Abstract
Background
Myocardial perfusion (MP) SPECT is a well-established method for diagnosing cardiac disease, yet its radiation risk poses safety concern. This study aims to apply and evaluate the use of Pix2Pix generative adversarial network (Pix2Pix GAN) in denoising low dose MP SPECT images.
Methods
One hundred male and female patients with different 99mTc-sestamibi activity distributions, organ and body sizes were simulated by a population of digital 4D Extended Cardiac Torso (XCAT) phantoms. Realistic noisy SPECT projections of full dose of 987 MBq injection and 16 min acquisition, and low dose ranged from 1/20 to 1/2 of the full dose, were generated by an analytical projector from the right anterior oblique (RAO) to the left posterior oblique (LPO) positions. Additionally, twenty patients underwent ~1,184 MBq 99mTc-sestamibi stress SPECT/CT scan were also retrospectively recruited for the study. For each patient, low dose SPECT images (7/10 to 1/10 of full dose) were generated from the full dose list mode data. Our Pix2Pix GAN model was trained with full dose and low dose reconstructed SPECT image pairs. Normalized mean square error (NMSE), structural similarity index (SSIM), coefficient of variation (CV), full-width-at-half-maximum (FWHM) and relative defect size differences (RSD) of Pix2Pix GAN processed images were evaluated along with a reference convolutional auto encoder (CAE) network and post-reconstruction filters.
Results
NMSE values of 0.0233±0.004 vs. 0.0249±0.004 and 0.0313±0.007 vs. 0.0579±0.016 were obtained on 1/2 and 1/20 dose level for Pix2Pix GAN and CAE in the simulation study, while they were 0.0376±0.010 vs. 0.0433±0.010 and 0.0907±0.020 vs. 0.1186±0.025 on 7/10 and 1/10 dose level in the clinical study. Similar results were also obtained from the SSIM, CV, FWHM and RSD values. Overall, the use of Pix2Pix GAN was superior to other denoising methods in all physical indices, particular in the lower dose levels in the simulation and clinical study.
Conclusions
The Pix2Pix GAN method is effective to reduce the noise level of low dose MP SPECT. Further studies on clinical performance are warranted to demonstrate its full clinical effectiveness.
Keywords: Generative adversarial network, Pix2Pix, low dose, myocardial perfusion (MP) SPECT, denoising
Introduction
Myocardial perfusion (MP) SPECT is a well-established method for diagnosing cardiac disease (1). In order to obtain high quality SPECT images for clinical usages, a higher dosage level of radioactive tracer is often desired as the detection efficiency of the gamma camera is usually low (~0.01%). However, higher dose SPECT procedures raise risks related health concerns with radiation. Gerber et al. (2) discussed the risks of ionizing radiation for MP SPECT. In addition, Faze et al. (3) indicated that MP SPECT is a significant source of ionizing radiation exposure, making it one of the largest contributions in terms of cumulative radiation dose. Thus, it is important to reduce the dosage level in MP SPECT scanning. On the other hand, the noise level is high in low dose SPECT images, leading to poor image quality which severely affects clinical diagnosis (4). The high noise problem is even more problematic in gated MP SPECT, as each gate only contains a part of the total detected counts, considerably affecting image quality (5,6).
There are existing methods to reduce the noise level and improve low dose SPECT image quality. Skiadopoulos et al. (7) applied a Butterworth post-processing filter on MP SPECT images to reduce noise, yet still preserving effective imaging details. Noubari et al. (8) evaluated wavelet-based denoising with optimal thresholding of the wavelet coefficients for SPECT images, improving the image quality and enhancing the diagnosis. Although these denoising methods can suppress the noise level of SPECT images, the resultant images are usually over-smoothed compared to the conventional full dose SPECT images.
Deep learning has been proven to be promising in image denoising (9), segmentation (10) and classification (11), showing better performance as compared to traditional methods when using an abundant training dataset. Among different tasks, deep learning-based denoising, mostly using convolutional neural network (CNN), has been applied for various PET studies and shown promising performance (12-15). Ramon et al. (16) evaluated CNN and a variant of CNN-based structure, i.e., convolutional auto encoder (CAE), on denoising low dose static SPECT and showed that CAE outperformed CNN in diagnostic accuracy, e.g., total perfusion deficit.
Recently, Zhou et al. (17) suggested that GAN can push the generator to study the inter-domain mapping, which can generate compelling target results compared to other deep learning methods. Wan et al. (18) adapted a joint GAN for reconstruction and segmentation of dual-tracer PET images and show improved results. Sundar et al. (19) employed a conditional GAN (cGAN) on motion correction for rigid body motion for total-body PET imaging. Islam et al. (20) proposed to use a deep convolutional GAN (DCGAN) for synthetic PET brain image generation of different stages of Alzheimer’s disease. Wang et al. (21) applied a cGAN on low dose brain PET denoising and showed the promising performance in normalized mean square error (NMSE), peak signal-to-noise ratio (PSNR) and standard uptake value (SUV) values as compared with U-net. To the best of our knowledge, there are limited works investigating cGAN in denoising MP SPECT images (22-23). In this paper, we aim to evaluate the effectiveness of the Pix2Pix GAN method in denoising low dose MP SPECT images as compared to post-reconstruction filtering and CAE methods in simulation and clinical study. We present the following article in accordance with the GRRAS reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-21-1042/rc).
Methods
Simulation dataset
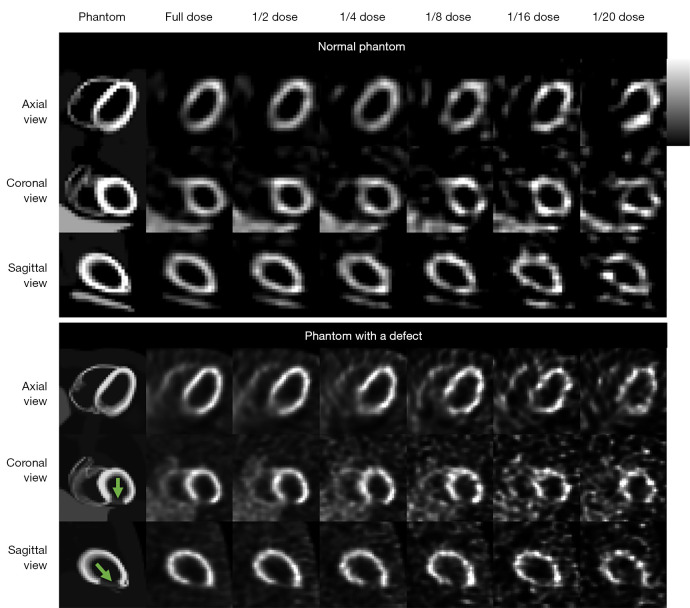
This study was performed using a population of 100 4D Extended Cardiac Torso (XCAT) digital phantoms as a virtual clinical trial (24). Fifty male and 50 female patients with normal cardiac motion, a large range of 99mTc-sestamibi distributions (25), respiratory motion amplitude (11.0–37.4 mm), cardiac wall thickness (7.7–11.6 mm), organ and body sizes (26) were simulated. Thirty of the 100 phantoms were modeled with heart defects additionally. The defects circumference ranged from 30° to 70°, length ranged from 30 to 70 mm, uptake ranged from 20% to 70% of the normal cardiac uptake. A rotation-based analytical projector (27) which has been verified with Monte Carlo methods (28) was used to model a clinical dual-head SPECT system with low-energy high-resolution (LEHR) collimators (5,29), modelling attenuation, scatter and spatially variant geometric collimator response functions. Sixty-four projections (30) with a matrix size of 128×114 and a voxel size of 0.442 cm were simulated from the right anterior oblique (RAO) to the left posterior oblique (LPO) with various noise levels. In this study, we simulated the clinical realistic noise level with count rate based on a 987 MBq injection and 16 min acquisition with a total projection count of 34.27 M (5). The projection count was scaled to the 1/20 to 1/2 of the original total count for each noise level correspondingly. Poisson noise was modeled to the scaled projections to produce a series of low dose noisy SPECT images as shown in Figure 1. The detector-collimator response, scatter and attenuation were modelled in the projection generation. Projections were then reconstructed by the 3D ordered subset expectation maximization (OS-EM) algorithm with 12 iterations and 4 subsets with attenuation, model-based scatter (31) and detector-collimator response corrections. The reconstructed SPECT image size was 128×128×114 with a voxel size of 0.442 cm.
Figure 1.
Sample two 4D XCAT phantoms and their corresponding reconstruction images with different dose levels used in this study (green arrows indicate cardiac defects). XCAT, Extended Cardiac Torso.
Clinical dataset
Twenty anonymized patients (Table 1) who underwent 99mTc-sestamibi stress SPECT/CT scan on a clinical SPECT/CT system (GE NM/CT 870 CZT) were retrospectively enrolled in this study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) under the approval of Institutional Review Board of Taipei Veterans General Hospital, and individual consent for this retrospective analysis was waived. They were injected with ~1,184 MBq 99mTc-sestamibi. Prior to the SPECT scan, a helical CT scan [120 kVp, smart mA (10–150 mA), 0.375 cm thickness] was acquired in the heart region for attenuation correction in SPECT reconstruction with a matrix of 512×512. Sixty projections were acquired through 180° from the RAO to LPO positions. The original acquisition time for each projection was 10 s. The photopeak energy window was centered at 140.5 keV with a 20% width and the scatter window was centered at 120 keV with a 10% width. The clinical data were reconstructed by the OS-EM algorithm with 5 iterations and 4 subsets, incorporating CT based attenuation and dual energy window scatter corrections. The reconstruction matrix size was 64×64×19 and the voxel size was 0.6096 cm.
Table 1. Demographic information for the patient study.
| Patient characteristics | Male | Female | Total |
|---|---|---|---|
| No. | 16 | 4 | 20 |
| Age (years) | 63.8±11.7 [45–79] | 66.0±3.9 [61–70] | 64.2±10.6 [45–79] |
| BMI (kg/m2) | 25.7±2.5 (19.23–30.12) | 24.1±3.7 (21.10–29.34) | 25.3±2.7 (19.23–30.12) |
| Cardiac defect | 7 | 2 | 9 |
BMI, body mass index.
We obtained a series of low dose SPECT images by reducing the projection acquisition time in list mode data, i.e., 10 s to 7, 5, 3, 2 and 1 s. Thus, dose levels of 7/10, 5/10, 3/10, 2/10 and 1/10 of the original full dose images were generated.
Pix2Pix generative adversarial network (Pix2Pix GAN)
The objective function V of the original GAN can be presented as:
| [1] |
where G and D are the generator and discriminator respectively. The G can produce a new sample G(z) with generative distribution PG through converting a random input drawn from a probability distribution, i.e., z~P(noise(z)), while x~Pdata indicates the true data distribution used in D. This generator G in GAN would be trained by “fighting against” with the discriminator D that attempts to differentiate between true samples with true distribution Pdata and generated samples with generative distribution PG.
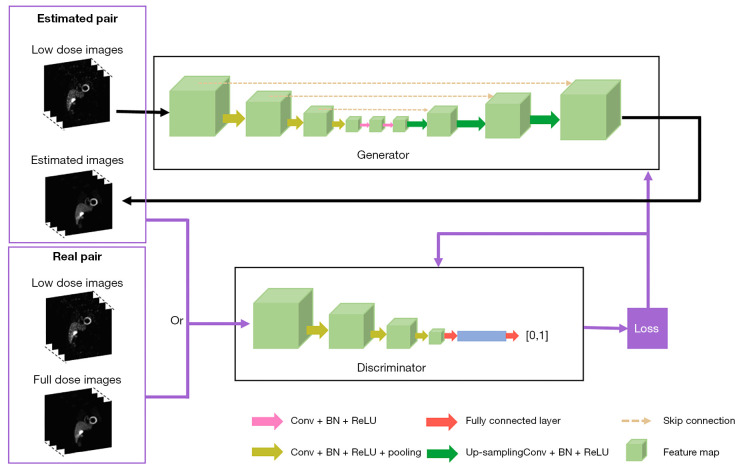
The 3D Pix2Pix GAN, a variant of cGAN, is implemented when the input of the original GAN generator is conditioned with additional information instead of the random noise vector (21), such as 3D low dose SPECT images in this study (Figure 2) to generate an estimated 3D full dose SPECT image. Pairing with the original 3D low dose image, the discriminator compares the estimated paired images with the real paired images. Dropout was applied in the generator of the proposed model. The calculated loss would be applied for tuning the generator and discriminator simultaneously. Specifically, the objective function VPix2Pix GAN of Pix2Pix GAN is defined as:
Figure 2.
The diagram of the Pix2Pix GAN used in this study. Conv, convolution; BN, batch normalization; ReLU, rectified linear unit; GAN, generative adversarial network.
| [2] |
where x~Plow(x) indicates low dose SPECT images, while y~Pfull(y) represents the corresponding full dose SPECT images. The discriminator loss, defined as cross-entropy as in Eq. [2], was used for adjusting the discriminator weights. While comparing to the common L2 loss, L1 loss (21,32) can enforce low-frequency correctness Eq. [3] and encourage less blurring. Therefore, the generator is thus trained by minimizing the difference, i.e., mean absolute error (MAE) between denoised images and reference images:
| [3] |
Then, our final objective function can be expressed as:
| [4] |
where λ=100 is an adjustable parameter (32) used to control the balance between VPix2Pix GAN(G,D) and VL1(G).
For comparison, 3D CAE (16) was also implemented in this study. Unlike Pix2Pix GAN, CAE has a symmetric encoder-decoder structure without a discriminator. Similarly, the low dose SPECT images was input into the encoding layers and the decoding layers will reconstruct it to the denoised output images. The CAE is trained by minimizing a MAE cost function between denoised images and their corresponding full dose images.
The 3D Pix2Pix GAN and 3D CAE were implemented using TensorFlow (33) and ran on a NVIDIA GeForce RTX 2080Ti GPU. The input images were normalized to a range of 0–1 for training. For validation, the Adam optimizer (34) was applied to optimize this proposed model with various numbers of layer and filter for each layer at 1/8 dose level on simulation and 3/10 dose level on clinical SPECT reconstructed images. Specifically, the number of layers varied as 2, 3, 4 and 5, while the number of filters within each layer varied as 8, 16, 24, 32 and 40 for Pix2Pix GAN and CAE.
Experiments
Determination of optimal parameters for the conventional post-reconstruction filtering
The 3D Gaussian filter (35) can be defined as follows:
| [5] |
where the x, y, z indicate the coordinate of a given image I. The σ denotes the standard deviation of the Gaussian distribution, which can control the smoothing performance on a given image I. Here, we evaluated the use of a 3D Gaussian filter with different σ, i.e., from 0.1 to 2.5 voxels with an interval of 0.1 voxel, to denoise different low dose SPECT images.
The 3D Butterworth filter can be expressed as follows:
| [6] |
where C represents a given image I, Ck is the cut-off frequency, the order of the Butterworth filter, n, controls the roll-off of Ck. Here, we fixed the order n=4 (36) for MP SPECT, while evaluate Ck from 0.11 to 0.35 cycle/voxel with an interval of 0.01 cycle/voxel for denoising different low dose SPECT images. The NMSE values on the filtered reconstructed images were assessed. This optimization was based on average results from 20 testing phantoms and 20 patients respectively.
Evaluation of denoising methods between Pix2Pix GAN, CAE and post-reconstruction filtering on the phantom dataset
Conventional denoising methods, i.e., Butterworth filter and Gaussian filter with optimal parameters determined in the previous section, and deep learning denoising methods, i.e., 3D CAE and 3D Pix2Pix GAN, were applied for different low dose SPECT images on the simulation dataset. For the deep learning methods, the full dose SPECT images were used as the training reference. The 3D CAE/Pix2Pix GAN network was trained to estimate the denoised images for each specific low dose SPECT dataset (1/2, 1/4, 1/8, 1/16 and 1/20 of full dose).
Among the 100 phantoms, SPECT imaging data of 70 phantoms were randomly selected for training, while 10 phantoms were used for validation and 20 phantoms were used for testing. The training time for 3D Pix2Pix GAN and CAE was ~3.5 and 3 h for each dose level respectively.
Evaluation of denoising methods between Pix2Pix GAN, CAE and post-reconstruction filtering on the clinical dataset
We also applied the Pix2Pix GAN method on the clinical dataset and compared it with traditional filtering methods and 3D CAE. For CAE and Pix2Pix GAN, the full dose images, i.e., SPECT images with 10 s acquisition time/prj were used as the training reference to denoise different low dose clinical SPECT images (7/10, 5/10, 3/10, 2/10 and 1/10 of full dose). In addition, we performed a 10-fold cross-validation on the clinical dataset to evaluate CAE approach and our proposed Pix2Pix GAN approach on each low dose level. Specifically, for each round of evaluation, 16 patients were selected for training, 2 patients were used for validation, while 2 patients were applied for testing. Therefore, this process was repeated 10 times and all 20 patient datasets were tested and averaged for the final results. The training time for Pix2Pix GAN and CAE was ~1 h and 50 min for each dose level.
Data analysis
MAE (21) was used to select the best hyper-parameters from the validation dataset using the full dose images as reference:
| [7] |
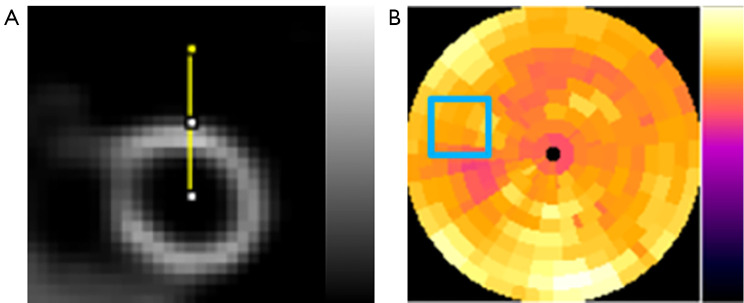
In both simulation and clinical study, to evaluate the accuracy and precision for the voxel error of the denoised images, NMSE and SSIM for a 3D volume-of-interest (VOI; N=28×28×28), covering the whole heart (Figure 3A) were computed on the denoised images as compared to the original full dose SPECT reconstructed images.
Figure 3.
The ROIs used for data analysis in this study. (A) 3D VOI chosen from denoised SPECT images for measuring NMSE and SSIM. The yellow line indicated the profile drawn across the cardiac wall for measuring the FWHM. (B) The CV value measuring from the selected ROI (blue box) on the polar plots. ROI, region-of-interest; VOI, volume-of-interest; NMSE, normalized mean square error; SSIM, structural similarity index; FWHM, full-width-at-half-maximum; CV, coefficient of variation.
| [8] |
| [9] |
where xd represents the voxel value in denoised images, xFD represents the voxel values in the corresponding full dose images, μd and μFD are the mean values of denoised images and full dose images respectively, σd and σFD are the standard deviations and σd,FD is the cross-covariance for denoised images and full dose images respectively. The constants C1 and C2 were set to be 0.01 and 0.02 (37).
To evaluate the noise level of the denoised SPECT images, coefficient of variation (CV) was measured on a 2D uniform region-of-interest (ROI; 20×20) drawn on the polar plots to assess the noise reduction (Figure 3B) for the denoised reconstructed images, avoiding defects and motion artifact regions based on the original phantoms and patients’ clinical records.
| [10] |
where m is 400 voxels for the selected ROI, μ is the mean voxel value of the chosen ROI, j is the voxel index and x is the voxel count value.
In order to evaluate the degree of image blurring, full-width-at-half-maximum (FWHM) was measured on the image profiles drawn across the left ventricle wall (Figure 3A) of the denoised images, original full dose and low dose images.
For phantoms with defects, the defect size on the polar plots of various denoised images were measured and compared to that of the full dose polar plots to calculate the relative defect size difference (RSD):
| [11] |
where SFD is the pixel number of the defect from the full dose polar plot, Sd indicates the pixel number of the defect from different denoised polar plots, calculated by the thresholding method according to the actual defect uptake ratios.
We performed a statistical analysis using a paired t-test with Bonferroni correction (SPSS, IBM Corporation, Armonk, NY, USA). A P value <0.05 is regarded as statistically significant.
Results
Filter parameters optimization
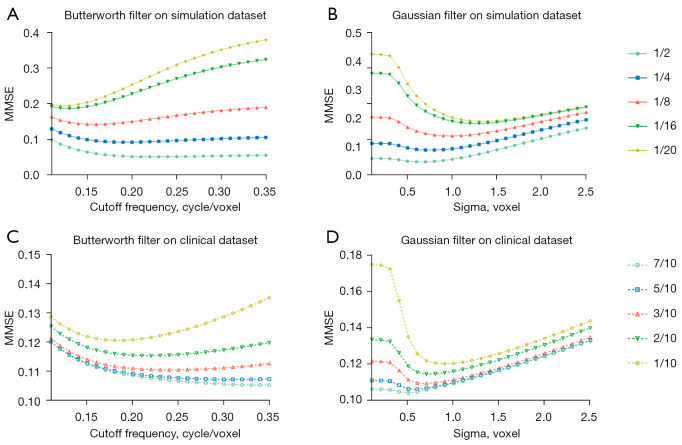
The NMSE results of applying various standard deviation in the Gaussian filter and cutoff frequency in the Butterworth filter for SPECT images at different dose levels are shown in Figure 4. For the phantom dataset, the selected cutoff frequency for Butterworth filter is 0.22, 0.19, 0.16, 0.13 and 0.12 cycle/voxel and the standard deviation for Gaussian filter is 0.7, 0.8, 1.0, 1.4 and 1.5 voxels for denoising images at 1/2, 1/4, 1/8, 1/16 and 1/20 of the original full dose level (Figure 4A,4B). For the clinical dataset, the optimal cutoff frequency for Butterworth filter is 0.33, 0.30, 0.24, 0.22 and 0.18 cycle/voxel, while the optimal standard deviation for Gaussian filter is 0.5, 0.6, 0.7, 0.7 and 0.9 voxel for denoising low dose MP SPECT images at 7/10, 5/10, 3/10, 2/10 and 1/10 of the full dose level (Figure 4C,4D).
Figure 4.
The NMSE curves with various Ck from 0.11 to 0.35 cycle/voxel in the Butterworth filter and various σ from 0.1 to 2.5 voxels in the Gaussian filter, for 1/2–1/20 dose level of full dose SPECT reconstructed images on the phantom dataset (A,B) and for 7/10–1/10 dose level of full dose SPECT reconstructed images on the clinical dataset (C,D). NMSE, normalized mean square error.
Network validation
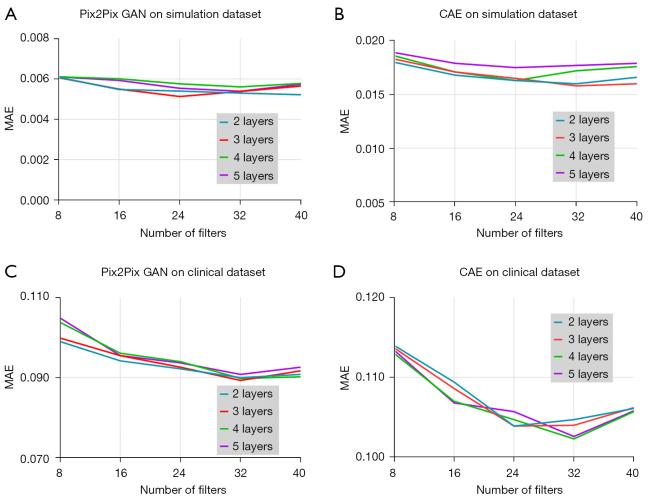
Figure 5 shows the MAE results obtained with different numbers of layer and filter of Pix2Pix GAN and CAE from the validation dataset with 1/8 dose level in the simulation and 3/10 dose level in the clinical study respectively. For the simulation study, the layer and filter number for the lowest MAE value was 3 and 24 for Pix2Pix GAN, and 3 and 32 for CAE respectively. For the clinical study, the layer and filter number for the lowest MAE value was 3 and 32 for Pix2Pix GAN, and 4 and 32 for CAE respectively. Thus, these network hyper-parameters were used for the latter study.
Figure 5.
MAE values obtained using different numbers of layer and filter for training the (A) Pix2Pix GAN and (B) CAE model in the simulation study; (C) Pix2Pix GAN and (D) CAE in the clinical study from the validation dataset respectively. GAN, generative adversarial network; MAE, mean absolute error; CAE, convolutional auto encoder.
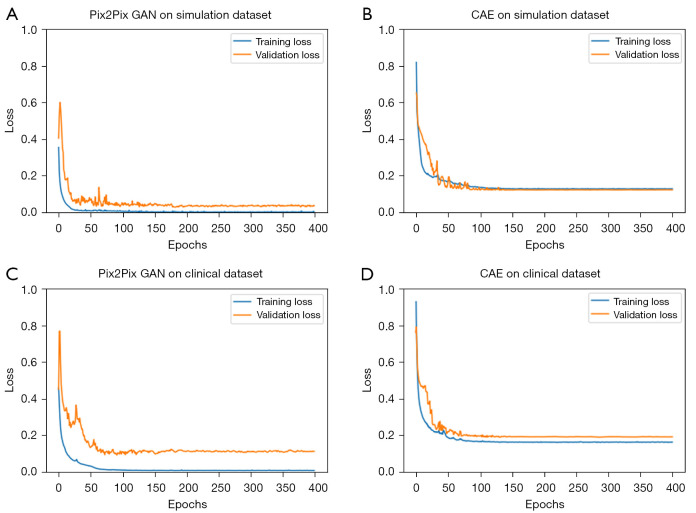
Figure 6 shows the loss versus epoch curves of Pix2Pix GAN and CAE obtained with the above optimized hyper-parameters. It can be observed that the loss function converges after 200 epochs in general.
Figure 6.
The loss versus epochs curves obtained from the optimized (A) Pix2Pix GAN and (B) CAE in the simulation study; (C) Pix2Pix GAN and (D) CAE in the clinical study. GAN, generative adversarial network; CAE, convolutional auto encoder.
Evaluation of denoising method between Pix2Pix GAN, CAE and post-reconstruction filtering on the phantom dataset
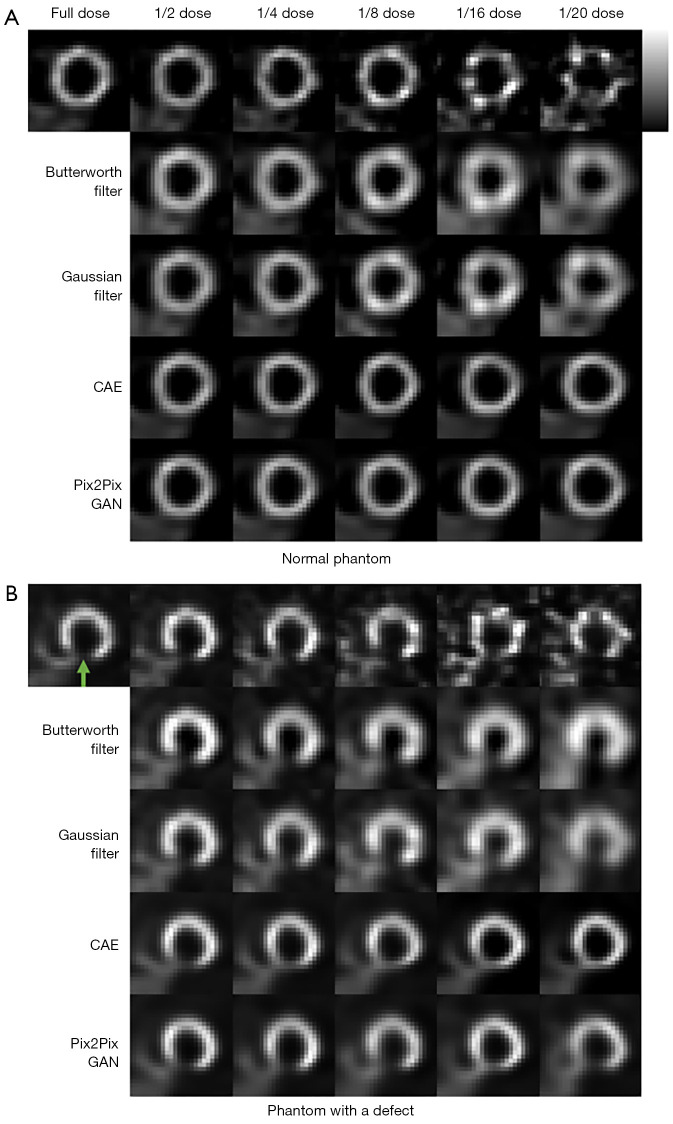
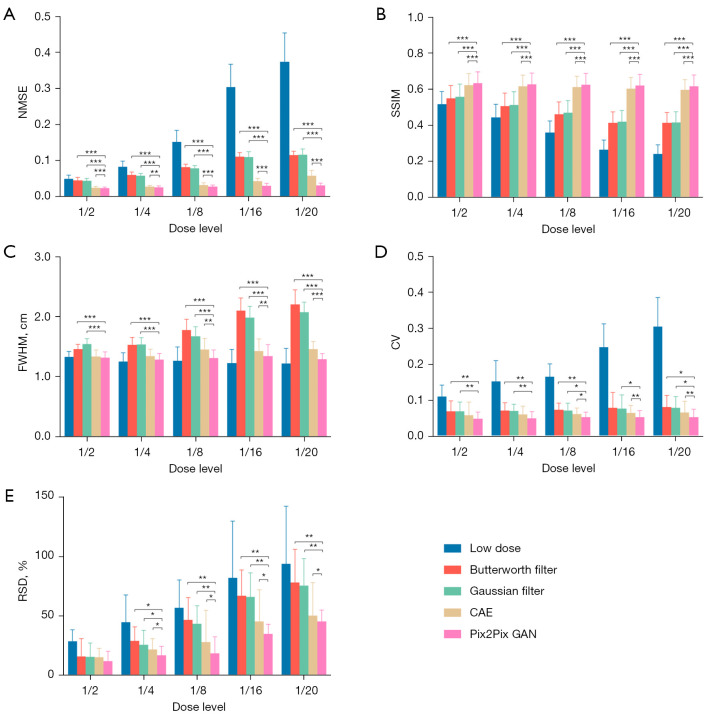
The sample denoising results of Pix2Pix GAN, optimized conventional filters and CAE for a range of low dose MP SPECT images for a normal male phantom and an abnormal female phantom are shown in Figure 7. The average NMSE, SSIM, FWHM, CV and RSD results of our simulation dataset for various dose levels are shown in Figure 8.
Figure 7.
Simulated MP SPECT images with various dose levels of (A) a sample normal male phantom and (B) a sample abnormal female phantom processed under different denoising methods. The green arrow indicates the defect location. CAE, convolutional auto encoder; GAN, generative adversarial network; MP, myocardial perfusion.
Figure 8.
The average (A) NMSE, (B) SSIM, (C) FWHM, (D) CV and (E) RSD measurements for different denoising methods for the simulation dataset. Error bars indicate the standard deviation of the respective indices for 20 phantoms. *, P<0.05; **, P<0.01; ***, P<0.001. NMSE, normalized mean square error; SSIM, structural similarity index; FWHM, full-width-at-half-maximum; CV, coefficient of variation; RSD, relative defect size differences; CAE, convolutional auto encoder; GAN, generative adversarial network.
For the accuracy and precision measurement for the voxel error of denoised images, i.e., NMSE and SSIM, the difference among different denoising methods is generally smaller in lower noise level as compared to the higher noise level, and Pix2Pix GAN shows the best results (P value <0.01). The deep learning methods show better performances compared to conventional filters, which can be observed in most of the indices. In addition, the results of Pix2Pix GAN are superior to those of CAE (P value <0.01). The results of CAE and Pix2Pix GAN model are more similar to the full dose images. However, small artifacts are observed on the heart wall for various denoising methods for images with higher noise levels particularly on 1/20 of the full dose images (Figure 7). The use of Pix2Pix GAN is superior to other denoising methods in all physical indices (Figure 8), while other methods substantially affect the defect quantification as compared to the Pix2Pix GAN method (Figure 8E). The CAE method masks the heart defects to some extent (Figure 7B), leading to inferior quantitative results as compared to the Pix2Pix GAN method, especially on higher dose levels (P value <0.05 on dose levels from 1/4 to 1/20) (Figure 8E). With the use of Pix2Pix GAN, dose reduction on lower dose levels (1/16 or 1/20) seems feasible with less artifacts observed and shows minimum quantitative errors among all methods.
Evaluation of denoising method between Pix2Pix GAN, CAE and post-reconstruction filtering on the clinical dataset
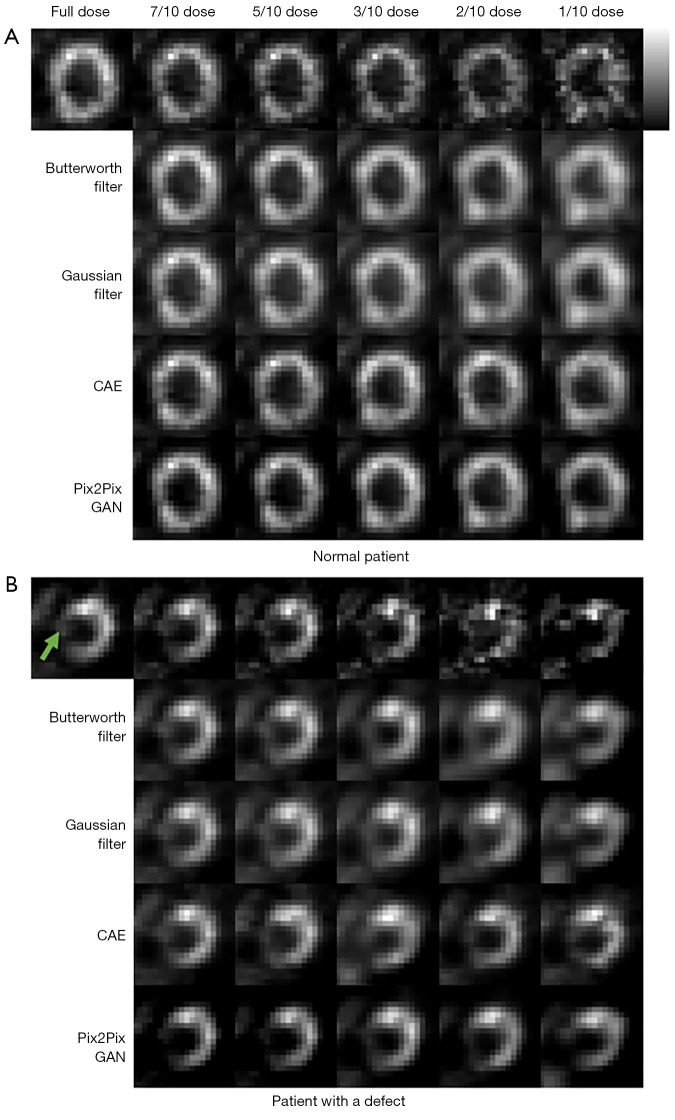
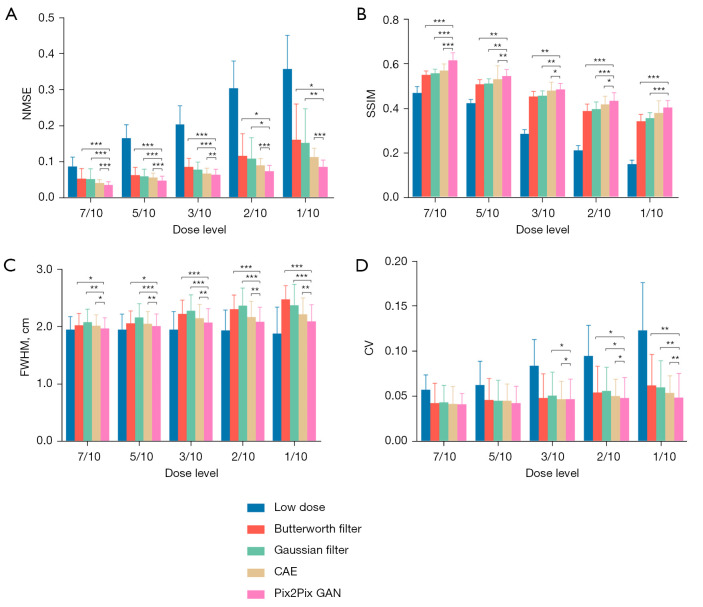
Figure 9 shows the sample full dose, low dose and denoised reconstructed images on two sample patients. The Pix2Pix GAN processed images are similar to the full dose images with less image blurring as compared to filtered images and CAE processed images at different low dose levels. The quantitative results at different dose levels on twenty patients are shown in Figure 10, which are consistent with those of the simulation dataset (Figure 8). Pix2Pix GAN method shows better performance as compared to other methods for all indices.
Figure 9.
Clinical MP SPECT images with various noise levels of (A) a sample normal patient (male, age =49 years, BMI =25.5 kg/m2) and (B) a sample patient (male, age =76 years, BMI =19.2 kg/m2) with a known cardiac defect (green arrow) processed under different denoising methods. CAE, convolutional auto encoder; GAN, generative adversarial network; MP, myocardial perfusion; BMI, body mass index.
Figure 10.
The average (A) NMSE, (B) SSIM, (C) FWHM and (D) CV measurements for different denoising methods for the clinical dataset. Error bars indicate the standard deviation of the respective indices for 20 patients. *, P<0.05; **, P<0.01; ***, P<0.001. NMSE, normalized mean square error; SSIM, structural similarity index; FWHM, full-width-at-half-maximum; CV, coefficient of variation; CAE, convolutional auto encoder; GAN, generative adversarial network.
Discussion
The filter optimization results suggest that the optimal parameters for different dose levels are different. The lower dose SPECT images would need a stronger filter. The filter parameters vary considerably between the simulation and the clinical study since the low dose levels are different (1/2–1/20 of full dose for simulation and 7/10–1/10 of full dose for clinical) in our study. For a series of low dose MP SPECT images, the Pix2Pix GAN denoising method significantly improves the image quality based on the physical indices (Figures 7-10) with less artifacts observed among all methods. Evaluation on lower dose level is not feasible on the clinical datasets as 1 s acquisition time/projection is the lowest setting available on the specific scanner used in this study. Further dose reduction from the proposed Pix2Pix GAN method is potentially feasible if a lower dose level can be achieved in other models of clinical scanners.
Various heart defects exist in real clinical patient studies, thus 30 of the 100 phantoms were modeled with heart defects to further evaluate if our proposed Pix2Pix GAN would affect the defect quantification on the simulation study. Our average results based on ten abnormal phantoms showed that the RSD values using the Pix2Pix GAN model is closer to the full dose SPECT comparing with the original low dose SPECT input, other conventional filtering methods and the CAE model (P value <0.05 on dose levels from 1/4 to 1/20), indicating the proposed methodology is effective and robust for hearts with defects. However, we found that all denoising methods would potentially “mask out” the relatively small defect especially in lower dose levels (1/8–1/20) in this study. Further improvement and evaluation on defect quantification using deep learning techniques with more clinical relevant indices is warranted.
We also applied the Pix2Pix GAN model to the clinical dataset with normal patients as well as patients with defects. Although the number of training patients was 16, the results are consistent with the simulations. The use of more clinical patient data for training is expected to further improve the training performance with lower bias, though increase of computational time is expected. We did not calculate the RSD in the clinical study since the actual defect size and uptake ratio were not known.
Comparing with the conventional Gaussian and Butterworth filtering methods and CAE model, our proposed Pix2Pix GAN can substantially reduce the noise level of low dose SPECT, while still preserving high quality SPECT images based on the physical indices. When comparing to other neural networks such as CAE approach in image denoising, GAN has two different networks with adversarial training method and a large variety of functions can be incorporated into the model (38). The Pix2Pix GAN, a derivative model of GAN, applies additional information, e.g., low dose MP SPECT images in this study instead of only random data as input to control over the characteristics of images that are being generated (39), further approaching the true distribution of data. In this study, we implemented the Pix2Pix GAN generator with a U-net architecture. The effectiveness of this design has been confirmed in many computer vision applications. The input of paired low and full dose MP SPECT images for the discriminator for training in this study, could provide more auxiliary information and the validity of this structure has also been confirmed in various works (32). For testing, pairing with corresponding full dose images is not required in the proposed Pix2Pix GAN method. Recently, Bradshaw et al. proposed the pipelines on best practices for AI algorithm development in nuclear medicine and this study follows most of the pipelines (40).
Our results showed that the injection dose can be substantially reduced for MP SPECT, leading to the concomitant reduced radiation absorbed dose to the patients. The reduced injection dose can be traded for a shorter acquisition time and increased patient throughput. The reduction of dose and acquisition time is particularly important for pediatric patients, as the lifetime risk from dose radiation for children was higher than adults (41) and sedation demand can be lowered respectively. Besides, for single or dual gating SPECT or PET, as the detecting counts are divided into more number of bins and the noise level would further increase (42), denoising in each bin for better motion estimation is another research topic that is currently under investigation in our group (23). Moreover, our preliminary results have shown that further dose reduction is feasible by applying Pix2Pix GAN on projection domain as compared to the reconstruction domain on the phantom dataset (43). Further evaluations on denoising on both domains using more patient data are warranted to investigate the lowest achievable dose limit for MP SPECT.
In the simulation, projection number and voxel size were based on previous clinical studies (30) using a conventional NaI clinical SPECT system for cross references, while the actual clinical datasets were acquired by a new CZT SPECT/CT scanner. However, our results on two different types of scanners and acquisition protocols were consistent. Another main limitation in this study is that we only considered the physical indices measurement. Existing literatures have shown that evaluation of denoising methods with physical metrics such as NMSE and SSIM are not reflective of performance on clinical tasks (44-46). Whether the lowest dose level could be achieved by deep learning-based denoising method is unknown without the evaluation of the actual clinical diagnostic index, e.g., ROC study of defect detectability (47-49). Further studies on clinical performance are warranted.
Conclusions
In this study, we proposed and evaluated a deep learning approach, i.e., 3D Pix2Pix GAN model to reduce the noise level for simulated and clinical low dose MP SPECT images. Pix2Pix GAN denoising is superior to conventional post-reconstruction filtering and CAE approaches, especially for higher noise levels based on the physical quantitative measurements. Further task-based evaluation based on more clinical data is warranted to demonstrate the full clinical effectiveness of the proposed method.
Acknowledgments
The authors would like to thank Mr. Qi Zhang and Ms. Yingqing Lyu for assisting data analysis. The authors also would like to thank Dr. Eric Frey from Johns Hopkins University for providing the rotation-based analytical projector program.
Funding: This work was supported by a research grant from the National Natural Science Foundation of China (NSFC), China (No. 81922080).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Institutional Review Board of Taipei Veterans General Hospital, and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Reporting Checklist: The authors have completed the GRRAS reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-21-1042/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-21-1042/coif). GSPM serves as an unpaid editorial board member of Quantitative Imaging in Medicine and Surgery. The other authors have no conflicts of interest to declare.
References
- 1.Buck AK, Nekolla S, Ziegler S, Beer A, Krause BJ, Herrmann K, Scheidhauer K, Wester HJ, Rummeny EJ, Schwaiger M, Drzezga A. SPECT/CT. J Nucl Med 2008;49:1305-19. 10.2967/jnumed.107.050195 [DOI] [PubMed] [Google Scholar]
- 2.Gerber TC, Gibbons RJ. Weighing the risks and benefits of cardiac imaging with ionizing radiation. JACC Cardiovasc Imaging 2010;3:528-35. 10.1016/j.jcmg.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 3.Fazel R, Krumholz HM, Wang Y, Ross JS, Chen J, Ting HH, Shah ND, Nasir K, Einstein AJ, Nallamothu BK. Exposure to low-dose ionizing radiation from medical imaging procedures. N Engl J Med 2009;361:849-57. 10.1056/NEJMoa0901249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin M, Niu X, Qi W, Yang Y, Dey J, King MA, Dahlberg S, Wernick MN. 4D reconstruction for low-dose cardiac gated SPECT. Med Phys 2013;40:022501. 10.1118/1.4773868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang D, Pretorius PH, Ghaly M, Zhang Q, King MA, Mok GSP. Evaluation of different respiratory gating schemes for cardiac SPECT. J Nucl Cardiol 2020;27:634-47. 10.1007/s12350-018-1392-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang D, Sun J, Pretorius PH, King M, Mok GSP. Clinical evaluation of three respiratory gating schemes for different respiratory patterns on cardiac SPECT. Med Phys 2020;47:4223-32. 10.1002/mp.14354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skiadopoulos S, Karatrantou A, Korfiatis P, Costaridou L, Vassilakos P, Apostolopoulos D, Panayiotakis G. Evaluating image denoising methods in myocardial perfusion single photon emission computed tomography (SPECT) imaging. Meas Sci Technol 2009;20:104023. 10.1088/0957-0233/20/10/104023 [DOI] [Google Scholar]
- 8.Noubari HA, Fayazi A, Babapour F. De-noising of SPECT images via optimal thresholding by wavelets. Annu Int Conf IEEE Eng Med Biol Soc 2009;2009:352-5. 10.1109/IEMBS.2009.5332777 [DOI] [PubMed] [Google Scholar]
- 9.Xue H, Zhang Q, Zou S, Zhang W, Zhou C, Tie C, Wan Q, Teng Y, Li Y, Liang D, Liu X, Yang Y, Zheng H, Zhu X, Hu Z. LCPR-Net: low-count PET image reconstruction using the domain transform and cycle-consistent generative adversarial networks. Quant Imaging Med Surg 2021;11:749-62. 10.21037/qims-20-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianconi F, Fravolini ML, Pizzoli S, Palumbo I, Minestrini M, Rondini M, Nuvoli S, Spanu A, Palumbo B. Comparative evaluation of conventional and deep learning methods for semi-automated segmentation of pulmonary nodules on CT. Quant Imaging Med Surg 2021;11:3286-305. 10.21037/qims-20-1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Zhou L, Wang M, Shao C, Shi L, Yang S, Zhang Z, Feng M, Shan F, Liu L. Combination of generative adversarial network and convolutional neural network for automatic subcentimeter pulmonary adenocarcinoma classification. Quant Imaging Med Surg 2020;10:1249-64. 10.21037/qims-19-982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui J, Gong K, Guo N, Wu C, Meng X, Kim K, Zheng K, Wu Z, Fu L, Xu B, Zhu Z, Tian J, Liu H, Li Q. PET image denoising using unsupervised deep learning. Eur J Nucl Med Mol Imaging 2019;46:2780-9. 10.1007/s00259-019-04468-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto F, Ohba H, Ote K, Teramoto A, Tsukada H. Dynamic PET image denoising using deep convolutional neural networks without prior training datasets. IEEE Access 2019;7:96594-603.
- 14.Gong K, Guan J, Liu CC, Qi J. PET Image Denoising Using a Deep Neural Network Through Fine Tuning. IEEE Trans Radiat Plasma Med Sci 2019;3:153-61. 10.1109/TRPMS.2018.2877644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan S, Zhu YM. Full-Dose PET Image Estimation from Low-Dose PET Image Using Deep Learning: a Pilot Study. J Digit Imaging 2019;32:773-8. 10.1007/s10278-018-0150-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramon AJ, Yang Y, Pretorius PH, Johnson KL, King MA, Wernick MN. Improving Diagnostic Accuracy in Low-Dose SPECT Myocardial Perfusion Imaging With Convolutional Denoising Networks. IEEE Trans Med Imaging 2020;39:2893-903. 10.1109/TMI.2020.2979940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou L, Schaefferkoetter JD, Tham IWK, Huang G, Yan J. Supervised learning with cyclegan for low-dose FDG PET image denoising. Med Image Anal 2020;65:101770. 10.1016/j.media.2020.101770 [DOI] [PubMed] [Google Scholar]
- 18.Wan Y, Ye H, Liu H. Deep-learning based joint estimation of dual-tracer PET image activity maps and clustering of time activity curves. SPIE Medical Imaging, 2021. [Google Scholar]
- 19.Sundar LS, Iommi D, Spencer B, Wang Q, Cherry S, Beyer T, Badawi R. Data-driven motion compensation using cGAN for total-body [18F] FDG-PET imaging. J Nucl Med 2021;62:35. [Google Scholar]
- 20.Islam J, Zhang Y. GAN-based synthetic brain PET image generation. Brain Inform 2020;7:3. 10.1186/s40708-020-00104-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Yu B, Wang L, Zu C, Lalush DS, Lin W, Wu X, Zhou J, Shen D, Zhou L. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage 2018;174:550-62. 10.1016/j.neuroimage.2018.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q, Sun J, Mok GSP. Low dose SPECT image denoising using a generative adversarial network. International Conference on Medical Imaging with Deep Learning (MIDL), 2019. [Google Scholar]
- 23.Sun J, Zhang Q, Du Y, Zhang D, Pretorius PH, King MA, Mok GSP. Dual gating myocardial perfusion SPECT denoising using a conditional generative adversarial network. Med Phys 2022. doi: . 10.1002/mp.15707 [DOI] [PubMed] [Google Scholar]
- 24.Abadi E, Segars WP, Tsui BMW, Kinahan PE, Bottenus N, Frangi AF, Maidment A, Lo J, Samei E. Virtual clinical trials in medical imaging: a review. J Med Imaging (Bellingham) 2020;7:042805. 10.1117/1.JMI.7.4.042805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghaly M, Du Y, Fung GS, Tsui BM, Links JM, Frey E. Design of a digital phantom population for myocardial perfusion SPECT imaging research. Phys Med Biol 2014;59:2935-53. 10.1088/0031-9155/59/12/2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D, Ghaly M, Mok GSP. Interpolated CT for attenuation correction on respiratory gating cardiac SPECT/CT - A simulation study. Med Phys 2019;46:2621-8. 10.1002/mp.13513 [DOI] [PubMed] [Google Scholar]
- 27.Frey EC, Tsui BMW. A practical projector-backprojector modeling attenuation, detector response, and scatter for accurate scatter compensation in SPECT. Conference Record of the 1991 IEEE Nuclear Science Symposium and Medical Imaging Conference. Santa Fe, NM, USA: IEEE, 1991. [Google Scholar]
- 28.Frey EC, Tsui BMW. A practical method for incorporating scatter in a projector-backprojector for accurate scatter compensation in SPECT. IEEE Transactions on Nuclear Science 1993;40:1107-16. 10.1109/23.256720 [DOI] [Google Scholar]
- 29.Chen J, Li Y, Luna LP, Chung HW, Rowe SP, Du Y, Solnes LB, Frey EC. Learning fuzzy clustering for SPECT/CT segmentation via convolutional neural networks. Med Phys 2021;48:3860-77. 10.1002/mp.14903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pretorius PH, Johnson KL, Dahlberg ST, King MA. Investigation of the physical effects of respiratory motion compensation in a large population of patients undergoing Tc-99m cardiac perfusion SPECT/CT stress imaging. J Nucl Cardiol 2020;27:80-95. 10.1007/s12350-017-0890-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frey EC, Tsui BMW. A new method for modeling the spatially-variant, object-dependent scatter response function in SPECT. 1996 IEEE Nuclear Science Symposium. Conference Record. Anaheim, CA, USA: IEEE, 1996. [Google Scholar]
- 32.Isola P, Zhu JY, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Honolulu, HI, USA: IEEE, 2017. [Google Scholar]
- 33.Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Kudlur M, Levenberg J, Monga R, Moore S, Murray DG, Steiner B, Tucker P, Vasudevan V, Warden P, Wicke M, Yu Y, Zheng X. TensorFlow: a system for large-scale machine learning. In: Proceedings of the 12th USENIX conference on Operating Systems Design and Implementation (OSDI'16). Berkeley, CA, USA: USENIX, 2016. [Google Scholar]
- 34.Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv 2014. arXiv:1412.6980.
- 35.Lyra M, Ploussi A. Filtering in SPECT Image Reconstruction. Int J Biomed Imaging 2011;2011:693795. 10.1155/2011/693795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vakhtangandze T, Hall DO, Zananiri FV, Rees MR. The effect of Butterworth and Metz reconstruction filters on volume and ejection fraction calculations with 99Tcm gated myocardial SPECT. Br J Radiol 2005;78:733-6. 10.1259/bjr/25662922 [DOI] [PubMed] [Google Scholar]
- 37.Shiri I, AmirMozafari Sabet K, Arabi H, Pourkeshavarz M, Teimourian B, Ay MR, Zaidi H. Standard SPECT myocardial perfusion estimation from half-time acquisitions using deep convolutional residual neural networks. J Nucl Cardiol 2021;28:2761-79. 10.1007/s12350-020-02119-y [DOI] [PubMed] [Google Scholar]
- 38.Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial nets. In: Proceedings of the 27th International Conference on Neural Information Processing Systems. Cambridge, MA, USA: MIT Press, 2672-80. [Google Scholar]
- 39.Wolterink JM, Kamnitsas K, Ledig C, Išgum I. Deep learning: Generative adversarial networks and adversarial methods. In: Handbook of Medical Image Computing and Computer Assisted Intervention. Cambridge, MA, USA: Academic Press, 2020. [Google Scholar]
- 40.Bradshaw TJ, Boellaard R, Dutta J, Jha AK, Jacobs P, Li Q, Liu C, Sitek A, Saboury B, Scott PJH, Slomka PJ, Sunderland JJ, Wahl RL, Yousefirizi F, Zuehlsdorff S, Rahmim A, Buvat I. Nuclear Medicine and Artificial Intelligence: Best Practices for Algorithm Development. J Nucl Med 2022;63:500-10. 10.2967/jnumed.121.262567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gelfand MJ, Lemen LC. PET/CT and SPECT/CT dosimetry in children: the challenge to the pediatric imager. Semin Nucl Med 2007;37:391-8. 10.1053/j.semnuclmed.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 42.Zhang Q, Zhang D, Mok GSP. Comparison of Different Attenuation Correction Methods for Dual Gating Myocardial Perfusion SPECT/CT. IEEE Transactions on Radiation Plasma Medical Sciences 2019;3:565-71. 10.1109/TRPMS.2019.2899066 [DOI] [Google Scholar]
- 43.Mok GSP, Zhang Q, Sun J, Du Y. Comparison of Projection-based and Reconstruction-based Low Dose SPECT Image Denoising Using a Conditional Generative Adversarial Network. 2020 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). Boston, MA, USA: IEEE, 2020. [Google Scholar]
- 44.Li K, Zhou W, Li H, Anastasio MA. Task-based performance evaluation of deep neural network-based image denoising. SPIE Medical Imaging, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Z, Rahman MA, Schindler T, Gropler R, Laforest R, Wahl R, Jha A. AI-based methods for nuclear-medicine imaging: Need for objective task-specific evaluation. J Nucl Med 2020;61:575. [Google Scholar]
- 46.Jha AK, Myers KJ, Obuchowski NA, Liu Z, Rahman MA, Saboury B, Rahmim A, Siegel BA. Objective Task-Based Evaluation of Artificial Intelligence-Based Medical Imaging Methods:: Framework, Strategies, and Role of the Physician. PET Clin 2021;16:493-511. 10.1016/j.cpet.2021.06.013 [DOI] [PubMed] [Google Scholar]
- 47.Frey EC, Gilland KL, Tsui BM. Application of task-based measures of image quality to optimization and evaluation of three-dimensional reconstruction-based compensation methods in myocardial perfusion SPECT. IEEE Trans Med Imaging 2002;21:1040-50. 10.1109/TMI.2002.804437 [DOI] [PubMed] [Google Scholar]
- 48.Gifford HC, King MA, de Vries DJ, Soares EJ. Channelized hotelling and human observer correlation for lesion detection in hepatic SPECT imaging. J Nucl Med 2000;41:514-21. [PubMed] [Google Scholar]
- 49.Gifford HC, Wells R, King MA. A comparison of human observer LROC and numerical observer ROC for tumor detection in SPECT images. IEEE Transactions on Nuclear Science 1999;46:1032-7. 10.1109/23.790820 [DOI] [Google Scholar]