Abstract
The peptidases of thermophilic lactic acid bacteria have a key role in the proteolysis of Swiss cheeses during warm room ripening. To compare their peptidase activities toward a dairy substrate, a tryptic/chymotryptic hydrolysate of purified β-casein was used. Thirty-four peptides from 3 to 35 amino acids, including three phosphorylated peptides, constitute the β-casein hydrolysate, as shown by tandem mass spectrometry. Cell extracts prepared from Lactobacillus helveticus ITG LH1, ITG LH77, and CNRZ 32, Lactobacillus delbrueckii subsp. lactis ITG LL14 and ITG LL51, L. delbrueckii subsp. bulgaricus CNRZ 397 and NCDO 1489, and Streptococcus thermophilus CNRZ 385, CIP 102303, and TA 060 were standardized in protein. The peptidase activities were assessed with the β-casein hydrolysate as the substrate at pH 5.5 and 24°C (conditions of warm room ripening) by (i) free amino acid release, (ii) reverse-phase chromatography, and (iii) identification of undigested peptides by mass spectrometry. Regardless of strain, L. helveticus was the most efficient in hydrolyzing β-casein peptides. Interestingly, cell extracts of S. thermophilus were not able to release a significant level of free proline from the β-casein hydrolysate, which was consistent with the identification of numerous dipeptides containing proline. With the three lactic acid bacteria tested, the phosphorylated peptides remained undigested or weakly hydrolyzed indicating their high intrinsic resistance to peptidase activities. Finally, several sets of peptides differing by a single amino acid in a C-terminal position revealed the presence of at least one carboxypeptidase in the cell extracts of these species.
Thermophilic lactobacilli such as Lactobacillus helveticus or Lactobacillus delbrueckii along with Streptococcus thermophilus constitute essential lactic starters in Swiss-type cheese (400,000 tons of Emmentaler produced per year worldwide). The three species have a key role in the acidification step and in cheese proteolysis (13). Adequate proteolysis is essential for acceptable quality of the ripened cheese. Whole caseins are first hydrolyzed by milk protease (plasmin) and/or rennet (chymosin and pepsin) into large and intermediate-sized peptides. The proteinases and peptidases from starters subsequently hydrolyze them in small peptides and amino acids, which are known to be aroma precursors. In the three species considered, peptidases are intracellular enzymes released upon lysis in the curd, where they remain active for several weeks (14, 40, 41). Because of their predominant role in proteolysis, studies on peptidases of thermophilic lactobacilli and S. thermophilus have expanded greatly in recent years (5, 18, 35, 36). Several aminopeptidases, dipeptidases, and peptidases specific to proline-containing peptides (prolidase, prolinase, X-prolyl-dipeptidyl aminopeptidase, and prolyl aminopeptidase) were isolated, cloned, and sequenced (20); only the general aminopeptidase(s) and the X-prolyl-dipeptidyl-aminopeptidase of thermophilic lactobacilli were shown to be essential to cheese flavor (27, 33). To date, no carboxypeptidase has been isolated. The role of each species in cheese proteolysis is still unclear since their peptidases were mainly characterized on synthetic substrates. Few reports compared the overall potential of the different species; all results showed that peptidase activities are generally highest in L. helveticus strains (17, 36).
The objective of this work was to better assess how L. helveticus, L. delbrueckii, and S. thermophilus contribute to proteolysis in Swiss cheese in order to improve the choice of starters and to control this essential process. For that purpose, we compared the intracellular peptidase potentials of these species, using a dairy substrate and the pH and temperature used for warm room ripening. The substrate chosen was a tryptic/chymotryptic hydrolysate of β-casein, mimicking the initial proteolysis as proposed by Lemée et al. (24). We characterized it by tandem mass spectrometry (MS-MS) and then subjected it to the action of cell extracts, allowing quantitative and qualitative analyses of the peptidase activities.
MATERIALS AND METHODS
Strains and culture conditions.
Origins of the strains used are summarized in Table 1. L. helveticus, L. delbrueckii subsp. lactis, and L. delbrueckii subsp. bulgaricus were stored at −80°C in MRS broth (Difco, Sparks, Md.) (7), and S. thermophilus was kept in M17 broth (Difco) supplemented with 5% (wt/vol) lactose (38). The media were supplemented with 15% glycerol prior to freezing. For the propagation of strains, media without glycerol were used, and growth at 37°C was monitored by the optical density at 650 nm. (OD650).
TABLE 1.
Origins of strains used
| Species | Strain | Origin |
|---|---|---|
| Lactobacillus helveticus | ITG LH1 | Industrial strain |
| ITG LH77 | Industrial strain | |
| CNRZ 32 | Artisanal lactic starter of Gruyère de Comté, France | |
| Streptococcus thermophilus | CNRZ 385 | |
| CIP 102303T | Pasteurized milk | |
| TA 060a | Industrial strain | |
| L. delbrueckii subsp. lactis | ITG LL14 | Industrial strain |
| ITG LL51 | Industrial strain | |
| L. delbrueckii subsp. bulgaricus | CNRZ 397 | Starter of yoghurt |
| NCDO 1489 | Bulgarian yoghurt |
Kindly supplied by Texel, Dangé St Romain, France.
Preparation of cell extracts.
Cells were collected during the exponential growth phase (OD650 = 1) by centrifugation (7,000 × g, 20 min, 7°C) and washed twice with cold distilled sterile water (15% of the initial culture volume). The pellets were stored 24 h at −18°C before being resuspended in cold sterile water (6% of initial culture volume) and submitted to a precooled French press apparatus at 138 MPa for 3 min (one run). Suspensions were centrifuged at 4,000 × g at 4°C for 30 min to eliminate unbroken cells. The supernatants representing cell extracts were filtered (0.45-μm-pore-size Sartorius filter) and stored at −18°C until used. Protein content was assayed according to Bradford method (Bio-Rad S.A., Ivry-sur Seine, France), using bovine serum albumin (Sigma, Saint Quentin, Fallavier, France) as a standard. Depending on the strain, the protein content was between 1 and 5 mg per ml of cell extract.
Assay of peptidase activity.
The lyophilized β-casein hydrolyzate used as the substrate was obtained as described by Lemée et al. (24). Briefly, pure β-casein (kindly supplied by EURIAL Poitouraine, Nantes, France) (23) was dissolved in sterile distilled water at a final concentration of 10 g/liter. Hydrolysis was performed using a mixture of trypsin and chymotrypsin (enzyme/substrate ratio, 1/1,000 [wt/wt]). After 3 h of incubation at 37°C, the β-casein was completely converted to peptides. Both enzymes were inactivated by heating at 80°C for 20 min, and the hydrolysate was lyophilized and stored at 4°C.
Five hundred fifty micrograms of protein of thawed cell extract adjusted to 500 μl with sterile distilled cold water was added to 3.5 ml of β-casein hydrolysate solution (0.8 mg of β-casein hydrolysate per ml in 20 mM ammonium acetate buffer [pH 5.5]), sterilized by filtration (0.22-μm-pore-size filter), and incubated at 24°C. Samples were collected at different times. Enzyme activity was stopped by one of two methods, depending on the nature of subsequent analysis: (i) by adding 1% (vol/vol) trifluoroacetic acid (TFA) or (ii) by adding 80% (vol/vol) absolute ethanol and incubating the mixture for 6 h at 4°C to eliminate precipitated proteins and large peptides by centrifugation (5,000 × g, 15 min, 4°C). After inactivation, samples were stored at −18°C until use. Controls were made without cell extract and with heat-treated (100°C, 20 min) cell extracts. For each strain, two independent assays were carried out.
Free amino acids.
Free amino acids were quantified using the cadmium ninhydrin reagent by the method of Baer et al. (2), with methionine as a standard. Triplicate samples at each time, inactivated by ethanol treatment, were thawed at 4°C. Fifty microliters of each sample was mixed with 550 μl of distilled water, and 1.25 ml of Cd-ninhydrin solution was added. Free amino acids were also analyzed by cation-exchange chromatography with an automatic amino acids analyzer (Pharmacia LKB Alpha Plus; Amersham Pharmacia Biotech Europe GmbH, Orsay, France) as previously described by Spackman et al. (37), using lithium citrate buffer for elution. Before analysis, samples inactivated with 1% TFA were thawed at room temperature and centrifuged (5,000 × g, 15 min, 4°C). The supernatant was recovered, and sulfosalicylic acid (30 mg/ml) was added. The solution was then incubated for 1 h at 4°C and centrifuged (5,000 × g, 15 min, 4°C). The free amino acids were assayed in the supernatant fraction.
Amino acids compositions of the β-casein peptides were determined after acid hydrolysis in sealed tubes with 6 N HC1 at 110°C for 24 h (6). Hydrolysates were evaporated to dryness in vacuum over KOH pellets, washed twice with distilled water, and analyzed with the amino acid analyzer described above but using sodium instead of lithium citrate for elution.
Peptides.
Peptides were separated by reverse-phase high-pressure liquid chromatography (RP-HPLC) on a Waters (Milford, Mass.) 625LC system, using solvent A (0.106% TFA in MilliQ water) and solvent B (0.100% TFA, 80% acetonitrile, 20% MilliQ water). The samples were analyzed on a Nova-Pack C18 column (39- by 150-mm inside diameter 4-μm particle size, and 60-Å pore size; Waters, Saint Quentin en Yvelines, France) with a linear gradient from 0 to 55% solvent B for 60 min followed by 55 to 80% solvent B for 2 min, at a flow rate of 0.8 ml/min at 40°C. The eluted peaks were detected by absorbance at 214 nm using a photodiode array detector spectrometer (Waters 991 series). Before injection, samples inactived with TFA were thawed at room temperature and centrifuged (10,000 × g, 15 min, 4°C); 50 μl of the supernatant were directly injected into the column.
MS.
Mass spectra were recorded on a PE Sciex Api III Plus triple-quadruple mass spectrometer (Sciex, Thornhill, Ontario, Canada) equipped with an atmospheric pressure ionization source. The masses of the peptides were determined by HPLC–electrospray ionization (ESI)-MS analysis as follows. Online with the separation by RP-HPLC, a postcolumn flow splitter was used to introduce 10% of the HPLC eluate into the mass spectrometer (25). The ion source voltage was set at 4.8 kV, and the orifice voltage was set at 80 V. The quadruple mass analyzer (Q3) was scanned to an m/z range of 200 to 2,000 Da, with a step size of 0.3 Da and a dwell time of 0.5 ms per step. The molecular masses were determined from these data by using the software supplied by Sciex (Biomultiview 1.2). The amino acid sequence of each peptide was determined by MS-MS as follows. The peptides collected after RP-HPLC separation were continuously infused with a pump model 22 (Harvard, South Natick, Mass.) at a flow rate of 3 μl/min. Collision-actived dissociation experiments were performed with argon, and the collision energies were set around 30 eV in accordance with the molecular mass and the charge state of the parents ions (29). The resolution was set to pass a 2-Da window around the parent ion on the Q1 and was adjusted a unit m/z for analysis of the product ions on the Q3.
RESULTS
Characterization of the β-casein hydrolysate.
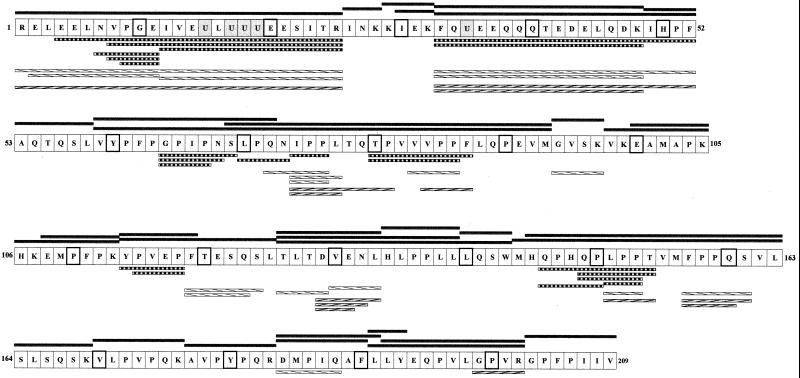
RP-HPLC coupled with MS-MS showed that the tryptic/chymotryptic β-casein hydrolysate contained 34 peptides from 3 to 35 amino acids (373.2 to 3,864.4 Da) including three phosphorylated peptides [Arg1-Arg25], [Phe33-Lys48], and [Phe33-Phe52] (Table 2). Figure 1 shows the complete sequence of the β-casein (209 amino acids) as well as the 34 peptides obtained in the hydrolysate. Two iterations of the tryptic/chymotryptic hydrolysis led to the same 34 peptides, showing the reproducible preparation of this substrate. The bonds hydrolyzed corresponded to those predicted in the review by Pelissier (32), with two additional sites: Leu125-Thr126 and Met144-His145. The amino acid composition was determined after complete acid hydrolysis of the β-casein peptides. The total amino acid content, 5.9 mM, was 96% of the expected theoretical value (6.1 mM).
TABLE 2.
Characteristics of the 34 peptides contained in the tryptic/chymotryptic hydrolysate of β-caseina
| Measured mass (Da; mean ± SD) | Sequence assignment (β-casein amino acids) | Peak no. |
|---|---|---|
| 373.40b | 26–28 | 2 |
| 388.40b | 30–32 | 3 |
| 389.30b | 94–97 | 1 |
| 407.20 | 191–193 | 12 |
| 512.30b | 49–52 | 13 |
| 516.50b | 29–32 | 4 |
| 532.40b | 140–143 | 10 |
| 645.50b | 100–105 | 5 |
| 646.40 | 53–58 | 6 |
| 648.50b | 164–169 | 4 |
| 688.70b | 134–139 | 21 |
| 741.50b | 203–209 | 22 |
| 747.50b | 108–113 | 14 |
| 750.50b | 114–119 | 16 |
| 779.60b | 170–176 | 10 |
| 820.40b | 184–190 | 18 |
| 829.70b | 177–183 | 8 |
| 872.80 | 98–105 | 7 |
| 903.50b | 126–133 | 15 |
| 933.80b | 184–191 | 22 |
| 1,012.94b | 106–113 | 11 |
| 1,270.49 ± 0.99 | 192–202 | 16 |
| 1,383.14 ± 0.07 | 191–202 | 19 |
| 1,396.19 ± 0.56 | 114–125 | 17 |
| 1,565.69 ± 0.15 | 59–72 | 20 |
| 1,574.99 ± 0.29 | 126–139 | 23 |
| 2,062.00b | 33–48 | 9 |
| 2,089.60b | 126–143 | 25 |
| 2,150.80b | 145–163 | 22 |
| 2,281.18 ± 0.98 | 144–163 | 22 |
| 2,556.98 ± 0.28 | 33–52 | 17 |
| 2,742.18 ± 0.29 | 69–93 | 24 |
| 3,123.23 ± 0.07 | 1–25 | 18 |
| 3,864.78 ± 0.56 | 59–93 | 25 |
The mass of each peptide was determined by RP-HPLC–ESI-MS. Sequence assignment was done from the masses, and the sequences were all verified by MS-MS. The peak numbers correspond to those indicated in the RP-HPLC profile of the β-casein peptides in Fig. 5.
Singly charged molecular ions (no SD).
FIG. 1.
β-Casein sequence (209 amino acids; one-letter amino acid code). Phosphorylated serines are indicated by the letter “U” and highlighted in grey; every 10th amino acid is framed in bold. The 34 initial peptides of the hydrolysate are represented in black rectangles above the β-casein sequence. After 72 h of hydrolysis, nonhydrolyzed peptides were identified by MS-MS and are presented below the sequence with different patterns, depending on the cell extract used: ▥ for L. helveticus ITG LH1, ▧ for L. delbrueckii subsp. lactis ITG LL14, and for S. thermophilus TA 060.
Kinetics of β-casein peptide hydrolysis by cell extracts.
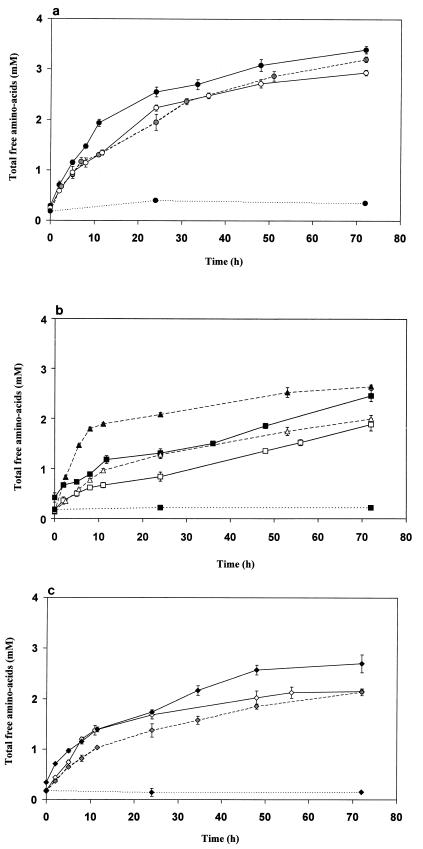
The release of free amino acids (Fig. 2) showed that all strains were able to degrade β-casein peptides at the pH (5.5) and temperature (24°C) used. No release of amino acids was noted in the absence of cell extracts or with heat-treated extracts, indicating no subsequent degradation of the β-casein hydrolysate by residual tryptic or chymotryptic activity.
FIG. 2.
Time course of hydrolysis of the β-casein hydrolysate
by cell extracts of the following strains: (a) L. helveticus
ITG LH1 (—●—), ITG LH77 (○), and CNRZ 32 (---
---), (b)
L. delbrueckii subsp. lactis ITG LL14 (—■—)
and ITG LL51 (□), and L. delbrueckii subsp.
bulgaricus NCDO 1489 (▴) and CNRZ 397 (▵); and (c)
S. thermophilus CNRZ 385 (—⧫—), TA 060 (◊), and CIP
(Pasteur Institut Collection, Paris, France) 102303
(--- ---). The enzyme reactions
were performed for 72 h at 24°C in 20 mM acetate ammonium buffer
(pH 5.5) with 137.5 μg of cell extract proteins per ml for 0.7 mg of
lyophilized β-casein hydrolysate per ml. Blanks (⋯●⋯,
⋯⧫⋯, and ⋯▴⋯) were assayed with the same conditions
but with boiled cell extracts.
---). The enzyme reactions
were performed for 72 h at 24°C in 20 mM acetate ammonium buffer
(pH 5.5) with 137.5 μg of cell extract proteins per ml for 0.7 mg of
lyophilized β-casein hydrolysate per ml. Blanks (⋯●⋯,
⋯⧫⋯, and ⋯▴⋯) were assayed with the same conditions
but with boiled cell extracts.
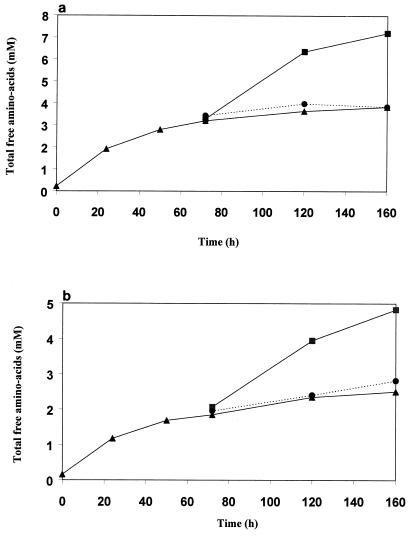
The time course of hydrolysis exhibited a high initial rate which decreased drastically after 8 to 10 h of incubation. After 72 h of incubation, no significant increase of free amino acids was observed for most of the strains. This can be explained by either peptidase inactivation or resistant peptides. The addition at 72 h of fresh β-casein hydrolysate led to new hydrolysis with a similar high initial rate, showing that the peptidases were still active (Fig. 3). Thus, the decreased rate was due to peptides resistant to hydrolysis. This was further confirmed by the addition at 72 h of fresh cell extracts; in this case, no increase of amino group was noted (Fig. 3).
FIG. 3.
Hydrolysis of the β-casein hydrolysate by cell extracts (▴) of L. helveticus ITG LH77 (a) and S. thermophilus CIP 102303 (b): effect of the addition at 72 h of fresh β-casein hydrolysate (■) or fresh cell extracts (●).
The initial rate and final extent were species and strain dependent. L. helveticus showed the most efficient peptidase activity regardless of strain, with a mean initial rate of 0.12 mM/h (±0.01 [standard deviation {SD}], depending on the strain) and final amount of 2.93 ± 0.25 mM free amino acids. These values were, respectively, 0.10 ± 0.02 mM/h and 2.1 ± 0.13 mM for the S. thermophilus strains. L. delbrueckii showed the largest intraspecies variability: L. delbrueckii subsp. bulgaricus NCDO (National Collection of Dairy Organisms) 1489 exhibited a higher initial rate than all of the L. helveticus strains studied, but the final content of free amino acids was lower. L. delbrueckii subsp. lactis (Institut Technique du Gruyère, Gruyère, France) ITG LL51 and ITG LL14 and L. delbrueckii subsp. bulgaricus CNRZ (Centre National de la Recherche Zootechnique collection, Institut National de la Recherche Agronomique, Jouy-en-Josas, France) 397 had the lowest initial rate (0.064 ± 0.01 mM/h) and the lowest final content (only 1.86 ± 0.15 mM free amino acids after 72 h).
Analysis of amino acids released from β-casein peptides by cell extracts.
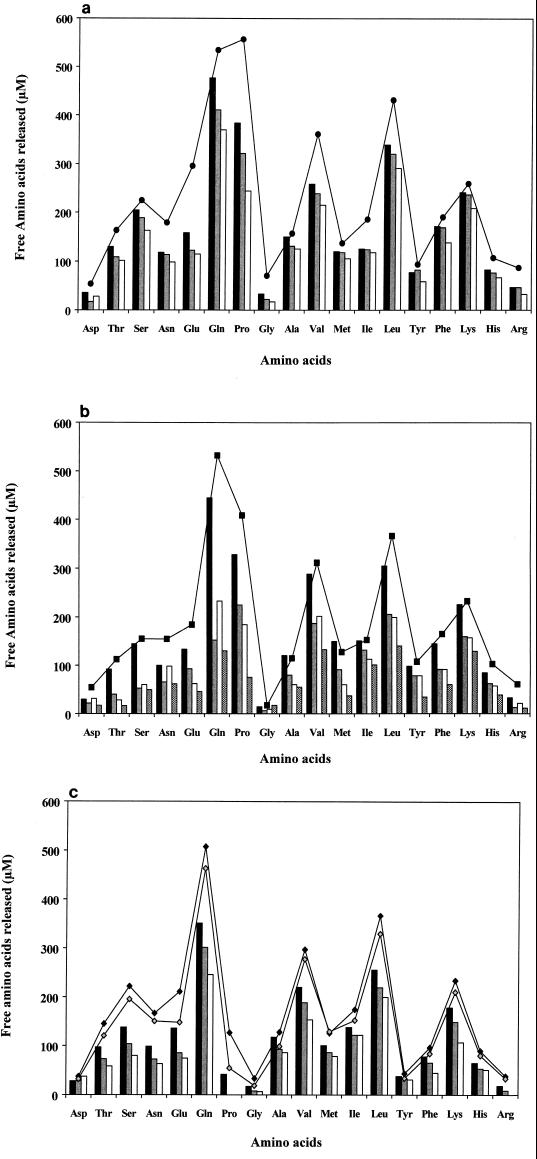
At zero time, free amino acids represented less than 2.5% of the total amino acid content. After 24 h, 42 to 53% was released by L. helveticus cell extracts depending on the strain; corresponding values were 24 to 36% for S. thermophilus and 20 to 48% for L. delbrueckii. After 72 h of incubation, cell extracts of L. helveticus ITG LH1, S. thermophilus CNRZ 385 and TA 060, and L. delbrueckii subsp. lactis ITG LL14 were able to liberate 69, 44, and 57% of the total amino acid content, respectively. These results are in complete agreement with the kinetics presented in Fig. 2 in terms of relative peptidase efficiencies of the strains. The profiles shown in Fig. 4 were qualitatively similar for L. helveticus, L. delbrueckii subsp. lactis, and L. delbrueckii subsp. bulgaricus, with the following predominant amino acids: Gln, Pro, Leu, Val, and Lys. Again the species L. delbrueckii exhibited the highest variation from strain to strain. Interestingly, the cell extracts of S. thermophilus were not able to release free proline from the β-casein peptides, except for the very low amount detected for strain CNRZ 385 (4% of the total proline content).
FIG. 4.
Free amino acids released after 24 h by hydrolysis
of β-casein peptides by cell extracts of the following strains: (a)
L. helveticus ITG LH1
(&cjs3744;), CNRZ 32
( ), and ITG LH77
(▯); (b) L.
delbrueckii subsp. bulgaricus NCDO 1489
(&cjs3744;), and CNRZ 397
(
), and ITG LH77
(▯); (b) L.
delbrueckii subsp. bulgaricus NCDO 1489
(&cjs3744;), and CNRZ 397
( and L. delbrueckii subsp. lactis ITG
LL14 (▯), and ITG
LL51 (
); and; (c)
S. thermophilus CNRZ 385
(&cjs3744;), TA 060
(
and L. delbrueckii subsp. lactis ITG
LL14 (▯), and ITG
LL51 (
); and; (c)
S. thermophilus CNRZ 385
(&cjs3744;), TA 060
( ), and CIP 102303
(▯). Curves indicate
values obtained after 72 h of incubation for L.
helveticus ITG LH1 (a; ●), L. delbrueckii subsp.
lactis ITG LL14 (b; ▪), and S.
thermophilus CNRZ 385 (♦), and TA 060
(
), and CIP 102303
(▯). Curves indicate
values obtained after 72 h of incubation for L.
helveticus ITG LH1 (a; ●), L. delbrueckii subsp.
lactis ITG LL14 (b; ▪), and S.
thermophilus CNRZ 385 (♦), and TA 060
( ) (c).
) (c).
Chromatographic analysis of peptides released from β-casein peptides by cell extracts.
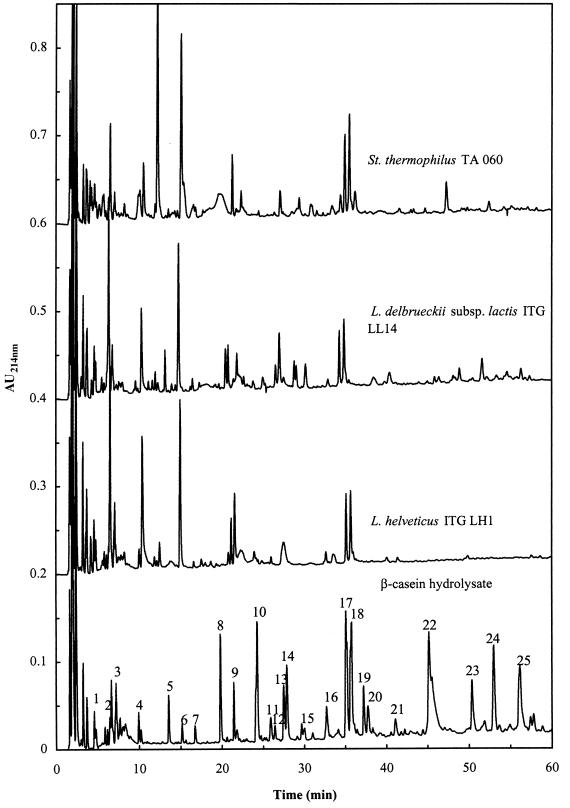
Following the addition of cell extracts, the peptide profile at time zero was identical to that for β-casein hydrolysate alone. After 24 h, the profiles were species dependent; therefore, only one profile per species is reported in Fig. 5. The peptides with a retention time higher than 38 min were completely hydrolyzed by L. helveticus; concomitantly the area of the peaks with retention times between 22 and 38 min decreased and new peaks with retention times of less than 22 min appeared. The hydrolysis of peptides by cell extracts of S. thermophilus and L. delbrueckii was less extensive than that observed with L. helveticus. These results are in agreement with the kinetics of β-casein hydrolysis. Surprisingly, three peaks, at retention times of 22, 35, and 37 min, appeared to be particularly resistant to the peptidase activity of the three species.
FIG. 5.
RP-HPLC profiles of β-casein peptides at time zero and after incubation for 24 h at 24°C with cell extracts of L. helveticus ITG LH1, L. delbrueckii subsp. lactis ITG LL14, and S. thermophilus TA 060.
Identification by MS-MS of peptides undigested by cell extracts.
After 72 h of incubation of the β-casein hydrolysate with extracts from L. helveticus ITG LH1, S. thermophilus TA 060, and L. delbrueckii subsp. lactis ITG LL14, several peptides remained undigested. They were identified by MS-MS and are indicated in Fig. 1, except for some dipeptides in the case of S. thermophilus (PP, GP, MP, YP, and LP), for which several locations in the β-casein sequence were possible.
Some specific features are noteworthy. The phosphorylated peptides [Phe33- Lys48] and [Phe33-Phe52] were not hydrolyzed by the three species. The phosphorylated peptide [Arg1-Arg25] was not hydrolyzed by S. thermophilus but could be degraded by an endopeptidase activity of thermophilic lactobacilli, followed by aminopeptidase processing in the case of L. helveticus. The cleavage at the peptide bond [Glu11-Ile12] reflected an endopeptidase activity. Peptides shorter than 12 amino acids, particularly from the N- and C-terminal regions of the β-casein, were completely hydrolyzed by all of the cell extracts. The short peptides located in the central region of the β-casein sequence, [Gly94-Lys97], [Tyr114-Pro119], and [Thr126-Leu133], were more or less hydrolyzed depending on the species. The long peptides (longer than 12 amino acids) rich in proline residues and containing in particular a Pro-Pro sequence, i.e, [Pro75-Pro76], [Pro85-Pro86], [Pro152-Pro153] and [Pro158-Pro159] were only partially hydrolyzed; the one exception is the sequence [Pro136-Pro137] included in the peptides [Thr126-Leu139], [Thr126-Trp143], and [His134-Leu139]. The prolyl aminopeptidase (PepIP) is the only enzyme able to hydrolyze this bond. This enzyme is active mainly on di- and tripeptides (15) and PepIP from L. helveticus is active on tetrapeptides (28), implying the preliminary involvement of endopeptidases and/or aminopeptidases in forming the long peptides. PepIP is not synthesized in S. thermophilus, explaining the numerous dipeptides containing proline identified by MS-MS (Pro-Pro, Gly-Pro, Met-Pro, Tyr-Pro, and Leu-Pro). The presence in L. helveticus of undigested di- and tripeptides containing Pro-Pro, [Ile74-Pro76] and [Leu150-Pro152], was surprising considering its high PepIP activity and could indicate an inhibition by some peptides released. Finally, the presence of a carboxypeptidase in L. helveticus and in S. thermophilus was strongly supported by the presence of the two sets of peptides differing by a single amino acid in a C-terminal position: [Gly64-Pro69], [Gly64-Asn68], and [Gly64-Ser67] in L. helveticus and [Asp129-Leu133], [Asp129-Asn132], and [Asp129-Glu131] in S. thermophilus.
DISCUSSION
This work presents new insight into the peptidase activity of various lactic acid bacteria by using a dairy substrate, a hydrolysate of β-casein. The cell extracts of 10 strains belonging to thermophilic starters, L. helveticus, L. delbrueckii, and S. thermophilus hydrolyzed efficiently most of the β-casein peptides, at a pH and temperature close to cheese ripening conditions.
Phosphorylated peptides were shown to be much more resistant than other peptides to hydrolysis by peptidases from thermophilic lactic acid bacteria, in particular to the aminopeptidases, providing the first experimental evidence for a hypothesis previously proposed (13, 22). This is in agreement with the numerous phosphorylated peptides identified by MS in various cheese aqueous extracts or cheese juices at the end of the ripening (Grana, Emmentaler, Cheddar, etc.) (12, 26, 34). The phosphorylated N-terminal part of β-casein is also very resistant to the cell surface proteinases of lactococci (20, 21). This mechanism of resistance is not dependent on the number of phosphorylated residues: the fragment [Phe33-Lys48] with one phosphorylated residue is resistant to all peptidases. By contrast, the fragment [Arg1-Arg25] with four phosphorylated residues can be partially hydrolyzed by endopeptidases. Several endopeptidases have already been purified from L. helveticus, L. delbrueckii subsp. bulgaricus, and S. thermophilus (3, 10, 35).
The accumulation of phosphorylated peptides raised the question of bacterial phosphatases. To our knowledge, little is known in this regard. Acid phosphatase activity was detected in Lactococcus lactis and L. delbrueckii subsp. bulgaricus (22), but under our conditions, this activity may not be maintained at an efficient rate. It cannot be ruled out that the degradation of the phosphorylated peptides is controlled by a mechanism involving bacterial phosphatases.
Another original result was the identification of peptides with successive removal of C-terminal amino acids in L. helveticus and S. thermophilus which strongly suggested the presence of at least one carboxypeptidase in lactic acid bacteria. This carboxypeptidase is not able to remove a proline in the C-terminal position, which suggests that no carboxypeptidase P is synthesized by these species. The presence of a carboxypeptidase has been reported for Lactobacillus casei (8, 9) and L. helveticus (36), but this point was controversial because of the synthetic substrate used. Carboxypeptidase activity was also recently suspected from the identification of peptides in Emmentaler juice at the end of the ripening (26). Some of the β-casein peptides produced in this work could constitute valuable substrates for further purification of this carboxypeptidase.
Some species-specific features can also be highlighted from that work. L. helveticus exhibited the highest activity regardless of strain, in agreement with previous data obtained with synthetic substrates or cheese models (9, 36). This supports the idea of the essential role of L. helveticus in Swiss cheese proteolysis and highlights the importance of its efficient lysis for the release of peptidases in curd. Lysis of L. helveticus was shown to occur during cold room ripening (40), to be strain dependent, and to have an obvious impact on the final extent of proteolysis (41). L. delbrueckii exhibited a large strain dependence with a threefold difference in free amino acid content between the less efficient and the more efficient strain. This should be considered in selecting strains of L. delbrueckii, particularly when it is used in Swiss cheese without L. helveticus. Interestingly, no strain of S. thermophilus was able to release significant amounts of free proline from the β-casein peptides. The peptidases of S. thermophilus were recently reviewed by Rul and Monnet (35). In lactic acid bacteria, the major peptidase activity specific of proline-containing peptides corresponds to PepX, releasing dipeptides, X-Pro, from the N terminus of peptides. A prolidase (PepQ), specific for X-Pro dipeptides, is then required to release free proline and has been purified from several lactococci and lactobacilli (4, 11, 30). Another means of proline release would involve a PepIP that removes proline in an N-terminal position. PepIP has been purified and the corresponding gene has been cloned from various lactobacilli including L. helveticus and L. delbrueckii (1, 16, 19, 31, 42). The absence of PepIP in S. thermophilus (35) and a weak PepQ activity can explain the inability of S. thermophilus to release free proline from β-casein peptides. With regard to the accumulating di- and tripeptides containing Pro-Pro sequences in the presence of L. helveticus extracts, PepIP activity may possibly be controlled by inhibitory peptides, resulting from peptidase action toward β-casein. This hypothesis is supported by the inhibitory effect of peptides derived from the sequence 58–72 against PepN, PepX, and endopeptidase activities (39). Swiss cheeses are known to be especially rich in free proline, which is associated with the sweet flavor. As S. thermophilus cannot release it, the amount of free proline will directly depend on the activity from the thermophilic lactobacilli peptidases (PepIP and PepQ) and on the efficiency of their release through lysis (41). This result can be of great technological value in terms of starter selection.
REFERENCES
- 1.Atlan D, Gilbert C, Blanc B, Portalier R. Cloning, sequencing and characterization of a PepIP gene encoding a proline iminopeptidase from Lactobacillus delbrueckii subsp. bulgaricusCNRZ 397. Microbiology. 1994;140:527–535. doi: 10.1099/00221287-140-3-527. [DOI] [PubMed] [Google Scholar]
- 2.Baer A, Riba I, Meyer J, Bütikofer U. Microplate assay of free amino acids in Swiss cheeses. Lebensm Wiss Technol. 1996;29:58–62. [Google Scholar]
- 3.Bockelman W, Hoppe-Seyler T, Heller K. Purification and characterization of an endopeptidase from Lactobacillus delbrueckii subsp. bulgaricusB14. Int Dairy J. 1996;6:1167–1180. [Google Scholar]
- 4.Booth M, Jennings V, Fhaolain I, O'Cuinn G. Prolidase activity of Lactococcus lactis ssp. cremorisAM2: partial purification and characterization. J Dairy Res. 1990;57:245–254. [Google Scholar]
- 5.Christensen J E, Dudley E G, Pederson J A, Steele J L. Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Leeuwenhoek. 1999;76:217–246. [PubMed] [Google Scholar]
- 6.Davies M G, Thomas A J. An investigation of hydrolytic techniques for the amino acid analysis of foodstuffs. J Sci Food Agric. 1973;24:1505–1540. doi: 10.1002/jsfa.2740241208. [DOI] [PubMed] [Google Scholar]
- 7.De Man J-C, Rogosa M, Sharpe E. A medium for the cultivation of lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 8.El Abboudi M, El Soda M, Pandian S, Barreau M, Trépanier G, Simard R E. Peptidase activities in debittering and nondebittering strains of lactobacilli. Int Dairy J. 1991;1:55–64. [Google Scholar]
- 9.El Soda M, Desmazeaud M J. Les peptide-hydrolases des lactobacilles du groupe Thermobacterium. I. Mise en évidence de ces activités chez Lactobacillus helveticus, L. acidophilus, L. lactis et L. bulgaricus. Can J Microbiol. 1982;28:1181–1188. [PubMed] [Google Scholar]
- 10.Fenster K, Parkin K, Steele J. Characterization of a thiol-dependent endopeptidase from Lactobacillus helveticusCNRZ 32. J Bacteriol. 1997;179:2529–2533. doi: 10.1128/jb.179.8.2529-2533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez-Espla M D, Martin-Hernandez M C, Fox P F. Purification and characterization of a prolidase from Lactobacillus casei subsp. caseiIFPL 731. Appl Environ Microbiol. 1997;63:314–316. doi: 10.1128/aem.63.1.314-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferranti P, Itolli E, Barone F, Malorni G, Garro G, Laezza P, Chianese L, Migliaccio F, Stingo V, Addeo F. Combined high resolution chromatographic techniques (FPLC and HPLC) and mass spectrometry-based identification of peptides and proteins in Grana Padano cheese. Lait. 1997;77:683–697. [Google Scholar]
- 13.Fox P, McSweeney P L H. Proteolysis in cheese during ripening. Food Rev Int. 1996;12:457–459. [Google Scholar]
- 14.Gagnaire V, Lortal S, Leonil J. Free active peptidases are evidenced in Emmental juices extracted before the warm room ripening. J Dairy Res. 1998;65:119–128. [Google Scholar]
- 15.Gilbert G, Atlan D, Blanc B, Portalier R. Proline iminopeptidase from Lactobacillus delbrueckii subsp. bulgaricusCNRZ 397: purification and characterization. Microbiology. 1994;140:537–542. doi: 10.1099/00221287-140-3-537. [DOI] [PubMed] [Google Scholar]
- 16.Habibi-Najafi M, Lee B. Purification and characterization of proline iminopeptidase from Lactobacillus casei subsp. caseiLLG. J Dairy Sci. 1995;78:251–259. [Google Scholar]
- 17.Hickey M W, Hillier A J, Jago G R. Peptidase activities in lactobacilli. Aust J Dairy Technol. 1983;38:118–123. [Google Scholar]
- 18.Kalantzopoulos G, Tsakalidou E, Manalopoulou E. Proteinase, peptidase and esterase activities of cell free extracts from wild strains of Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus salivarius subsp. thermophilusisoled from traditional Greek yogurt. J Dairy Res. 1990;62:601–610. [Google Scholar]
- 19.Klein J, Schmidt U, Plapp R. Cloning, heterologous expression and sequencing of a novel proline iminopeptidase gene, PepI, from Lactobacillus delbrueckii subsp. lactisDSM7290. Microbiology. 1994;140:1133–1139. doi: 10.1099/13500872-140-5-1133. [DOI] [PubMed] [Google Scholar]
- 20.Kunji E R S, Mierau I, Hagting A, Poolman B, Konings W N. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:187–221. doi: 10.1007/BF00395933. [DOI] [PubMed] [Google Scholar]
- 21.Kunji E, Fang G, Jeronimus-Stratingh C, Bruins A, Poolman B, Konings W. Reconstruction of the proteolytic pathway for use of β-casein by Lactococcus lactis. Mol Microbiol. 1998;17:1107–1118. doi: 10.1046/j.1365-2958.1998.00769.x. [DOI] [PubMed] [Google Scholar]
- 22.Kyriakidis S M, Sakellaris G, Sotiroudis T G. Is protein phosphorylation a control mechanism for the degradation of caseins by lactic acid bacteria? The detection of an extracellular acid phosphatase. Lett Appl Microbiol. 1993;16:295–298. [Google Scholar]
- 23.Le Magnen, C., and J.-J. Maugas. January 1992. Method and device for obtaining beta casein. Patent WO92/00017.
- 24.Lemée R, Gagnaire V, Maubois J-L. Strain variability of the cell-free proteolytic activity of dairy propionibacteria towards β-casein peptides. Lait. 1998;78:227–240. [Google Scholar]
- 25.Léonil J, Mollé D, Gaucheron F, Arpino P, Guénot P, Maubois J-L. Analysis of major bovine milk proteins by on-line high-performance liquid chromatography and electrospray ionization-mass spectrometry. Lait. 1995;75:193–210. [Google Scholar]
- 26.Léonil J, Gagnaire V, Mollé D, Maubois J-L. Symposium on Quality and Microbiology of Traditional and Raw Milk Cheeses. Dijon, France: Office for Official Publications of the European Communities; 1998. Casein breakdown in Emmental cheese and its relation with the release of phosphopeptides in aqueous phase during ripening; p. 344. [Google Scholar]
- 27.Meyer J, Spahni A. Influence of X-prolyl-dipeptidylaminopeptidase of Lactobacillus delbrueckii subsp. lactis on proteolysis and taste of Swiss Gruyère cheese. Milchwissenschaft. 1998;53:449–453. [Google Scholar]
- 28.Miyakawa H, Hashimoto T, Nakamura T, Ishibashi N, Shimamura S, Igoshi K. Purification and characterization of a prolyl aminopeptidase from Lactobacillus helveticusLHE-511. Milchwissenschaft. 1994;49:615–619. [Google Scholar]
- 29.Mollé D, Morgan F, Bouhallab S, Léonil J. Selective detection of lactolated peptides in hydrolysates by liquid chromatography electrospray tandem mass spectrometry. Anal Biochem. 1998;259:152–161. doi: 10.1006/abio.1998.2636. [DOI] [PubMed] [Google Scholar]
- 30.Morel F, Frot-Coutaz J, Aubel D, Portalier R, Atlan D. Characterization of a prolidase from Lactobacillus delbrueckii subsp. bulgaricusCNRZ 397 with an unusual regulation of biosynthesis. Microbiology. 1999;145:437–446. doi: 10.1099/13500872-145-2-437. [DOI] [PubMed] [Google Scholar]
- 31.Morel F, Gilbert C, Geourjon C, Frot-Coutaz J, Portalier R, Atlan D. The prolyl aminopeptidase from Lactobacillus delbrueckii subsp. bulgaricusbelongs to the α/β hydrolase fold family. Biochim Biophys Acta. 1999;1429:501–505. doi: 10.1016/s0167-4838(98)00264-7. [DOI] [PubMed] [Google Scholar]
- 32.Pelissier J-P. Protéolyse des caseines. Sci Aliments. 1984;4:1–35. [Google Scholar]
- 33.Prost F, Chamba J-F. Effect of aminopeptidases activity of thermophilic lactobacilli on Emmental cheese characteristics. J Dairy Sci. 1994;77:24–33. [Google Scholar]
- 34.Roudot-Algaron F, Le Bars D, Kerhoas L, Einhorn J, Gripon J-C. Phosphopeptides from Comtécheese: nature and origin. J Food Sci. 1994;59:544–547. [Google Scholar]
- 35.Rul F, Monnet V. Presence of additional peptidases in Streptococcus thermophilus CNRZ 302 compared to Lactococcus lactis. J Appl Microbiol. 1997;82:695–704. doi: 10.1046/j.1365-2672.1997.00185.x. [DOI] [PubMed] [Google Scholar]
- 36.Sasaki M, Bosman B W, Tan P S T. Comparison of proteolytic activities in various lactobacilli. J Dairy Res. 1995;62:601–610. doi: 10.1017/s0022029900031332. [DOI] [PubMed] [Google Scholar]
- 37.Spackman D H, Stein W H, Moore S. Automatic recording apparatus for use in the chromatography of amino-acids. Anal Chem. 1958;30:1190–1206. [PubMed] [Google Scholar]
- 38.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tobiassen R, Stepaniak L, Sorhaug T. Screening for differences in the proteolytic systems of Lactococcus, Lactobacillus, and Propionibacterium. Z Lebensm Unters Forsch A. 1997;204:273–278. [Google Scholar]
- 40.Valence F, Richoux R, Thierry A, Palva A, Lortal S. Autolysis of Lactobacillus helveticus and Propionibacterium freundenreichiiin Swiss cheese: first evidence by using species specific lysis markers. J Dairy Res. 1998;65:609–620. [Google Scholar]
- 41.Valence F, Deutsch S-M, Richoux R, Gagnaire V, Lortal S. Autolysis and related proteolysis in Swiss cheese for two Lactobacillus helveticusstrains. J Dairy Res. 2000;67:261–271. doi: 10.1017/s0022029900004118. [DOI] [PubMed] [Google Scholar]
- 42.Varmanen P, Vesanto E, Steele J, Palva A. Characterization and expression of the pepN gene encoding a general aminopeptidase from Lactobacillus helveticus. FEMS Microbiol Lett. 1994;124:315–320. doi: 10.1111/j.1574-6968.1994.tb07302.x. [DOI] [PubMed] [Google Scholar]