Abstract
Background
The registration of computed tomography (CT) and cone-beam computed tomography (CBCT) plays a key role in image-guided radiotherapy (IGRT). However, the large intensity variation between CT and CBCT images limits the registration performance and its clinical application in IGRT. In this study, a learning-based unsupervised approach was developed to address this issue and accurately register CT and CBCT images by predicting the deformation field.
Methods
A dual attention module was used to handle the large intensity variation between CT and CBCT images. Specifically, a scale-aware position attention block (SP-BLOCK) and a scale-aware channel attention block (SC-BLOCK) were employed to integrate contextual information from the image space and channel dimensions. The SP-BLOCK enhances the correlation of similar features by weighting and aggregating multi-scale features at different positions, while the SC-BLOCK handles the multiple features of all channels to selectively emphasize dependencies between channel maps.
Results
The proposed method was compared with existing mainstream methods on the 4D-LUNG data set. Compared to other mainstream methods, it achieved the highest structural similarity (SSIM) and dice similarity coefficient (DICE) scores of 86.34% and 89.74%, respectively, and the lowest target registration error (TRE) of 2.07 mm.
Conclusions
The proposed method can register CT and CBCT images with high accuracy without the needs of manual labeling. It provides an effective way for high-accuracy patient positioning and target localization in IGRT.
Keywords: Image registration, computed tomography (CT), cone-beam computed tomography (CBCT), image-guided radiotherapy (IGRT), deep-learning, neural network
Introduction
In clinical practice, physicists usually make treatment plans based on computed tomography (CT) images, and then verify patient positions in the treatment room based on cone-beam computed tomography (CBCT) images. Due to positioning errors and anatomy changes, there are certain offsets between the real position and the planned position of target in treatment room. Therefore, registration between CT and CBCT images is important that it can provide the offset of treatment site when patient is on the treatment couch, which is crucial for the success of modern radiotherapy.
Traditional registration methods usually employed iterative optimization algorithms to search for the best displacement vector (1). These include elastic models (2,3), deformable registration via attribute matching and mutual-saliency weighting (DRAMMS) (4), statistical parametric mapping (SPM) (5), Demons algorithm (6), standard symmetric normalization (SyN) (2,7), large diffeomorphic distance metric mapping (LDDMM) (8), diffeomorphic registration using B-splines (9). These algorithms are mostly time-consuming due to the large amount of iterations in data manipulation.
Recently several deep-learning-based deformable image registration methods were proposed (10). They roughly fall into three categories, i.e., supervised registration, semi-supervised registration, and unsupervised registration. Supervised registration requires the deformation field generated by traditional algorithms as a teaching signal (11-13), or the images transformed by the deformation field as training samples (14). The semi-supervised registration method deals with the problem of the insufficient number of medical image labels and uses part of the data as the supervising information (15). In order to make the best use of the unlabeled image data, several unsupervised registration networks have been proposed. In (16), a method based on Generative Adversarial Network (GAN) is developed, which replace the similarity function with a discriminator. Compared with GAN, U-Net-like architecture is easier to converge with fewer data. For example, VoxelMorph (17) and VTN (18) have achieved excellent results in medical image registration. These unsupervised registration networks are similar to U-Net. However, due to the inherent locality of convolutional operations, the architecture of convolutional networks is often limited in modeling explicit long-range spatial relationships present in images (19).
Although U-Net-like frameworks overcome this limitation, the influence from distant voxels decays rapidly as the number of convolutional layers deepens (20). There is a large voxel difference between CT and CBCT, so we expect the convolutional network to see long-range relationships between voxels in the two images. Therefore, some researchers hope to improve this problem through attention mechanism. For multi-modal image registration tasks, the required accuracy is hardly met due to the large variation of image intensity. Therefore, several researchers introduced the attention mechanism to assist convolution in the cross-modal registration problem. The cross-attention mechanism was used to realize the information exchange between multi-modal images (21,22). The convolutional block attention module (CBAM) was integrated into VTN (23). The spatial attention was introduced to further enhance the prominent areas of the feature map (24). And the binary spatial attention module (BSAM) was introduced to the spatial information extraction in the jump connection (25). Compared with the conventional deep learning network, the network with attention mechanism achieved better results.
In this study, an unsupervised dual attention network (UDAN) is proposed for CT-CBCT image registration. The U-Net-like encoder and decoder are used to extract and fuse features. Besides, a dual attention module (DAM) is placed between the encoder and decoder. DAM mainly consists of two parts: a scale-aware position attention block (SP-BLOCK) and a scale-aware channel attention block (SC-BLOCK). Dilated convolution and residual structure are first used to obtain multi-scale position and multi-scale channel features, respectively. Then these features are weighted using a non-linear combination to weaken the influence of intensity differences between the two image modalities.
This paper is organized as follows. In section Methods, details of the proposed unsupervised dual attention registration network are introduced. In section Results, performance of our method and the other six methods, i.e., ANTs (7), ELASTIX (26), B-spline (9), VTN (18), VoxelMorph (17), and CycleMorph (27) are summarized. In addition, the ablation studies of the proposed method are also reported. In section Discussion, the advantages and disadvantages of the proposed method are discussed. We present the following article in accordance with the MDAR checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-21-1194/rc).
Methods
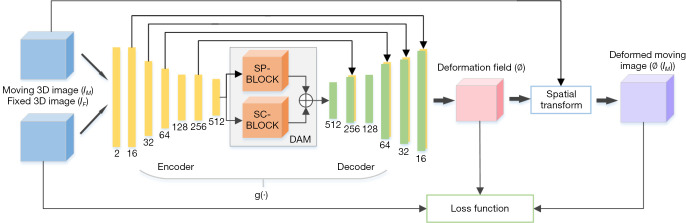
The overall network structure is shown in Figure 1. First, the CT and CBCT images are passed to the encoder, DAM, and decoder. Then, the deformation field is generated and the input CT image is warped using the spatial transformation function. Next, the similarity between the transformed CT image and the CBCT image is calculated as the loss function. Finally, the similarity and regularization terms of the deformation field are back-propagated to update the network parameters.
Figure 1.
Overview of the registration network. DAM, dual attention module; SP-BLOCK, scale-aware position attention block; SC-BLOCK, scale-aware channel attention block; g(·), the network for computing the deformation field ∅, including Encoder and Decoder.
Assuming there are a moving image IM and a fixed image IF. In a d-dimensional space Ω, the goal of deformable image registration is to find optimal parameters ϕ to align IM and IF. For IM and IF, ϕ can be obtained via a network g(·),
| [1] |
As shown in Figure 1, the input of the network is the concatenation of IM and IF, the output of network is the deformation field ϕ in dimension of 3×48×512×512. In the encoder, six consecutive convolution layers with kernels size of 3×3 are used to extract different levels of information, and an activation function ReLU is used after each convolutional layer. The dimension of the encoder output is 512×3×32×32. Then these outputs are sent to DAM for attention calculation. In the decoder, the convolution layer with kernels size of 3×3 followed by ReLU is used again to process feature mapping. The feature maps of each layer of the encoder are spliced into the feature maps of the symmetrical layer in the encoder.
Dual attention module
DAM is introduced to adaptively utilize images’ high-dimensional features with different scales. To better integrate the features obtained by DAM and improve efficiency, DAM is placed between the encoder and the decoder. DAM consists of two parts, SP-BLOCK and SC-BLOCK. A detailed introduction of these two parts is given below.
SP-BLOCK
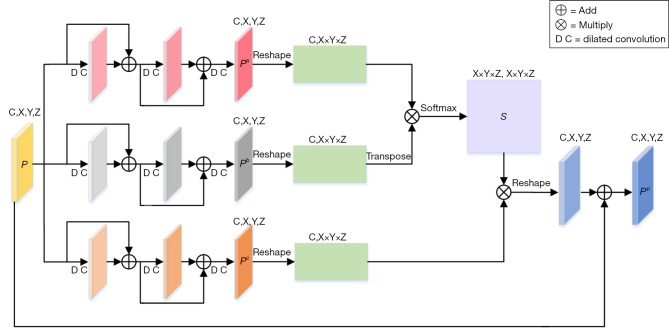
The SP-BLOCK enhances the correlation of similar features by weighting and aggregating multi-scale features at different positions. The structural details of SP-BLOCK are shown in Figure 2. Given a feature map , inspired by the success of the residual structure (28), three discriminative consecutive residual blocks are used to obtain Pa, Pb and Pc with multi-scale position information. The sizes of C, X, Y, and Z are 512, 3, 32, and 32, respectively. Moreover, in addition to the necessary layer jump connections in each residual block, dilated convolution (29) with different dilation rates is added. The dilation rates in the three-layer residual blocks are 1, 2 and 3, respectively. This structure can expand the receptive field without introducing additional parameters, obtain information of different scales, and enhance the network’s ability to perceive different scales. Then Pa, Pb and Pc are reshaped to (C,X×Y×Z). Given Pa and Pb, a position attention map, , is obtained via a Softmax operation,
Figure 2.
The detailed architecture of SP-BLOCK. DC, dilated convolution; SP-BLOCK, scale-aware position attention block.
| [2] |
where sji measures the correlation from ith position to jth position. Next, the matrix product of Pc and S is made, and reshape the result to (C,X,Y,Z). Finally, the output of SP-BLOCK is obtained,
| [3] |
where α is a scale parameter. It is initially set to 0 and gradually increased. As shown in Eq. [3], for each position, a weighted sum of the features from all positions, i.e., the multi-scale global context, is selectively integrated into the multi-scale features.
SC-BLOCK
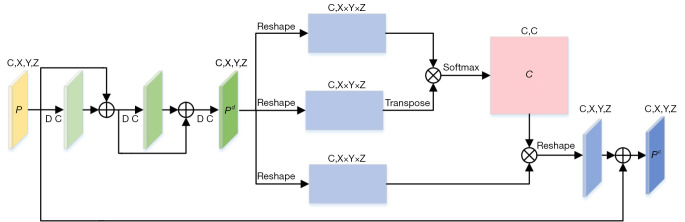
The SC-BLOCK acquires multi-scale channel features, then weights these channel features to selectively enhance the correlation between different channels. The structural details of SC-BLOCK are shown in Figure 3. Unlike the SP-BLOCK, the consecutive residual block is executed only once for P because the channel attention took the dependence between channels into account. We denote the output of the consecutive residual block as Pd, which is reshaped into (C,X×Y×X). The channel attention map is obtained by performing matrix multiplication between Pd and the transpose of Pd,
Figure 3.
The detailed architecture of SC-BLOCK. DC, dilated convolution; SC-BLOCK, scale-aware channel attention block.
| [4] |
where cji measures the correlation from ith channel to jth channel. After that, the matrix product of the transformed Pd and C is obtained in the dimension of (C,X×Y×X). Finally, the output of SC-BLOCK is calculated,
| [5] |
where β is a scale parameter. Eq. [5] shows that each channel’s features are the weighted sum of the global channel features and the original input of the multi-scale features. It allows the network to integrate the features of different channels with consideration of the structural relationship among channels.
Spatial transformation function
The spatial transformation function is used to warp IM to ϕ(IM) (30), which allows the evaluation of the similarity between ϕ(IM) and IF. Denoting the voxel of IM as p, the voxel ϕ(p) after transformation can be achieved. Since image values are only defined at integer positions, linear interpolation is performed based on eight neighboring voxels:
| [6] |
where N(ϕ(p)) represent the neighbors of ϕ(p).
Loss function
The loss function consists of two parts: Lsim, which penalizes the difference between a fixed image and a moving image after the deformation field, and Lsmooth, which penalizes the local spatial variation of the deformation field. Lsim is the normalized cross-correlation (NCC) (17), and its definition is:
| [7] |
where and are normalized images with mean intensities removed, and ϕ(IM) is the image transformed by the deformation field. The regularization term is used to punish the excessive deformation and the unsmooth deformation field, the definition is:
| [8] |
The overall loss function is:
| [9] |
where λ represents the regularization term coefficient.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Ethics Committee of The Affiliated Cancer Hospital of Zhengzhou University, Henan Cancer Hospital. And the informed consent was waived in this retrospective study.
Experiments data
The 4D-lung dataset was used for the test (31,32). This data set includes four-dimensional fan beam CT (4D-FBCT) and cone-beam CT (4D-CBCT), which were obtained during chemotherapy of 20 patients with locally advanced non-small-cell lung cancer during radiotherapy. The planned CT image was obtained using a 16-slice spiral CT scanner (Brilliance Big Bore, Philips Medical Systems), with a slice thickness of 3 mm and an axial resolution of 512×512 (in-plane spacing approximately 1 mm). The CBCT image was obtained on a commercial CBCT system (On-Board ImagerTM, Varian Medical Systems, Inc.), and the size was 512×512×50. Two operations were performed before CT and CBCT image registration:
Pair the data. All images were resampled to the same pixel spacing and slice thickness. The slice thickness was 2.34 mm and pixel spacing was 1.16 mm. The field of view (FOV) of the CT irradiation area was larger than that of CBCT, the FOV of CT was cropped to match the FOV of CBCT.
Align the data (18). The lung in both CT and CBCT images were aligned. Rigid registration was performed using ANTs (7) to reduce the excessive deformation.
Experiment settings
To evaluate the effectiveness of UDAN, two experiments were performed (The code of the network structure and the pre-trained model can be found in https://github.com/Eric-Hu88/UDAN). The first experiment was to compare our method with three traditional widely-used deformable registration algorithms [ANTs (7), ELASTIX (26), B-Spline non-rigid registration (9)], and three deep-learning-base deformable registration algorithms [VoxelMorph (17), VTN (18) and CycleMorph (27)]. In the second experiment, the effectiveness of SC-BLOCK and SP-BLOCK was investigated by six variants of UDAN:
Baseline: UDAN’s backbone network without dual attention module;
Baseline with P-BLOCK: UDAN’s backbone network with position attention block (P-BLOCK) (33);
Baseline with C-BLOCK: UDAN’s backbone network with channel attention block (C-BLOCK) (33);
Baseline with SP-BLOCK: UDAN’s backbone network with SP-BLOCK;
Baseline with SC-BLOCK: UDAN’s backbone network with SC-BLOCK;
UDAN: The proposed registration network.
P-BLOCK and C-BLOCK were simplified versions of SP-BLOCK and SC-BLOCK that did not use dilated convolution and residual structure.
Implementation details
CT and CBCT images of total 20 patients were used. Due to the limited number of patients in the dataset, directly using a portion of the data as a validation set will reduce the generalization of the model. In the process of cross-validation, 50% patients were used for training and 25% patients were used for validation to tune hyper-parameters, 50% of patients for training and 25% of patients for validation were synthesized into the final training set, then trained the model by the proposed network with pre-tuned hyper-parameters and the remaining 25% of patients were used for testing. The total amount of data was 86 pairs of CT and CBCT images. The number of slices per one set of CT and CBCT images was 48. The network was trained for 1,000 epochs. ADAM optimizer with the initial learning rate of 4×10−4 was used and multiplied by 0.5 after 400 epochs. Regularization parameter λ was set to 0.9. The networks were trained and tested on a computer cluster equipped with NVIDIA Tesla V100 GPU with 32 GB of memory.
Evaluation metric
Following previous study (17,18), five metrics, dice similarity coefficient (DICE), target registration error (TRE), structural similarity index (SSIM), TIME, and negative Jacobian percentage were used to evaluate the registration performance. They are:
Dice similarity coefficient (DICE): experienced radiologists manually delineated the position of tumor in the data and calculated the DICE coefficient of the tumor before and after registration.
Target registration error (TRE) of anatomical: for each pair of CT and CBCT data, a professional radiologist manually marked around 23 significant anatomical landmarks, which were distributed throughout the lungs.
Structural similarity index (SSIM): the structural similarity between the registered image and the fixed image.
TIME: the time spent on registering a pair of CT and CBCT images.
Negative Jacobian percentage |Jϕ|≤0(%): in the deformation field generated by the network, areas with negative Jacobian determinants were considered folding, and the percentage of the area to the whole was the negative Jacobian percentage (34).
Results
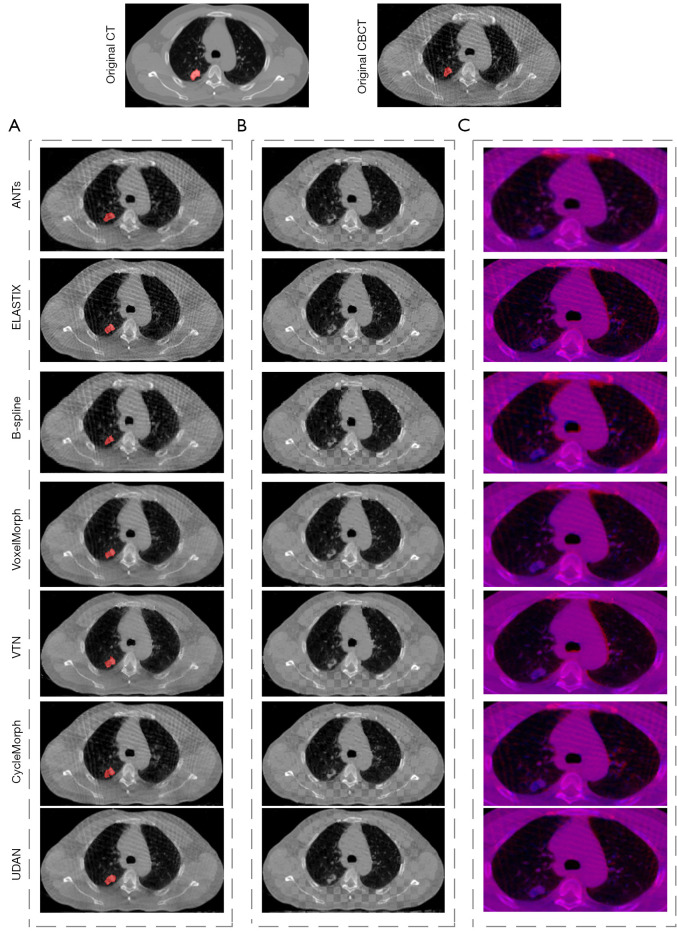
Table 1 showed the results of performance comparison between our method and the six existing methods. Our method achieved 2.07 mm on TRE, 86.34% on SSIM, and 89.74% on DICE. Compared to the six existing methods, our method obtained the best performance on four metrics. Figure 4A showed the visual comparison between our method and the six existing methods, red regions indicated the warped tumor labels in CBCT after registration, it can be seen that the proposed UDAN yields the tumor label that was closer to the CT tumor label. The checkerboard overlaps between the CT image and the CBCT image were provided in Figure 4B. Apparently, the proposed UDAN reduced the dislocation of organ boundaries and bone structure compared to the other six existing methods. Also, the registered image pairs were fused and presented in Figure 4C. The R channel was the CT image and presented in red color, while the B channel was the deformed CBCT image and presented in blue color. When the two images match perfectly, a purple image was displayed. In the example shown in Figure 4C, the results of ANTs (7), Elastix (26), and B-splines (9) displayed several red or blue areas. Compared with traditional algorithms, the VoxelMorph (17), VTN (18), and CycleMorph (27) algorithms based on deep learning achieved excellent results. However, our method was more competitive for the details of the parts where artifacts exist. CycleMorph maintained a low percentage of negative Jacobian using cyclic consistency, but did not show better performance in the other metrics. Paired t-tests indicated that the decrease of TRE, the increases of DICE and SSIM in our method were statistically significant (P<0.05) compared to those of the other methods tested. Although the B-Spline algorithm achieved the lowest negative Jacobian percentage, it was worse in the other metrics.
Table 1. Performance comparison of the proposed UDAN with six mainstream deformable image registration methods.
| Method | TRE (mm) ↓ | SSIM (%) ↑ | DICE (%) ↑ | TIME (s) ↓ | |Jϕ|≤ 0(%) ↓ |
|---|---|---|---|---|---|
| Affine | 4.27 | 68.17 | 64.38 | – | – |
| ANTs | 2.32 | 74.43 | 79.27 | 1123 | 0.16 |
| ELASTIX | 2.45 | 73.56 | 75.24 | 265 | 0.09 |
| B-spline | 3.27 | 69.49 | 70.49 | 943 | 0.06 |
| VTN | 2.12 | 85.48 | 88.61 | 16 | 0.12 |
| VoxelMorph | 2.17 | 85.34 | 87.94 | 14 | 0.34 |
| CycleMorph | 2.14 | 85.73 | 87.81 | 23 | 0.16 |
| UDAN | 2.07 | 86.34 | 89.74 | 18 | 0.28 |
The best results are shown bold font. The line of affine is the result of rigid registration before the application of deformable registration. UDAN, unsupervised dual attention network; TRE, target registration error; SSIM, structural similarity; DICE, dice similarity coefficient; |Jϕ|≤ 0(%), areas with negative Jacobian determinant are considered folding, the folded area as a percentage of the total area.
Figure 4.
The comparison of registration results between our method and the other mainstream methods. (A) The registered images, where the red area is the CBCT tumor segmentation label deformed by the deformation field; (B) the checkerboard display of registered images; (C) the red and blue channel overlay of the registered images, zoomed in on the lungs and parts of the surrounding area. CT, computed tomography; CBCT, cone-beam computed tomography.
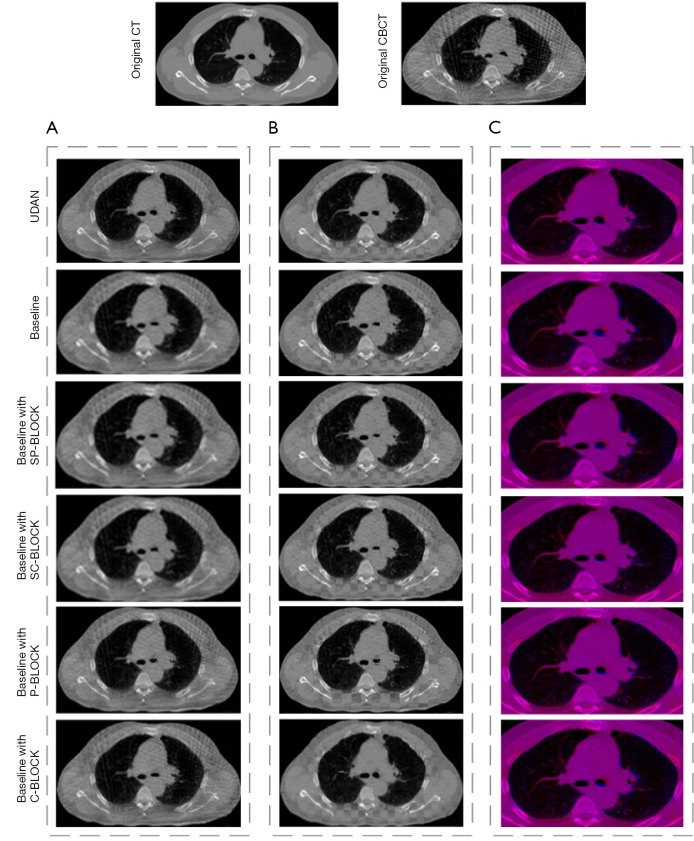
The effect of several important elements in our framework was investigated through a set of ablation experiments. Table 2 listed the results of the comparison between the proposed method and its five variants. The best scores were shown in boldface. When using the proposed UDAN, TRE, SSIM, and DICE reached the highest level. TIME increased but was close to the average. As shown in Figure 5, with the application of the DAM, the dislocation of organ boundaries and bone structure was reduced in Figure 5B, and red and blue areas were less in Figure 5C. In addition, we also performed paired t-tests for different variants. The results showed that when the complete DAM module was added to the baseline network, the TRE, DICE, and SSIM indicators achieved the best results with statistical significance. The negative Jacobiavn percentage indicator was not the best, but its value was second to the best.
Table 2. Performance comparison of the proposed UDAN with its six variations.
| Method | TRE (mm) ↓ | SSIM (%) ↑ | DICE (%) ↑ | TIME (s) ↓ | |Jϕ|≤ 0(%) ↓ |
|---|---|---|---|---|---|
| Baseline | 2.25 | 83.92 | 86.54 | 14 | 0.34 |
| Baseline with P-BLOCK | 2.21 | 84.27 | 87.15 | 17 | 0.29 |
| Baseline with C-BLOCK | 2.09 | 84.72 | 87.56 | 16 | 0.31 |
| Baseline with SP-BLOCK | 2.13 | 85.13 | 87.43 | 16 | 0.25 |
| Baseline with SC-BLOCK | 2.11 | 85.08 | 88.57 | 17 | 0.33 |
| UDAN | 2.07 | 86.34 | 89.74 | 18 | 0.28 |
The best results are shown bold font. TRE, target registration error; SSIM, structural similarity; DICE, dice similarity coefficient; |Jϕ|≤ 0(%), areas with negative Jacobian determinant are considered folding, the folded area as a percentage of the total area; P-BLOCK, position attention block; SP-BLOCK, scale-aware position attention block; SC-BLOCK, scale-aware channel attention block; UDAN, unsupervised dual attention network.
Figure 5.
The comparison of registration results among UDAN variants. (A) The registered images; (B) the checkerboard display of registered images; (C) the red and blue channel overlay of the registered images, zoomed in on the lungs and parts of the surrounding area. CT, computed tomography; CBCT, cone-beam computed tomography; UDAN, unsupervised dual attention network.
Discussion
In this paper, we have proposed an unsupervised registration network with a dual attention module. Compared with the other six existing methods, the slice junction of our method was relatively smooth, and there were fewer dislocation boundaries. While presented in the overlay of the red and blue channels, the proposed method displayed more purple areas than the other six existing methods. In summary, our method provided high accuracy in registering CT and CBCT images in the tested cases.
With the introduction of SC-BLOCK and SP-BLOCK, the registration accuracy is improved apparently. SP-BLOCK and SC-BLOCK are the enhanced versions of P-BLOCK and C-BLOCK that use dilated convolution and residual structure. By using dilated convolutions with different dilation rates, the high-dimension feature receptive field is gradually increased and the search space of feature matching is reduced. This can effectively alleviate the problem of inaccurate feature matching caused by the different voxel intensities between CT and CBCT images. When all blocks are added to the network, the registration accuracy reaches the best value.
High-dimension features usually contain rich detailed information. For the complex high-dimension features, DAM is effective to process them. DAM not only takes both low-dimension features and high-dimension detailed information into account but also processes the semantic information of the high-level features while maintaining the integrity of the low-level location information. DAM weights features in different levels which makes them adaptively match the corresponding structural information in fixed and moving images.
There are certain problems in the current study. One is the limited data used for training. In future work, we plan to take advantage of unsupervised registration and make more use of unlabeled clinical data. Another problem is that the output could be affected considerably by the quality of CBCT images. The registration accuracy of the proposed model with high-quality CBCT images is better than that with low-quality CBCT images. We plan to improve the quality of CBCT images by preprocessing them while preserving the anatomical structure of CBCT. Enhancing the image quality of CBCT might be a feasible way to obtain higher registration accuracy.
The proposed method is not limited to the CT and CBCT medical image registration. It would be applied in other clinical registration tasks, such as 4DCT registration with automatically detected landmarks. Also, it would be beneficial to compare our proposed method with the advanced deep-learning based methods proposed in the other medical image processing fields (35). In the future, comparing the proposed method with various advanced registration methods and testing it in other clinical scenarios will be our next work.
Conclusions
An unsupervised CT-CBCT medical image registration model for CT and CBCT images is developed and evaluated based on publicly available data. The proposed method shows an apparent advantage over the existing methods. The introduction of SP-BLOCK and SC-BLOCK in the DAM can enhance the capability of capturing high-level semantic information and low-level spatial information in CT and CBCT image pairs. It provides an effective way to accelerate the automated image registration between CT and CBCT images in routine clinics.
Acknowledgments
Thanks to William Rochelle, PhD, from National Institutes of Health (NIH), who edited the English text of a draft of this manuscript.
Funding: This work was supported by National Natural Science Foundation of China (No. 11975312); Anhui Provincial Natural Science Foundation of China (No. 1908085J25); National Natural Science Foundation of China (No. 61902104) and Anhui Provincial Natural Science Foundation (No. 2008085QF295).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Ethics Committee of The Affiliated Cancer Hospital of Zhengzhou University, Henan Cancer Hospital. And the informed consent was waived in this retrospective study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-21-1194/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-21-1194/coif). RH and TL report that a patent under consideration constitutes a conflict of interest [patent application number: CN202110637086.9 (pending)]. All authors report grant from National Natural Science Foundation, grant from Anhui Provincial Natural Science Foundation of China, grant from National Natural Science Foundation of China, and grant from Anhui Provincial Natural Science Foundation, during the conduct of the study. The authors have no other conflicts of interest to declare.
References
- 1.Sotiras A, Davatzikos C, Paragios N. Deformable medical image registration: a sur-vey. IEEE Trans Med Imaging 2013;32:1153-90. 10.1109/TMI.2013.2265603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bajcsy R, Kovačič S. Multiresolution elastic matching. Computer Vision, Graphics, and Image Processing 1989;46:1-21. 10.1016/S0734-189X(89)80014-3 [DOI] [Google Scholar]
- 3.Sridharan R, Dalca AV, Fitzpatrick KM, Cloonan L, Kanakis A, Wu O, Furie KL, Rosand J, Rost NS, Golland P. Quantification and Analysis of Large Multimodal Clinical Image Studies: Application to Stroke. Multimodal Brain Image Anal (2013) 2013;8159:18-30. 10.1007/978-3-319-02126-3_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ou Y, Sotiras A, Paragios N, Davatzikos C. DRAMMS: Deformable registration via attribute matching and mutual-saliency weighting. Med Image Anal 2011;15:622-39. 10.1016/j.media.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashburner J, Friston KJ. Voxel-based morphometry--the methods. Neuroimage 2000;11:805-21. 10.1006/nimg.2000.0582 [DOI] [PubMed] [Google Scholar]
- 6.Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: efficient non-parametric image registration. Neuroimage 2009;45:S61-72. 10.1016/j.neuroimage.2008.10.040 [DOI] [PubMed] [Google Scholar]
- 7.Avants BB, Tustison N, Song G. Advanced normalization tools (ANTS). Insight J 2009;2:1-35. [Google Scholar]
- 8.Khan AR, Wang L, Beg MF. FreeSurfer-initiated fully-automated subcortical brain segmentation in MRI using Large Deformation Diffeomorphic Metric Mapping. Neuroimage 2008;41:735-46. 10.1016/j.neuroimage.2008.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanai T, Kadoya N, Ito K, Onozato Y, Cho SY, Kishi K, Dobashi S, Umezawa R, Matsushita H, Takeda K, Jingu K. Evaluation of accuracy of B-spline transfor-mation-based deformable image registration with different parameter settings for thoracic images. J Radiat Res 2014;55:1163-70. 10.1093/jrr/rru062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haskins G, Kruger U, Yan P. Deep learning in medical image registration: a survey. Machine Vision and Applications 2020;31:1-18. 10.1007/s00138-020-01060-x [DOI] [Google Scholar]
- 11.Fan J, Cao X, Yap PT, Shen D. BIRNet: Brain image registration using du-al-supervised fully convolutional networks. Med Image Anal 2019;54:193-206. 10.1016/j.media.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao X, Yang J, Zhang J, Wang Q, Yap PT, Shen D. Deformable Image Registration Using a Cue-Aware Deep Regression Network. IEEE Trans Biomed Eng 2018;65:1900-11. 10.1109/TBME.2018.2822826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teng X, Chen Y, Zhang Y, Ren L. Respiratory deformation registration in 4D-CT/cone beam CT using deep learning. Quant Imaging Med Surg 2021;11:737-48. 10.21037/qims-19-1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokooti H, de Vos B, Berendsen F, Lelieveldt BP, Išgum I, Staring M. editors. Non-rigid image registration using multi-scale 3D convolutional neural networks. Inter-national conference on medical image computing and computer-assisted intervention. Cham: Springer, 2017. [Google Scholar]
- 15.Sedghi A, Luo J, Mehrtash A, Pieper S, Tempany CM, Kapur T, Mousavi P, Wells III WM. Semi-supervised deep metrics for image registration. arXiv preprint arXiv: 180401565 2018.
- 16.Fan J, Cao X, Wang Q, Yap PT, Shen D. Adversarial learning for mono- or mul-ti-modal registration. Med Image Anal 2019;58:101545. 10.1016/j.media.2019.101545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV. VoxelMorph: A Learning Framework for Deformable Medical Image Registration. IEEE Trans Med Imaging 2019. [Epub ahead of print]. doi: . 10.1109/TMI.2019.2897538 [DOI] [PubMed] [Google Scholar]
- 18.Zhao S, Lau T, Luo J, Chang EI, Xu Y. Unsupervised 3D End-to-End Medical Image Registration With Volume Tweening Network. IEEE J Biomed Health Inform 2020;24:1394-404. 10.1109/JBHI.2019.2951024 [DOI] [PubMed] [Google Scholar]
- 19.Luo W, Li Y, Urtasun R, Zemel R. Understanding the effective receptive field in deep convolutional neural networks. Advances in Neural Information Processing Systems 2016;29:4898-906. [Google Scholar]
- 20.Li S, Sui X, Luo X, Xu X, Liu Y, Goh R. Medical image segmentation using squeeze-and-expansion transformers. arXiv preprint arXiv:210509511 2021. 10.24963/ijcai.2021/112 10.24963/ijcai.2021/112 [DOI]
- 21.Song X, Guo H, Xu X, Chao H, Xu S, Turkbey B, Wood BJ, Wang G, Yan P. editors. Cross-Modal Attention for MRI and Ultrasound Volume Registration. International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer, 2021. [Google Scholar]
- 22.Sun J, Liu C, Li C, Lu Z, He M, Gao L, Lin T, Sui J, Xie K, Ni X. CrossModalNet: exploiting quality preoperative images for multimodal image registration. Phys Med Biol 2021;66:175002. 10.1088/1361-6560/ac195e [DOI] [PubMed] [Google Scholar]
- 23.Cai G, Wang J, Chi Z. An Improved Convolutional Neural Network for 3D Unsuper-vised Medical Image Registration. 2020 IEEE 6th International Conference on Computer and Communications (ICCC), 2020: 1908-14. [Google Scholar]
- 24.Li S, Ma Y, Wang H. 3D Medical Image Registration Based on Spatial Attention. 2020 The 4th International Conference on Video and Image Processing; 2020: 98-103. [Google Scholar]
- 25.Zhu Y, Zhou Z, Liao G, Yuan K. A novel unsupervised learning model for dif-feomorphic image registration. Proc SPIE 11596, Medical Imaging 2021: Image Processing, 115960M (15 February 2021).
- 26.Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. elastix: a toolbox for inten-sity-based medical image registration. IEEE Trans Med Imaging 2010;29:196-205. 10.1109/TMI.2009.2035616 [DOI] [PubMed] [Google Scholar]
- 27.Kim B, Kim DH, Park SH, Kim J, Lee JG, Ye JC. CycleMorph: Cycle consistent un-supervised deformable image registration. Med Image Anal 2021;71:102036. 10.1016/j.media.2021.102036 [DOI] [PubMed] [Google Scholar]
- 28.Chen H, Dou Q, Yu L, Qin J, Heng PA. VoxResNet: Deep voxelwise residual net-works for brain segmentation from 3D MR images. Neuroimage 2018;170:446-55. 10.1016/j.neuroimage.2017.04.041 [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Zhang W, Shen W, Li N, Chen Y, Li S, Chen B, Guo S, Wang Y. Automatic segmentation of the cardiac MR images based on nested fully convolutional dense network with dilated convolution. Biomedical Signal Processing and Control 2021;68:102684. 10.1016/j.bspc.2021.102684 [DOI] [Google Scholar]
- 30.Jaderberg M, Simonyan K, Zisserman A, Kavukcuoglu K. Spatial transformer net-works. Advances in Neural Information Processing Systems 2015;28:2017-25. [Google Scholar]
- 31.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013;26:1045-57. 10.1007/s10278-013-9622-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hugo GD, Weiss E, Sleeman WC, Balik S, Keall PJ, Lu J, Williamson JF. A longitu-dinal four-dimensional computed tomography and cone beam computed tomography dataset for image-guided radiation therapy research in lung cancer. Med Phys 2017;44:762-71. 10.1002/mp.12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu J, Liu J, Tian H, Li Y, Bao Y, Fang Z, Lu H, editors. Dual attention network for scene segmentation. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition; 2019. [Google Scholar]
- 34.Mok TC, Chung A. Fast symmetric diffeomorphic image registration with convolu-tional neural networks. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2020: 4643-4652. [Google Scholar]
- 35.Hering A, Hansen L, Mok TC, Chung A, Siebert H, Häger S, Lange A, Kuckertz S, Heldmann S, Shao W. Learn2Reg: comprehensive multi-task medical image regis-tration challenge, dataset and evaluation in the era of deep learning. arXiv preprint arXiv:211204489 2021. [DOI] [PubMed]