Abstract
Purpose of Review:
The purpose of this review is to provide an update regarding the gut barrier and its involvement with chronic diseases, as well as to review biomarkers for identification of gut barrier integrity. This review is timely and relevant as our knowledge is increasing regarding the role of the gut microbiome and the gut barrier in health and disease.
Recent findings:
This review provides an overview of: the gut barrier, which is complex and comprised of the mucus layer and the intestinal apical junctional protein complex; the gut microbiome in its relation to regulating the integrity of the gut barrier; select acute and chronic conditions that are known to be associated with gut dysbiosis and impaired gut integrity or “leaky gut”; and current means for identifying loss in gut barrier integrity.
Summary:
Many chronic conditions are associated with gut dysbiosis and systemic inflammation. Identifying whether the gut barrier is compromised in these conditions could help to inform potential therapeutics as a means to correct losses in gut barrier integrity and mitigate associated medical conditions.
Keywords: gut barrier, tight junction proteins, gut microbiome, butyrate, leaky gut
Introduction:
There is emerging knowledge regarding the interrelationship between the gut microbiome, gut barrier and many acute and chronic diseases. The gut microbiome and the host intestinal cells communicate with each other in order to maintain homeostasis, which includes supporting an intact gut barrier. Many chronic diseases are associated with gut dysbiosis and systemic inflammation. Systemic inflammation that is associated with gut dysbiosis is presumed to stem from gut microbial byproducts entering into systemic circulation as a consequence of a compromise in the integrity of the intestine. Identifying individuals that may be at risk for gut permeability utilizing specific biomarkers is of high interest as this may assist to inform potential therapeutics which could correct gut barrier disruption and mitigate disease. The aim of this review is to provide an overview of the gut barrier and potential biomarkers used to identify its compromise.
Overview of the Gut Barrier
The physical gut barrier is comprised of a mucus layer and a single layer of intestinal epithelial cells (IEC). (Fig 1) On the apical surface of the epithelium lies a single layer of mucus in the small intestine and a double layer in the colon. Produced by the goblet cells, the mucus is composed primarily of water and glycosylated mucins which form a gel-like substance overlaying the epithelium.[1] Other secretory cells of the epithelium release various molecules into the mucus layer including anti-microbial peptides, trefoil factor 3, and others that regulate the immune response and promote epithelial repair.[2–4] Beneath the mucus, the IECs are adhered to one another via the apical junctional complex forming another physical barrier of the epithelium. Sealing the paracellular space, the junctional protein complex includes tight junctional proteins (e.g., zonula occludens, claudins, occludin), adherens junctions (e.g., zonula adherens), and desmosomes (e.g., macula adherens).[5] The apical junctional complex is also regulated by a peri-junctional actomyosin ring associated with myosin light chain kinase (MLCK). Overall, the tight junctional proteins regulate the permeability of the epithelium and upon their disassembly, pathogen and pathogenic byproducts can translocate from the gut lumen into systemic circulation.[6]
Figure 1: The Intestinal Barrier.
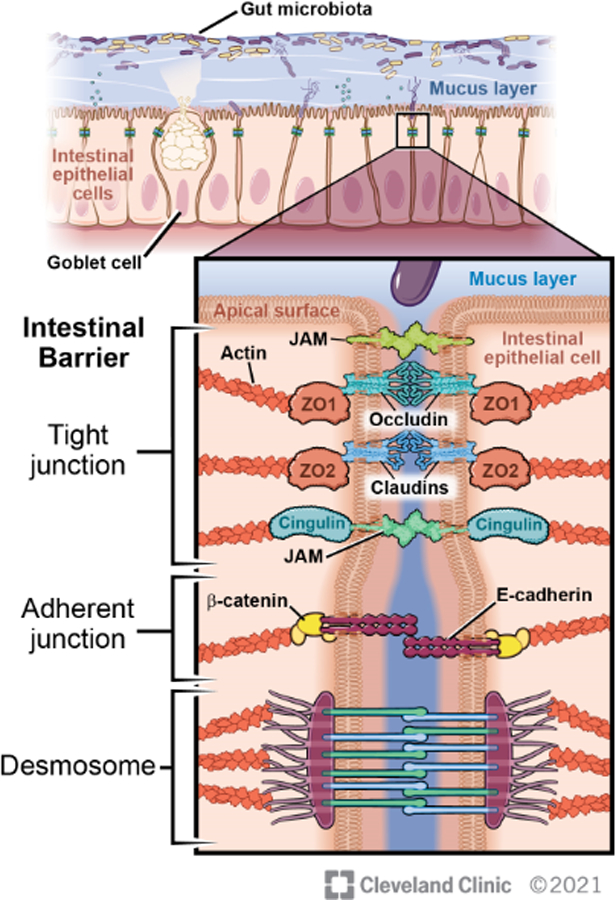
The intestinal barrier is comprised of a physical barrier which includes a mucus layer and intestinal epithelial cells. The intestinal epithelial cells are joined together by the apical junctional complex which seals the paracellular space and prevents intestinal permeability. The gut microbiome regulates both the mucus layer and the apical junctional complex. Illustration by David Schumick, BS, CMI. Reprinted with the permission of the Cleveland Clinic Center for Medical Art & Photography © 2021. All Rights Reserved
The Gut Microbiome and Gut Barrier
Within the gut lumen resides trillions of microbes including bacteria, fungi/yeasts, viruses, and protozoa. Microbial densities increase from the proximal to distal gut where the numbers of microbes can reach 1012 colony forming units (CFU) in the colon. The host and the microbiome have a mutualistic relationship in that each party benefits from the presence of the other in healthy conditions. The host supplies the microbiome a safe niche to reside and a steady food supply. The gut microbiome supports the host in digestion, fermentation, and synthesis of important vitamins, enzymes, and other metabolites, as well as provides protection against pathogens through multiple mechanisms.[7]
In the absence of disease, the gut microbiome supports the gut barrier through many mechanisms, one of which is the development and support of the mucus layer. Studies in germ-free mice support that in the absence of microbes, mucin-generating goblet cells are reduced and release of the mucus layer from the goblet cells is compromised.[8] Mucus detachment, an important step for small intestinal homeostasis, requires the enzyme meprinβ, and this enzyme is dependent upon the gut microbiome for its activation.[9] The thickness and permeability of the mucus layer is also regulated by the gut microbiome.[10] Through its production of beneficial metabolites, the gut microbiome also supports the tight junctional proteins in the intestinal epithelium. In particular, the short-chain fatty acid (SCFA) butyrate, which is generated by gut microbial fermentation of dietary fibers, has been shown to regulate the intestinal tight junctional proteins and intestinal mucus production. In vitro studies demonstrate butyrate protects the functional integrity of the epithelium as demonstrated by mitigating the losses in trans epithelial resistance (TER) and permeability to fluorescein-isothiocyanate (FITC)-dextran induced by endotoxin or ethanol exposure.[11] Multiple mechanistic effects of butyrate on the intestinal epithelial barrier have been demonstrated including induction of AMP-activated protein kinase activity, reduced activation of NLRP3 inflammasome and autophagy due to butyrate’s histone deacetylase activity.[12] The hypoxia-inducible factor (HIF), a transcription factor that coordinates barrier function, was shown to be dependent upon microbial-generated butyrate.[13]
Diseases with Associated Gut Barrier Disruption and Gut Dysbiosis
About 2500 years ago Hippocrates hypothesized that “all disease begins in the gut”. Though this idea has not come fully to fruition, our understanding of the role the gut barrier plays in a variety of diseases is rapidly expanding. Technological advancements such as genome wide association studies and next generation sequencing are revolutionizing our understanding of both the genetic and environmental factors that underlay the pathology of many diseases related to the intestinal barrier. Interactions between the gut microbiome and IEC are incredibly dynamic. Thus, the causes and consequences of gut dysbiosis are broad-ranging and often not fully elucidated. There are diseases in which breakdown of the intestinal barrier, or “leaky gut” is a primary pathology such as in gastric ulcers or inflammatory bowel disease (IBD). There are also diseases in which changes of the intestinal barrier occurs secondarily to other pathology such as intestinal ischemia from hypotension or thromboembolism.
Various medical therapies and conditions can result in gut dysbiosis. Antibiotic therapy not only targets pathogenic microbes but also gut commensals which allow for the overgrowth of opportunistic pathogens, such as Clostridioides difficile. Surgical intestinal resection that removes the ileocecal valve, results in short bowel syndrome, or manipulates architecture of the intestine (e.g., gastric bypass surgery) can promote small intestinal bacterial overgrowth. Moreover, there are many non-gastrointestinal diseases that have associated evidence of gut dysbiosis in which it is not yet known to be the cause or effect of the disease. Included are autoimmune diseases (e.g., ankylosing spondylitis (AS), rheumatoid arthritis), metabolic diseases (obesity, type 2 diabetes mellitus), cardiovascular disease, cancers, neurologic diseases (e.g., autism, attention deficit hyperactivity disorder, multiple sclerosis), and hepatic diseases (non-alcoholic steatohepatitis (NASH), non-alcoholic fatty liver disease (NAFLD)). The variety of diseases involving gut barrier disruption is truly vast and beyond the scope of a single manuscript, however we highlight some clinically relevant topics below.
IBD is the result of complex interactions between genetic, immune, and environmental variables that culminate in a breakdown of the mutualistic relationship between the gut microbiome and barrier that begins at the mucus layer.[6, 14] The mucus layer in patients with IBD is characterized to have a reduction in beneficial butyrate-producing microbes, such as Faecalibacterium praunnitzii, and expansion in pathogenic Escherichia coli species, some of which can then invade the IEC intracellularly causing subsequent cell death.[15, 16] Butyrate, along with other SCFAs, has various beneficial anti-inflammatory and immunomodulating effects.[17] SCFAs serve as an energy source for colonocytes, and importantly exert action on the host through G-protein coupled receptors and inhibition of histone deacetylase activity which results in epigenetic changes.[17] Indeed, patients with AS have reduced levels of butyrate metabolism.[18] Interestingly, the relationship between IBD with rheumatoid arthritis and ankylosing spondylitis has been known for over 100 years to the extent that they were once considered a single disease.[19] Patients with AS also exhibit an increased abundance of Proteobacteria, Bacteroidetes, Enterobacteriaceae, and Prevotella and a decrease in SCFA-producing F. prausnitzii and Eubacterium halli.[18, 20–23] This relationship with the microbiome though is not fully explained. Other research has shown that mice with increased human leukocyte antigen (HLA)-B27 fail to develop spondyloarthritis when in a sterile environment, however this changes when commensal bacteria are introduced into their microbiome.[24] Beyond arthritis, IBD is associated with a variety of other autoimmune diseases including primary sclerosing cholangitis, type 1 diabetes (T1D), and even multiple sclerosis, all of which exhibit their own microbial alterations.[25–29]
Patients with T1D have an increased abundance of Bacteroides species and reduced abundance of beneficial SCFA-producing bacteria.[30] Testing NOD mouse models that develop spontaneous autoimmune T1D, it has been suggested that gut barrier disruption accelerates development of T1D by increasing activation of pancreatic draining lymph diabetogenic CD8+ T cells.[31] In patients with obesity, gut microbial diversity shifts to increase the abundance of Firmicutes at the expense of Bacteroidetes.[32] Gut dysbiosis has been proposed as a possible cause of obesity. A study in which the gut microbiota of human identical twins discordant for obesity was transplanted into germ-free mice found that mice only gained increased body mass if they received the microbiota from the obese twin.[33] Interestingly, when mice transplanted with the obese twin’s microbiota cohabited with the lean mice, this prevented the obese phenotype from developing as a result of invasion with Bacteroidetes from the lean mice microbiome.[33] NAFLD and NASH often co-present with obesity in humans and exhibit similar dysbiotic trends. Both obesity and NASH microbiomes are heavily represented by Prevotella species as compared to healthy individuals.[34] Patients with NAFLD and NASH are also more likely to have increased intestinal permeability via disruption of tight junction proteins that occurs after initial hepatic injury.[35] Interestingly, intestinal permeability through direct exposure of endotoxemia from lipopolysaccharide of gram negative microbes has also been suggested as playing role in the pathogenesis of NASH.[36]
Biomarkers for the Appearance and Progression of Gut Barrier Compromise
In the context of this review, biomarkers for failure of the gut barrier are considered to be the detection of or changes in a biological product or process that are indicative of an increase in gut permeability. Additionally, we sought to highlight biomarkers that are associated directly with the gut barrier structure or function. While a variety of other biological activities are under investigation for their use in monitoring gut function, the ones highlighted in the following sections are most reliable in identifying changes in gut barrier function. Markers can be host-derived or produced by opportunistic pathogenic microbes, and will be discussed in three categories:
Primary – Changes in the production of junctional protein components, or their assembly into functional exclusionary junctions, leading to increased intestinal permeability.
Secondary – Detection of or changes in a biological component or process that would only be present if gut integrity has already been compromised. For example elevated levels of an endotoxin in the circulatory entering by way of a compromised gut barrier.
Increased Risk – Changes in a biological process or component that would increase the likelihood of cell junctional failure in the immediate future.
Together these three categories could be thought of as a manner to track the progression of intestinal permeability, from initial risk factors to the pathological end results of such a condition. Individually each marker provides an indication that failure of the gut barrier may have occurred, but multiple markers, either from the same category or multiple, may be necessary to confidently identify intestinal permeability as the primary cause for their appearance. Table 1 shows the various metabolic functions and products that will be discussed for each category.
Table 1:
Summary of Biomarkers for Gut Barrier Integrity
| Category | Biomarker | Advantages | Disadvantages |
|---|---|---|---|
| Primary | Loss of junctional proteins (zonulins, claudins, occludin, etc.) | IHC allows visualization of barrier disruption Can be quantified by western blot or RT-PCR |
Disagreement over which proteins are diagnostic and what concentration constitutes loss of function |
| Differential Sugar Method | Direct measurement of gut barrier function | Need for multiple urine time point collections. Different time points needed to asses small intestine permeability vs entire intestine | |
| Secondary | Endotoxin in circulatory system | Quantitative analysis available | Most widely accepted test (LAL) is currently being phased out |
| Presence of LPS Binding Protein and CD14 | Direct evidence of antigen translocation | No consistent protocol has been established for clinicians | |
| Presence of Intestinal Fatty Acid Binding Protein | Measurement of tissue destruction that would indicate Gut failure | Can be present due to other causes of epithelial damage and infection | |
| Increased Risk | Decreased SCFA and Butyrate Levels | Directly promotes the function of gut barrier | Absence isn’t a definitive marker or loss of function |
| Microbiome Diversity | Associated with a wide variety of diseases. | “Healthy” ratios have not been established. Microbial populations fluctuate based on a number of variables besides health |
Primary Indicators
As multiple junctional proteins maintain intestinal integrity to prevent the translocation of pathogenic material across the epithelial barrier, the loss of one or several of these components can compromise barrier function. In this capacity, these proteins may serve as direct markers for gut barrier disruption. Several molecular biology techniques can be utilized to identify epithelial barrier integrity. Histological analyses employing immunostaining for antigens against epithelial barrier proteins in intestinal tissue, obtained either surgically or endoscopically, allows for visual analysis of the epithelial barrier proteins. (Fig 2) Additionally, intestinal tissue samples can be quantitatively analyzed for messenger RNA (mRNA) and protein levels of junctional proteins by real-time polymerase chain reaction (RT-PCR) or immunoblotting, respectively. Zonulin, a human analogue of zonula occludens toxin produced by Vibrio cholorae, is a protein secreted by intestinal epithelial cells in in response to the presence of certain bacteria and environmental triggers. When bound to the epithelial cells, this inflammatory protein causes various components of tight junctions to disassemble, leading to a loss in their function. Zonulin protein can be assessed in both serum and stool samples. While these characteristics allow for elevated levels of zonulin protein to potentially act as a biomarker of such diseases, recent studies have called in to question whether or not changes in zonulin protein concentration consistently equates with barrier loss. Thus, while these techniques may assist in identifying potential loss in the intestinal barrier integrity, they do not indicate whether the barrier function is altered.
Figure 2: Immunostaining of Tight Junctional Proteins in Normal Mouse Proximal Colon.
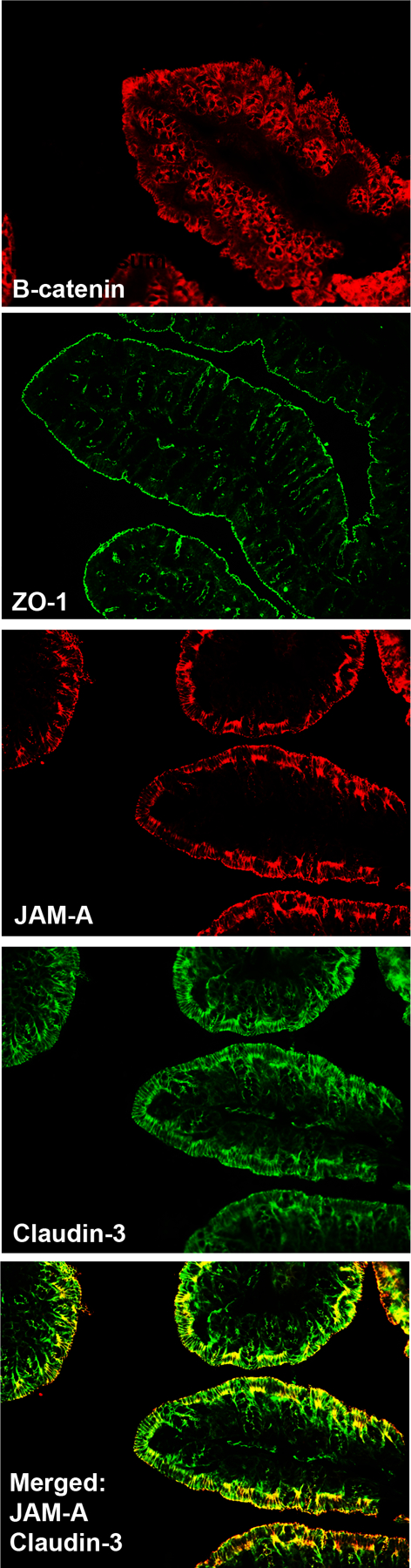
Proximal colon was dissected from a C57BL/6 mouse and immunostained for antibodies against the tight junctional proteins: β-catenin, zonulen occludin-1 (ZO-1), JAM-A, and Claudin-3.
Use of the “differential sugar” method can inform whether there is a functional loss in the epithelial barrier.[37–39] In this research method, a mixture of sugars that are absorbed from different regions of the gastrointestinal tract are ingested orally, then excreted in the urine unchanged. For example:lactulose, a non-absorbable disaccharide comprised of galactose and fructose, has limited ability to passively diffuse across an intact intestinal mucosa due to its large molecular size, and is metabolized in the colon by the gut microbiota into monosaccharides and then volatile fatty acids. Undigested mannitol, a sugar alcohol, is capable of crossing an intact intestinal mucosa, entering into circulation and being excreted in urine unchanged. Thus, lactulose and mannitol are markers of intestinal permeability via paracellular and transcellular pathways, respectively making the lactulose to mannitol ratio a marker of small bowel permeability. Therefore, an intact intestinal barrier will exhibit a lower lactulose:mannitol ratio, while a compromised intestinal barrier will allow for the passage of lactulose, and thus have a higher lactulose:mannitol ratio. While lactulose and mannitol are commonly used, a variety of other sugars may be used as both the absorbable and non-absorbable saccharide, such as L- rhamnose and sucralose respectively.
Secondary Indicators
Secondary indicators can be considered biomarkers that appear after the loss of gut barrier function, and may serve as a signal that primary indicators may also be present. An example of this is the appearance of endotoxin or other pathogenic compound derived from a gut microbe in circulation (e.g., endotoxemia). This event would occur as a consequence of gut permeability allowing the endotoxin to translocate from the intestinal lumen into the blood stream. Endotoxins, mostly lipopolysaccharides (LPS) derived from the outer membrane of gram negative bacteria, have been associated with the onset of various disease states.[40–42] Endotoxin levels can be measured using the Limulus Amebocyte Lysate assay (LAL), though there is currently and effort to replace it with the Recombinant Factor C test due to LAL potentially harming the horseshoe crab population since their blood is a primary reagent in the assay. Host immune responses to increased gut permeability may also be used as a marker for gut permeability. For instance, LPS binding protein captures and delivers LPS to cluster of differentiation 14 (CD14) positive immune cells which then secrete CD14 as part of the immune response. These functions allow the elevated presence to LPS binding protein and secretory CD14 to serve as secondary indicators for LPS translocation due to increased intestinal permeability.[40, 43–48] Finally, intestinal fatty acid binding protein has been suggested as another biomarker due to its release in the circulatory system in response to intestinal tissue damage, and its presence can be detected and quantified in plasma using ELISA.
Markers of Increased Risk
The final biomarker category are those that indicate a patient is at an increased risk of gut barrier disruption. While these markers are not directly indicative of increased gut permeability, they may serve as a signal that a potential problem and prophylactic intervention may be indicated. The most commonly discussed marker in this category is butyrate levels. As butyrate is known to regulate the intestinal barrier via multiple mechanisms, depleted luminal butyrate levels coincide with impaired intestinal integrity.[11, 49] Therefore, butyrate levels could be thought of as a biomarker, with reduced levels signaling conditions favoring an increased likelihood of gut permeability. In a broader sense, the composition of the microbiome as a whole could be seen as a biomarker. For instance a reduction in Firmicutes, one of the primary phyla in the gut, could be seen as a marker for gut health with a higher presence of Firmicutes generally being suggested as favorable due to a majority of butyrate-producing bacteria belonging to this phyla.[50–52] Unfortunately, a definitive “healthy” microbiome has not been firmly established since composition can be affected by a wide variety of factors.[53–55]
Gut permeability could be considered a biomarker itself for other conditions. For example, Clostridioides difficile infection leads to tissue destruction by way of toxins produced by the pathogen.[56–59] Consequential damage to the intestinal lining may present with increased intestinal permeability. While interesting, most gastrointestinal infections have superior diagnostic markers, such as presence of the inciting pathogen in the stool by culture or PCR, or detection of characteristic compounds such as toxins or fecal calprotectin.
Conclusion
As knowledge regarding the relationship between the gut barrier and its disassembly with various disease states continues to emerge, it is important to understand and recognize indicators of gut barrier disruption. While current biomarkers of gut barrier disruption are in their infancy, rapid technological development futuristically will expand our knowledge regarding direct and indirect biomarkers that originate from the gut microbiome and its metabolome for early detection of gut barrier integrity. These findings will assist in early identification of an impaired gut barrier. Additionally, targeted investigations may expand our knowledge to potential mechanisms of an impaired gut barrier. Thus, this will help identify new biomarkers of gut function, as well as inform new therapeutics to target the gut barrier and mitigate disease.
Key points:
There is crosstalk between the gut microbiome and the host intestine that maintains homeostasis including an intact gut barrier.
The physical gut barrier is comprised of a mucus layer and apical junctional complex which seals the paracellular space between intestinal epithelial cells.
Many acute and chronic conditions are associated with a breakdown of the intestinal barrier, often termed “leaky gut”.
Currently there are several methods used to identify gut integrity, but these have many limitations and better biomarkers are needed.
Acknowledgements:
We would like to thank Dave Schumick, Cleveland Clinic Art Department, for his expertise in developing Figure 1.
Financial Support:
This work was supported in part by the National Institutes of Health, United States. (R01AA028043– 01A1 to GAMC)
Footnotes
Conflicts of interest: The authors have no conflicts of interest.
References
- 1.Melhem H, Regan-Komito D, Niess JH. Mucins Dynamics in Physiological and Pathological Conditions. Int J Mol Sci 2021;22(24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehmann D, Wendler J, Koeninger L, Larsen IS, Klag T, Berger J, et al. Paneth cell α-defensins HD-5 and HD-6 display differential degradation into active antimicrobial fragments. Proc Natl Acad Sci U S A 2019;116(9):3746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelaseyed T, Hansson GC. Membrane mucins of the intestine at a glance. J Cell Sci 2020;133(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann W Trefoil Factor Family (TFF) Peptides and Their Diverse Molecular Functions in Mucus Barrier Protection and More: Changing the Paradigm. Int J Mol Sci 2020;21(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckley A, Turner JR. Cell Biology of Tight Junction Barrier Regulation and Mucosal Disease. Cold Spring Harb Perspect Biol 2018;10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parikh K, Antanaviciute A, Fawkner-Corbett D, Jagielowicz M, Aulicino A, Lagerholm C, et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 2019;567(7746):49–55. [DOI] [PubMed] [Google Scholar]
- 7.Sędzikowska A, Szablewski L. Human Gut Microbiota in Health and Selected Cancers. Int J Mol Sci 2021;22(24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson Malin EV, Jakobsson Hedvig E, Holmén-Larsson J, Schütte A, Ermund A, Rodríguez-Piñeiro Ana M, et al. Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell Host & Microbe 2015;18(5):582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Werny L, Colmorgen C, Becker-Pauly C. Regulation of meprin metalloproteases in mucosal homeostasis. Biochim Biophys Acta Mol Cell Res 2022;1869(1):119158. [DOI] [PubMed] [Google Scholar]
- 10. Paone P, Cani PD. Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut 2020;69(12):2232–43. * This review highlights some of the most important advancements made in our understanding of the mucus barrier.
- 11. Siddiqui MT, Cresci GAM. The Immunomodulatory Functions of Butyrate. J Inflamm Res 2021;14:6025–41. * This paper succintly presents the most up to date experimental models and human studies which explore therapeutic potential of butyrate supplementation.
- 12.Feng Y, Wang Y, Wang P, Huang Y, Wang F. Short-Chain Fatty Acids Manifest Stimulative and Protective Effects on Intestinal Barrier Function Through the Inhibition of NLRP3 Inflammasome and Autophagy. Cell Physiol Biochem 2018;49(1):190–205. [DOI] [PubMed] [Google Scholar]
- 13.Wang RX, Henen MA, Lee JS, Vögeli B, Colgan SP. Microbiota-derived butyrate is an endogenous HIF prolyl hydroxylase inhibitor. Gut Microbes 2021;13(1):1938380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham DB, Xavier RJ. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 2020;578(7796):527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018;67(1):108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamali Dolatabadi R, Feizi A, Halaji M, Fazeli H, Adibi P. The Prevalence of Adherent-Invasive Escherichia coli and Its Association With Inflammatory Bowel Diseases: A Systematic Review and Meta-Analysis. Front Med (Lausanne) 2021;8:730243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Wang J, He T, Becker S, Zhang G, Li D, et al. Butyrate: A Double-Edged Sword for Health? Adv Nutr 2018;9(1):21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou C, Zhao H, Xiao XY, Chen BD, Guo RJ, Wang Q, et al. Metagenomic profiling of the pro-inflammatory gut microbiota in ankylosing spondylitis. J Autoimmun 2020;107:102360. [DOI] [PubMed] [Google Scholar]
- 19.Ashrafi M, Kuhn KA, Weisman MH. The arthritis connection to inflammatory bowel disease (IBD): why has it taken so long to understand it? RMD Open 2021;7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Han R, Zhang X, Fang G, Chen J, Li J, et al. Fecal microbiota in patients with ankylosing spondylitis: Correlation with dietary factors and disease activity. Clin Chim Acta 2019;497:189–96. [DOI] [PubMed] [Google Scholar]
- 21.Li M, Dai B, Tang Y, Lei L, Li N, Liu C, et al. Altered Bacterial-Fungal Interkingdom Networks in the Guts of Ankylosing Spondylitis Patients. mSystems 2019;4(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klingberg E, Magnusson MK, Strid H, Deminger A, Ståhl A, Sundin J, et al. A distinct gut microbiota composition in patients with ankylosing spondylitis is associated with increased levels of fecal calprotectin. Arthritis Res Ther 2019;21(1):248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harkins P, Burke E, Swales C, Silman A. ‘All disease begins in the gut’-the role of the intestinal microbiome in ankylosing spondylitis. Rheumatol Adv Pract 2021;5(3):rkab063. * Ankylosing spondylitis is a chronic, complex and debilitating arthropathy with a strong genetic predisposition. Alterations in the gut microbiome are implicated in disease pathogenesis. Disregulation of the intestinal tight junction proteins and increased intestinal permeability are present in AS patients and first-degree relatives. Targeting the gut microbiome may be a means to mitigate AS and this study presents the msot comprehensive review of our current understanding between the microbiome and AS..
- 24.Rath HC, Herfarth HH, Ikeda JS, Grenther WB, Hamm TE Jr., Balish E, et al. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J Clin Invest 1996;98(4):945–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bar Yehuda S, Axlerod R, Toker O, Zigman N, Goren I, Mourad V, et al. The Association of Inflammatory Bowel Diseases with Autoimmune Disorders: A Report from the epi-IIRN. J Crohns Colitis 2019;13(3):324–9. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y, Musco H, Simpson-Yap S, Zhu Z, Wang Y, Lin X, et al. Investigating the shared genetic architecture between multiple sclerosis and inflammatory bowel diseases. Nat Commun 2021;12(1):5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halling ML, Kjeldsen J, Knudsen T, Nielsen J, Hansen LK. Patients with inflammatory bowel disease have increased risk of autoimmune and inflammatory diseases. World J Gastroenterol 2017;23(33):6137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang EA, Han K, Chun J, Soh H, Park S, Im JP, et al. Increased Risk of Diabetes in Inflammatory Bowel Disease Patients: A Nationwide Population-based Study in Korea. Journal of clinical medicine 2019;8(3):343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Losurdo G, Brescia IV, Lillo C, Mezzapesa M, Barone M, Principi M, et al. Liver involvement in inflammatory bowel disease: What should the clinician know? World J Hepatol 2021;13(11):1534–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018;562(7728):589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee AS, Gibson DL, Zhang Y, Sham HP, Vallance BA, Dutz JP. Gut barrier disruption by an enteric bacterial pathogen accelerates insulitis in NOD mice. Diabetologia 2010;53(4):741–8. [DOI] [PubMed] [Google Scholar]
- 32.Castaner O, Goday A, Park YM, Lee SH, Magkos F, Shiow STE, et al. The Gut Microbiome Profile in Obesity: A Systematic Review. Int J Endocrinol 2018;2018:4095789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013;341(6150):1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwimmer JB, Johnson JS, Angeles JE, Behling C, Belt PH, Borecki I, et al. Microbiome Signatures Associated With Steatohepatitis and Moderate to Severe Fibrosis in Children With Nonalcoholic Fatty Liver Disease. Gastroenterology 2019;157(4):1109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luther J, Garber JJ, Khalili H, Dave M, Bale SS, Jindal R, et al. Hepatic Injury in Nonalcoholic Steatohepatitis Contributes to Altered Intestinal Permeability. Cell Mol Gastroenterol Hepatol 2015;1(2):222–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sookoian S, Pirola CJ. Liver tissue microbiota in nonalcoholic liver disease: a change in the paradigm of host-bacterial interactions. Hepatobiliary Surg Nutr 2021;10(3):337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Khan MR, Faubion WA, Dyer R, Singh R, Larson JJ, Absah I. Role of Lactulose Rhamnose Permeability Test in Assessing Small Bowel Mucosal Damage in Children with Celiac Disease. Glob Pediatr Health 2020;7:2333794x20969278. * Autoimmune disorders, such as celiac disease, are increasing and linked with genetic and environmental factors, such as the gut microbiome. This was one of the few prospective studies that evaluated the mucosal integrity of pediatric pateints with celiac disease and proposed a noninvasive test of muscoal healing. Current standards of care include invasive tests such as endoscopy with biopsies and this proposed technique may be allow for an alternative in the future. The lactulose/rhamnose was permeability test was studied to determine if gut permeability was linked with celiac disease-induced small bowel damage as a means to assess mucosal healing in children. When compared to siblings and age-matched controls, the L/R ratio was associated with the celiac disease group comoared with controls. There were no differences between L/R in siblings vs. controls.
- 38.Musa MA, Kabir M, Hossain MI, Ahmed E, Siddique A, Rashid H, et al. Measurement of intestinal permeability using lactulose and mannitol with conventional five hours and shortened two hours urine collection by two different methods: HPAE-PAD and LC-MSMS. PLOS ONE 2019;14(8):e0220397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sequeira IR, Lentle RG, Kruger MC, Hurst RD. Standardising the Lactulose Mannitol Test of Gut Permeability to Minimise Error and Promote Comparability. PLOS ONE 2014;9(6):e99256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ghosh SS, Wang J, Yannie PJ, Ghosh S. Intestinal Barrier Dysfunction, LPS Translocation, and Disease Development. Journal of the Endocrine Society 2020;4(2):bvz039. * The “leaky gut” relationship between disease and LPS translocation has been a topic of immense scientific and public interest. This manuscript synthesizes the most current scientific understanding of LPS translocation and the development of disease. It also highlights the development of potential therapies for barrier dysfunction associated diseases. Understanding of these complex pathologic mechanisms could be the key to developing the next generation of treatment for various disease.
- 41.Tulkens J, Vergauwen G, Van Deun J, Geeurickx E, Dhondt B, Lippens L, et al. Increased levels of systemic LPS-positive bacterial extracellular vesicles in patients with intestinal barrier dysfunction. Gut 2020;69(1):191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vatanen T, Kostic Aleksandar D, d’Hennezel E, Siljander H, Franzosa Eric A, Yassour M, et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 2016;165(4):842–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sikora M, Stec A, Chrabaszcz M, Waskiel-Burnat A, Zaremba M, Olszewska M, et al. Intestinal Fatty Acid Binding Protein, a Biomarker of Intestinal Barrier, is Associated with Severity of Psoriasis. J Clin Med 2019;8(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin L, Yuan F, Chen C, Wu J, Gong R, Yuan G, et al. Degradation Products of Polydopamine Restrained Inflammatory Response of LPS-Stimulated Macrophages Through Mediation TLR-4-MYD88 Dependent Signaling Pathways by Antioxidant. Inflammation 2019;42(2):658–71. [DOI] [PubMed] [Google Scholar]
- 45.Ciesielska A, Matyjek M, Kwiatkowska K. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell Mol Life Sci 2021;78(4):1233–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho SSC, Keenan JI, Day AS. The Role of Gastrointestinal-Related Fatty Acid-Binding Proteins as Biomarkers in Gastrointestinal Diseases. Dig Dis Sci 2020;65(2):376–90. [DOI] [PubMed] [Google Scholar]
- 47.Goswami P, Sonika U, Moka P, Sreenivas V, Saraya A. Intestinal Fatty Acid Binding Protein and Citrulline as Markers of Gut Injury and Prognosis in Patients With Acute Pancreatitis. Pancreas 2017;46(10):1275–80. [DOI] [PubMed] [Google Scholar]
- 48.Kupčinskas J, Gedgaudas R, Hartman H, Sippola T, Lindström O, Johnson CD, et al. Intestinal Fatty Acid Binding Protein as a Marker of Necrosis and Severity in Acute Pancreatitis. Pancreas 2018;47(6):715–20. [DOI] [PubMed] [Google Scholar]
- 49.Cresci GA, Glueck B, McMullen MR, Xin W, Allende D, Nagy LE. Prophylactic tributyrin treatment mitigates chronic-binge ethanol-induced intestinal barrier and liver injury. J Gastroenterol Hepatol 2017;32(9):1587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheridan PO, Martin JC, Lawley TD, Browne HP, Harris HMB, Bernalier-Donadille A, et al. Polysaccharide utilization loci and nutritional specialization in a dominant group of butyrate-producing human colonic Firmicutes. Microbial Genomics 2016;2(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 2014;5(2):e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott KP, Martin JC, Duncan SH, Flint HJ. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol Ecol 2014;87(1):30–40. [DOI] [PubMed] [Google Scholar]
- 53.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell 2014;159(4):789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016;352(6285):565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Goffau MC, Fuentes S, van den Bogert B, Honkanen H, de Vos WM, Welling GW, et al. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia 2014;57(8):1569–77. [DOI] [PubMed] [Google Scholar]
- 56.Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, Li L, et al. Defining the Roles of TcdA and TcdB in Localized Gastrointestinal Disease, Systemic Organ Damage, and the Host Response during Clostridium difficile Infections. mBio 2015;6(3):e00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leslie JL, Huang S, Opp JS, Nagy MS, Kobayashi M, Young VB, et al. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect Immun 2015;83(1):138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fletcher JR, Pike CM, Parsons RJ, Rivera AJ, Foley MH, McLaren MR, et al. Clostridioides difficile exploits toxin-mediated inflammation to alter the host nutritional landscape and exclude competitors from the gut microbiota. Nat Commun 2021;12(1):462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaban L, Chen Y, Fasciano AC, Lin Y, Kaplan DL, Kumamoto CA, et al. A 3D intestinal tissue model supports Clostridioides difficile germination, colonization, toxin production and epithelial damage. Anaerobe 2018;50:85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]


