Abstract
A nutrient deprivation-induced locus in Sinorhizobium meliloti strain 1021 was identified by use of a Tn5-luxAB reporter gene transposon. The tagged locus is comprised of two open reading frames (ORFs) designated ndiA and ndiB for nutrient deprivation-induced genes A and B. Comparison of the deduced amino acid sequences of both ndiA and ndiB to the protein databases failed to reveal similarity to any known genes. The expression of the ndi locus was found to be induced by carbon and nitrogen deprivation, osmotic stress, and oxygen limitation and during entry into stationary phase. To identify regulatory components involved in the control of ndi gene expression, a second round of mutagenesis was performed on the primary ndiB::Tn5-luxAB-tagged strain (C22) with transposon Tn1721. A double-mutant strain was obtained that lacked ndi locus transcriptional activity under all of the inducing conditions tested. The Tn1721-tagged gene showed a high degree of similarity to tryptophan-rich sensory protein TspO from Rhodobacter sphaeroides, as well as to mitochondrial benzodiazepine receptor pK18 from mammals. Induction of the ndi::Tn5-luxAB reporter gene fusion was restored under all inducing conditions by introducing the tspO coding region, from either S. meliloti or R. sphaeroides, in trans. Furthermore, it was found that, in addition to tspO, fixL, which encodes the sensor protein of an oxygen-sensing two-component system, is required for full expression of the ndi locus, but only under low oxygen tension.
In nature, bacterial growth is restricted by a wide variety of environmental factors. One factor of particular importance is the lack of essential nutrients. In most natural settings, at least some essential nutrients are limiting; therefore, negligible growth or dormancy is the more typical physiological state of bacteria (24). Understanding how bacteria are able to monitor, sense, and respond to their environment in this nutrient-deprived state is fundamental to our understanding of microbial biology and ecology. Some bacteria, such as Bacillus and Myxococcus spp., sporulate when challenged with environmental stresses (11, 14). However, the majority of bacteria do not appear to differentiate into these stress-resistant forms. Research on primarily Escherichia coli, Salmonella typhimurium, and Vibrio spp. has shown that under starvation conditions, these nonsporulating bacteria enter into a specific genetic program which results in the generation of a survival state supporting the persistence of the species until conditions improve (17, 19, 28, 35).
Research on the starvation survival of bacteria whose natural habitat is soil, such as Pseudomonas or Rhizobium spp., has been relatively limited thus far (see Discussion). Soil is generally a harsh, oligotrophic environment (31, 40, 44). Nutrient deprivation and oxygen limitation may represent the most prevailing stress conditions for bacteria that persist in this environment. Although a small amount of organic matter is present in most soils, the bulk of it is in a recalcitrant form, such as lignin or humus (1). Furthermore, in soils where utilizable carbon is available, the lack of inorganic nutrients such as phosphorus or nitrogen may limit growth. Rhizosphere soil has been reported to be a somewhat less oligotrophic environment than bulk soil, due to the presence of plant root exudates as readily utilizable nutrients. However, even in the rhizosphere, bacterial growth and activity are generally limited to the short periods when these exudates are available (18). It is likely that bacteria, which have evolved within the soil and rhizosphere, have developed distinct mechanisms for persisting in a nutrient-limited soil environment.
We have been investigating gene expression during nutrient deprivation in an indigenous soil bacterium, Sinorhizobium meliloti. This bacterium is a capable of establishing a symbiosis with the legume alfalfa (Medicago sativa) during which a new specialized organ is formed, the nitrogen-fixing root nodule. These nodules provide the proper physiological conditions for the bacteria to survive in the absence of competing microflora and to reduce atmospheric dinitrogen to ammonia, which is then assimilated by the plant (reviewed in references 34 and 41). The symbiotic properties of rhizobia have been thoroughly investigated (34), yet little is known about how these bacteria are able to persist in their free-living state in the soil and rhizosphere. Previously, we reported the isolation of 33 S. meliloti strains with Tn5-luxAB reporter gene fusions induced by deprivation of carbon, nitrogen, or both (23). Here we report further characterization of one of these strains (C22) that harbors a transcriptional fusion induced by oxygen, nitrogen, or carbon deprivation; by osmotic stress; and during entry into postexponential stationary phase. The locus containing this fusion has been designated ndi for nutrient deprivation induced. In addition, we describe the identification of an S. meliloti gene encoding a homologue of the tryptophan-rich sensory protein TspO from Rhodobacter sphaeroides which is involved in regulating the expression of the ndi locus.
MATERIALS AND METHODS
Media and growth conditions.
S. meliloti strains were grown on or in rich TY medium (4) or defined minimal GTS medium (23) at 28°C, as indicated. GTS-N is GTS medium lacking nitrogen. GTS-C is GTS medium lacking carbon. GTS+NaCl is GTS containing NaCl (400 mM). GTS+sucrose is GTS containing sucrose (30%). E. coli strains were grown on rich medium (Luria-Bertani medium) at 37°C. Antibiotics were added at various concentrations. S. meliloti received streptomycin at 250 μg ml−1, kanamycin at 200 μg ml−1, tetracycline at 10 μg ml−1, and spectinomycin at 50 μg ml−1. E. coli received ampicillin at 100 μg ml−1, kanamycin at 50 μg ml−1, spectinomycin at 100 μg ml−1, and tetracycline at 5 μg ml−1.
Survival during stationary phase.
Strains 1021, C22, 12,C-4, and R-C22 were grown in GTS medium containing limiting amounts of carbon (0.025% glucose). Triplicate cultures (25 ml) were grown in 125-ml flasks with shaking at 28°C for 21 days. Viable cell counts of each culture were determined at regular intervals by plating a dilution series (five replicates) on TY medium containing the appropriate antibiotics. The data were recorded as CFU per milliliter.
Nodulation experiments.
S. meliloti mutant strains C22 and 12,C-4 were screened for the symbiotic phenotype by inoculation on alfalfa (M. sativa) seedling roots and for the ability to fix nitrogen as described previously (22).
Substrate utilization experiments.
To test strains for the ability to utilize or oxidize a variety of different carbon sources, GN Biolog Microplates from BiOLOG (Hayward, Calif.) were used as described by the manufacturer.
DNA manipulations and plasmid constructions.
The strains and plasmids used in this study are described in Table 1. Plasmid DNA for restriction analysis and DNA sequencing was prepared using Wizard minipreps from Promega (Madison, Wis.). Chromosomal DNA was isolated from S. meliloti strains as described by de Bruijn et al. (6). All enzymes for DNA manipulations were purchased from Boehringer Mannheim (Indianapolis, Ind.) or New England Biolabs (Beverly, Mass.). Restriction enzyme digests and ligations were carried out as described by Sambrook et al. (32). Probes were labeled with [α-32P]dATP using a random primer kit (Boehringer Mannheim) and following the manufacturer's instructions. Plasmids were introduced into E. coli hosts by electroporation and into S. meliloti strains via triparental conjugation (5).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristics and (or) derivations | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | supE44 lacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 8a |
| S. meliloti | ||
| 1022 | Smr; wild type | 20a |
| C22 | Kmr; S. meliloti 1021 ndiB::Tn51063a | 23 |
| 12,C-4 | Tcr Kmr; C22 tspO::Tn1721 | This study |
| R-C22 | Tcr; S. meliloti 1021 tspO::Tn1721 | This study |
| 5001 | ntrC::Tn5; Smr Kmr | 37 |
| Plasmids | ||
| pRK2013 | Kmr; triparental mating helper, ColE1 replication, Tra+ | 6a |
| pJOE105 | Tcr suicide vector, contains transposon Tn1721 | 32a |
| pTB93F | Ptrp-GFP-S65T in pMB393; Spr | 8 |
| pBluescript II SK | Apr; cloning and sequencing vector | Stratagene, La Jolla, Calif. |
| pLAFR1 | Broad host range; Tcr IncP Mob+ Tra− | 7a |
| pC22 | Kmr; EcoRI transposon recovery from strain C22 | This study |
| pC22lux | Spr Kmr; entire 12.2-kb EcoRI fragment of pC22 inserted into pTB93 | This study |
| pBSR | Apr; HindIII transposon Tn1721 recovery from strain 12,C-4 | This study |
| pLtspO | Tcr; clone containing tspO locus from pLAFR1 S. meliloti library | 6 |
| pTtspO | Spr; ppuMI-SalI fragment of pLtspO containing tspO ORF from S. meliloti inserted into pTB93 | This study |
| pUI1126 | Tcr; pRK415 containing tspO under control of PrrnB | 48 |
| pRstspO | Spr; BamHI-KpnI fragment containing tspO gene from plasmid pUI1126 inserted into pTB93 | This study |
Two plasmids (pTtspO and pRstspO) were constructed for complementation studies with double-mutant strain 12,C-4. To construct plasmid pTtspO, we replaced the gfp coding region in plasmid pTB93F (8) with an S. meliloti tspO-like open reading frame (ORF) containing a PpuMI-SalI fragment derived from plasmid pLtspO. Expression of the S. meliloti tspO-like ORF in this construct is controlled by the constitutive trp promoter of pTB93F. To construct plasmid pRstspO, we replaced the gfp coding region in plasmid pTB93F (8) with a BamHI-KpnI fragment containing the R. sphaeroides tspO gene from plasmid pUI1126 (48). Expression of R. sphaeroides tspO in this construct is also under the control of the constitutive trp promoter of pTB93F.
Transposon mutagenesis.
Tn1721 transposon mutants were generated by triparental conjugation (5; A. Milcamps, P. Struffi, and F. J. de Bruijn, submitted for publication) using E. coli strain DH5α carrying plasmid pJOE105 (donor; Tcr) or plasmid pRK2013 (helper; Kmr) and the S. meliloti insertion mutant C22 (recipient). Three thousand transposon mutants were isolated, and single colonies were purified, grown in liquid TY medium, and stored in microtiter plates at −80°C. The resulting double-mutant strains were screened for luciferase activity on agar plates under the inducing conditions specified using a Hamamatsu Photonic System model C1966-20 (Photonic Microscopy) as described by Milcamps et al. (23).
Isolation of the Tn1721-tagged locus and genetic techniques.
The Tn1721-tagged locus from double-mutant strain 12,C-4 was isolated by creating a partial genomic library in pBluescript II KS, using 12,C-4 genomic HindIII fragments between 4.0 and 5.0 kb, and screening this library via colony hybridization using a probe corresponding to the first 350 bp of the 5′ end of Tn1721. This clone was designated pBSR. In order to obtain additional flanking sequence information, a pLAFR1 genomic library of S. meliloti 1021 (6) was probed with pBSR to isolate the wild-type genomic clone of the S. meliloti tspO locus. The obtained pLAFR1 genomic clone was designated pLtspO.
Phage φM12 was used for general transduction as described by Finan et al. (7) to reconstruct tspO::Tn1721 as a single mutation. This strain is designated R-C22.
Induction and quantitative measurement of luciferase activity.
To evaluate expression under various stress conditions, strain C22 or strains harboring pC22lux were grown in TY broth with the appropriate antibiotics for 36 h, subcultured in triplicate into GTS or TY broth (1:100 dilution), and grown overnight to early exponential growth phase (optical density between 0.1 and 0.3). The cultures were maintained under aeration for experiments evaluating luxAB gene expression during stationary phase or constantly bubbled with a mixture of 1% oxygen in nitrogen to test the effect of microaerophilic conditions on expression. Alternatively, the cells were centrifuged at room temperature and the pellets were resuspended in regular or modified GTS medium for examination of expression under nutrient deprivation conditions or osmotic stress. All cultures were incubated in a rotory shaker at 28°C, and luciferase activity and cell density were measured at the designated time points.
Luciferase activity was determined with a luminometer (model TD-20e; Turner Designs, Sunnyvale, Calif.) by mixing 140 μl of bacterial culture with 10 μl of n-decyl aldehyde (Sigma, St. Louis, Mo.) and immediately starting the analysis. The aldehyde solution was prepared in water (0.1%, vol/vol) and vortexed for 10 min to form an emulsion. Photons were counted for 20 s, and data was recorded as light units (LU). The assay was performed in triplicate on each culture. Calibration of the luminometer by the method of Hastings and Weber (10) was used to determine that 1 LU equaled approximately 1.4 × 107 photons.
To quantitatively determine the luminescence of carbon-limited and therefore energy-starved cells, a 500-μl aliquot of culture was mixed with an equal portion of complete GTS medium and vortexed briefly. This mixture was incubated at room temperature for 15 min, and a 140-μl aliquot was used for luciferase activity testing as described above.
Since oxygen is required for luciferase activity, the cells that were exposed to low oxygen tension (1%) were aerated briefly before testing by pelleting a 750-μl aliquot of culture and resuspending the pellet in an equal amount of fresh GTS medium. This sample was subsequently immediately tested for luciferase activity as described above.
DNA sequence analysis.
Sequence analysis of DNA fragments carried in Bluescript vectors was performed using standard primers and primer walking strategies. The sequencing was carried out at the DNA sequencing facility at Michigan State University. Initial DNA sequence analysis was carried out using the Sequencher software package (Gene Code Corporation, Ann Arbor, Mich.). ORFs were identified by analyzing the DNA sequence with a codon preference program based on codon usage for S. meliloti (C. Halling, University of Chicago, Chicago, Ill.) and by examining the DNA sequence for start codons and Shine-Dalgarno motifs around translational start sites indicated by the codon usage program. Possible promoter regions were identified by searching for characteristic motifs in the DNA sequence using the Predict Promoter for Prokaryotes program (29a, 29b). Database searches were conducted through the National Center for Biotechnology Information (NCBI) web page using the Gapped BLAST program (2). Alignments of deduced amino acid sequences were obtained using the pileup program of the GCG software package (Genetics Computer Group, Madison, Wis.).
Nucleotide sequence accession numbers.
The DNA sequences obtained in this study have been submitted to GenBank under accession numbers AF178441 (ndi locus) and AF179401 (tspO locus).
RESULTS
S. meliloti strain C22 carries a Tn5-luxAB insertion in a novel locus.
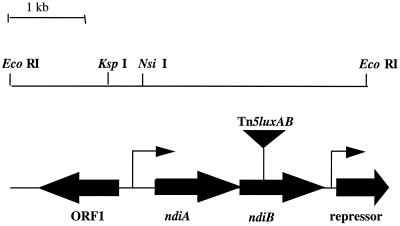
Tn5-luxAB insertion mutant C22 was isolated in a previous screen for S. meliloti strains carrying luxAB reporter gene fusions induced under nutrient deprivation conditions (23). The tagged locus in strain C22 was cloned by excision from the genome as an EcoRI fragment (EcoRI does not cleave within Tn5-luxAB), self-ligated, and electroporated into E. coli (DH5α). The resulting plasmid (pC22) is self-replicating due to the presence of oriV within the Tn5-luxAB transposon (46). The sequence of the DNA flanking the Tn5-luxAB insertion site was determined using unique outward reading primers corresponding to the left and right ends of the transposon, as described previously (23). Additional sequence in the region was determined by primer walking. DNA sequence analysis of the region flanking the Tn5-luxAB insertion (4.7 kb) predicted four ORFs (Fig. 1). The site of insertion in strain C22 was found to be located near the middle of an ORF that, upon comparison of the deduced amino acid sequence to the NCBI protein databases, failed to reveal significant similarity to any known protein. This ORF has been designated ndiB for nutrient deprivation-induced gene B. The predicted translational stop codon (TGA) of the ORF just upstream of ndiB, which has been designated ndiA, and the predicted start (ATG) site of ndiB were found to overlap (GAAGGAGAGACTGCAATGAA). Therefore, it is possible that these two genes are translationally coupled and constitute an operon. In addition, comparison of the deduced amino acid sequence of ndiA also showed no similarity to any sequence in the NCBI protein databases; therefore, both ndiA and ndiB appear to be novel.
FIG. 1.
Map of the Tn5-luxAB-tagged locus from strain C22. Insertion site of Tn5-luxAB is indicated (by an inverted triangle), along with selected restriction sites used for cloning, predicted ORFs (➞), and putative promoter regions ( ). The presumptive direction of transcription is indicated by arrows.
Two strategies were used to delimit the predicted promoter region of the ndiAB locus. First, plasmid pC22 was digested with EcoRI and the entire fragment was cloned into broad-host-range vector pTB93 (pC22lux) and subsequently introduced into S. meliloti strain 1021. Induction of luciferase activity during oxygen, nitrogen, or carbon deprivation; osmotic stress; and stationary phase was observed in the strain containing plasmid pC22lux (data not shown), indicating that the complete ndi promoter region is contained within the 4.7-kb fragment. In addition, the DNA sequence analysis program Promoter Predictions for Prokaryotes (http://www.fruitfly.org /seq_tools/promoter.html) predicts a putative promoter region (score, 0.91) approximately 275 bp upstream of ndiA (Fig. 1).
An ORF encoding a protein with a high degree of similarity (31% identity, 46% similarity) to various repressor proteins, such as PurR and LacI (33), was found adjacent to and downstream (194 bp) of the ndiAB genes, along with a putative promoter region (score, 0.98) identified by the Predict Promoter Program (Fig. 1). It is highly similar to the consensus (−35, −10) promoter element of E. coli (9). It should be noted that within the 4.7-kb sequence analyzed, the promoter region upstream of ndiA and this promoter region were the only two promoters predicted by the program.
In addition, a divergent ORF was predicted at the 5′ end of the putative ndiAB operon, containing a region with significant similarity (41% identity, 60% similarity) to the gene osmY from E. coli, which encodes an osmotically induced periplasmic protein whose function is unknown (51, 52).
The ndiB::Tn5-luxAB gene fusion in strain C22 is induced by O2, N, and C deprivation; by osmotic stress; and during stationary phase.
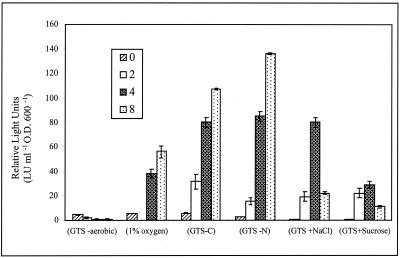
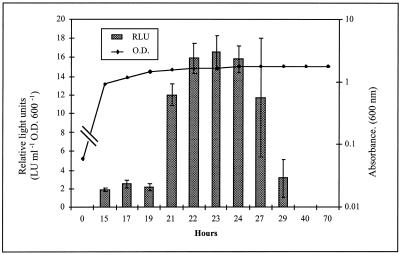
The expression of the chromosomal ndiB::Tn5-luxAB fusion harbored by strain C22 was quantitatively examined by measuring luminescence (relative LU [RLU]) under a variety of growth conditions. The ndi promoter was found to be active at low levels throughout exponential growth (≅3 RLU) and induced 12- to 27-fold after 4 h of carbon deprivation, nitrogen deprivation, or osmotic stress and at reduced oxygen tension (Fig. 2). In addition, an eightfold induction of the ndiB::Tn5-luxAB fusion was observed as the cells of strain C22 entered stationary phase in complex TY medium (Fig. 3). This expression profile was also observed in stationary-phase cells grown in defined GTS medium (data not shown). The observed luxAB reporter gene expression pattern persisted for approximately 8 h after entry into stationary phase. However, since luciferase activity is dependent on the energy status of the cell due to the requirement for reducing equivalents (reduced flavin mononucleotide) to catalyze the bioluminescence reaction (21), the persistence of expression during later stages of stationary phase (after 24 h) could not be quantitatively evaluated.
FIG. 2.
Strain C22 ndi::Tn5-luxAB expression under oxygen (1% oxygen), carbon (GTS-C), and nitrogen (GTS-N) deprivation and under osmotic stress (GTS plus NaCl or sucrose). Luciferase activity was determined at 2-h intervals for 8 h. Luminescence values are defined as described in Materials and Methods. Data are presented as the mean ± the standard deviation (n = 9). O.D. 600, optical density at 600 nm.
FIG. 3.
Strain C22 ndi::Tn5-luxAB expression during entry into postexponential stationary phase in complex TY medium. Cultures were grown in triplicate for 72 h. Luciferase activity and absorbance were determined at the specified intervals. Data are presented as the mean ± the standard deviation (n = 9). O.D. 600, optical density at 600 nm.
A homologue of tspO regulates expression of the ndi locus.
In order to identify genes involved in the regulation of the ndi locus, strain C22 (ndiB::Tn5-luxAB) was mutagenized with a second transposon (Tn1721) as described in Materials and Methods and a library of 3,000 double mutants was screened for altered patterns of ndiB::Tn5-luxAB expression. One double mutant, designated strain 12,C-4, was found in which the ndiB::Tn5-luxAB reporter gene fusion was found to be no longer expressed under any of the previously determined inducing conditions. Southern hybridization analysis of strain 12,C-4 confirmed that this strain contained a single Tn1721 insertion and that the insertion site did not fall within the initial Tn5-luxAB-tagged locus (data not shown).
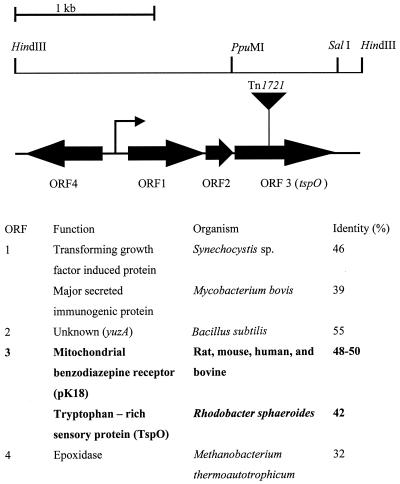
The corresponding Tn1721-tagged locus from double-mutant strain 12,C-4 was isolated as described in Materials and Methods. The structure and DNA sequence of the region surrounding the transposon were determined (Fig. 4). The site of insertion was found to be in a gene encoding a protein with a high degree of similarity to the 18-kDa outer membrane component (pK18) of the mammalian mitochondrial benzodiazepine receptor (≅45% identity, 65% similarity) (20), as well as to an outer membrane oxygen sensor protein (TspO) of R. sphaeroides (42% identity, 67% similarity) (48–50).
FIG. 4.
Map of the Tn1721-tagged locus from double-mutant strain 12,C-4 with indication of the Tn1721 insertion site (inverse triangle), the putative promoter region ( ), selected restriction sites, predicted ORFs (➞), protein sequence similarities, the organism with which the similarity was found, and percent identity. The presumptive direction of transcription is indicated by arrows.
Upstream (53 bp) of the tspO-like gene, a 225-bp ORF (ORF2) was found with a high degree of similarity (55% identity) to the protein encoded by the yuzA gene from Bacillus subtilis (accession no. Z99120). This protein, whose function is unknown, is approximately the same size as the predicted protein in ORF2 (78 amino acids; Fig. 4). Upstream of ORF2 (29 bp) was found another ORF (ORF1), in the same transcriptional direction, which shares a high degree of similarity with a gene encoding a transforming growth factor-induced protein from Synechocystis sp. (46% identity) (15) and a major secreted protein MBP70 precursor from Mycobacterium bovis (39% identity; accession no. A37195). Both of these genes encode secreted proteins that affect eukaryotic gene expression.
A putative promoter region was identified by computer analysis just upstream of ORF1 (score, 0.94), and a divergent ORF (ORF4) upstream of this putative promoter region was identified with similarity to a gene encoding an epoxidase from Methanobacterium thermoautotrophicum (32% identity and 50% similarity) (accession no. AAB85164). In addition, downstream of the tspO-like gene, a divergent ORF encoding a protein with low similarity (31% identity and 46% similarity) to a Tra protein from Streptomyces lividans plasmid pIJ101 (accession no. P22409) was found (data not shown). Thus, the sequence analysis predicts that three ORFs (ORF1, ORF2, and ORF3, which encodes TspO) constitute a Tn1721-tagged operon.
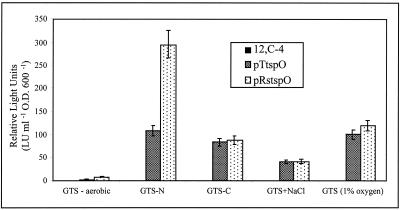
To determine if the dark phenotype of double-mutant strain 12,C-4 was exclusively due to the mutation in the tspO-like gene, a plasmid (pTtspO) which constitutively expresses the coding region of only the tspO-like ORF was introduced into strain 12,C-4 via conjugation for complementation studies. This region was found to restore induction of the ndi locus under all of the previously tested induction conditions (Fig. 5). In addition, the tspO gene from R. sphaeroides was also found to restore proper induction patterns when provided in trans (pRstspO) and expressed under the control of the same constitutive promoter (Fig. 5).
FIG. 5.
Complementation of double-mutant strain 12,C-4 with pTtspO (S. meliloti) and pRstspO (R. sphaeroides) under all inducing conditions. Data for pTtspO is after 6 h, and that for pRstspO is after 8 h of exposure to inducing conditions. Double-mutant strain 12,C-4 is not responsive (0.0 RLU) under any of the conditions tested. A strain 12,C-4 pTB93 vector-only control also showed no expression, as expected (data not shown). Luminescence values are defined as described in Materials and Methods. Data are presented as the mean ± the standard deviation (n = 9). O.D. 600, optical density at 600 nm.
FixL is involved in regulating expression of the ndi locus.
As described above, expression of the Tn5-luxAB fusion in strain C22 is induced by nitrogen and oxygen deprivation (Fig. 2). Two regulatory pathways have been described in S. meliloti that are involved in the control of nitrogen deprivation and microaerobically induced genes, namely, the ntr system, consisting of the ntrA (30) and ntrBC (37) genes, and the fixLJ system (3), respectively. In order to evaluate the involvement of these signaling pathways in the control of ndiB expression, plasmid pC22lux was introduced into fixL::Tn5-233 (J. Trzebiatowki, unpublished data) and ntrC::Tn5 (37) mutant strains of S. meliloti 1021 and luciferase activity was monitored over time under inducing conditions. A similar reporter gene expression profile was found in the wild-type and ntrC mutant backgrounds during nitrogen deprivation (data not shown), indicating that ntrC is not required for ndiB expression during nitrogen deprivation. However, a much lower level of luciferase expression was found under oxygen-limiting conditions in the fixL mutant strain than in the wild-type background (Table 2). To further evaluate this observation, a double chromosomal (S. meliloti 1021 fixL::Tn5-233 ndiB::Tn5-luxAB) mutant was created by transducing the ndi::Tn5-luxAB insertion into S. meliloti 1021 fixL::Tn5-233. This strain (fixL::Tn5-233 ndiB::Tn5-luxAB) was examined for luciferase activity under oxygen-limiting conditions. A decreased level of ndiB::Tn5-luxAB expression during oxygen deprivation was observed, compared to that found in the wild-type background (Table 2), confirming the data obtained with strains carrying the plasmid-borne fusion. Therefore, it appears that fixL is required for full induction of ndiB::Tn5-luxAB reporter gene expression under low oxygen tensions. Furthermore, ndiB-luxAB reporter gene expression patterns were found to be unaltered in the fixL mutant under all of the other inducing conditions tested (carbon and nitrogen deprivation and osmotic stress) (data not shown). Therefore, fixL involvement in the regulation of the ndi locus appears to be specific to sensing and/or responding to low oxygen tension.
TABLE 2.
Plasmid pC22lux and chromosomal ndi::Tn5-luxAB expression in S. meliloti 1021 wild-type and tspO and fixL mutant strains after exposure to low oxygen tension for 8 ha
| Regulatory mutant background | Plasmid pC22lux
|
Chromosomal
ndi::Tn5lux
|
||||
|---|---|---|---|---|---|---|
| 0 h | 8
h
|
0 h | 8 h
|
|||
| Aerobic | 1% O2 | Aerobic | 1% O2 | |||
| Wild type | 493 ± 19.7 | 344 ± 10.7 | 2,632 ± 158.0 | 5.2 ± 0.25 | 0.4 ± 0.0 | 55.8 ± 5.0 |
| fixL::Tn5 | 883 ± 35.2 | 116 ± 4.5 | 1,064 ± 49.7 | 11 ± 0.9 | 0.0 | 8.8 ± 0.72 |
| tspO::Tn1721 | 344 ± 6.9 | 101 ± 2.0 | 453 ± 12.9 | 0.0 | 0.0 | 0.0 |
Luminescence values are defined as described in Materials and Methods. Data are presented as the mean ± the standard deviation (n = 9).
Luciferase activity is dependent on the energy status of the cell. Therefore, it was possible that the observed decrease in ndiB::Tn5-luxAB reporter gene expression in the fixL and tspO mutant backgrounds could be the result of a general effect of the physiology of the cells on luciferase activity. To examine this question, a control strain (CV2) harboring a constitutively expressed Tn5-luxAB fusion in a gene whose function is unknown (23) was used. Double-mutant strains CV2:tspO::Tn1721 and CV2: fixL::Tn5-233 were created by φM12 transduction and examined for luciferase activity under low oxygen tension. Inactivation of the fixL or tspO gene did not affect the luciferase expression of the Tn5-luxAB reporter gene (data not shown). Therefore, expression of the ndi locus, rather than luciferase activity per se, appears to be dependent on fixL and tspO.
Phenotypic analysis of mutant strains.
The growth rate and survival characteristics of strains C22, 12,C-4, and R-C22 were examined. These strains, as well as wild-type strain 1021, were grown in minimal GTS medium, and viable cell counts of each culture were determined at regular intervals for 21 days. All three strains were found to have growth rates similar to that of the wild-type strain. Furthermore, a decrease in viable counts was not observed during stationary phase, indicating that these mutants are not impaired in the ability to persist under starvation conditions (data not shown). In addition, the ability to utilize or oxidize a wide variety of different carbon sources was screened using the GN Biolog Microplates. All three strains had utilization patterns similar to that demonstrated by the wild-type strain (data not shown). Therefore, these mutants do not appear to be deficient in carbon utilization.
The abilities of the C22 and 12,C-4 mutants to nodulate and to fix nitrogen symbiotically were examined in a plant infection test. Both strains were able to nodulate the plant with equivalent numbers of nodules, and the nodules demonstrated levels of nitrogenase activity comparable to that of plants infected with the wild-type strain. Therefore, ndiB and tspO do not appear to be absolutely required for the formation and function of the symbiotic association.
DISCUSSION
Rhizobia provide a unique model system with which to investigate environmental control of gene expression in a bacterium indigenous to soil. These bacteria persist in the free-living state in bulk soil or in an endosymbiotic state in plants. In order to do so, rhizobia must survive under stressful conditions in soil, competitively sense and utilize signaling molecules and growth-promoting nutrients excreted from the plant into the rhizosphere, and adapt to the homeostatic environment within the host plant tissue. It is evident that these three distinct modes of existence require a high degree of physiological adaptability and mechanisms to sense changes in environmental parameters.
The nutrient deprivation responses in rhizobia are just beginning to be investigated. Recently, evidence for a general starvation response in Rhizobium leguminosarum similar to that found in E. coli and Vibrio sp. has been reported (38) and Uhde et al. (39) have identified S. meliloti mutants that are affected in stationary-phase survival (39). In addition, phosphate stress-induced genes in S. meliloti (36), as well genes expressed during carbon or nitrogen deprivation, have been reported (23) and starvation-induced changes in chemotaxis, motility, and flagellation have also been reported (42).
Although S. meliloti starvation or stress response research has only just begun, there is great potential for rapid progress in this well-studied organism. Many of the environmental parameters bacteria encounter in soil, such as low oxygen tensions, nitrogen and carbon limitation, and osmotic changes, are also experienced in planta, either during the infection process or after symbiosis is established. Consequently, a number of genes have been identified during studies on symbiosis that are involved in both physiological adaptations in the free-living state and legume infection (26). These include functions required for the synthesis of cytochrome complexes, amino acids, nucleotides, and chaperonins (GroEL), as well as utilization of different carbon and energy sources (26). It is likely that these genes play a role in the starvation or stress response of rhizobia; however, further studies are required to determine this.
As indicated earlier, upstream of ndiA is a divergent ORF with similarity to an osmY-encoded protein whose function is unknown. Interestingly, this gene has also been found to be induced by carbon starvation (43) and during stationary phase (13, 16), in addition to osmotic stress (52), in E. coli. It should be noted, however, that the similarity is restricted to the last 71 amino acids of the protein encoded by osmY. The first 130 amino acids of osmY do not show similarity to the ORF. In addition, an ORF was identified adjacent to and downstream of the ndi locus which is predicted to encode a protein with striking similarity to transcriptional repressor proteins and it is possible that this protein is involved in controlling expression of the ndi locus. The high background levels of luciferase activity exhibited by cells carrying plasmid pC22lux, even in a tspO mutant background (Table 2), may indicate titration of repressor molecules resulting in a higher level of expression. However, additional experiments are required to determine if this putative regulatory protein is involved in controlling expression of the ndi locus.
To our knowledge, this is the first report that a tspO-like gene in S. meliloti is involved in signal transduction. Recently, however, this same locus was identified by Oke and Long (27) in a screen for S. meliloti genes expressed predominantly in the nodule during the intermediate stages of nodule development. As stated earlier, the environmental parameters used to evaluate gene expression in this study are likely to be found in both the free-living state and in planta, so it is not surprising that the same locus should be identified under these distinctly different screens. Their studies found that plants infected with cells containing a disruption in nex-18 (this gene correlates with ORF1 in Fig. 4) resulted in an altered symbiotic phenotype in that a mixture of Fix+ and Fix− nodules were elicited by the plants infected with this mutant strain. It is therefore evident that this putative operon is expressed during the intermediate stages of nodule development and that ORF1 of this putative operon is important for symbiosis (27). However, further studies are required to determine the function and role of this locus in the development of the symbiotic association.
The TspO outer membrane receptor does not appear to be ubiquitous in nature. Homologues of pK18/TspO have been found in vertebrates, invertebrates, plants, and some yeasts; however, they have not been observed in the majority of the genomes of prokaryotes whose DNA sequencing has been completed. Since a member of the alpha subdivision of purple bacteria is the likely source of the endosymbiont that gave rise to the mammalian mitochondrion (45), the finding of a pK18 orthologue in R. sphaeroides, a member of this subdivision, has been of great interest. Moreover, Yeliseev et al. (50) have demonstrated that the rat pK18 gene could complement a TspO-deficient strain of R. sphaeroides, indicating functional homology. The finding of a pK18/TspO homologue in S. meliloti is also intriguing. Members of the genus Sinorhizobium also belong to the alpha subdivision of purple bacteria; therefore, it is not surprising that similar proteins are retained in these organisms. However, since only a few prokaryotes contain this protein, it may be evolutionarily significant that mitochondria, members of the alpha subdivision of purple bacteria that are the likely source of the endosymbiont that gave rise to the mammalian mitochondrion, and S. meliloti, an endosymbiont, contain this protein.
In Rhodobacter spp., TspO is located in the outer membrane and is associated with the major outer membrane porins. In the mitochondrion, the data indicate that pK18 is also localized to the outer membrane and is associated with the voltage-dependent anion channel (20). Moreover, both the TspO and pk18 complexes bind and transport dicarboxylic tetrapyrrole intermediates of the heme biosynthetic pathway (20, 48). This has been proposed as the likely mechanism by which TspO regulates gene expression in Rhodobacter (47, 49). Although it seems counterintuitive that synthesis of porphyrins would occur during nutrient deprivation, it has been reported that under a variety of conditions, such as iron deficiency, starvation for heme, or oxygen limitation, bacterial cells produce and excrete porphyrins (12, 25, 29). We propose that heme itself or an intermediate of the heme biosynthetic pathway acts as an effector molecule or triggers the synthesis of another effector molecule that binds to a repressor, which then inhibits transcription of the ndi locus. When the concentration of porphyrins or putative effector molecules is decreased, the repressor can no longer inhibit transcription and expression of the ndi locus is restored. It is possible that in the TspO mutant, the concentrations of porphyrins remains high under physiologically stressful conditions; therefore, there is constant repression of the ndi locus.
In addition, FixL appears to be required for full induction of the ndi locus under low oxygen tension. This control may be imposed directly through the response regulator FixJ, or the control may be an indirect effect; that is, FixL may control porphyrin biosynthesis in response to oxygen tensions and thereby indirectly regulate expression of the ndi locus when the oxygen tension is low. However, TspO appears to be epistatic to FixL, since no expression is observed in the tspO mutant, even though FixL is still present.
The mechanism by which TspO in Rhodobacter controls the transport of porphyrins and the nature of the signal that TspO senses remains to be determined. In addition, further investigations are necessary to determine if these two related TspO homologues in S. meliloti and Rhodobacter spp. actually function in a similar manner and respond to the same signals. Nonetheless, these two proteins are involved in regulating gene expression and are likely to provide a new and important way to think about signal transduction in prokaryotes.
ACKNOWLEDGMENTS
This work was supported by NSF STC grant DEB9120006 from the Center of Microbial Ecology and grant DE-FG02-91ER20021 from the Department of Energy.
We thank Jodi Trzebiatowski for providing us with fixL::Tn5-233 and for many helpful discussions and Sam Kaplan for generously providing us with a variety of tspO constructs. We also thank Julie Hines for technical assistance.
REFERENCES
- 1.Alexander M. Non biodegradable and other recalcitrant molecules. Biotechnol Bioeng. 1973;15:611–647. [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zheng Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batut J, Boistard P. Oxygen control in Rhizobium. Antonie van Leeuwenhoek. 1994;66:129–150. doi: 10.1007/BF00871636. [DOI] [PubMed] [Google Scholar]
- 4.Beringer J E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 5.de Bruijn F J, Rossbach S. Transposon mutagenesis. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 387–405. [Google Scholar]
- 6.de Bruijn F J, Rossbach S, Schneider M, Ratet P, Messmer S, Szeto W W, Ausubel F M, Schell J. Rhizobium meliloti1021 has three differentially regulated loci involved in glutamine biosynthesis, none of which is essential for symbiotic nitrogen fixation. J Bacteriol. 1989;171:1673–1682. doi: 10.1128/jb.171.3.1673-1682.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finan T M, Hartwieg E, LeMieux K, Bergman K, Walker G C, Signer E R. General transduction in Rhizobium meliloti. J Bacteriol. 1984;159:120–124. doi: 10.1128/jb.159.1.120-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Friedman A M, Long S R, Brown S E, Buikema W I, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobiummutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 8.Gage D J, Bobo T, Long S R. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa) J Bacteriol. 1996;178:7159–7166. doi: 10.1128/jb.178.24.7159-7166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Hanahan D. Studies on transformation of Escherichia coliwith plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 9.Harley C B, Reynolds R P. Analysis of E. colipromoter. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hastings J W, Weber G. Total quantum flux of isotropic sources. J Opt Soc Am. 1963;53:1410–1415. [Google Scholar]
- 11.Hecker M, Schumann W, Volker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 12.Hendry G S, Jordan D C. Coproporphyrin excretion by Rhizobium meliloti. Can J Microbiol. 1969;15:242–244. doi: 10.1139/m69-043. [DOI] [PubMed] [Google Scholar]
- 13.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 14.Kaiser D. Control of multicellular development: Dictyostelium and Myxococcus. Annu Rev Genet. 1986;20:539–566. doi: 10.1146/annurev.ge.20.120186.002543. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystissp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 16.Lange R, Barth M, Hengge-Aronis R. Complex transcriptional control of the ςs-dependent stationary-phase-induced and osmotically regulated osmY (csi-5) gene suggests novel roles for Lrp, cyclic AMP (cAMP) receptor protein-cAMP complex, and integration host factor in stationary-phase response of Escherichia coli. J Bacteriol. 1993;175:7910–7917. doi: 10.1128/jb.175.24.7910-7917.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loewen P C, Hu B, Strutinsky J, Sparling R. Regulation in the rpoS regulon of Escherichia coli. Can J Microbiol. 1998;44:707–717. doi: 10.1139/cjm-44-8-707. [DOI] [PubMed] [Google Scholar]
- 18.Lynch J M, Whipps J M. Substrate flow in the rhizosphere. Plant Soil. 1990;129:1–10. [Google Scholar]
- 19.Matin A, Auger E A, Blum P H, Schultz J E. Genetic basis of starvation survival in nondifferentiating bacteria. Annu Rev Microbiol. 1989;43:293–316. doi: 10.1146/annurev.mi.43.100189.001453. [DOI] [PubMed] [Google Scholar]
- 20.McEnery M W, Snowman A M, Trifiletti R R, Snyder S H. Isolation of the mitochondrial benzodiazapine receptor: association with the voltage-dependent anion channel and the adenine nucleotide carrier. Proc Natl Acad Sci USA. 1992;89:3170–3174. doi: 10.1073/pnas.89.8.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Meade H M, Long S R, Ruvkun G B, Brown S E, Ausubel F M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meighen E A, Dunlap P V. Physiological, biochemical and genetic control of bacterial bioluminescence. Adv Microb Physiol. 1993;34:1–67. doi: 10.1016/s0065-2911(08)60027-2. [DOI] [PubMed] [Google Scholar]
- 22.Milcamps A, de Bruijn F J. Identification of a novel, nutrient deprivation induced gene (hmgA) in Sinorhizobium meliloti, involved in tyrosine degradation. Microbiology. 1999;145:935–947. doi: 10.1099/13500872-145-4-935. [DOI] [PubMed] [Google Scholar]
- 23.Milcamps A, Ragatz D M, Lim P, Berger K A, de Bruijn F J. Isolation of carbon and nitrogen deprivation induced loci of Sinorhizobium meliloti by Tn5-luxABmutagenesis. Microbiology. 1998;144:3205–3218. doi: 10.1099/00221287-144-11-3205. [DOI] [PubMed] [Google Scholar]
- 24.Morita R Y. Bioavailability of energy and the starvation state. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 1–23. [Google Scholar]
- 25.Nakayashiki T, Inokuchi H. Effects of starvation for heme on the synthesis of porphyrins in Escherichia coli. Mol Gen Genet. 1997;255:376–381. doi: 10.1007/s004380050509. [DOI] [PubMed] [Google Scholar]
- 26.Niner B M, Hirsch A M. How many Rhizobium genes, in addition to nod, nif/fix, and exo, are needed for nodule development and function? Symbiosis. 1998;24:51–102. [Google Scholar]
- 27.Oke V, Long S R. Bacterial genes induced within the nodules during the Rhizobium-legume symbiosis. Mol Microbiol. 1999;32:837–849. doi: 10.1046/j.1365-2958.1999.01402.x. [DOI] [PubMed] [Google Scholar]
- 28.Ostling J, Holmquist L, Flardh K, Svenblad B, Jouper-Jaan A, Kjelleberg S. Starvation and recovery of Vibrio. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 103–123. [Google Scholar]
- 29.Pronk A F, Stigter J, Stouthamer A H, de Bruijn F J, Boogerd F C. Coproporphyrin excretion by Azorhizobium caulinodansunder micro-aerobic conditions. Antonie van Leeuwenhoek. 1998;74:245–251. doi: 10.1023/a:1001734626121. [DOI] [PubMed] [Google Scholar]
- 29a.Reese MG, Eeckman adn F H. New neural network algorithms for improved eukaryotic promoter site recognition. In: Venter J C, Doyle D, editors. Genome science and technology. 1, no.1. Proceedings of the Seventh International Genome Sequencing and Analysis Conference, Hilton Head, S.C. 1995. p. 45. [Google Scholar]
- 29b.Reese M G, Harris N L, Eeckman F H. Large scale sequencing specific neural networks for promoter and spliced site recognition, poster 737–738. In: Hunter L, Klein T E, editors. Biocomputing: Proceedings of the 1996 Pacific Symposium. Singapore: World Scientific Publishing Co.; 1996. [Google Scholar]
- 30.Ronson C W, Nixon B T, Albright L M, Ausubel F M. Rhizobium meliloti ntrA (rpoN) gene is required for diverse metabolic functions. J Bacteriol. 1987;169:2424–2431. doi: 10.1128/jb.169.6.2424-2431.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roszak D B, Colwell R R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 32a.Schoeffl R, Arnold W, Puhler A, Altenbuchner J, Schmitt R. The tetracycline resistance transposons Tn1721 and Tn1771have three 38-base pair repeats and generate five-base pair direct repeats. Mol Gen Genet. 1981;181:87–94. doi: 10.1007/BF00339010. [DOI] [PubMed] [Google Scholar]
- 33.Schumacher M A, Choi K Y, Zalkin H, Brennan R G. Crystal structure of the LacI member, PurR, bound to DNA: minor groove binding by α helices. Science. 1994;266:763–770. doi: 10.1126/science.7973627. [DOI] [PubMed] [Google Scholar]
- 34.Spaink H P, Kondorosi A, Hooykaas P J J. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Press; 1998. [Google Scholar]
- 35.Spector M P. The starvation-stress response (SSR) of Salmonella. Adv Microb Physiol. 1998;40:233–279. doi: 10.1016/s0065-2911(08)60133-2. [DOI] [PubMed] [Google Scholar]
- 36.Summers M L, Elkins J G, Elliot B A, McDermott T R. Expression and regulation of phosphate stress inducible genes in Sinorhizobium meliloti. MPMI. 1998;11:1094–1101. doi: 10.1094/MPMI.1998.11.11.1094. [DOI] [PubMed] [Google Scholar]
- 37.Szeto W W, Nixon B T, Ronson C W, Ausubel F M. Identification and characterization of the Rhizobium meliloti ntrC gene: R. melilotihas separate regulatory pathways for activation of nitrogen fixation genes in free-living and symbiotic cells. J Bacteriol. 1987;169:1423–1432. doi: 10.1128/jb.169.4.1423-1432.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorne S H, Williams H D. Adaptation to nutrient starvation in Rhizobium leguminosarumbv. phaseoli: analysis of survival, stress resistance, and changes in macromolecular synthesis during entry to and exit from stationary phase. J Bacteriol. 1997;179:6894–6901. doi: 10.1128/jb.179.22.6894-6901.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhde C, Schmidt R, Jording D, Selbitschka W, Puhler A. Stationary-phase mutants of Sinorhizobium melilotiare impaired in stationary-phase survival or in recovery to logarithmic growth. J Bacteriol. 1997;179:6432–6440. doi: 10.1128/jb.179.20.6432-6440.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Elsas J D, van Overbeek L S. Bacterial responses to soil stimuli. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 55–77. [Google Scholar]
- 41.van Rhijn P, Vanderleyden J. The Rhizobium-plant symbiosis. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei X, Bauer W D. Starvation-induced changes in motility, chemotaxis and flagellation of Rhizobium meliloti. Appl Environ Microbiol. 1998;64:1708–1714. doi: 10.1128/aem.64.5.1708-1714.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weichart D, Lange R, Henneberg N, Hengge-Aronis R. Identification and characterization of stationary phase-inducible genes in Escherichia coli. Mol Microbiol. 1993;10:407–420. [PubMed] [Google Scholar]
- 44.Williams S T. Oligotrophy in soil: fact or fiction? In: Fletcher M, Floodgate G, editors. Bacteria in the natural environment: the effect of nutrient conditions. New York, N.Y: Academic Press; 1985. pp. 81–100. [Google Scholar]
- 45.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolk C P, Cai Y, Panoff J-M. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc Natl Acad Sci USA. 1991;88:5355–5359. doi: 10.1073/pnas.88.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeliseev A A, Kaplan S. A novel mechanism for the regulation of photosynthesis gene expression by the TspO outer membrane protein of Rhodobacter sphaeroides2.4.1. J Biol Chem. 1999;274:21234–21243. doi: 10.1074/jbc.274.30.21234. [DOI] [PubMed] [Google Scholar]
- 48.Yeliseev A A, Kaplan S. A sensory transducer homologous to the mammalian peripheral-type benzodiazepine receptor regulates photosynthetic membrane complex formation in Rhodobacter sphaeroides2.4.1. J Biol Chem. 1995;270:21167–21175. doi: 10.1074/jbc.270.36.21167. [DOI] [PubMed] [Google Scholar]
- 49.Yeliseev A A, Kaplan S. TspO of Rhodobacter sphaeroides: a structural and functional model for the mammalian peripheral benzodiazepine receptor. J Biol Chem. 2000;275:5657–5667. doi: 10.1074/jbc.275.8.5657. [DOI] [PubMed] [Google Scholar]
- 50.Yeliseev A A, Krueger K E, Kaplan S. A mammalian mitochondrial drug receptor functions as a bacterial “oxygen” sensor. Proc Natl Acad Sci USA. 1997;94:5101–5106. doi: 10.1073/pnas.94.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yim H H, Brems R L, Villarejo M. Molecular characterization of the promoter of osmY, an rpoS-dependent gene. J Bacteriol. 1994;176:100–107. doi: 10.1128/jb.176.1.100-107.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yim H H, Villarejo M. osmY, a new hyperosmotically inducible gene, encodes a periplasmic protein in Escherichia coli. J Bacteriol. 1992;174:3637–3644. doi: 10.1128/jb.174.11.3637-3644.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]