Abstract
Continual development in instrumental and analytical techniques have aided in establishing rigorous connections between protein glycosylation and human illness. These illnesses, such as various forms of cancer, are often associated with poor prognoses, prompting the need for more comprehensive characterization of the glycoproteome. While innovative instrumental and computational strategies have largely benefited glycoproteomic analyses, less attention is given to benefits gained through alternative, optimized chromatographic techniques. Porous graphitic carbon (PGC) chromatography has gained considerable interest in glycomics research due to its mobile phase flexibility, increased retention of polar analytes and improved structural elucidation at higher temperatures. PGC has yet to be systematically compared against or in tandem with standard reversed phase liquid chromatography (RPLC) in high-throughput bottom-up glycoproteomics experiments, leaving the potential benefits unexplored. Performing comparative analysis of single and biphasic separation regimes at a range of column temperatures illustrates complementary advantages for each method. PGC separation is shown to selectively retain shorter, more hydrophilic glycopeptide species, imparting higher average charge, and exhibiting greater microheterogeneity coverage for identified glycosites. Additionally, we demonstrate that liquid-phase separation of glycopeptide isomers may be achieved through both single and biphasic PGC separations, providing a means towards facile, multidimensional glycopeptide characterization. Beyond this, we demonstrate how utilization of multiple separation regimes and column temperatures can aid in profiling the glycoproteome in tumorigenic and aggressive prostate cancer cells. RAW MS proteomics and glycoproteomics datasets have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD024196 (10.6019/PXD024196) and PXD024195, respectively.
Keywords: Glycopeptides, Glycoproteomics, Proteomics, Porous Graphitic Carbon, Mass Spectrometry
Graphical Abstract
Introduction
Glycoproteins, a unique class of post-translationally modified biomolecules, have long been an area of dedicated investigation within proteomic research communities. The high degree of modification complexity (1) has revealed the numerous roles glycoproteins play in physiological processes such as cell trafficking (2–4), signaling pathways (5–7) and protein folding (8–10), among others. Aided by continual improvements in mass spectrometry (MS)-based biomolecule discovery and identification, researchers have turned their attention towards understanding the relationship between these protein modifications and human disease. The dynamic nature of modification site occupancy and microheterogeneity (11) has established connections between glycoproteins and neurodegenerative diseases (12–17), autoimmune disorders (18–20) and numerous forms of cancer (21–23).
The growing relevance of the human glycoproteome has prompted analytical innovation, stimulating curiosity in strategies best suited for detecting, identifying, and characterizing glycoproteins. Specific to bottom-up glycoproteomic investigations, glycopeptide enrichment strategies and instrument capabilities are often the areas of most intense focus. Numerous novel enrichment strategies (24–26) have been developed to compensate for the low abundance of glycopeptides within complex proteolytic mixtures, while novel dissociation techniques (27–30) are pursued to provide more effective single-run sequencing of peptide backbones. Beyond this, ion separation regimes are often the primary focus when seeking to provide structural information of diverse glycan modifications (31–33).
Though these analytical developments have provided unique avenues towards more comprehensive glycoproteomic analysis, the experiments validating their efficacy almost exclusively rely on traditional reversed-phase liquid chromatography (RPLC). As there has been no extensive utilization of alternative separation regimes, with the exception of hydrophilic interaction chromatography (HILIC), the potential benefits and complements offered through novel chromatography have been left uncharacterized. Porous graphitic carbon (PGC) has shown increased utility in recent years for glycan identification and characterization (34–37) experiments but has not been evaluated in high-throughput glycopeptide analyses. As Zhu et al. (38) recently demonstrated the utility of PGC in elucidating glycopeptide modification structure, it inspired us to ask the question to what extent PGC may be employed to deepen the coverage of the human glycoproteome and complement the robust strategies that currently exist.
Presented here is a systematic comparison of proteome and glycoproteome profiling enabled through traditional RPLC C18, PGC, and biphasic RPLC-PGC separation regimes. We build on current literature that highlights the differences in analyte retention at elevated column temperatures, illustrating the impact it has in complex glycopeptide profiling experiments. This study brings to light the complementary proteome coverage provided by PGC, selectively retaining those shorter, more hydrophilic analytes that go unretained in RPLC analyses. PGC is also shown to provide greater coverage for glycan microheterogeneity and enable higher charge states on the retained hydrophilic glycopeptides. Interestingly, we also demonstrate both PGC and biphasic separation regimes provide higher isomeric resolution as column temperature increases, indicating potential utility in characterization experiments. Finally, we examine two complex, human-derived prostate cancer cell lines, demonstrating the utility of complementary separation regimes and characterizing the benefits and drawbacks of altering column temperatures in complex glycopeptide profiling experiments.
Materials and methods
Reagents and Materials
Bovine fetuin (F3004), bovine ribonuclease B (R7884), and bovine a1-Acid Glycoprotein (G3643) standards, as well as 1,4-dithiothreitol (DTT, D9779), 2-iodoacetamide (IAA, I6125), and methanol (MeOH, 34860) were purchased from MilliporeSigma (Burlington, MA). Urea (U15), tris-base (BP152), hydrochloric acid (A144SI), chloroform (C297), and acetonitrile (ACN, A998) were purchased from Fisher Scientific (Waltham, MA). Trypsin was purchased from Promega (V5113, Madison, WI). Capillary tubing (1068150019) was purchased from PolyMicro. RPLC packing material (4451GP) was purchased from Osaka Soda Co. (Osaka, Japan). PGC packing material was obtained by extracting stationary phase from PGC guard columns (35003-014001) purchased from Thermo Fisher Scientific (Waltham, MA). PolyHYDROXYETHYL A packing material was purchased from PolyLC (Columbia, MD). Materials for preparing frits were purchased from Next Advance (Troy, NY).
Pancreatic Cancer Cell (PANC1) Culture
The commercial pancreatic cancer cell lines PANC-1 obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) were cultured and maintained in DMEM:F12 (Hyclone, GE Healthcare Life Sciences, Logan, Utah, USA) containing 10% fetal bovine serum (FBS) (Gibco, Origin: Mexico), 1% penicillin-streptomycin solution (Gibco, Life Technologies Corporation, Grand Island, NY, USA). Cells cultured in a 37°C moisture incubator supplied with 5% CO2. PANC-1 cultures between passage 3 and 15 were used for all experiments. Cells at 70%−90% confluence were trypsinized using 0.25% trypsin EDTA solution (Corning, Mediatech, Inc., Manassas, VA, USA). Cell suspension was centrifuged at 800g for 5 minutes and the medium was discarded. Cells were resuspended in phosphate-buffered saline (PBS) (Corning, Mediatech, Inc., Manassas, VA, USA) and washed 3 times with PBS, flash frozen in dry ice, and stored at −80°C.
Prostate Cancer Cell (BCaP, LNCaP) Culture
BPH1 to cancer progression (BCaP) cells were produced by Ricke and colleagues and have been described previously (39–41) and lymph node carcinoma of the prostate (LNCaP) (42) cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cell lines were grown and maintained in phenol-free DMEM/Ham’s F-12 (Gibco) supplemented with 5% fetal bovine serum (HyClone) and 1% penicillin-streptomycin solution (Gibco). T175 culture flasks were placed in an incubator at 5% CO2 and 98% humidity. Cells were grown to 90% confluency, washed with 1X phosphate-buffered saline (Cytiva) and harvested using a cell scraper. Cell pellets were washed twice using phosphate-buffered saline and stored at −80°C for subsequent processing.
Protein Extraction and Digestion
Evaluating various preparation protocols, an adapted FASP approach was chosen for extraction and digestion of proteins from PANC1, LNCaP, BCaP-NT1, and BCaP-T10 cell lines. This strategy, which does not provide targeted membrane protein enrichment, proved to enable highest recovery of secretory glycoproteins and access to a small number of heavily glycosylated (43–45) membrane-bound species. Dedicated strategies are required to target membrane proteins and may be pursued in future analyses. Briefly, cells were resuspended in and washed with PBS, centrifuging at 14,000 rcf between washes. Cells were then resuspended in lysis buffer (8M Urea, protease inhibitor, 20mM HEPES) and lysed via ultrasonication. Cell debris was removed, and proteins were reduced and alkylated with centrifuge filtering between each step. Proteins were digested with trypsin (1:50) overnight at 37°C and desalted via reverse phase desalting cartridges. Volume was reduced under vacuum and concentration of peptides was estimated via Pierce Peptide Assay. Standard glycoproteins were reduced alkylated and digested similarly; a detailed preparation workflow is described in the electronic supplemental information.
Glycopeptide Enrichment
The enrichment procedure is fully detailed in the electronic supplemental information. Briefly, 200μL pipette tips were blocked with 3mg sterile cotton, and loaded with a hydrated polyHYDROXYETHYL-A resin. Weight of beads is adjusted according to the mass of peptides used. Peptide mixture was added and allowed to bind through repeated centrifugation. Non-specific peptides were washed away using 80% ACN+0.1% TFA with enriched glycopeptides eluted in 10% ACN+5% FA. Prior to analysis, glycopeptides were dried under vacuum and resuspended in LC/MS-grade water+0.1% FA.
LC-MS Parameters
Samples were analyzed on an Thermo Scientific QE-HF mass spectrometer coupled with a NanoUltimate 3000 chromatography stack. Follow-up analyses were performed on an Thermo Scientific Fusion Lumos with the same chromatography system. Custom capillary columns were fabricated and coupled as described (electronic supplemental information, Figure S1) and temperatures were controlled by a 30 cm pencil column heater (Phoenix S&T). Flow rate was established for each column by achieving a stable pressure drop of 450 bar at 30°C and was held constant as temperature was varied (C18: 475μL/min, PGC: 750μL/min, BP: 525μL/min). Using water+0.1%FA and ACN+0.1%FA as buffers A and B LC gradients were held consistent between columns: a trapping phase from 0–16 minutes at 3% B moving to 40% at minute 100, 75% B at from minutes 102.5–105, 97% B from minutes 105.1–113, and 3% B from minutes 115–125.
Mass spectrometry settings were configured to balance profiling depth with mass resolution. Using a full MS/dd-MS2 method, MS1 settings were as follows: polarity, positive; default charge state, 2; microscans, 1; resolution, 60K; AGC target, 1e6; maximum injection time, 250 ms; m/z range, 150–1500; spectrum type, centroid. MS2 settings: microscans, 1; resolution, 15K; AGC target, 2e5; maximum AGC 8e3; maximum injection time, 120 ms; loop count, 15; isolation window, 2 m/z, isolation offset, 0 m/z; fixed first mass, 100.0 m/z; collision energy, stepped HCD (25, 35, 45); spectrum type, centroid; dynamic exclusion, 30s; rejected charge states, 1+, 8+ >8+.
Data Analysis and Availability
Preliminary results from analysis of PANC1 cell lysate digest were identified in PEAKS X (Bioinformatics Solutions Inc.) searching against the UniProt reviewed human proteome. Glycopeptide data was annotated via Byonic (Protein Metrics). Standard glycopeptides were run against a custom database of UniProt protein sequences while LNCaP and BCaP samples were searched against the human proteome. The parameters used for PEAKS X and Byonic searching are described in the electronic supplemental information.
RAW MS data for proteomics and glycoproteomics datasets have been deposited to the ProteomeXchange Consortium via the PRIDE (46) partner repository with the dataset identifier PXD024196 (10.6019/PXD024196) and PXD024195, respectively. Peptide, glycopeptide, and protein identifications are also provided in Supplemental Tables S1–6. Byonic output files are available at https://figshare.com/account/home#/projects/98459. Custom data scraping, analysis, calculations, and visualization was done primarily in Python using the Altair (47) library with additional components done in R, D3, and vega-lite. All code is available at https://github.com/lingjunli-research/pgc-parallel-comparison.
Results and Discussion
While PGC is noted to be amenable to an array of solvent systems, a key benefit is its compatibility with typical water/ACN buffer systems. In hand with the principle of contrasting retention mechanisms compared to RPLC, this indicates a facile means of complementary proteomic/glycoproteomic analysis and multidimensionality compared to organic-based separations such as HILIC. Though a multidimensional C18-PGC strategy was reported for pronase-digested glycopeptides (48), the profiling depths of these two separation regimes have not yet been compared for complex proteomic or glycoproteomic analyses.
We address this knowledge gap through comparative analyses of complex proteolytic and enriched glycopeptide mixtures across three custom-fabricated nanoflow separation regimes run at varying temperatures (Figure 1). Columns were prepared as described (electronic supplemental information) and coupled to a Q-Exactive-HF nano-electrospray source as shown (Figure S1). As a note, 3μm diameter packing materials are used for each stationary phase as, at the time of writing, this was the smallest available PGC packing material. Smaller diameter RPLC material may be employed to increase liquid-phase resolution and is under evaluation for future research.
Figure 1.
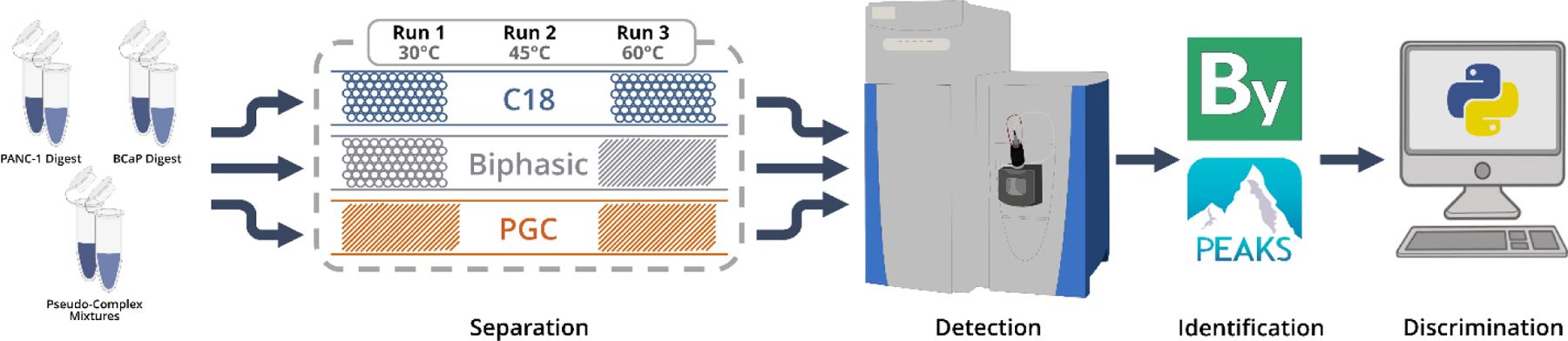
General workflow utilized throughout the experiment. Samples were run sequentially at different temperatures (Run 1: 30°C, Run 2: 45°C, Run 3: 60°C). Data was collected on a Thermo Q-Exactive-HF orbitrap mass sepctrometer, with peptide and glycopeptide annotations provided by PEAKS and Byonic prior to custom data analysis.
Proteomic Performance
To evaluate the performance in shotgun-like proteomic investigations, proteins were extracted from pancreatic cancer cells (PANC1), digested with trypsin, and subjected to LC-MS/MS at varying temperatures (30°C, 45°C, 60°C). RPLC demonstrated the best overall performance in peptide and protein identifications, followed by PGC and biphasic (BP) separations (Figure 2A, S2, Table S1). While temperature variations had negligible effect on BP performance, both C18 and PGC experiments displayed noticeable improvements in number of identified species when run at 45°C; this observation is consistent with later analyses and is discussed within. Across all temperatures, our results produced the expected overlap in highly abundant identified species, though RPLC and PGC demonstrate the most distinct differences in number of unique identifications (Figure 2B). This observation supports the idea of complementary proteome access enabled through PGC separation, indicating more holistic coverage may be obtained when incorporated alongside RPLC. We further investigated the species found to be unique in RPLC or PGC separations to establish any preferential retention of subcellular species. Our data demonstrate that there are noticeable differences in the most prevalent subcellular locations and cellular compartments associated with proteins unique to each separation regime (Figure S3, Table S2). While this finding supports the gain of complementary proteome access through respective separations, it must be considered carefully. Rather than definitively conclude that each separation regime targets specific protein subclasses, these data only present trends that may be further defined after in-depth discovery proteomics experiments.
Figure 2.
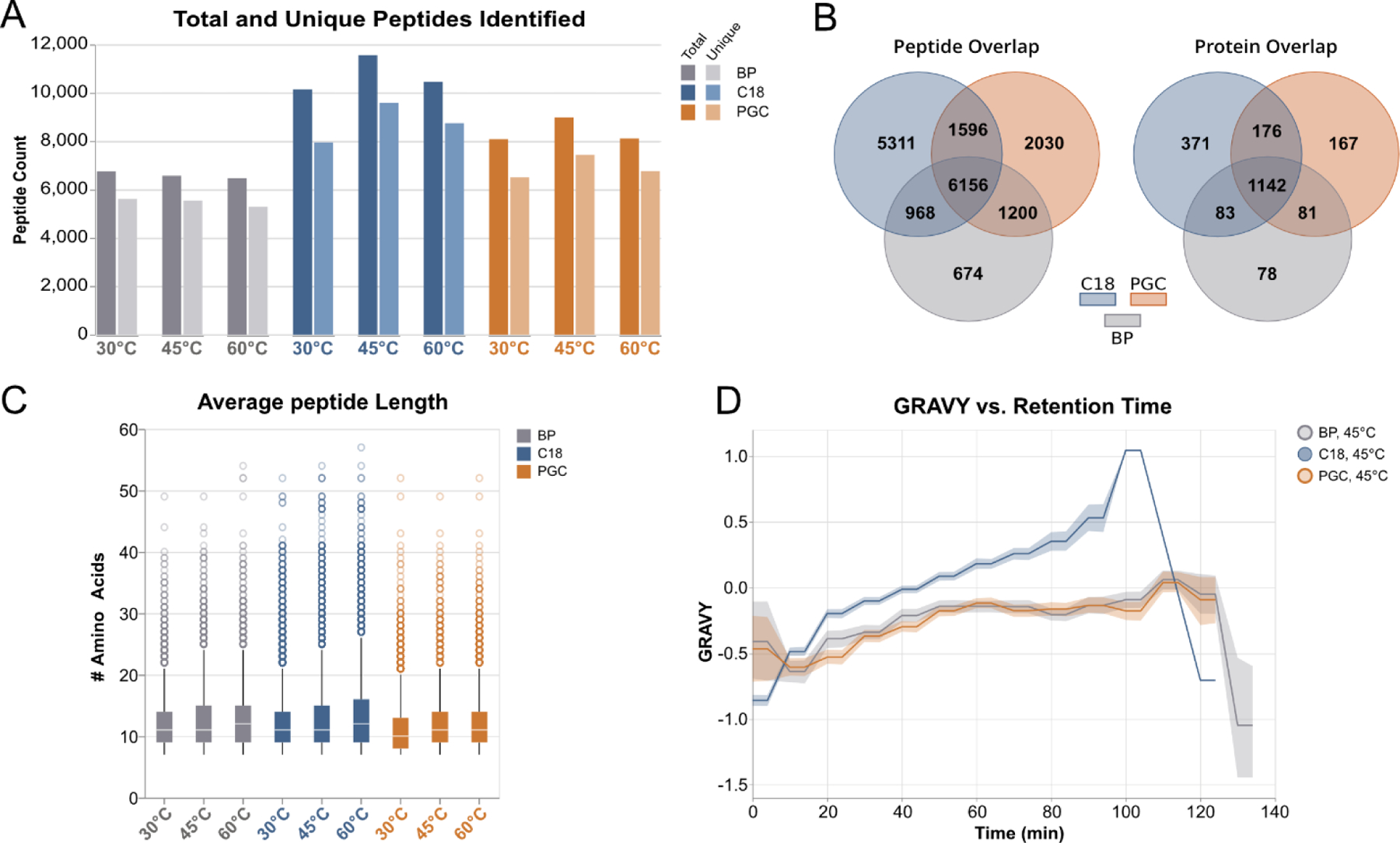
High-level overview of identified peptides resulting from analysis of PANC1 cell lysate digests. A) Comparison of total and unique peptides identified between all columns at varying temperatures. B) Overlap of identified peptides (left) and identified proteins (right), demonstrating the clear distinction in the species retained in PGC and RPLC separations. C) Comparison of identified peptide lengths. D) Hydrophilicity of identified peptides as a function of time, demonstrating the complementary retention between PGC and RPLC. Lesser values are associated with hydrophilic character while greater values are associated with hydrophobic character. The line represents average GRAVY score, shaded area represents 95% confidence interval.
Hypothesizing that differences in peptide character are the primary factor in driving this complementary proteome access, we first evaluated the distribution of retained peptide lengths. Considering the high number of peptides identified in each experiment (between 6,000 and 11,000) it is not surprising the median peptide lengths are largely conserved across all experiments (Figure 2C). However, at all temperatures, PGC demonstrates the lowest median length, a more concise interquartile range, and shorter maximums and outlier species. More importantly is the evaluation of hydrophilic character of retained peptides identified in each experiment. Using GRAVY (grand average of hydropathy) score to infer hydrophilic character, RPLC separations retained those peptide species with the greatest overall hydrophobic character at all gradient timepoints, as evidenced by the higher, positive GRAVY scores (Figure 2D). Conversely, PGC separations yield peptides with overall hydrophilic character (lower, more negative GRAVY scores). Noting intermediate trend of biphasic separations compared to the two higher performing regimes, this knowledge, combined with the presence of unique peptide identifications in this separation strategy, indicates that further optimization of biphasic separations could provide synergistic retention and identification. The utility of biphasic regimes is further expanded in glycopeptide analyses, as seen below.
To establish the reliability of our custom separation strategies, it was imperative we evaluate inter-run reproducibility. Our follow-up replicate analyses of each separation regime (Table S3) established high reproducibility in peptide and protein identifications (Figure S4), as well as confirmed the trends seen in peptide length and hydropathy (Figure S5). In total, these results point toward two notable conclusions. First, PGC separations can be successfully employed in high-throughput analyses of tryptic proteolytic mixtures. As previous experiments have been confined to analyses of small peptides - often using sequential or non-specific digestion - this observation stands alone as the first of its kind and implies high utility in proteomic analyses. Second, our results demonstrate the complementary access to the human proteome that is granted when PGC separations are employed. Whereas PGC selectively retains those shorter, more hydrophilic peptides, as well as being amenable to standard LC mobile phases, this regime presents a useful approach to proteome profiling that may be readily implemented in research settings where RPLC is employed. As well, greater benefit may be found in multidimensional chromatography experiments and instances where fractionation is used to reduced sample complexity.
Trends in Glycopeptide Identification
To understand the performance of these complementary separation regimes in glycopeptide identification and characterization experiments, we performed HILIC enrichment on three glycopeptide-rich samples: one mixture of standard glycoprotein digests and two BPH1 to cancer progression (BCaP) cell line digests. The former sample was employed to provide a robust, reproducible dataset that may be validated in other research settings, while the latter two are meant to serve as real, complex glycoproteomic mixtures. Within our analyses, we sought to profile the characteristic differences of glycopeptides retained in RPLC, PGC, and biphasic separations while evaluating the extent to which incorporation of PGC separations would benefit glycoproteomic workflows, expand microheterogeneity profiling, and offer benefits in structural elucidation.
After compiling annotated glycopeptides identified in all samples, we again observed a greater number of total and unique glycopeptides in RPLC experiments with PGC as the next best performer (Figure 3A), with these two regimes also displaying the greatest number of unique glycopeptides across all experiments (Figure 3B). Comparing glycopeptide identifications when sample constituents and column temperature are held constant confirms the difference in retention mechanism is the primary cause of unique identifications (Figure 3C, S6). However, our data also confirms that that there are temperature-dependent differences in glycopeptide identification (Figure 3C, S7), an observation further validated in subsequent analyses (Figure S8, Tables S4–5). Knowing this, we then sought to understand what differences exist between glycopeptides retained on each stationary phase. As a note, we again observe a noticeable improvement in analyte identification when RPLC and PGC columns are operated at 45°C compared to other conditions, which is discussed below.
Figure 3.
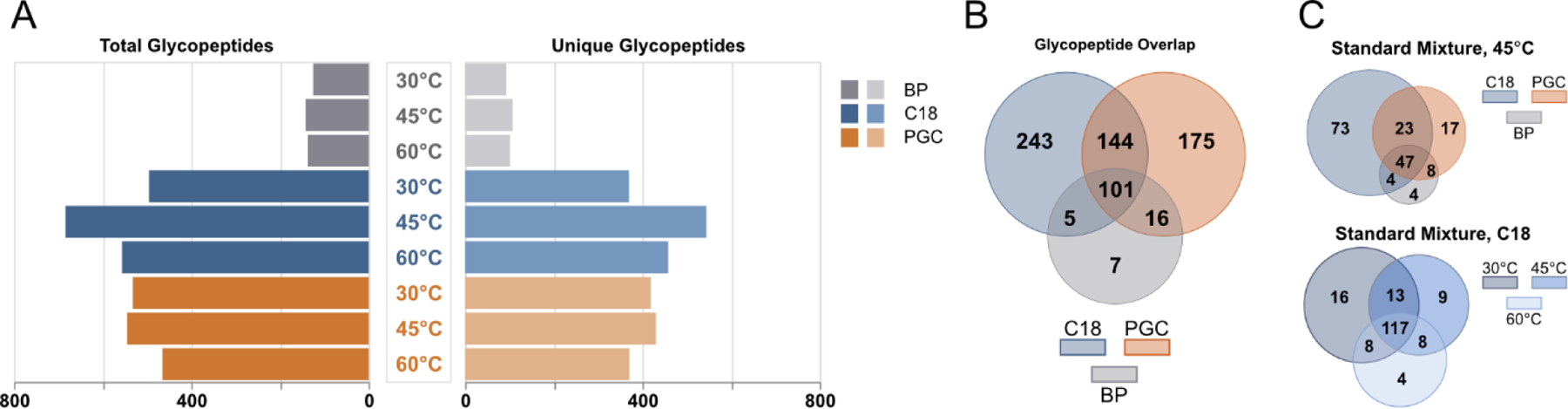
Overview of glycopeptides identified between all separation regimes. A) Comparison of total (left) and unique (right) glycopeptides identified across all samples, shown according to separation strategy and column temperature. B) Overlap of all glycopeptides identified in each separation phase across all samples. C) (top) Overlap of glycopeptides identified in the standard mixture when column temperature is held constant and column is varied, (bottom) overlap of glycopeptides from standard mixture when separation regime is constant and temperature is varied.
Peptide-Level Differences in Retained Glycopeptides
To illuminate the peptide-level differences existing between RPLC and PGC separations, we consolidated the dataset to contain only those glycopeptides that were identified in a single separation regime. Given the hydrophobicity-driven separation mechanism of RPLC, we had anticipated this regime would vastly outpace the others in number of unique glycopeptide backbones. In contrast to this hypothesis, holistic and sample-specific data (Figure 4A, S9, Table S6) revealed comparable numbers of unique species in RPLC and PGC with the former regime identifying only slightly more peptide backbones. Additionally, RPLC-specific glycopeptides were found to be significantly longer than those identified in PGC (p=1.44E−32, Wilcoxin rank-sum) and notably different from those in BP experiments (p=0.102, Wilcoxin rank-sum) while simultaneously providing a greater distribution of charge states (Figure 4A–B). These stark discrepancies in RPLC experiments result from a higher prevalence of basic amino acid residues, due in large part to missed cleavage sites.
Figure 4.
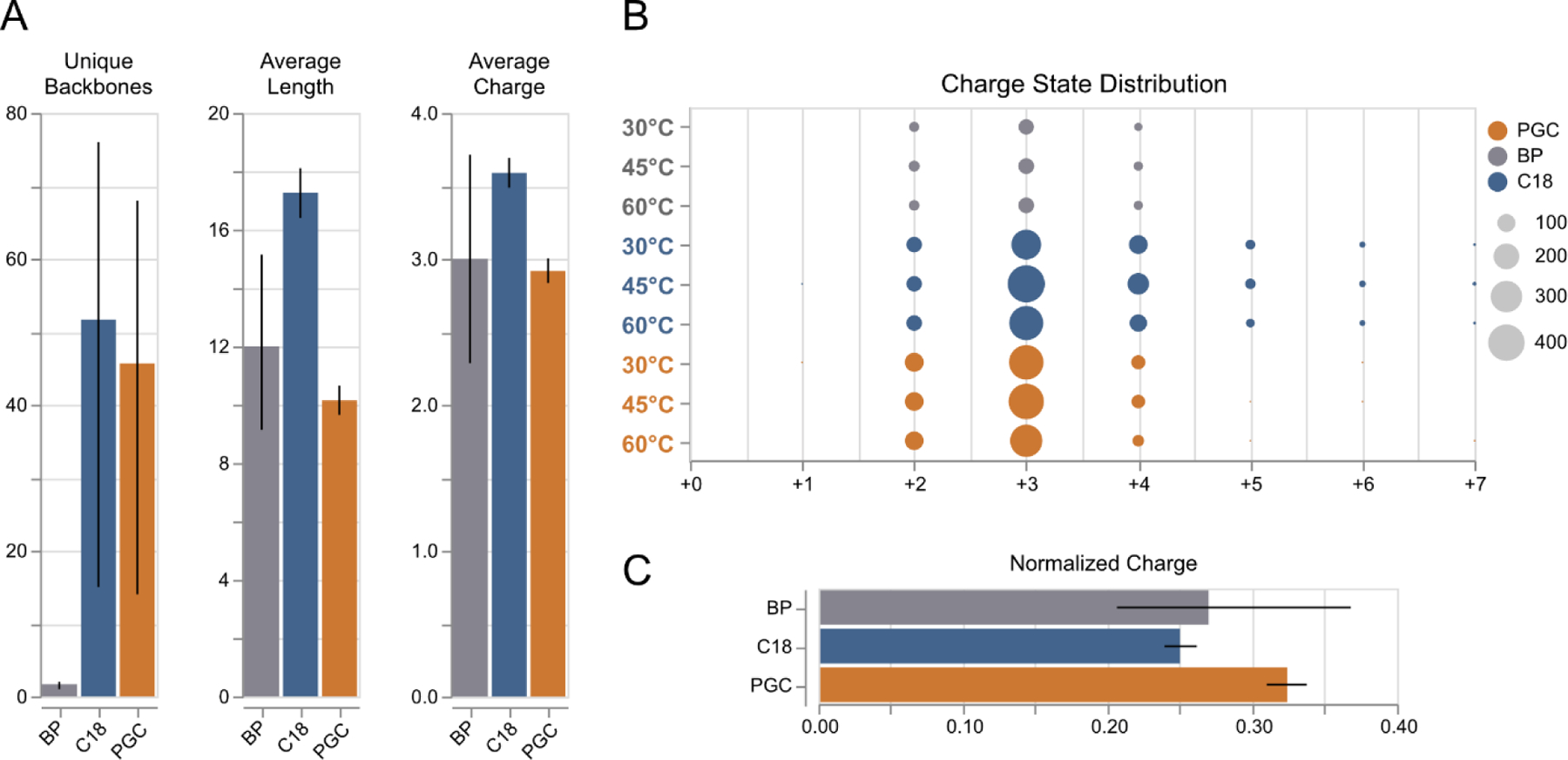
Peptide level differences between glycopeptides identified across all experiments. A) Distribution of number unique backbones identified, average glycopeptide length, and average glycopeptide charge between all separation regimes. Error bars: 95% confidence inteval, n=3 (each sample). B) Distribution of glycopeptide charge states between all separations and temperatures. C) Normalized charge per glycopeptide, shown as a function of average charge/amino acid residue. Error bars: 95% confidence inteval, n=3 (each sample).
These observations must be carefully considered in the context of glycoproteomics. Our data suggest that RPLC separations provide the highest identification rates of glycopeptides, agreeing with the general utility of this technique across current literature. However, RPLC separations very clearly benefit from instances where the hydrophobic character of the peptide outpaces the often-dominating hydrophilicity of the glycan; such is the case for long peptides and those with missed cleavage sites. These results highlight that the incorporation of PGC separations into existing experiments directly complements, rather than detracts from current glycoproteomics methodologies. As each regime provides meaningful access to unique components of the glycoproteome, these two regimes may be leveraged in tandem to increase glycoproteomic coverage.
Furthermore, the differences in charge state distribution across all experiments may be further contextualized for potential benefits and drawbacks. Within glycoproteomic analyses, longer peptides with higher charge states could be considered beneficial in some analytical applications as they may be leveraged for modification site localization through electron transfer dissociation (ETD) and directly combats traditional limitations in ionization efficiency of glycopeptides. However, this can also result in precursor reporting signal being distributed across several channels, lowering overall signal intensity, and potentially limiting precursor selection in data-dependent analyses. Interestingly, glycopeptides identified in PGC analyses display a consolidated charge state distribution, primarily displaying 2+ and 3+ ions at rates higher than or equal to those seen in RPLC analyses. Normalizing this charge to the length of the peptide backbone reveals that PGC-specific glycopeptides demonstrated higher charge density among all separation regimes (Figure 4B, S9). This increased charge density displayed in short, hydrophilic analytes is a direct consequence of not selectively retaining those hydrophobic species that traditionally outcompete during ionization. As such, we may conclude that for applications where charge-mediated fragmentation is desired, RPLC separations are more beneficial as they will retain longer, highly charged glycopeptides. At the same time, PGC separations may find utility in applications where researchers wish to remedy ionization inefficiency hydrophilic analytes – the need for which is exacerbated when considering glycan-level differences of retained glycopeptides.
Glycan-Level Differences
Turning to the differences exhibited at the glycan level, the benefits of PGC separations become more pronounced. Topically, PGC analyses retained the broadest array of unique glycan structures, only falling behind RPLC in the standard glycopeptide sample (Figure 5A, S10). Interestingly, when comparing the extent of glycosylation (inferred from the number of monosaccharide residues within each glycan structure), initial observations revealed that PGC retained smaller glycans compared to other separation regimes, though this discrepancy is confined to the standard peptide sample and is likely due to microheterogeneity differences at retained glycosites.
Figure 5.
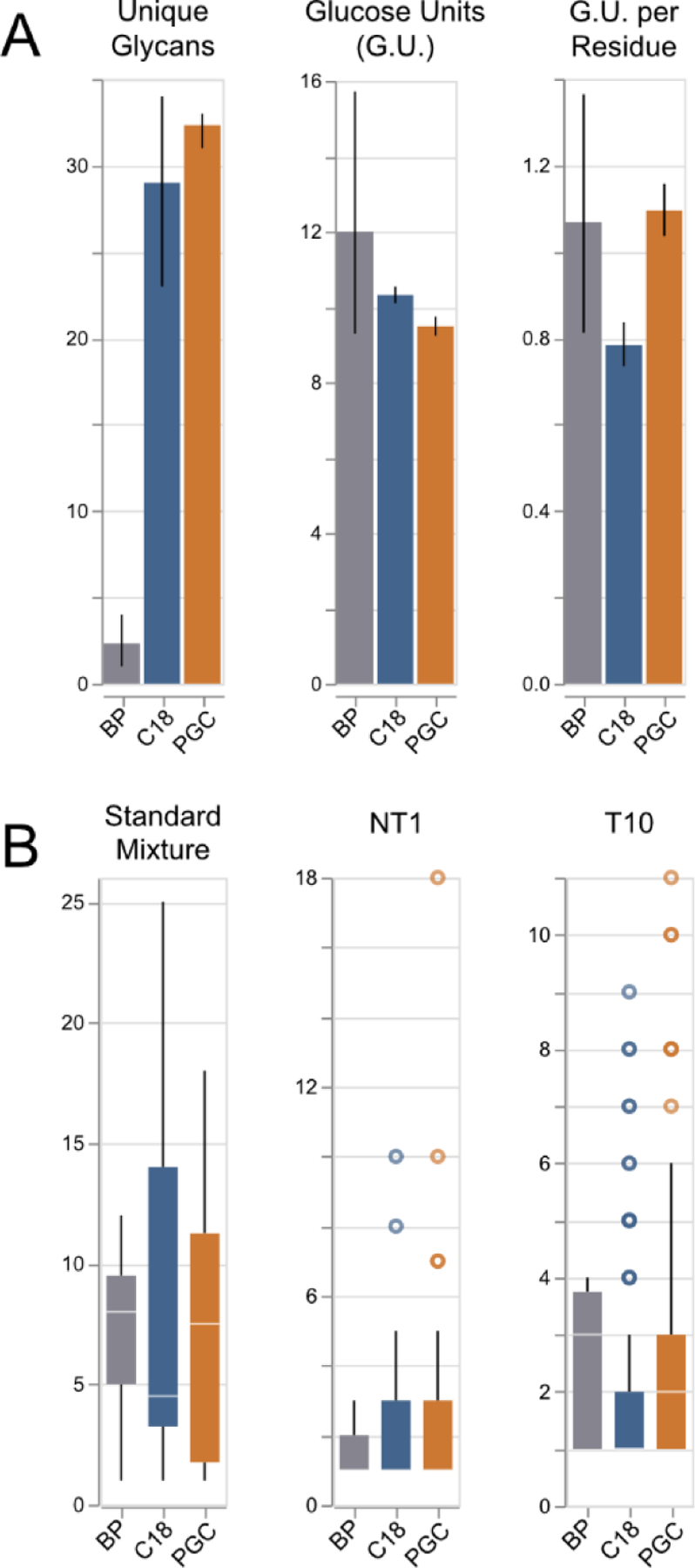
A) Distribution of # unique glycans, glucose units per glycan, and glucose units per amino acid residue. Error bars: 95% confidence inteval, n=3 (each sample). B) Boxplots displaying the number of glycans associated with retained peptide bacbones. Note: for samples “NT1” and “T10”, the median values of RPLC are found to be 1, making them difficult to view here.
Across all experiments, we did not observe notable bias towards any glycopeptide type, as all identifications were found to be exclusively high-mannose or complex. More notable however, is the relative size of the glycan in relation to the peptide backbone. Across all samples, PGC-specific peptides displayed the highest monosaccharide-to-amino-acid ratio, indicating the character of these glycopeptides is largely dominated by the large, hydrophilic glycan. Rearticulating the above-mentioned observation that PGC selectively retains glycopeptides with shorter, more hydrophilic backbones, this revelation further emphasizes the utility of PGC separations in accessing those portions of the glycoproteome that would traditionally go undetected in RPLC analysis.
As a primary goal in glycoprotein profiling investigations is to characterize site microheterogeneity, it is of topical concern to evaluate the number of glycans associated with each identified peptide backbone. Across all experiments, PGC regularly outpaced the other separations, providing a higher median number of glycans per glycosite, with higher maximums found in all but one sample (Figure 5B). This observation further illustrates the value of PGC separation in glycoproteomic analyses as it not only accesses those portions of the glycoproteome not found in RPLC analyses, but it also provides improved microheterogeneity profiling depth. This reality may be further leveraged to provide unparalleled glycoproteome profiling, if used alongside established methods of sequential or non-specific enzymatic digestion to provide a broad array of PGC-compatible glycopeptides.
Improved Isomeric Resolution
While analytical methods capable of assigning glycan location and composition occupy a large portion of glycoproteomics investigations, facile structural elucidation remains on the outset of widespread investigation. Previous studies have detailed the ability of PGC to provide separation of isomeric glycans at higher temperatures (34, 38, 49) postulating the expanded glycan morphology that results from increased temperature interacts more readily with the stationary phase, improving liquid-phase resolution. Though this PGC-based approach was recently demonstrated in collections of tryptic/chymotryptic glycopeptides (38), it has never been explored in complex samples that display high diversity in peptide sequence and glycan composition.
Similar to these previous results, our studies show that isomeric glycopeptides can be resolved through PGC separations with benefits often being further pronounced at higher temperatures (Figure 6, S11–12). However, as previous studies utilized commercial columns of shorter length (L=10cm, i.d.=75μm), we predicted our longer stationary phase would impart greater resolution of constitutional isomers prior to MS. Contrary to this hypothesis, a subset of glycopeptides that were partially or fully resolved in PGC experiments also displayed comparable resolution in BP separations, with some more well-resolved in the latter regime (Figure S13). This realization, in tandem with the poorer demonstrated performance of BP in glycopeptide retention indicates a novel approach may be to couple higher resolution RPLC stationary phases (L=15cm, d.p.=1.7μm) with a 15cm PGC stationary component. This method may provide a more effective means of deep profiling and structural elucidation.
Figure 6.
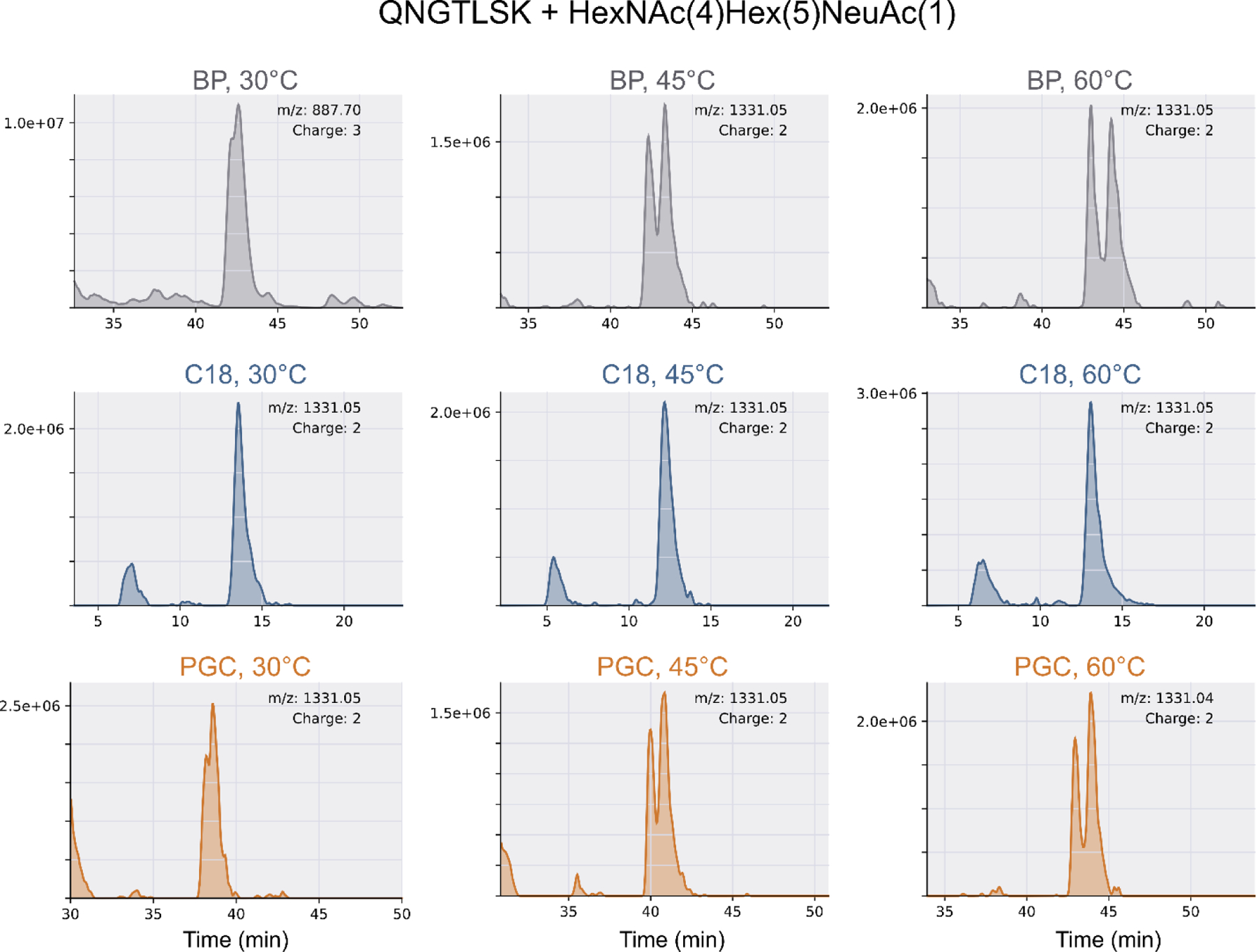
Extract ion chromatogram (EIC) of a representative glycopeptide (QN*GTLSK + HexNAc(4)Hex(5)NeuAc(1)), which was identified on all columns at all temperatures. As the retention mechanism of RPLC is driven by hydrophobic interactions, the lack of any liquid-phase resolution is expected. However, both a longer (L=30cm) and short (L=15cm) PGC stationary phases were shown to provide adequate liquid phase resolution of isomeric glycopeptides. Note, the 3+ charge state in BP, 30°C has been manually confirmed as correct.
In addition, a survey of those species with altered liquid phase retention yields two putative trends. The less surprising observation is that isomeric resolution is greater for those containing high mannose and complex glycans with unequal antennae, especially those found to be singly sialylated. Second, as liquid-phase separation was more readily achieved in glycopeptides from our standard protein mixture and in species identified on all separation regimes, we hypothesize this observation is concentration-dependent. This is also supported in theory. Considering the unequal distribution between major and minor glycan conformations, it is logical to assume some minor species may not be preserved after extensive sample handling.
Taken together, our results serve to further emphasize the potential benefits of incorporating PGC separation strategies into routine glycoproteomics investigations. Whereas previous studies often employ more expensive commercial columns – many of which are no longer available for purchase – our study highlights the facility of custom fabrication and customization of analytical columns capable of discerning structural information. Comprehensive structural characterization is not a primary goal of many bottom-up glycoproteomics experiments, we present a low-cost, facile means of deepening glycoproteome coverage that may be further developed to provide substantial gains in biological information.
Glycoproteome Profiling
Though the results have thus far demonstrated the marked differences in proteome access provided through PGC and RPLC separations, we must assess any potential bias in glycopeptide retention presented in PGC analysis and evaluate the potential impact that higher temperature plays on glycoproteome profiling. Focusing on the two complex samples, BPH1 to cancer progression (BCaP) NT1 and T10 cell lines (50), it is immediately noticeable that high mannose glycans present the highest proportion of glycan species identified (Figure 7). As this observation is shared between RPLC and PGC separations, and because analyses of our standard mixture deviates from this trend (Figure S14), we can hypothesize this is not a result of preferential retention of high mannose glycans and is instead related to higher prevalence of these species in the samples analyzed. Future quantitative investigations, performed across numerous cell lines, are needed to confirm and validate this hypothesis.
Figure 7.
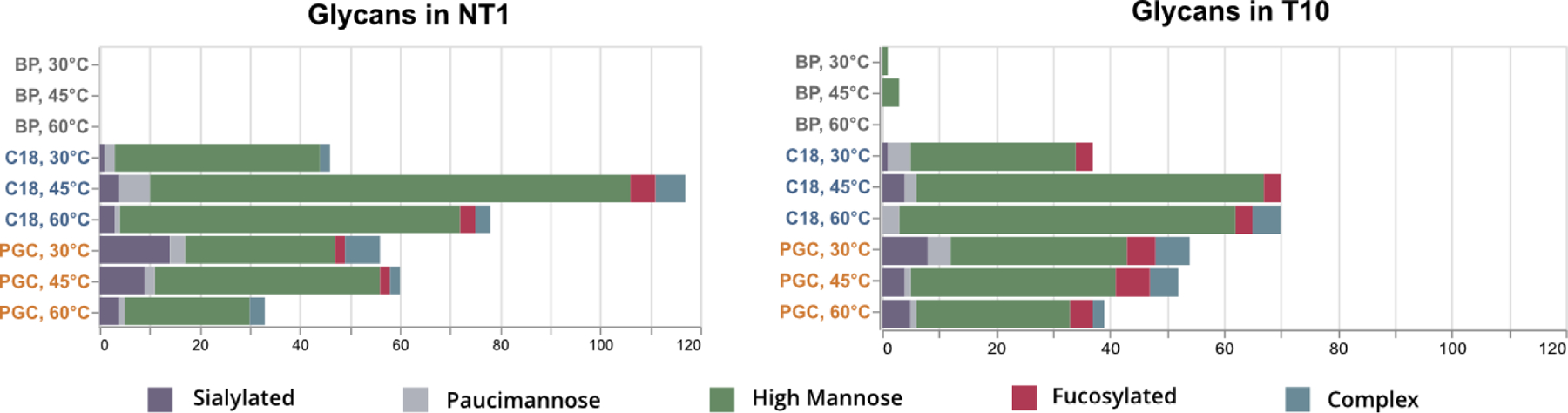
Distribution of glycan types identified within prostate cancer cell lines NT1 and T10. Glycan class (i.e., High Mannose, Complex, etc.) are colored to highlight the changing quantities of glycans as a function of column temperature.
Mammalian N-glycoproteins are known to be dominated by complex glycans, with high mannose modifications being considered “immature” within the synthetic pathway (1). Recently, a growing number of studies have revealed high abundances of high mannose glycans in cancer cells (51, 52) with these moieties noted as being involved in cell-cell interactions (53, 54). Of note, a recent study revealed that extended high mannose glycans directly contribute to bile duct cancer metastasis, noting the importance of specific cell surface glycosylation (55). Importantly, this study notes the importance of terminal α−1,2 mannose residues, bolstering the importance of providing structural elucidation in glycoproteomic investigations. Based on our results that demonstrate resolution of isomeric glycopeptides in BP and PGC separations, we anticipate this type of analysis would provide a novel path toward in-depth, targeted glycoproteomics. As the importance given to glycan isomers continues to grow, and the ability to easily distinguish these species without specialized instrumentation becomes invaluable, the incorporation of a PGC separation component will play a vital role in future investigations.
With respect to the observed prevalence of high mannose glycan modifications, we also observe an obvious difference in the number of species identified at 45°C versus those at any other temperature. As this trend was also conserved in both RPLC and PGC experiments, we surveyed the composition of high mannose glycans identified at all temperatures and found no significant disparities at the peptide level suggesting a cause for increased identifications (Figure S15–16).
Speculating the increase of high mannose glycopeptides may be due to species only being retained at 45°C, extracting the masses of identified glycopeptides demonstrated that most precursors could be identified across all temperatures, and that very few glycopeptides displayed temperature-dependent retention. A more consistent observation, however, is that different column temperatures result in different MS1 peak heights/areas, likely impacting their selection in data-dependent experiments. The prevailing trend is for species to exhibit highest median peak heights at 45°C, either increasing or decreasing in magnitude at 60°C (Figure S17). We did not observe notable differences in MS2 fragmentation across unique temperatures (Figure S18), indicating MS1 level differences are likely driving altered identification of glycopeptides. More intensive analysis is needed to determine the exact cause of this observation, though we hypothesize that 45°C may provide a more optimal balance between droplet desolvation and liquid-phase resolution, resulting in higher identification rates overall.
One potential concern of raising column temperatures is the additional energy provided to species prior to ionization. A consistent observation in both the complex and standard glycopeptide samples is that lower temperatures were more successful in identifying sialylated glycans (Figure 7, S14). In general, we observed a decay in reporting signal as temperature increased (Figure S19) though a wider collection of sialylated species may be needed to confirm the universality of this trend. However, it cannot be ignored that in a small number of cases, little or no MS1 signal could be observed for sialylated glycopeptides at 60°C. Considering the labile nature of glycosidic bonds, it is possible the increased energy prompted premature dissociation of glycosidic bonds and therefore has direct adverse effects on identification. These hypotheses are currently under investigation.
These potential limitations notwithstanding, our data demonstrate both RPLC and PGC separations are benefited by moderate increases in running temperatures. Raising column temperature even higher may result in better isomeric resolution, as suggested by previous studies, but may cause undesired signal decay. This tradeoff must be carefully balanced depending on experimental objectives.
Conclusions
Taken together, our results demonstrate the incorporation of a PGC stationary phase grants complementary, though not orthogonal, access to the proteome and glycoproteome. While traditional RPLC separation regimes provide the greatest overall retention of peptides and glycopeptides, our results demonstrate RPLC preferentially retain longer, more hydrophobic species within solution. In contrast, PGC expands overall sample coverage by retaining those smaller, more hydrophilic peptides and glycopeptides that would otherwise go unidentified. Furthermore, we demonstrate that the glycopeptides retained by PGC have a more consolidated charge state distribution than those in RPLC experiments, generating more charge per length of peptide regardless of dominant hydrophilic character. As well, PGC separations provided greater microheterogeneity profiling depth by identifying more glycans per retained peptide. Our results also build on previous studies that show the capacity for liquid-phase separation of isomeric glycopeptides at higher temperatures. However, our results show a long PGC stationary phase is not exclusively necessary to provide improved liquid-phase separation. This knowledge indicates the combination of high-resolution RPLC stationary components in tandem with a PGC stationary phase could provide a useful strategy in high-throughput characterization studies. Finally, we demonstrate that running both RPLC and PGC separations at higher temperatures provides altered peptide and glycopeptide identification rates, with the most optimal temperature shown to be 45°C. In summary, PGC enables pronounced benefits in proteomic and glycoproteomic analyses and provides a means towards facile, high-throughput characterization. As such, this regime is sure to exhibit utility in applications targeting unique post-translational modifications and as a separation strategy for structural interrogation.
Supplementary Material
Acknowledgments
The authors thank Dr. Qinying Yu, Danqing Wang and Dr. Dustin Frost in the Li Research Group for their helpful suggestions in the areas of glycopeptide enrichment and custom LC separations This work was funded in part by the National Institutes of Health (NIH) grants RF1 AG052324, U01CA231081, and R01 DK071801. The QE-HF Orbitrap instrument was generously provided by Thermo Fisher Scientific. LL would like to acknowledge NIH grant support R21AG065728, NCRRS10RR029531, and S10OD025084, a Pancreas Cancer Pilot grant from the University of Wisconsin Carbone Cancer Center (233-AAI9632), as well as a Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Distinguished Chair Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
Conflicts of interest/Competing interests: The authors declare no competing interests.
References
- 1.Varki A Essentials of glycobiology Third edition. ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2017. xxix, 823 pages p. [Google Scholar]
- 2.Sperandio M, Gleissner CA, Ley K. Glycosylation in immune cell trafficking. Immunol Rev. 2009;230(1):97–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hobbs SJ, Nolz JC. Regulation of T Cell Trafficking by Enzymatic Synthesis of O-Glycans. Frontiers in Immunology. 2017;8(600). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sackstein R, Merzaban JS, Cain DW, Dagia NM, Spencer JA, Lin CP, et al. Ex vivo glycan engineering of CD44 programs human multipotent mesenchymal stromal cell trafficking to bone. Nature Medicine. 2008;14(2):181–7. [DOI] [PubMed] [Google Scholar]
- 5.Dong Y, Li W, Gu Z, Xing R, Ma Y, Zhang Q, et al. Inhibition of HER2-Positive Breast Cancer Growth by Blocking the HER2 Signaling Pathway with HER2-Glycan-Imprinted Nanoparticles. Angew Chem Int Ed Engl. 2019;58(31):10621–5. [DOI] [PubMed] [Google Scholar]
- 6.Xuan L, Luan G, Wang Y, Lan F, Zhang X, Hao Y, et al. MicroRNAs regulating mucin type O-glycan biosynthesis and transforming growth factor β signaling pathways in nasal mucosa of patients with chronic rhinosinusitis with nasal polyps in Northern China. Int Forum Allergy Rhinol. 2019;9(1):106–13. [DOI] [PubMed] [Google Scholar]
- 7.Au - Bagavant H, Au - Trzeciak M, Au - Papinska J, Au - Biswas I, Au - Dunkleberger ML, Au - Sosnowska A, et al. A Method for the Measurement of Salivary Gland Function in Mice. JoVE. 2018(131):e57203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaffey PK, Guan X, Wang X, Ruan Y, Li Y, Miller SG, et al. Quantitative Effects of O-Linked Glycans on Protein Folding. Biochemistry. 2017;56(34):4539–48. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y-S, Guo X-Y, Hirata T, Rong Y, Motooka D, Kitajima T, et al. N-Glycan–dependent protein folding and endoplasmic reticulum retention regulate GPI-anchor processing. Journal of Cell Biology. 2017;217(2):585–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuribara T, Totani K. Structural insights into N-linked glycan-mediated protein folding from chemical and biological perspectives. Current Opinion in Structural Biology. 2021;68:41–7. [DOI] [PubMed] [Google Scholar]
- 11.Čaval T, Heck AJR, Reiding KR. Meta-heterogeneity: evaluating and describing the diversity in glycosylation between sites on the same glycoprotein. Molecular & Cellular Proteomics. 2021:100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Losev Y, Frenkel-Pinter M, Abu-Hussien M, Viswanathan GK, Elyashiv-Revivo D, Geries R, et al. Differential effects of putative N-glycosylation sites in human Tau on Alzheimer’s disease-related neurodegeneration. Cellular and Molecular Life Sciences. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Losev Y, Paul A, Frenkel-Pinter M, Abu-Hussein M, Khalaila I, Gazit E, et al. Novel model of secreted human tau protein reveals the impact of the abnormal N-glycosylation of tau on its aggregation propensity. Scientific Reports. 2019;9(1):2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frenkel-Pinter M, Shmueli MD, Raz C, Yanku M, Zilberzwige S, Gazit E, et al. Interplay between protein glycosylation pathways in Alzheimer’s disease. Science Advances. 2017;3(9):e1601576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Videira PAQ, Castro-Caldas M. Linking Glycation and Glycosylation With Inflammation and Mitochondrial Dysfunction in Parkinson’s Disease. Frontiers in Neuroscience. 2018;12(381). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson JW, Sorum AW, Hsieh-Wilson LC. Deciphering the Functions of O-GlcNAc Glycosylation in the Brain: The Role of Site-Specific Quantitative O-GlcNAcomics. Biochemistry. 2018;57(27):4010–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reindl M, Waters P. Myelin oligodendrocyte glycoprotein antibodies in neurological disease. Nature Reviews Neurology. 2019;15(2):89–102. [DOI] [PubMed] [Google Scholar]
- 18.Stack JR, Madigan A, Helbert L, Dunne E, Gardiner EE, Andrews RK, et al. Soluble glycoprotein VI, a specific marker of platelet activation is increased in the plasma of subjects with seropositive rheumatoid arthritis. PLOS ONE. 2017;12(11):e0188027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salem D, Subang R, Kuwana M, Levine JS, Rauch J. T cells from induced and spontaneous models of SLE recognize a common T cell epitope on β2-glycoprotein I. Cell Mol Immunol. 2019;16(8):685–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rauch J, Salem D, Subang R, Kuwana M, Levine JS. β2-Glycoprotein I-Reactive T Cells in Autoimmune Disease. Frontiers in Immunology. 2018;9(2836). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mereiter S, Balmaña M, Campos D, Gomes J, Reis CA. Glycosylation in the Era of Cancer-Targeted Therapy: Where Are We Heading? Cancer Cell. 2019;36(1):6–16. [DOI] [PubMed] [Google Scholar]
- 22.Holst S, Belo AI, Giovannetti E, van Die I, Wuhrer M. Profiling of different pancreatic cancer cells used as models for metastatic behaviour shows large variation in their N-glycosylation. Scientific Reports. 2017;7(1):16623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ideo H, Kondo J, Nomura T, Nonomura N, Inoue M, Amano J. Study of glycosylation of prostate-specific antigen secreted by cancer tissue-originated spheroids reveals new candidates for prostate cancer detection. Scientific Reports. 2020;10(1):2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riley NM, Bertozzi CR, Pitteri SJ. A Pragmatic Guide to Enrichment Strategies for Mass Spectrometry-based Glycoproteomics. Molecular & Cellular Proteomics. 2021:100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue Y, Xie J, Fang P, Yao J, Yan G, Shen H, et al. Study on behaviors and performances of universal N-glycopeptide enrichment methods. Analyst. 2018;143(8):1870–80. [DOI] [PubMed] [Google Scholar]
- 26.Qing G, Yan J, He X, Li X, Liang X. Recent advances in hydrophilic interaction liquid interaction chromatography materials for glycopeptide enrichment and glycan separation. TrAC Trends in Analytical Chemistry. 2020;124:115570. [Google Scholar]
- 27.Riley NM, Malaker SA, Driessen MD, Bertozzi CR. Optimal Dissociation Methods Differ for N- and O-Glycopeptides. Journal of Proteome Research. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riley NM, Hebert AS, Westphall MS, Coon JJ. Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis. Nature Communications. 2019;10(1):1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Escobar EE, King DT, Serrano-Negrón JE, Alteen MG, Vocadlo DJ, Brodbelt JS. Precision Mapping of O-Linked N-Acetylglucosamine Sites in Proteins Using Ultraviolet Photodissociation Mass Spectrometry. Journal of the American Chemical Society. 2020;142(26):11569–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei J, Tang Y, Ridgeway ME, Park MA, Costello CE, Lin C. Accurate Identification of Isomeric Glycans by Trapped Ion Mobility Spectrometry-Electronic Excitation Dissociation Tandem Mass Spectrometry. Analytical Chemistry. 2020;92(19):13211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pathak P, Baird MA, Shvartsburg AA. High-Resolution Ion Mobility Separations of Isomeric Glycoforms with Variations on the Peptide and Glycan Levels. Journal of the American Society for Mass Spectrometry. 2020;31(7):1603–9. [DOI] [PubMed] [Google Scholar]
- 32.Mookherjee A, Guttman M. Bridging the structural gap of glycoproteomics with ion mobility spectrometry. Current Opinion in Chemical Biology. 2018;42:86–92. [DOI] [PubMed] [Google Scholar]
- 33.Chen Z, Glover MS, Li L. Recent advances in ion mobility–mass spectrometry for improved structural characterization of glycans and glycoconjugates. Current Opinion in Chemical Biology. 2018;42:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou S, Huang Y, Dong X, Peng W, Veillon L, Kitagawa DAS, et al. Isomeric Separation of Permethylated Glycans by Porous Graphitic Carbon (PGC)-LC-MS/MS at High Temperatures. Analytical Chemistry. 2017;89(12):6590–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei J, Tang Y, Bai Y, Zaia J, Costello CE, Hong P, et al. Toward Automatic and Comprehensive Glycan Characterization by Online PGC-LC-EED MS/MS. Analytical Chemistry. 2020;92(1):782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashwood C, Lin C-H, Thaysen-Andersen M, Packer NH. Discrimination of Isomers of Released N- and O-Glycans Using Diagnostic Product Ions in Negative Ion PGC-LC-ESI-MS/MS. Journal of the American Society for Mass Spectrometry. 2018;29(6):1194–209. [DOI] [PubMed] [Google Scholar]
- 37.Ashwood C, Waas M, Weerasekera R, Gundry RL. Reference glycan structure libraries of primary human cardiomyocytes and pluripotent stem cell-derived cardiomyocytes reveal cell-type and culture stage-specific glycan phenotypes. Journal of Molecular and Cellular Cardiology. 2020;139:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu R, Huang Y, Zhao J, Zhong J, Mechref Y. Isomeric Separation of N-Glycopeptides Derived from Glycoproteins by Porous Graphitic Carbon (PGC) LC-MS/MS. Analytical Chemistry. 2020;92(14):9556–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu TT, Ewald JA, Ricke EA, Bell R, Collins C, Ricke WA. Modeling human prostate cancer progression in vitro. Carcinogenesis. 2018;40(7):893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vellky JE, Ricke EA, Huang W, Ricke WA. Expression and Localization of DDX3 in Prostate Cancer Progression and Metastasis. The American Journal of Pathology. 2019;189(6):1256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vellky JE, McSweeney ST, Ricke EA, Ricke WA. RNA-binding protein DDX3 mediates posttranscriptional regulation of androgen receptor: A mechanism of castration resistance. Proceedings of the National Academy of Sciences. 2020;117(45):28092–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horoszewicz JS, Leong SS, Kawinski E, Karr JP, Rosenthal H, Chu TM, et al. LNCaP Model of Human Prostatic Carcinoma. Cancer Research. 1983;43(4):1809–18. [PubMed] [Google Scholar]
- 43.Chandler KB, Leon DR, Kuang J, Meyer RD, Rahimi N, Costello CE. N-Glycosylation regulates ligand-dependent activation and signaling of vascular endothelial growth factor receptor 2 (VEGFR2). Journal of Biological Chemistry. 2019;294(35):13117–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandler KB, Costello CE. Glycomics and glycoproteomics of membrane proteins and cell-surface receptors: Present trends and future opportunities. Electrophoresis. 2016;37(11):1407–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin C-Y, Lee C-H, Chuang Y-H, Lee J-Y, Chiu Y-Y, Wu Lee Y-H, et al. Membrane protein-regulated networks across human cancers. Nature Communications. 2019;10(1):3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–d50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.VanderPlas J, Granger B, Heer J, Moritz D, Wongsuphasawat K, Satyanarayan A, et al. Altair: Interactive Statistical Visualizations for Python. The Open Journal; 2018. p. 1057. [Google Scholar]
- 48.Stavenhagen K, Plomp R, Wuhrer M. Site-Specific Protein N- and O-Glycosylation Analysis by a C18-Porous Graphitized Carbon–Liquid Chromatography-Electrospray Ionization Mass Spectrometry Approach Using Pronase Treated Glycopeptides. Analytical Chemistry. 2015;87(23):11691–9. [DOI] [PubMed] [Google Scholar]
- 49.Zhou S, Dong X, Veillon L, Huang Y, Mechref Y. LC-MS/MS analysis of permethylated N-glycans facilitating isomeric characterization. Analytical and Bioanalytical Chemistry. 2017;409(2):453–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu TT, Ewald JA, Ricke EA, Bell R, Collins C, Ricke WA. Modeling human prostate cancer progression in vitro. Carcinogenesis. 2019;40(7):893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Leoz ML, Young LJ, An HJ, Kronewitter SR, Kim J, Miyamoto S, et al. High-mannose glycans are elevated during breast cancer progression. Mol Cell Proteomics. 2011;10(1):M110.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lattová E, Skřičková J, Hausnerová J, Frola L, Křen L, Ihnatová I, et al. N-Glycan profiling of lung adenocarcinoma in patients at different stages of disease. Modern Pathology. 2020;33(6):1146–56. [DOI] [PubMed] [Google Scholar]
- 53.Brown Chandler K E.Costello C, E.Rahimi N Glycosylation in the Tumor Microenvironment: Implications for Tumor Angiogenesis and Metastasis. Cells. 2019;8(6):544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.An HJ, Gip P, Kim J, Wu S, Park KW, McVaugh CT, et al. Extensive Determination of Glycan Heterogeneity Reveals an Unusual Abundance of High Mannose Glycans in Enriched Plasma Membranes of Human Embryonic Stem Cells*. Molecular & Cellular Proteomics. 2012;11(4):M111.010660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park DD, Phoomak C, Xu G, Olney LP, Tran KA, Park SS, et al. Metastasis of cholangiocarcinoma is promoted by extended high-mannose glycans. Proceedings of the National Academy of Sciences. 2020;117(14):7633–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


