Abstract
Background:
Routine clinical data from clinical charts are indispensable for retrospective and prospective observational studies and clinical trials. Their reproducibility is often not assessed. We developed a prostate cancer-specific database for clinical annotations and evaluated data reproducibility.
Methods:
For men with prostate cancer who had clinical-grade paired tumor–normal sequencing at a comprehensive cancer center, we performed team-based retrospective data collection from the electronic medical record using a defined source hierarchy. We developed an open-source R package for data processing. With blinded repeat annotation by a reference medical oncologist, we assessed data completeness, reproducibility of team-based annotations, and impact of measurement error on bias in survival analyses.
Results:
Data elements on demographics, diagnosis and staging, disease state at the time of procuring a genomically characterized sample, and clinical outcomes were piloted and then abstracted for 2,261 patients (with 2,631 samples). Completeness of data elements was generally high. Comparing to the repeat annotation by a medical oncologist blinded to the database (100 patients/samples), reproducibility of annotations was high; T stage, metastasis date, and presence and date of castration resistance had lower reproducibility. Impact of measurement error on estimates for strong prognostic factors was modest.
Conclusions:
With a prostate cancer-specific data dictionary and quality control measures, manual clinical annotations by a multidisciplinary team can be scalable and reproducible. The data dictionary and the R package for reproducible data processing are freely available to increase data quality and efficiency in clinical prostate cancer research.
Keywords: prostate cancer, clinical data, electronic health record, reproducibility, open source software
1. Background
Clinical data have a central role in any clinical research study. In prostate cancer, data elements often include demographics, cancer characteristics at diagnosis, time-updated information on the disease course, and clinical outcomes such as metastasis and survival. Defining which elements are measured and how has been recognized as critical for the success of clinical trials, leading to standardized definitions for metastatic castration-resistant prostate cancer by the Prostate Cancer Working Group.1
Based on the premise that high-quality clinical data coupled with genomic profiling could identify predictive and prognostic genomic alterations,2 large-scale data extraction efforts from medical records are underway, such as Project GENIE3 and others.4 How well such pan-cancer approaches capture elements relevant to prostate cancer is unclear, as is the reproducibility of manual clinical annotations by investigators at medical centers.
A key source of clinical data is the medical record. Many studies are hospital-based observational studies that entirely rely on information from the medical record. Even prospective observational studies and clinical trials have the medical record as the sole source for key data elements, such as Gleason score, prostate-specific antigen, and staging. Data are distributed across narrative reports or structured data sources and are often internally discordant.5 With notable exceptions,6 it is often not reported from what sources, how, and by whom clinical data are collected for research and how they are prepared for analysis.
In this study, we designed, piloted, and implemented a clinical database for prostate cancer research (Fig. 1). We describe and share prostate cancer-specific data elements for manual curation and a software pipeline to preprocess, recode, and deidentify the resulting dataset for analyses. We also report results from a reproducibility study using this framework.
Figure 1.
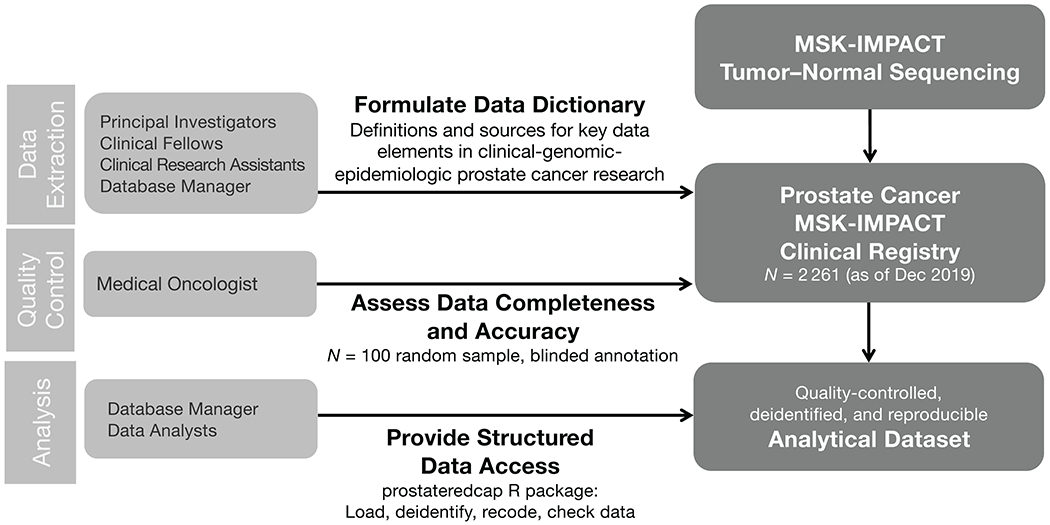
Workflow in a clinical database for prostate cancer research coupled with genomic testing.
2. Methods
2.1. Design and Implementation of a Clinical Database for Prostate Cancer Research
The clinical research database was designed for data from all men with prostate cancer who had provided written informed consent for an institutional review board-approved study of tumor–normal genomic profiling through MSK-IMPACT.7,8
First, we designed data elements applicable to prostate cancer research, led by a board-certified medical oncologist and adapting Prostate Cancer Working Group 3 recommendations1 as much as necessary for data retrieval from the medical record. Data elements were designed to be useful for prostate cancer research, without reference to data capture models3,9–11 existing in late 2017, and they were not intended for interoperability across other tumor types.
The four data categories for each patient are demographics/at-diagnosis characteristics (“baseline form”); information about genomically profiled specimens (“sample form”); outcome data (“freeze form”); and lines of therapy (“treatment form”). Nearly all data elements are structured data, predominantly binary or categorical selections from predefined lists. Numeric and date values are captured through single-line text fields, for which data formats are recommended by written instructions (“enter PSA in ng/ml”). For each data element, the source hierarchy is defined. Brief instructions address common questions, how missing data should be coded, and whether incomplete or discordant data need to be escalated for review.
Second, we implemented preliminary data definitions in a Research Electronic Data Capture (REDCap) database.12 (The database software is exchangeable.) We then piloted data extraction. After a set of 20 patient records, we revised data elements, source hierarchy, and instructions based on feasibility and an informal assessment of reproducibility by a clinician. For example, biochemical recurrence was removed from the data dictionary, given feasibility challenges. A further pilot with 80 records followed, after which the data dictionary was finalized (examples are in Table 1).
Table 1.
Domains and 5 example input data elements of the clinical database of prostate cancer, and derived data elements by the prostateredcap R package.
| Input data elements for clinical database |
Derived analytical dataset |
|||||
|---|---|---|---|---|---|---|
| Data element | Type | Source hierarchy | Instructions | Data element | Type | Source |
| Baseline form: Patient and tumor characteristics at initial diagnosis | ||||||
| Date of birth | Text1 | Automated pull from medical record | Answer Format: MM/DD/YYYY | (removed) | ||
| Date of initial diagnosis | Text1 | 1. Initial consultation note: MD-reported date of first biopsy showing prostate cancer 2. Initial consultation note: other MD-reported date of assumed diagnosis of prostate cancer, if treatment started outside without initial biopsy |
Answer Format: MM/DD/YYYY ○ Enter the date to the greatest level of granularity available. Use format “MM/YYYY” for month/year only and format “YYYY” for year only. ○ Flag for resolution if unable to find any approximate date. |
Age at diagnosis (age_dx) | Continuous value; rounded to 0.1 years | Interval between date of birth and date of initial diagnosis |
| Clinical N stage (regional lymph node metastases) | Categorical: 0 / 1 / X | 1. Initial Consultation note. 2. First GU Oncology follow-up note, particularly if the Initial Consultation note mentioned that outside records were incomplete at that time. |
○ Enter ‘X’ if unknown. ○ If N stage at diagnosis is mentioned, but it is not documented if this is clinical or path staging, enter as clinical N stage at diagnosis. ○ If note only describes names of positive lymph nodes, code as N1 for these regional lymph node stations: pelvic, hypogastric, obturator, internal iliac, external iliac, sacral. Code as M1a for all other positive lymph nodes (including common iliac). |
Clinical N stage (clin_n) | Binary: TRUE/ FALSE; can be missing | Clinical N stage |
| Other data elements: patient ID, race, ethnicity, smoking status at diagnosis, date of initial prostate biopsy, sum Gleason at diagnosis (biopsy), primary Gleason pattern at diagnosis, secondary Gleason pattern at diagnosis, histology at diagnosis, PSA at diagnosis, clinical T stage, clinical M stage, primary therapy, sum Gleason at prostatectomy, primary Gleason pattern at prostatectomy, secondary Gleason pattern at prostatectomy, pathologic T stage, pathologic N stage | ||||||
|
| ||||||
| Sample form: Characteristics of the genomically profiled sample | ||||||
| Sample tissue | Categorical: Prostate / Lymph node / Bone / Lung / Liver / Other soft tissue | Tumor sequencing report | ○ “Other soft tissue” only applies to distant metastases, not to local extension of the prostate tumor. ○ If unable to decide, flag for resolution. |
Sample tissue (tissue) | Categorical (same categories) | Sample tissue |
| Other data elements: patient ID, sample ID, date of collection, histology for sample, sample type, extent of disease at collection, sites of disease, volume of bone metastases at time of collection, continuous ADT | ||||||
|
| ||||||
| Outcome form: Clinical event data | ||||||
| Metastasis date | Text1 | 1. Oncology History of Last GU Oncology note. 2. Last Urology or Rad-Onc follow-up note. |
Answer Format: MM/DD/YYYY. Enter the date to the greatest level of granularity available. Use format “MM/YYYY” for month/year only and format “YYYY” for year only. Enter the date on which metastases were first detected. If M1 at diagnosis, enter diagnosis date. |
(removed) | – | Recoded as duration, e.g., diagnosis to metastasis |
| Other data elements: patient ID, freeze date, continuous ADT start date, castration resistance status and date, metastasis status, last MD visit date (censor date for castration resistance/metastasis), survival status and date of death/last contact | ||||||
|
| ||||||
| Treatment form: Lines of oncologic treatment | ||||||
| Data elements: patient ID, treatment name, start date, end date/last known treatment date/ongoing, reason for stop | ||||||
Dates are initially captured as text allow for incomplete but useful entries, such as a date of diagnosis as “03/2015” when the day of the month is unknown.
Third, we scaled data extraction and completed the data on all patients who had had MSK-IMPACT profiling for prostate cancer. The current manuscript describes patients included by December 2019. Weekly data capture “in real time” has since been implemented, adding patients with genomic profiling, currently MSK-IMPACT7 and MSK-ACCESS.13
Extraction was done by a team of clinical research study assistants who specifically support clinical research on genitourinary cancers and who underwent supervised training on prostate cancer data extraction by research study managers with supervision by clinicians. Clinical subspecialty fellows (urology, radiation oncology) collaborated on extractions, as did a medical student with a background as a research study assistant.
2.2. Quality Control and Data Processing
We addressed data quality and reproducibility during two key steps, data entry and data processing. During data entry, questions on data elements were flagged as queries in order to open issues on specific data fields of an individual patient/sample record, route them to colleagues, and track completion. Queries were resolved by discussion between research study assistants or escalated to project leaders.
Raw data entered in the database, even if largely in structured fields, require substantial processing. Steps include, but are not limited to: (1) recoding of many categorical variables (e.g., the many combinations of Gleason patterns are collapsed to five Gleason grade groups for analyses); (2) imputation of date variables (e.g., “03/2015” should be converted into an appropriate date format for the mid-point of March 2015); (3) calculation of time intervals (e.g., a sequencing date of April 12, 2015 and a death date of June 12, 2016 correspond to 14.0 months of follow-up for overall survival from the time of sequencing); (4) creation of time-varying covariates (e.g., castration-resistance status at the time of genomic sequencing, based on the occurrence and date of castration resistance); (5) removal of protected health information that is required for the preceding steps (e.g., exact date of cancer diagnosis); (6) assessment for internal consistency (e.g., if stage is “M1,” the date of developing metastases cannot be months after diagnosis).
Manual data processing in a spreadsheet program like Microsoft Excel, as we suspect is frequently done, is time-intensive, introduces human error, and is, by definition, not reproducible. Instead, we developed the “prostateredcap” package for the free R statistical software. The package handles data processing starting with a labeled comma-separated file exported from REDCap, data de-identification, and consistency checks. The latter step flagged approximately 10% of all records for missingness in required data elements or internal discrepancies, most of which were fixable. The output dataset with data elements recommended for analysis (see Table 1 for examples) is suitable for statistical analyses and merging with, e.g., molecular data.
2.3. Reproducibility study
To assess the completeness and reproducibility of annotations, we conducted a nested quality control study based on 100 patients and tumor samples (one per patient), with 50 randomly selected samples from metastatic castration-sensitive disease and 50 randomly selected samples from metastatic castrate-resistant disease at the time of sample procurement. Blinded to the team-based annotations in the REDCap database, a board-certified medical oncologist reviewed the full medical record to re-extract data elements selected for the reproducibility study, without being limited to the narrow source hierarchies defined for the team-based annotation.
Completeness of data elements was expressed as proportions (percentages). Confidence intervals (CIs) for these and other proportions were score test-based.14 Dates that could be not reached because of censoring were excluded from denominators.
Reliability of annotations for binary variables (e.g., present/absent) was evaluated by comparing team-based annotations to the medical oncologist as the reference “gold standard.”
For categorical variables (e.g., Gleason pattern; T stage), we calculated the proportion of agreement between gold standard and team-based annotations as well as Cohen’s κ. Missing values were included as a separate category.
For date variables, we expressed the time difference between dates from team-based annotations and gold-standard annotations as median (2.5th, 97.5th percentile).
To evaluate the impact of measurement error on scientific inference, we compared inferential results from using team-based annotations to gold-standard annotations. For four strongly prognostic exposures measured at cancer diagnosis (age; prostate-specific antigen; primary treatment with androgen deprivation; Gleason score, per grade group), we quantified associations with three outcomes (castration resistance, metastasis, and death) using univariable Cox proportional hazards regression. These models for demonstration purposes on measurement error ignore late entry and are not suited for subject-matter inference.
3. Results
The prostate cancer clinical-genomic database was manually curated with clinical data on 2,261 men with prostate cancer (Table 2), including 2,631 genomically-profiled samples, on median 1 sample per person (maximum, 5). Men were diagnosed with prostate cancer between 1987 and 2019 (median year of diagnosis 2014) at a median age of 63 years (interquartile range 56–68, range 36–94). The first tumor sample per person was obtained on median 3 months after diagnosis (interquartile range 0–42) and underwent paired tumor–normal sequencing between 2014 and 2019. Survival follow-up after sequencing of the first sample, available on 2,204 men (97%), was on median 30 months (interquartile range, 16–46).
Table 2.
Selected patient and tumor characteristics for the full database (show the first sample per patient only; n = 2261), by disease extent at sample procurement, and the reproducibility study (n = 100), by gold-standard annotation or team-based annotation.1
| Entire Prostate Cancer Clinical Database2 |
Reproducibility Study |
|||||
|---|---|---|---|---|---|---|
| Localized | Regional nodes | Metastatic hormone-sensitive | Castration resistant | Gold standard | Team-based | |
|
|
||||||
| N | 759 | 393 | 624 | 469 | 100 | 100 |
|
| ||||||
| Age at sample (yr) | 63 (57, 69) | 63 (57, 69) | 66 (60, 72) | 70 (64, 76) | 68 (61, 73) | 68 (60, 73) |
| Unknown | 1 | |||||
|
| ||||||
| Diagnosis to sample (months) | 2 (0, 4) | 2 (1, 5) | 0 (0, 21) | 74 (31, 144) | 28 (2, 89) | 27 (2, 89) |
| Unknown | 1 | 0 | 0 | 0 | 0 | 0 |
|
| ||||||
| Self-reported race | ||||||
| Asian | 17 (2%) | 9 (2%) | 18 (3%) | 20 (5%) | 0 (0%) | 1 (1%) |
| Black | 54 (7%) | 29 (8%) | 50 (9%) | 38 (9%) | 6 (7%) | 6 (7%) |
| White | 646 (90%) | 336 (90%) | 515 (88%) | 379 (86%) | 82 (91%) | 83 (90%) |
| Other | 4 (1%) | 1 (0%) | 4 (1%) | 4 (1%) | 2 (2%) | 2 (2%) |
| Unknown | 38 | 18 | 37 | 28 | 10 | 8 |
|
| ||||||
| PSA at diagnosis | 6.4 | 9.2 | 20.6 | 11.1 | 12.8 | 13.1 |
| (ng/ml) | (4.6, 11.1) | (5.6, 18.7) | (7.2, 92.5) | (6.0, 40.6) | (6.4, 50.4) | (6.5, 53.6) |
| Unknown | 44 | 13 | 29 | 43 | 3 | 6 |
|
| ||||||
| Gleason score | ||||||
| <7 | 114 (16%) | 17 (4.5%) | 15 (3%) | 38 (9%) | 6 (6%) | 7 (8%) |
| 3+4 | 173 (24%) | 46 (12%) | 39 (7%) | 56 (14%) | 13 (14%) | 11 (12%) |
| 4+3 | 133 (18%) | 87 (23%) | 79 (14%) | 63 (16%) | 12 (12%) | 14 (15%) |
| 8 | 142 (20%) | 79 (21%) | 128 (23%) | 72 (18%) | 18 (19%) | 18 (20%) |
| 9–10 | 165 (23%) | 149 (39%) | 295 (53%) | 174 (43%) | 47 (49%) | 43 (46%) |
| Unknown | 32 | 15 | 68 | 66 | 4 | 7 |
|
| ||||||
| Stage N1 | 0 (0%) | 113 (32%) | 309 (59%) | 111 (32%) | 39 (42%) | 33 (40%) |
| Unknown | 0 | 35 | 98 | 126 | 7 | 18 |
|
| ||||||
| Stage (M) | ||||||
| 0 | 759 (100%) | 393 (100%) | 167 (27%) | 302 (65%) | 51 (52%) | 53 (53%) |
| 1 | 0 (0%) | 0 (0%) | 5 (1%) | 1 (0%) | ||
| 1a | 0 (0%) | 0 (0%) | 64 (10%) | 24 (5%) | 7 (7%) | 7 (7%) |
| 1b | 0 (0%) | 0 (0%) | 343 (55%) | 114 (25%) | 36 (36%) | 38 (38%) |
| 1c | 0 (0%) | 0 (0%) | 43 (7%) | 24 (5%) | 5 (5%) | 2 (2%) |
| Unknown | 0 | 0 | 2 | 4 | 1 | 0 |
|
| ||||||
| Disease extent | ||||||
| Prostate | 759 (100%) | Unknown3 | 307 (49%) | 105 (22%) | 70 (70%) | 45 (45%) |
| Distant lymph nodes | 0 (0%) | 0 (0%) | 339 (54%) | 267 (57%) | 60 (60%) | 63 (63%) |
| Bone | 0 (0%) | 0 (0%) | 456 (73%) | 347 (74%) | 73 (73%) | 73 (73%) |
| Liver | 0 (0%) | 0 (0%) | 26 (4%) | 81 (17%) | 10 (10%) | 10 (10%) |
| Lung | 0 (0%) | 0 (0%) | 78 (12%) | 68 (14%) | 15 (15%) | 12 (12%) |
| Other soft tissue | 0 (0%) | 0 (0%) | 35 (6%) | 65 (14%) | 15 (15%) | 12 (12%) |
Statistics are count (percent) or median (interquartile range).
Not shown are 16 patients with missing/unknown disease extent at biopsy (sample procurement) of their first sample.
For patients with disease in regional nodes, presence or absence of prostatic disease was not recorded but can be inferred from prior local therapy if needed.
In the reproducibility study (Table 2), the majority of the selected data elements were 100% complete (Fig. 2). Completeness ranged between 55% to 99% for elements of clinical TNM staging, self-reported race, biopsy Gleason score, and presence of variant histologies, both for the team-based annotation and the gold standard annotation.
Figure 2.
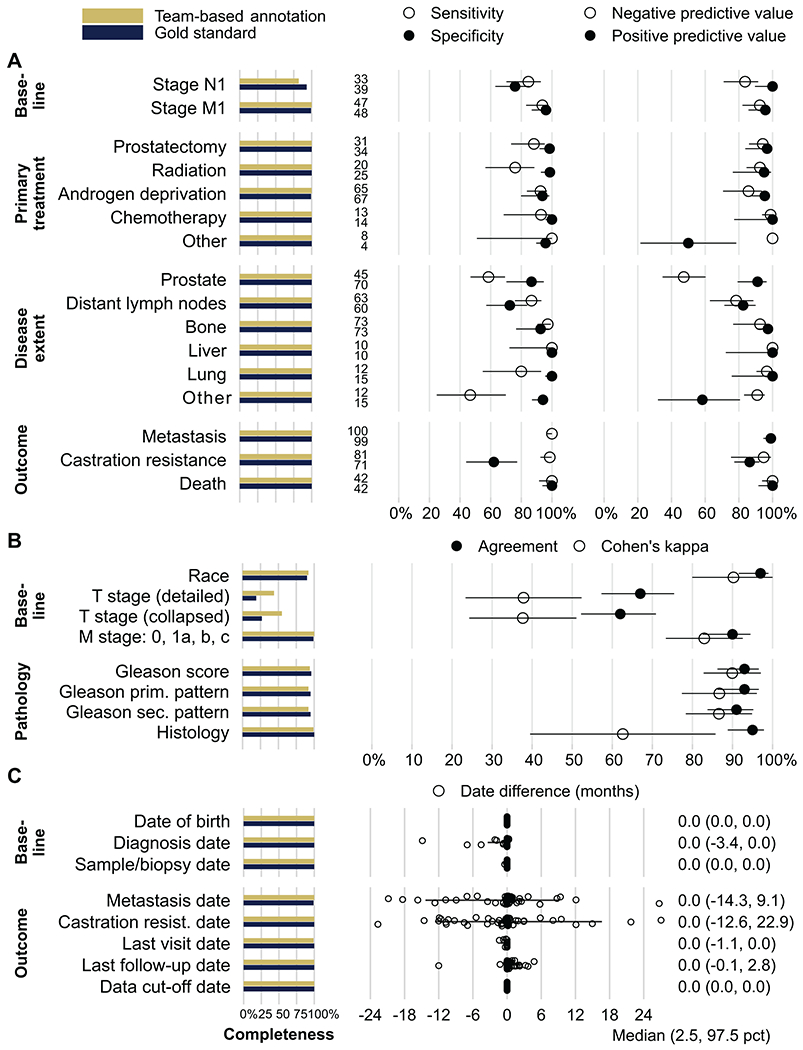
Results of the reproducibility study. The first panel shows completeness (in %) for each data element. A, Reproducibility for binary data elements: sensitivity, specificity, positive and negative predictive value (with 95% CI); and number of observations positive for each element (between the panels). B, Reproducibility for categorical data elements: agreement (team-based and gold-standard annotation gave the same value) and Cohen’s kappa (agreement corrected for agreement by chance; both with 95% CI). C, Reproducibility for date elements: difference between gold-standard and team-based annotation (individual patient’s data points). Positive values indicate that team-based annotations gave later dates than gold-standard annotations. Last visit date is the censor date for metastases and castration resistance; last follow-up date is the censor date for overall survival. Bars and values to the right are median difference (2.5th, 97.5th percentile).
To assess reproducibility of binary data elements, we first evaluated sensitivity and specificity, thus taking the perspective of the gold standard and indicating what proportions of patients with any given feature (e.g., nodal metastasis at diagnosis) present or absent were correctly recorded as such by the team-based annotation (Fig. 2A, middle panel). For 7 data elements, both sensitivity and specificity of the team-based annotations reached or exceeded 90%. The 9 data elements with lower reproducibility were nodal metastases at diagnosis (stage N1; sensitivity 85%; specificity 76%); primary treatments with any form of radiation therapy (sensitivity 88%) or prostatectomy (sensitivity 88%); presence of prostatic tumor tissue (sensitivity 59%), lung metastases (sensitivity 80%), and other soft-tissue metastases (sensitivity 47%) at sample procurement; and absence of lymph node metastases at sample procurement (specificity 72%). Finally, specificity for absence of castration resistance by end of follow-up was only modest (62%, 95% CI 44–77).
We then evaluated positive and negative predictive values, indicating the probability of features being present or absent if recorded as such in the team-based annotations. These estimates also incorporate feature prevalence and inform use of the team-based annotations when a gold standard is not available. With the exceptions of primary treatments as well as prostatic disease and other soft-tissue disease at sample procurement, predictive values were generally high (Fig. 2A, right panel).
For categorical data elements on baseline characteristics (Fig. 2B), including staging and histopathology, agreement between annotations was generally about 90%, with the exception of sub-categories of tumor (T) stage (agreement 67%, 95% CI 57–75). Agreement for T stage and variant histology was partially driven by chance, as indicated by lower Cohen’s κ (Fig. 2B), as many tumors had missing T stage and most were adenocarcinomas.
Dates of birth, diagnosis, sample procurement, and censor dates were very similar between team-based and gold standard annotations (Fig. 2C). The outcomes of metastasis and castration resistance showed notable date differences, even if without directional bias on average (median difference, 0 months). 95% of the time (in 95/100 patients), differences between team-based annotation for metastasis were between 14 months earlier and 9 months later than the gold-standard annotation; for castration resistance, 95% of date differences were between 13 months earlier and 23 months later.
To assess the impact of measurement error in team-based annotations, we quantified the association between four known strong prognostic factors and clinical outcomes: castration resistance, metastasis, and overall survival (order of decreasing measurement error). Hazard ratios for prognostic factors and overall survival did not differ between team-based or gold-standard annotations, as expected given the absence of measurement error for the outcome (Fig. 3). There were minor differences for metastasis, driven by date differences in when metastasis was recorded to have occurred. For castration resistance, for which team-based annotations had imperfect specificity and noticeable date differences, estimates using team-based annotations (e.g., hazard ratio per Gleason grade group of 1.91, 95% CI 1.52–2.40) were more noticeably, but still only slightly different from estimates using gold standard annotations (hazard ratio per Gleason grade group of 1.69, 95% CI 1.35–2.12).
Figure 3.
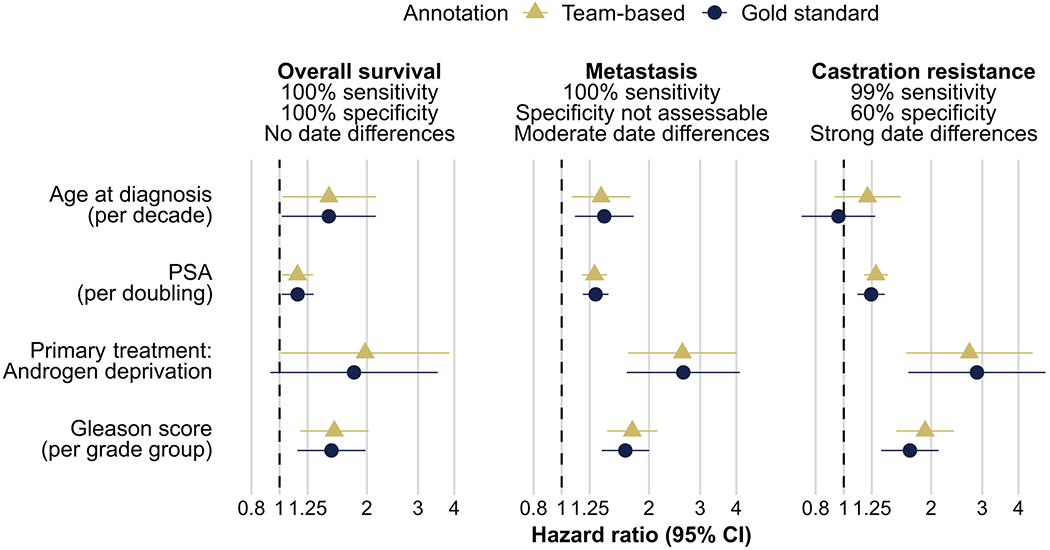
Impact of measurement error on scientific inference. Using team-based annotations (triangle) or gold-standard annotations (circle), hazard ratios for four selected prognostic factors (in rows: age at diagnosis; PSA; primary treatment androgen deprivation; and Gleason score) and three outcomes (in columns, by increasing reliability: castration resistance, metastasis, death) were estimated.
4. Discussion
The prostate cancer-specific clinical research database described here is notable for four key features: a data dictionary with a defined source hierarchy that was tested for feasibility; a data extraction pipeline that makes the conversion from medical record-derived raw data to an analyzable dataset a reproducible process; a reproducibility study that openly evaluates data quality in the setting that the database was implemented; and the provision of these tools to the scientific community for re-use.
Our undertaking was pragmatic. We intended to create a clinical research database that captured data elements essential in prostate cancer that could be linked with genomic profiling data. We relied on data captured during routine clinical practice. Data extraction had to be scalable to thousands of patient records without external funding, precluding desirable approaches such as blinded parallel annotation by more than one person. Earlier versions of the database have already been useful to assess genomic and clinical features in prostate cancer,15–17 as are similar databases.6,18,19 Unsurprisingly, for some data elements, reproducibility of annotations was suboptimal, including for data elements known to have low reproducibility, like tumor T stage.20 Besides suboptimal reproducibility, completeness of T stage for both annotations was low in the subset of patients selected for the reproducibility study, who all had metastatic disease. Outcome data can be imperfect, which highlights one challenge for establishing surrogate endpoints,21 with castration resistance or the date when metastases first occurred being examples in our study. Some data definitions that are consensus for clinical trials1 were not suitable, e.g., for castration resistance. Increasing reproducibility on these data elements would primarily require changing clinical care by mandating laboratory tests and imaging in regular intervals, as it is feasible in a clinical trial.
Importantly, while we considered annotations by a medical oncologist an alloyed gold standard, the reproducibility study merely assessed whether two investigators would come to the same annotation, given the same medical record (repeatability), and not a comparison with “truth” (validity). Even an annotation by two or more board-certified clinicians, possibly external to the local team as done frequently in clinical trials, would have the same limitation, because the true state of the patient may simply not be completely recorded or recordable in the medical record. Nevertheless, we believe that dedicated reproducibility studies like the current one should be done whenever data are collected for clinical research to help improve data quality and inform result interpretations.22 The data elements provided through our data dictionary, their source hierarchy, and how they are post-processed can and may need to be adapted to local needs. Feasibility, completeness, and reliability of data will differ depending on patient population, clinical setting, available data sources, annotation approach, team, and other factors. They should not be inferred from our estimates.
When researchers create and validate similar databases, we anticipate that the data dictionary, which can be directly uploaded into REDCap to create the database, and the data processing pipeline via the R package may be useful. Source hierarchies and data elements definitions will likely require local adaptation, because medical records differ between institutions. Different use cases of the data may prompt different levels of attention to specific data elements, possibly requiring changes in data definitions, source hierarchies, and the R code for data processing and quality control. For example, if T stage is only used as a descriptor of the study population, but not as an exposure or outcome, its lower reproducibility may not be a major concern. In contrast, in a previous study leveraging this database where radiographically-defined disease extent was a key exposure of interest,17 our study radiologist performed repeat annotations based on a review of scans, even though concordance of team-based annotations using radiology reports was high at 90%.
Aspects that we have found critical for success of our database were clinical knowledge about prostate cancer in the team; extensive pilot testing; modifications of the data dictionary until idiosyncrasies of specific medical record data elements were sufficiently addressed and clear hierarchies for potentially internally discordant data elements had been devised; structured approaches to re-reviewing challenging cases by other team members, including those with additional expertise (e.g., through “queries” in REDCap); and algorithm (code)-based data processing and quality control. Further, results from reproducibility studies should inform data usage and result interpretation. For example, result interpretations using the outcome of castration resistance need to be cautious, given its lower reproducibility. Data quality may even prompt not to collect certain data points of clinical interest if completeness or reliability are too low, as our pilot testing showed for biochemical recurrence.
Principled approaches to improving data quality are needed. A limitation of our work is that data definitions are not explicitly linked other data definitions, the NCI Thesaurus, or pan-cancer approaches. How well data could be integrated across institutions may be informed both by comparisons of data definitions of interest and by reproducibility studies that would best be conducted at each site, just as laboratories typically demonstrate performance on quality control samples at each site. How manual approaches to clinical data curation in prostate cancer compare to larger-scale, pan-cancer, or computer-assisted (“machine learning”) data extraction would be important to compare, as would be comparisons of such data to true gold standards.
5. Conclusions
With a prostate cancer-specific data dictionary and quality control measures, manual annotations of clinical data by a multidisciplinary team can be scalable and reproducible. The data dictionary and the R package should help increase data quality in clinical prostate cancer research.
Funding
This work was funded in part by the National Cancer Institute (1P01CA228696, to P.W. Kantoff; P30CA008748, Cancer Center Support Grant; P50CA092629, Prostate Cancer SPORE) and the Department of Defense (Early Investigator Research Award W81XWH-18-1-0330, to K.H. Stopsack; Physician Research Award W81XWH-17-1-0124, to W. Abida). D.E. Rathkopf, W. Abida, and K.H. Stopsack are Prostate Cancer Foundation Young Investigators. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Conflicts of Interest
M.J. Morris is an uncompensated consultant for Bayer, Advanced Accelerator Applications, Johnson and Johnson, Novartis, and Lantheus. He is a compensated consultant for Oric, Curium, Athenex, Exelexis, and Astra Zeneca. MSK receives funds for contracts for the conduct of clinical trials from Bayer, Advanced Accelerator Applications, Novartis, Corcept, Roche/Genentech, and Janssen.
D.E. Rathkopf is a consultant for Janssen, Genentech, AstraZeneca, Bayer, and Myovant Sciences, and has received research funding through her institution from Janssen Oncology, Medivation, Celgene, Tekeda, Millennium, Ferring, Novartis, Taiho Pharmaceutical, AstraZeneca, Genentech/Roche, TRACON Pharma, Bayer, and Phosplatin Therapeutics.
S.F. Slovin has received research support from Sanofi-Aventis, Novartis, Poseida, and the Prostate Cancer Foundation, and honoraria for advisory boards from Clovis, Janssen, Sanofi-Aventis, and PER.
D.C. Danila has received research support from the U.S. Department of Defense, American Society of Clinical Oncology, Prostate Cancer Foundation, Stand Up 2 Cancer, Janssen Research & Development, Astellas, Medivation, Agensys, Genentech, and CreaTV; he is a consultant for Angle LLT, Axiom LLT, Janssen Research & Development, Astellas, Medivation, Pfizer, Genzyme, and Agensys.
P.W. Kantoff reports the following disclosures for the last 24-month period: he has investment interest in ConvergentRx Therapeutics, Context Therapeutics LLC, DRGT, Placon, and Seer Biosciences; he is a company board member for ConvergentRx Therapeutics, Context Therapeutics LLC; he is a consultant/scientific advisory board member for Bavarian Nordic Immunotherapeutics, DRGT, GE Healthcare, Janssen, OncoCellMDX, Progenity, Seer Biosciences, and Tarveda Therapeutics; and he serves on data safety monitoring boards for Genentech/Roche and Merck.
W. Abida reports the following disclosures: he has received honoraria from CARET, Roche, Medscape, and Aptitude Health; is a consultant for Clovis Oncology, Janssen, MORE Health, ORIC Pharmaceuticals, and Daiichi Sankyo; he has received research funding through his institution from AstraZeneca, Zenith Epigenetics, Clovis Oncology, GlaxoSmithKline, ORIC Pharmaceuticals, and Epizyme; and he has had travel/accommodations/expenses paid by GlaxoSmithKline, Clovis Oncology, and ORIC Pharmaceuticals.
N.M. Keegan, S.E. Vasselman, E.S. Barnett, B. Nweji, E.A. Carbone, A. Blum, K.A. Autio, and K.H. Stopsack report no potential conflict of interest.
Data sharing
Data definitions to create the REDCap database, the prostateredcap R package, an overview of data elements recommended for analysis, and an example dataset are available at https://stopsack.github.io/prostateredcap.
References
- 1.Scher HI, Morris MJ, Stadler WM, et al. Trial Design and Objectives for Castration-Resistant Prostate Cancer: Updated Recommendations From the Prostate Cancer Clinical Trials Working Group 3. J Clin Oncol. 2016;34(12):1402–1418. doi: 10.1200/JCO.2015.64.2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mateo J, McKay R, Abida W, et al. Accelerating precision medicine in metastatic prostate cancer. Nat Cancer. 2020;1(11):1041–1053. doi: 10.1038/s43018-020-00141-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7(8):818–831. doi: 10.1158/2159-8290.CD-17-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singal G, Miller PG, Agarwala V, et al. Association of Patient Characteristics and Tumor Genomics With Clinical Outcomes Among Patients With Non-Small Cell Lung Cancer Using a Clinicogenomic Database. JAMA. 2019;321(14):1391–1399. doi: 10.1001/jama.2019.3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Lucadou M, Ganslandt T, Prokosch HU, Toddenroth D. Feasibility analysis of conducting observational studies with the electronic health record. BMC Med Inf Decis Mak. 2019;19(1):202. doi: 10.1186/s12911-019-0939-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh WK, Hayes J, Evan C, et al. Development of an integrated prostate cancer research information system. Clin Genitourin Cancer. 2006;5(1):61–66. doi: 10.3816/CGC.2006.n.019 [DOI] [PubMed] [Google Scholar]
- 7.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17(3):251–264. doi: 10.1016/j.jmoldx.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. doi: 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belenkaya R, Gurley MJ, Golozar A, et al. Extending the OMOP Common Data Model and Standardized Vocabularies to Support Observational Cancer Research. JCO Clin Cancer Inform. 2021;(5):12–20. doi: 10.1200/CCI.20.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goel AK, Campbell WS, Moldwin R. Structured Data Capture for Oncology. JCO Clin Cancer Inform. 2021;(5):194–201. doi: 10.1200/CCI.20.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guérin J, Laizet Y, Le Texier V, et al. OSIRIS: A Minimum Data Set for Data Sharing and Interoperability in Oncology. JCO Clin Cancer Inform. 2021;(5):256–265. doi: 10.1200/CCI.20.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Razavi P, Li BT, Brown DN, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med. 2019;25(12):1928–1937. doi: 10.1038/s41591-019-0652-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agresti A, Coull BA. Approximate is better than “exact” for interval estimation of binomial proportions. Am Stat. 1998;52(2):119–126. doi: 10.2307/2685469 [DOI] [Google Scholar]
- 15.Mota JM, Barnett E, Nauseef JT, et al. Platinum-Based Chemotherapy in Metastatic Prostate Cancer With DNA Repair Gene Alterations. JCO Precis Oncol. 2020;4:355–366. doi: 10.1200/po.19.00346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen B, Mota JM, Nandakumar S, et al. Pan-cancer Analysis of CDK12 Alterations Identifies a Subset of Prostate Cancers with Distinct Genomic and Clinical Characteristics. Eur Urol. 2020;78(5):671–679. doi: 10.1016/j.eururo.2020.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stopsack KH, Nandakumar S, Wibmer AG, et al. Oncogenic Genomic Alterations, Clinical Phenotypes, and Outcomes in Metastatic Castration-Sensitive Prostate Cancer. Clin Cancer Res. 2020;26(13):3230–3238. doi: 10.1158/1078-0432.CCR-20-0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koshkin VS, Patel VG, Ali A, et al. PROMISE: a real-world clinical-genomic database to address knowledge gaps in prostate cancer. Prostate Cancer Prostatic Dis. Published online August 6, 2021. doi: 10.1038/s41391-021-00433-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lubeck DP, Litwin MS, Henning JM, et al. The capsure database: a methodology for clinical practice and research in prostate cancer. Urology. 1996;48(5):773–777. doi: 10.1016/S0090-4295(96)00226-9 [DOI] [PubMed] [Google Scholar]
- 20.Reese AC, Sadetsky N, Carroll PR, Cooperberg MR. Inaccuracies in assignment of clinical stage for localized prostate cancer. Cancer. 2011;117(2):283–289. doi: 10.1002/cncr.25596 [DOI] [PubMed] [Google Scholar]
- 21.ICECaP Working Group, Sweeney C, Nakabayashi M, et al. The Development of Intermediate Clinical Endpoints in Cancer of the Prostate (ICECaP). J Natl Cancer Inst. 2015;107(12):djv261. doi: 10.1093/jnci/djv261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Smeden M, Lash TL, Groenwold RHH. Reflection on modern methods: five myths about measurement error in epidemiological research. Int J Epidemiol. 2020;49(1):338–347. doi: 10.1093/ije/dyz251 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data definitions to create the REDCap database, the prostateredcap R package, an overview of data elements recommended for analysis, and an example dataset are available at https://stopsack.github.io/prostateredcap.


