette C, Schippers B. Antagonistic effect of nonpathogenic Fusarium oxysporum Fo47 and pseudobactin 358 upon pathogenic Fusarium oxysporum f. sp. dianthi. Appl Environ Microbiol. 1993;59:74–82. doi: 10.1128/aem.59.1.74-82.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leong S A, Ditta G S, Helinski D R. Heme biosynthesis in Rhizobium: identification of a cloned gene coding for δ-aminolevulinic acid synthetase from Rhizobium meliloti. J Biol Chem. 1982;257:8724–8730. [PubMed] [Google Scholar]
- 29.Loper J E, Buyer J S. Siderophores in microbial interactions on plant surfaces. Mol Plant-Microbe Interact. 1991;4:5–13. [Google Scholar]
- 30.Loper J E, Ishimaru C A, Carnegie S R, Vanavichit A. Cloning and characterization of aerobactin biosynthesis genes of the biological control agent Enterobacter cloacae. Appl Environ Microbiol. 1993;59:4189–4197. doi: 10.1128/aem.59.12.4189-4197.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lösel D M. Functions of lipids: specialized roles in fungi and algae. In: Ratledge C, Wilkinson S G, editors. Micobial lipids. San Diego, Calif: Academic Press, Inc.; 1989. pp. 367–438. [Google Scholar]
- 32.Martin F N, Loper J E. Soilborne plant diseases caused by Pythium spp.: ecology, epidemiology, and prospects for biological control. Crit Rev Plant Sci. 1999;18:111–181. [Google Scholar]
- 33.Miller J H. Experiments in molecular biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 34.Miller V L, Mekalanos J J. A novel suicide vector and its uses in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mircetich S M. The role of Pythium in feeder roots of diseased and symptomless peach trees and in orchard soils in peach tree decline. Phytopathology. 1971;61:357–360. [Google Scholar]
- 36.Nelson E B. Biological control of Pythium seed rot and preemergence damping-off of cotton with Enterobacter cloacae and Erwinia herbicola applied as seed treatments. Plant Dis. 1988;72:140–142. [Google Scholar]
- 37.Nelson E B. Exudate molecules initiating fungal responses to seeds and roots. Plant Soil. 1990;129:61–73. [Google Scholar]
- 38.Nelson E B, Chao W L, Norton J M, Nash G T, Harman G E. Attachment of Enterobacter cloacae to hyphae of Pythium ultimum: possible role in the biological control of Pythium preemergence damping-off. Phytopathology. 1986;76:327–335. [Google Scholar]
- 39.Nelson E B, Hsu J S T. Nutritional factors affecting responses of sporangia of Pythium ultimum to germination stimulants. Phytopathology. 1994;84:677–683. [Google Scholar]
- 40.Osburn R M, Schroth M N. Effect of osmopriming sugar beet seed on exudation and subsequent damping-off caused by Pythium ultimum. Phytopathology. 1988;78:1246–1250. [Google Scholar]
- 41.Osburn R M, Schroth M N. Effect of osmopriming sugar beet seed on germination rate and incidence of Pythium ultimum damping-off. Plant Dis. 1989;73:21–24. [Google Scholar]
- 42.Paulitz T C. Biochemical and ecological aspects of competition in biological control. In: Baker R R, Dunn P E, editors. New directions in biological control: alternatives for suppressing agricultural pests and diseases. New York, N.Y: Alan R. Liss, Inc.; 1990. pp. 713–724. [Google Scholar]
- 43.Roberts D P, Sheets C J, Hartung J S. Evidence for proliferation of Enterobacter cloacae on carbohydrates in cucumber and pea spermosphere. Can J Microbiol. 1992;38:1128–1134. [Google Scholar]
- 44.Roeder D L, Collmer A. Marker-exchange mutagenesis of a pectate lyase isozyme gene in Erwinia chrysanthemi. J Bacteriol. 1985;184:51–56. doi: 10.1128/jb.164.1.51-56.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruttledge T R, Nelson E B. Extracted fatty acids from Gossypium hirsutum stimulatory to the seed-rotting fungus, Pythium ultimum. Phytochemistry. 1997;46:77–82. [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Simeoni L A, Lindsay W L, Baker R. Critical iron level associated with biological control of Fusarium wilt. Phytopathology. 1987;77:1057–1061. [Google Scholar]
- 48.Singleton L L. Storage of Pythium species on a wheat leaf: water medium. Phytopathology. 1986;76:1143. [Google Scholar]
- 49.Stanghellini M E. Spore germination, growth, and survival of Pythium in soil. Proc Am Phytopathol Soc. 1974;1:211–214. [Google Scholar]
- 50.Stanghellini M E, Hancock J G. Radial extent of the bean spermosphere and its relation to the behavior of Pythium ultimum. Phytopathology. 1971;61:165–168. [Google Scholar]
- 51.Stanghellini M E, Hancock J G. The sporangium of Pythium ultimum as a survival structure in soil. Phytopathology. 1971;61:157–164. [Google Scholar]
- 52.Tedla T, Stanghellini M E. Bacterial population dynamics and interactions with Pythium aphanidermatum in intact rhizosphere soil. Phytopathology. 1992;82:652–656. [Google Scholar]
- 53.van Dijk K, Nelson E B. Inactivation of seed exudate stimulants of Pythium ultimum sporangium germination by biocontrol strains of Enterobacter cloacae and other seed-associated bacteria. Soil Biol Biochem. 1998;30:183–192. [Google Scholar]
- 54.van Loon L C, Bakker P A H M, Pieterse C M J. Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol. 1998;36:453–483. doi: 10.1146/annurev.phyto.36.1.453. [DOI] [PubMed] [Google Scholar]
- 55.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 56.Weete J D. Lipid biochemistry of fungi and other organisms. New York, N.Y: Plenum Press; 1980. [Google Scholar]
- 57.Whipps J M. Developments in the biological control of soil-borne plant pathogens. Adv Bot Res. 1997;26:1–134. [Google Scholar]
- 58.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]