Abstract
The current perception of evolutionary relationships and the natural diversity of ammonia-oxidizing bacteria (AOB) is mainly based on comparative sequence analyses of their genes encoding the 16S rRNA and the active site polypeptide of the ammonia monooxygenase (AmoA). However, only partial 16S rRNA sequences are available for many AOB species and most AOB have not yet been analyzed on the amoA level. In this study, the 16S rDNA sequence data of 10 Nitrosomonas species and Nitrosococcus mobilis were completed. Furthermore, previously unavailable 16S rRNA sequences were determined for three Nitrosomonas sp. isolates and for the gamma-subclass proteobacterium Nitrosococcus halophilus. These data were used to revaluate the specificities of published oligonucleotide primers and probes for AOB. In addition, partial amoA sequences of 17 AOB, including the above-mentioned 15 AOB, were obtained. Comparative phylogenetic analyses suggested similar but not identical evolutionary relationships of AOB by using 16S rRNA and AmoA as marker molecules, respectively. The presented 16S rRNA and amoA and AmoA sequence data from all recognized AOB species significantly extend the currently used molecular classification schemes for AOB and now provide a more robust phylogenetic framework for molecular diversity inventories of AOB. For 16S rRNA-independent evaluation of AOB species-level diversity in environmental samples, amoA and AmoA sequence similarity threshold values were determined which can be used to tentatively identify novel species based on cloned amoA sequences. Subsequently, 122 amoA sequences were obtained from 11 nitrifying wastewater treatment plants. Phylogenetic analyses of the molecular isolates showed that in all but two plants only nitrosomonads could be detected. Although several of the obtained amoA sequences were only relatively distantly related to known AOB, none of these sequences unequivocally suggested the existence of previously unrecognized species in the wastewater treatment environments examined.
Chemolithoautotrophic ammonia-oxidizing bacteria (AOB) play a central role in the natural cycling of nitrogen by aerobically transforming ammonia to nitrite. From an anthropocentric point of view, the activity of AOB is considered to be both detrimental and beneficial. AOB oxidize urea and ammonia fertilizers to nitrite and, in conjunction with nitrite oxidizers which subsequently convert nitrite to nitrate, thus contribute to fertilizer loss from agricultural soils by producing compounds which are easily washed out or used as electron acceptors for denitrification (42). The former process is also responsible for significant pollution of water supplies with nitrite and nitrate. Furthermore, AOB can produce greenhouse gases (8, 74) and corrode, because of the produced acid, stonework and concrete (46). On the other hand, AOB activity is encouraged in wastewater treatment plants to reduce the ammonia content of sewage before discharge into the receiving waters (49). Reduction of ammonia releases into aquatic environments reduces the risk of local oxygen depletion, helps to prevent eutrophication (15), and protects aquatic life (6).
After the first reports on successful isolation of chemolithoautotrophic ammonia oxidizers at the end of the 19th century (14, 88), researchers have continued to investigate the diversity of AOB in natural and engineered environments by applying enrichment and isolation techniques. These efforts resulted in the description of 16 AOB species (27, 30, 32, 34, 84). Furthermore, DNA-DNA hybridization studies provided evidence for the existence of at least 15 additional species (30, 31, 67). However, low maximum growth rates and growth yields of AOB render cultivation-based analysis of their environmental diversity extremely time-consuming and tedious. Furthermore, all culture techniques are potentially selective and thus bear the risk of incomplete coverage of the actually existing bacterial diversity (5, 28, 79).
Comparative 16S rRNA sequence analyses of cultured AOB revealed that members of this physiological group are confined to two monophyletic lineages within the Proteobacteria. Nitrosococcus oceani (75, 84) is affiliated with the gamma-subclass of the class Proteobacteria, while members of the genera Nitrosomonas (including Nitrosococcus mobilis), Nitrosospira, Nitrosolobus, and Nitrosovibrio form a closely related grouping within the beta-subclass of Proteobacteria (17, 52, 67, 73, 76, 92). It has been suggested (17) and subsequently questioned (73) that the latter three genera should be reclassified in the single genus Nitrosospira.
The availability of 16S rRNA sequences also provided a basis for the development of cultivation-independent methods to investigate the diversity and community composition of these microorganisms in complex environments. PCR-mediated preferential amplification of AOB 16S rDNA and subsequent cloning and sequencing have been extensively applied to create phylogenetic inventories of various environments (7, 35, 37, 38, 44, 47, 50, 65, 87), which led to the recognition of seven 16S rRNA beta-subclass AOB sequence clusters. Recently, the battery of molecular tools to infer the presence of AOB in the environment has been supplemented by PCR primers for specific amplification of the ammonia monooxygenase structural gene amoA (22, 47, 56, 64). While environmental 16S rDNA and amoA libraries significantly extended our knowledge on the natural diversity of AOB, biases introduced by DNA extraction, PCR amplification, and cloning methods (10, 12, 51, 54, 71, 72, 90) blur quantitative information on the community composition. Furthermore, due to long-term stability of extracellular DNA and frequent passive dispersal of microbial cells over long distances, the detection of DNA from a certain AOB is inadequate to prove that this organism is part of the autochthonous microbial community. In contrast to PCR-based methods, quantitative information on AOB population structure and dynamics in the environment is obtainable via membrane or in situ hybridization techniques in combination with AOB-specific oligonucleotide probes (28, 40, 48, 61, 62, 80, 81). The latter approach also allows one to directly relate community structure with the morphology and spatial distribution of the detected organisms.
The application of molecular tools already provided exciting new insights into the diversity and community composition of AOB in various environments. However, incomplete coverage of cultured AOB in the current 16S rRNA and amoA data sets hampers the design and evaluation of specific primers and probes and renders it impossible to decide whether a novel environmentally retrieved 16S rRNA or amoA sequence represents a previously not cultured AOB or is identical to an already isolated AOB which is not yet included in the respective database. One goal of the present study was to complete the 16S rDNA and amoA sequence databases in regard to described AOB species. A thorough phylogenetic analysis including all available 16S rRNA and amoA sequences of AOB was conducted in order to establish robust phylogenetic frameworks for molecular surveys of the natural diversity of AOB. Furthermore, the specificity of all published AOB-specific 16S rRNA and amoA-targeting primers was reevaluated. These analyses helped to resolve several inconsistent results in the literature. Subsequently, the diversity of AOB occurring in wastewater treatment plants was analyzed by assigning more than 100 cloned amoA sequences from 11 nitrifying treatment plants to the established amoA framework.
MATERIALS AND METHODS
Pure cultures of AOB and sampled wastewater treatment plants.
Table 1 summarizes the AOB investigated in this study. AOB were cultured using the media and conditions described previously (30). Nitrosococcus sp. strains Nm 104 and Nm 107 were isolated from the industrial wastewater treatment plant Kraftisried by using the enrichment and isolation procedures (with 10 to 100 mM NH4Cl and 10 to 200 mM NaCl) described by Juretschko et al. (28). Samples of 11 different wastewater treatment plants were collected between 1997 and 1999 (Table 2).
TABLE 1.
Pure cultures of AOB used in this studya
| Organismb | Reference | Origin |
|---|---|---|
| Nitrosococcus halophilus Nc4T | 34 | Salt lagoon, Sardinia, Italy |
| Nitrosococcus mobilis Nc2T | 32 | North Sea, Harbour of Husum, Germany |
| Nitrosococcus sp. strain Nm 93 | 28 | Activated-sludge, rendering plant Kraftisried, Germany |
| Nitrosococcus sp. strain Nm 104 | This study | Activated-sludge, rendering plant Kraftisried, Germany |
| Nitrosococcus sp. strain Nm 107 | This study | Activated-sludge, rendering plant Kraftisried, Germany |
| Nitrosomonas aestuarii Nm36T | 30 | Brackish water, North Sea, Denmark |
| Nitrosomonas communis Nm2T | 30 | Soil, isle of Korfu, Greece |
| Nitrosomonas cryotolerans Nm55T | 27 | Kasitsna Bay, Alaska |
| Nitrosomonas europaea Nm50T, ATCC 25978 | 88, 91 | Soil, United States |
| Nitrosomonas halophila Nm1T | 30 | North Sea |
| Nitrosomonas marina Nm22T | 30 | Shell grit, great barrier reef, Australia |
| Nitrosomonas nitrosa Nm90T | 30 | Activated-sludge, chemical processing facility, Germany |
| Nitrosomonas oligotropha Nm45T | 30 | Soil, Hamburg, Germany |
| Nitrosomonas sp. strain Nm33 | 30 | Soil, Japan |
| Nitrosomonas sp. strain Nm41 | 30 | Soil, Leningrad, Russia |
| Nitrosomonas sp. strain Nm51, ATCC 25981 | 30, 87 | Seawater, off Peru |
| Nitrosomonas sp. strain Nm103 | 28 | Activated-sludge, rendering plant Kraftisried, Germany |
| Nitrosomonas ureae Nm10T | 30 | Soil, Sardinia, Italy |
AOB were obtained from the culture collection of the Institut für Allgemeine Botanik der Universität Hamburg, Mikrobiologische Abteilung, Germany.
T, type strain; ATCC, American Type Culture Collection.
TABLE 2.
Characteristics of 11 German nitrifying wastewater treatment plants analyzeda
| Type of treatment plant, location | System | PE | Sewage type |
|---|---|---|---|
| Semitechnical, Ingolstadt, SBBR1 | B | 1,800 | Concentrated sewage from sludge dewatering |
| Semitechnical, Ingolstadt, SBBR2 | B | 50 | Municipal |
| Semitechnical, Ingolstadt, BIOFOR1 | B | 500 | Municipal |
| Semitechnical, Ingolstadt, BIOFOR2 | B | 500 | Municipal |
| Full-scale, Poing | AS | 105,000 | Municipal |
| Full-scale, Munich I, Großlappen | AS | 1,200,000 | Municipal |
| Full-scale, Kraftisried | AS | 6,000 | Rendering plant effluent |
| Full-scale, Plattling | AS | 26,000 | Rendering plant effluent |
| Full-scale, Sünching, Plant A | AS | ND | Municipal |
| Full-scale, Sünching, Plant B | AS | ND | Industrial |
| Semitechnical, Stuttgart, trickling filter 1 | B | ND | Semisynthetic |
B, biofilm; AS, activated sludge; PE, population equivalent (1 PE = 60 g of biological oxygen demand d−1 [26]); SBBR, sequencing batch biofilm reactor; BIOFOR, biological fixed oxygen reactor ND, not determined.
DNA extraction.
AOB were harvested from 10 liters of exponentially growing cultures by continuous-flow centrifugation (20,000 × g, 400 ml min−1). Activated-sludge samples (2 ml each) were pelleted by centrifugation (5 min, 10,000 × g). Biofilm samples were detached from their substratum by swirling in a suitable volume of DNA extraction buffer (see below). After removal of the substratum, biofilm material was harvested by centrifugation (5 min, 10,000 × g). Total genomic DNA was extracted according to the following protocol. A 0.25-g (wet weight) pellet of each sample was resuspended in a 2-ml polypropylene tube with a screw top with 625 μl of DNA extraction buffer (100 mM Tris-HCl [pH 8.0], 100 mM sodium EDTA [pH 8.0], 100 mM sodium phosphate [pH 8.0], 1.5 M NaCl, 1% cetyltrimethylammonium bromide). After addition of 50 μl of enzyme mixture I (lysozyme [66,200 U mg−1; Fluka, Buchs, Switzerland], lipase type 7 [2,000 U mg−1; Sigma, Deisenhofen, Germany], pectinase [1,200 U mg−1; Roth, Karlsruhe, Germany], and β-glucuronidase [120,000 U mg−1; Sigma] each at 10 mg ml−1), the mixture was incubated for 30 min at 37°C. Subsequently, 50 μl of enzyme mixture II (proteinase K [20 U mg−1; Boehringer Mannheim], protease typ9 [1 U mg−1; Sigma], and pronase P [20,000 U mg−1; Serva, Heidelberg, Germany], each at 10 mg ml−1) was added and the mixture was incubated again for 30 min at 37°C. After addition of 75 μl of 20% sodium dodecyl sulfate and incubation at 65°C for 2 h, cell lysis was completed by addition of 600 μl of a mixture of phenol-chloroform-isoamyl alcohol (25:24:1) and 20 min of incubation at 65°C. After vortexing, the mixture was centrifuged for 10 min at 10,000 × g at room temperature. The aqueous phase was carefully transferred to a fresh tube, mixed with 1 volume of chloroform-isoamyl alcohol (24:1), and centrifuged for another 10 min at 10,000 × g. The aqueous phase was transferred to a fresh tube, and nucleic acids were precipitated by incubation with 0.6 volumes of isopropanol for 1 h at room temperature and subsequent centrifugation for 20 min at 10,000 × g. Pellets were washed with 1 ml of 70% ethanol, dried, and finally resuspended in 30 to 50 μl of elution buffer (10 mM Tris-HCl [pH 8.5]). The amount and purity of DNA were determined spectrophotometrically by determining the optical densities at 260 and 280 nm (58).
PCR amplification of the 16S rDNA.
Almost-complete 16S rDNA gene fragments (1,461 to 1,502 bp after deletion of the primer sequences) were amplified from pure cultures of AOB by using the 616V-630R primer pair as described previously (28). Positive controls containing purified DNA from Escherichia coli were included in all of the amplification sets along with negative controls (no DNA added). The presence and sizes of the amplification products were determined by agarose (1%) gel electrophoresis of the reaction product. Ethidium bromide stained bands were digitally recorded with a video documentation system (Cybertech, Hamburg, Germany).
PCR amplification of the amoA gene fragment.
For AOB of the beta-subclass of Proteobacteria, a 453-bp fragment (without primers) of the amoA gene was amplified from 100 ng of DNA by using the primers amoA-1F and amoA-2R (targeting positions 332 to 349 and 802 to 822 of the Nitrosomonas europaea amoA gene [56]) for PCR with a capillary cycler (Idaho Technology). A 507-bp amoA-amoB fragment was amplified from Nitrosococcus halophilus by using the newly designed primers amoA-3F (5′-GGT GAG TGG GYT AAC MG-3′, positions 295 to 310 of the amoA gene of Nitrosomonas europaea [45]) and amoB-4R (5′-GCT AGC CAC TTT CTG G-3′, positions 30 to 44 of the amoB gene of Nitrosococcus oceani C-107 [4]), which are complementary to target regions in the amoA and amoB genes of Nitrosococcus oceani and Nitrosococcus sp. strain C-113 [4]). Reaction mixtures containing 15 pM concentrations of each primer were prepared in accordance with the manufacturer's recommendations in a total volume of 50 μl by using 20 mM MgCl2 reaction buffer and 1.5 U of Taq polymerase (Promega, Madison, Wis.). Thermal cycling was carried out by an initial denaturation step at 94°C for 30 s, followed by 30 cycles of denaturation at 94°C for 15 s, annealing at 55 or 48°C (amoA-1F and amoA-2R at 55°C and amoA-3F and amoB-4R at 48°C) for 20 s, and elongation at 72°C for 40 s. Cycling was completed by a final elongation step at 72°C for 1 min.
Positive controls containing purified DNA from Nitrosomonas europaea Nm50 were included in all of the amplification sets along with negative controls (no DNA added). Examination of the amplification products was performed as described above.
Cloning, sequencing, and phylogeny inference.
amoA PCR products were ligated according to the manufacturer's recommendations into the cloning vector pCR2.1 supplied with the TOPO TA cloning kit (Invitrogen Corp., San Diego, Calif.). Nucleotide sequences were determined for both strands by the dideoxynucleotide method (59) by cycle sequencing of purified plasmid preparations (Qiagen, Hilden, Germany) with a Thermo Sequenase Cycle sequencing kit (Amersham, Little Chalfont, Buckinghamshire, United Kingdom) and an infrared automated DNA sequencer (Li-Cor, Inc., Lincoln, Nebr.) under conditions recommended by the manufacturers. Dye-labeled (IRD 800) M13-targeted sequencing primers were used. 16S rDNA PCR amplificates (approximately 80 to 100 ng) obtained from AOB pure cultures were sequenced directly using primers targeting conserved regions. The new 16S rRNA sequences were added to an alignment of about 18,000 homologous primary structures from bacteria using the alignment tool of the ARB program package (O. Strunk and W. Ludwig, http://www.biol.chemie.tu-muenchen.de/pub/ARB). Alignments were refined by visual inspection. Phylogenetic analyses were performed by applying distance-matrix, maximum-parsimony, and maximum-likelihood methods using the respective tools of the ARB and PHYLIP (Phylogeny Inference Package, version 3.57c; J. Felsenstein, Department of Genetics, University of Washington, Seattle) program packages and the fastDNAml program (39). The composition of the data sets varied with respect to the reference sequences and the alignment positions included. Variabilities of the individual alignment positions were determined using the ARB package and were used as criteria for removing or including variable positions for phylogenetic analyses.
The new amoA sequences were added to an ARB amoA sequence database which contains all publicly available amoA sequences. Deduced amino acid sequences were aligned using the editor GDE 2.2 (S. W. Smith, C. Wang, P. M. Gillevet, and W. Gilbert, Genetic Data Environment and the Harvard Genome Database, Genome Mapping and Sequencing, Cold Spring Harbor Laboratory) implemented in the ARB software package. Nucleic acid sequences were aligned according to the amino acid alignment. To construct phylogenetic trees based on amino acid alignments, protein distances were inferred by using a maximum-likelihood method implemented in the PROTDIST program, with the Dayhoff PAM 001 matrix as the amino acid replacement model. Trees were inferred from the distances by using FITCH with global rearrangements and randomized input order of species (PHYLIP, version 3.57c). In addition, protein maximum-likelihood (using the JTT-f amino acid replacement model, computer science monographs, no. 28, MOLPHY version 2.3; programs for molecular phylogenetics based on maximum likelihood, Institute of Statistics and Mathematics, Tokyo, Japan), protein parsimony (PHYLIP, version 3.57c), and neighbor–joining methods (using the Dayhoff PAM 001 matrix as amino acid replacement model and the respective tool in the ARB program package) were applied. To perform amoA phylogenetic analysis on the nucleotide level, filters were constructed which allowed exclusion of the third codon position for phylogenetic analysis. Nucleotide-level phylogenetic analyses were performed by applying distance-matrix, maximum-parsimony, and maximum-likelihood methods using the tools described above.
Bootstrap analysis for protein-level (AmoA) and nucleotide-level (amoA, 16S rRNA) phylogenetic analyses were performed for parsimony using the tool in the Phylogeny Inference Package PHYLIP (version 3.57c. Department of Genetics, University of Washington). For each calculation, 100 bootstrap resamplings were analyzed.
The terms nucleic acid similarity and amino acid similarity are used instead of nucleic acid identity and amino acid identity to indicate that, especially at variable positions, “false” identities (plesiomorphies) may result from multiple base changes during the course of evolution (41). It should be noted that the term amino acid similarity does not refer to chemical similarities in this context.
Nucleotide sequence accession numbers.
The sequences determined in this study are available in GenBank under accession no. AF272398 to AF272412 and AF272521 (amoA and AmoA sequences of reference strains); AF272426 to AF272520 and AF276464 to AF276499 (amoA and AmoA sequences of environmental clones); and AF272413 to AF272425, AF287297, and AF287298 (16S rDNA of reference strains). The amoA and AmoA sequences of Nitrosomonas halophila (AF272389) and Nitrosomonas nitrosa (AF272404) are identical with those recently published by Horz et al. (24) (AJ238541 and AJ238495).
RESULTS
AOB phylogeny inferred from 16S rRNA.
16S rDNA sequences (1,461 to 1,502 nucleotides) were determined for Nitrosomonas halophila, Nitrosomonas communis, Nitrosomonas ureae, Nitrosomonas marina, Nitrosomonas aestuarii, Nitrosomonas oligotropha, Nitrosomonas cryotolerans, Nitrosomonas nitrosa, Nitrosomonas sp. strain Nm33, and Nitrosomonas sp. strain Nm41. For these strains, only partial 16S rDNA sequences (209 to 1224 nucleotides) were published previously. Ambiguities and errors in the 16S rDNA sequence of Nitrosococcus mobilis Nc2 (17) were corrected. In addition, we determined almost-full-length 16S rDNA sequences (1,461 to 1,502 nucleotides) for Nitrosococcus halophilus (34), Nitrosomonas sp. strain Nm51 (30, 85), and two AOB strains (Nm104, Nm107) isolated in this study from the industrial wastewater treatment plant Kraftisried.
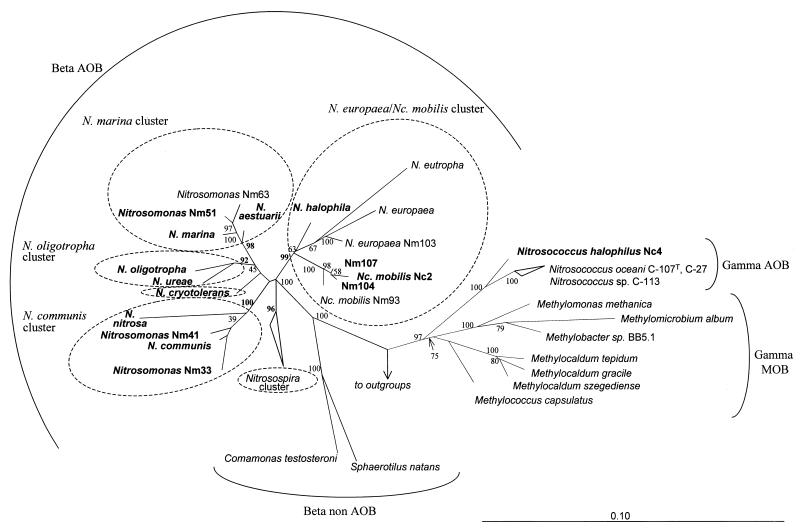
The 16S rDNA of Nitrosococcus halophilus showed the highest sequence similarity (95.6 and 95.7%) to the 16S rRNAs of the gamma-subclass AOB Nitrosococcus oceani strains C-107T (17, 91) and C-27 (17), respectively. These results confirm that Nitrosococcus halophilus should be considered a separate AOB species (34). The 16S rDNA sequences of all other AOB investigated were most similar to AOB sequences of the beta-subclass of Proteobacteria (Table 3). Phylogenetic trees for the 16S rDNA of AOB were estimated for data sets differing in regard to selection of outgroup organisms and number of variable positions included by distance, parsimony, and maximum-likelihood methods. Independent of the data set and method used, Nitrosococcus halophilus formed a monophyletic lineage together with Nitrosococcus oceani (strains C-107T and C-27) and Nitrosococcus sp. strain C-113 (4) within the gamma-subclass Proteobacteria while the other AOB analyzed formed a monophyletic grouping with the beta-subclass AOB (Fig. 1). Within the beta-subclass AOB, five stable clusters were revealed using the different treeing methods (Fig. 1). This clustering was also supported by high parsimony bootstrap values (92 to 100%). The nomenclature of the clusters was adopted from a study by Pommerening-Röser et al. (52). The first cluster comprised Nitrosomonas marina, Nitrosomonas aestuarii, together with two strains of a third species (30), Nitrosomonas sp. strain Nm63, and Nitrosomonas sp. strain Nm51 (Nitrosomonas marina cluster). The second cluster encompassed Nitrosomonas ureae and Nitrosomonas oligotropha (Nitrosomonas oligotropha cluster). Most but not all treeing analyses suggested that these two clusters formed a grouping to the exclusion of all other sequences. The third cluster was represented by Nitrosomonas europaea, Nitrosomonas eutropha, Nitrosomonas halophila, Nitrosococcus mobilis, and the isolates Nm104 and Nm107, which are most probably strains of Nitrosococcus mobilis (Nitrosomonas europaea-Nitrosococcus mobilis cluster). The fourth cluster allied Nitrosomonas nitrosa, Nitrosomonas communis, Nitrosomonas sp. strain Nm33, and Nitrosomonas sp. strain Nm41 (Nitrosomonas communis cluster). The fifth cluster contained all published Nitrosospira-like 16S rDNA sequences (Nitrosospira cluster). The phylogenetic position of Nitrosomonas cryotolerans and the specific branching order of the above-mentioned clusters varied dependently on the data set and treeing method used and could thus not unambiguously be resolved. In contrast to previous studies (17, 52, 73), phylogeny inference based on the more complete data set did not support that all nitrosomonads are more closely related with each other than with members of the Nitrosospira lineage (Fig. 1).
TABLE 3.
16S rRNA sequence similarities of beta-subclass AOBa
| Strain | % Sequence similarity
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Nitrosomonas communis cluster
|
Nitrosomonas marina cluster
|
Nitrosomonas oligotropha cluster
|
Nitrosomonas cryotolerans cluster (Nm55) |
Nitrosomonas europaea-Nitrosococcus mobilis cluster
|
Nitrosospira cluster
|
|||||||||||||||
| Nm2 | Nm33 | Nm41 | Nm90 | Nm22 | Nm63 | Nm51 | Nm36 | Nm45 | Nm10 | Nm50 | Nm57 | Nm1 | Nc2 | Nm104 | Nm107 | C128 | C71 | Nv12 | ||
| Nitrosomonas communis Nm2 | ||||||||||||||||||||
| Nitrosomonas sp. Nm33 | 98.2 | |||||||||||||||||||
| Nitrosomonas sp. Nm41 | 97.2 | 98.1 | ||||||||||||||||||
| Nitrosomonas nitrosa Nm90 | 94.9 | 95.3 | 94.6 | |||||||||||||||||
| Nitrosomonas marina Nm22 | 93.3 | 93.3 | 93.4 | 90.6 | ||||||||||||||||
| Nitrosomonas sp. Nm63 | 92.2 | 92.0 | 92.4 | 91.8 | 98.1 | |||||||||||||||
| Nitrosomonas sp. Nm51 | 93.3 | 93.2 | 93.4 | 90.6 | 98.9 | 98.8 | ||||||||||||||
| Nitrosomonas aestuarii Nm36 | 93.7 | 93.7 | 93.9 | 92.0 | 98.1 | 97.1 | 98.1 | |||||||||||||
| Nitrosomonas oligotropha Nm45 | 93.3 | 93.1 | 93.0 | 91.5 | 94.8 | 94.1 | 94.9 | 95.6 | ||||||||||||
| Nitrosomonas ureae Nm10 | 93.6 | 93.3 | 93.0 | 90.9 | 94.2 | 93.4 | 94.3 | 94.9 | 96.7 | |||||||||||
| Nitrosomonas cryotolerans Nm55 | 93.8 | 94.0 | 93.7 | 91.4 | 95.2 | 94.5 | 95.3 | 95.9 | 95.1 | 94.7 | ||||||||||
| Nitrosomonas europaea Nm50 | 92.9 | 93.2 | 92.4 | 90.8 | 92.4 | 91.8 | 92.5 | 92.6 | 92.3 | 91.6 | 93.4 | |||||||||
| Nitrosomonas eutropha Nm57 | 93.0 | 93.0 | 92.9 | 92.4 | 92.2 | 91.4 | 92.0 | 92.7 | 91.9 | 91.6 | 93.2 | 98.0 | ||||||||
| Nitrosomonas halophila Nm1 | 93.9 | 94.3 | 93.6 | 91.1 | 93.5 | 92.6 | 93.4 | 93.1 | 92.5 | 92.1 | 94.1 | 96.4 | 95.3 | |||||||
| Nitrococcus mobilis Nc2 | 92.4 | 92.4 | 91.9 | 89.5 | 93.0 | 92.6 | 93.2 | 93.2 | 92.9 | 92.1 | 93.7 | 95.0 | 94.1 | 94.9 | ||||||
| Nitrosomonas sp. Nm104 | 92.5 | 92.7 | 92.1 | 90.8 | 93.3 | 92.9 | 93.5 | 93.6 | 93.1 | 92.6 | 94.0 | 95.4 | 94.7 | 95.3 | 99.5 | |||||
| Nitrosomonas sp. Nm107 | 92.5 | 92.5 | 92.2 | 90.7 | 93.2 | 92.9 | 93.3 | 93.5 | 93.0 | 92.5 | 93.9 | 95.3 | 94.7 | 95.3 | 99.3 | 99.9 | ||||
| Nitrosospira sp. C128 | 94.0 | 93.7 | 93.1 | 91.9 | 93.4 | 92.3 | 93.3 | 93.7 | 92.5 | 92.6 | 95.3 | 92.0 | 92.1 | 93.8 | 92.0 | 92.5 | 92.5 | |||
| Nitrosospira multiformis C71 | 93.8 | 93.8 | 93.3 | 91.8 | 93.0 | 92.2 | 93.1 | 93.4 | 92.7 | 92.8 | 95.5 | 92.4 | 92.5 | 94.0 | 92.1 | 92.5 | 92.5 | 98.8 | ||
| Nitrosospira tenuis Nv12 | 93.5 | 93.3 | 93.0 | 92.2 | 93.0 | 91.6 | 92.7 | 93.4 | 92.1 | 92.5 | 95.1 | 92.1 | 92.1 | 93.6 | 91.7 | 92.1 | 92.2 | 98.8 | 98.3 | |
| Nitrosospira sp. NpAV | 93.7 | 93.7 | 93.5 | 93.2 | 93.8 | 92.8 | 93.4 | 94.2 | 92.4 | 93.0 | 96.1 | 93.2 | 92.3 | 94.4 | 92.4 | 92.8 | 92.9 | 99.0 | 98.9 | 98.5 |
The lowest sequence similarity within a cluster is in bold.
FIG. 1.
Phylogenetic 16S rRNA tree reflecting the relationships of AOB and several non-AOB reference organisms. The tree is based on results of neighbor-joining analysis using a 50% conservation filter for the Bacteria. An encompassing collection of organisms representing all major lineages of the Archaea and Bacteria were used as outgroups for treeing. The multifurcation connects branches for which a relative order could not be unambiguously determined by applying different treeing methods. Parsimony bootstrap values (100 replicates) for branches are reported. Missing bootstrap values indicate that the branch in question was not recovered in the majority of bootstrap replicates by the parsimony method. The bar indicates 10% estimated sequence divergence. MOB, methane-oxidizing bacteria.
AOB phylogeny inferred from amoA.
Partial (453 bp) amoA sequences were determined for Nitrosococcus mobilis Nc2, Nitrosococcus mobilis Nm93 (28), Nitrosomonas halophila, Nitrosomonas communis, Nitrosomonas ureae, Nitrosomonas marina, Nitrosomonas aestuarii, Nitrosomonas oligotropha, Nitrosomonas cryotolerans, Nitrosomonas nitrosa, Nitrosomonas europaea Nm103 (28), Nitrosomonas sp. strain Nm33, Nitrosomonas sp. strain Nm41, Nitrosomonas sp. strain Nm51, isolate Nm104, and isolate Nm107 after PCR amplification using the primers described by Rotthauwe et al. (56). Since these primers did not amplify an amoA fragment of Nitrosococcus halophilus, we exploited the complete amoA and amoB sequence of its closest known relative, Nitrosococcus oceani (4), for the design of the new PCR primer pair amoA-F3 and amoB-R4. These primers were successfully used to amplify the expected amoA and amoB fragment from Nitrosococcus halophilus. In accordance with the 16S rDNA phylogeny, nucleic acid similarities and amino acid similarities were highest between Nitrosococcus halophilus and Nitrosococcus oceani C-107 (77.8 and 82.5%) and Nitrosococcus sp. strain C-113 (77.6 and 81.0%). The amoA and AmoA sequences of the other AOB investigated showed highest sequence similarities and similarities to beta-subclass AOB (Table 4).
TABLE 4.
amoA and AmoA sequence similarities of beta-subclass AOBa
| Strain | % amoA (AmoA) sequence similarity
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
Nitrosomonas communis cluster
|
Nitrosomonas marina cluster
|
Nitrosomonas oligotropha cluster
|
|||||||
| Nm2 | Nm33 | Nm41 | Nm90 | Nm22 | Nm51 | Nm36 | Nm45 | Nm10 | |
| Nitrosomonas sp. Nm 33 | 86.2 (90.1) | ||||||||
| Nitrosomonas sp. Nm 41 | 85.3 (90.8) | 88.7 (90.8) | |||||||
| Nitrosomonas nitrosa Nm 90 | 80.9 (91.5) | 83.8 (90.8) | 86.5 (92.2) | ||||||
| Nitrosomonas marina Nm 22 | 73.3 (79.4) | 74.9 (81.6) | 73.7 (82.3) | 74 (81.6) | |||||
| Nitrosomonas sp. Nm 51 | 74.8 (81.4) | 75.2 (83.6) | 74 (84.3) | 75 (83.6) | 89 (97.2) | ||||
| Nitrosomonas aestuarii Nm 36 | 75.5 (80.7) | 76.7 (83.0) | 76.7 (83.6) | 74.8 (83.7) | 86.5 (97.2) | 88.5 (98.6) | |||
| Nitrosomonas oligotropha Nm 45 | 75.5 (79.4) | 78.6 (82.3) | 78.4 (83.0) | 76.7 (83.0) | 84.1 (93.7) | 82.8 (94.3) | 82.8 (95.0) | ||
| Nitrosomonas ureae Nm 10 | 74.5 (78.0) | 75.7 (79.4) | 76.4 (81.6) | 77.5 (83.0) | 81.9 (94.4) | 81.6 (94.3) | 84.1 (95.7) | 85.8 (93.7) | |
| Nitrosomonas cryotolerans Nm 55 | 74.5 (80.9) | 79.6 (83.7) | 76.7 (82.3) | 76.5 (83.0) | 79.2 (88.7) | 79.7 (88.7) | 80.4 (90.1) | 81.1 (90.8) | 80.4 (90.1) |
| Nitrosomonas europaea Nm 50 | 79.4 (89.4) | 80.8 (88.0) | 81.6 (90.1) | 80.4 (90.8) | 74 (80.1) | 71.8 (82.1) | 75 (82.3) | 75.2 (80.9) | 75.7 (82.3) |
| Nitrosomonas sp. Nm 103 | 78.8 (88.3) | 80 (87.0) | 81 (89.1) | 79.8 (89.9) | 73.5 (78.8) | 71.5 (80.9) | 74.2 (81.0) | 74.7 (79.6) | 75 (81.0) |
| Nitrosomonas eutropha Nm 57 | 80.1 (90.1) | 81.1 (88.7) | 81.1 (89.4) | 79.9 (88.0) | 74.3 (78.7) | 73 (80.7) | 76.2 (80.1) | 75 (80.1) | 73.5 (78.7) |
| Nitrosomonas halophila Nm 1 | 77.5 (87.2) | 79.4 (87.3) | 79.6 (88.7) | 77.5 (89.4) | 74.8 (78.7) | 71.3 (80.7) | 72.5 (80.9) | 76.7 (80.1) | 71.3 (79.4) |
| Nitrosococcus mobilis Nc2A | 75.5 (83.0) | 77.9 (84.5) | 78.1 (85.1) | 76.2 (87.3) | 70.1 (75.9) | 72.1 (77.9) | 73.3 (78.0) | 72.3 (77.3) | 72.5 (76.6) |
| Nitrosomonas sp. Nm 104 | 75.5 (83.0) | 77.9 (84.5) | 78.1 (85.1) | 76.2 (87.3) | 70.1 (75.9) | 72.1 (77.9) | 73.3 (78.0) | 72.3 (77.3) | 72.5 (76.6) |
| Nitrosomonas sp. Nm 107 | 75.5 (83.0) | 77.9 (84.5) | 78.1 (85.1) | 76.2 (87.3) | 70.1 (75.9) | 72.1 (77.9) | 73.3 (78.0) | 72.3 (77.3) | 72.5 (76.6) |
| Nitrosomonas sp. Nm 93 | 75.7 (83.0) | 78.1 (84.5) | 78.4 (85.1) | 76.5 (87.3) | 70.3 (75.9) | 72.3 (77.9) | 73.5 (78.0) | 72.1 (77.3) | 72.8 (76.6) |
| Nitrosospira sp. C128 | 69.1 (78.0) | 69.0 (78.9) | 69.7 (78.0) | 68.7 (79.6) | 74.0 (82.4) | 72.6 (83.0) | 72.0 (83.8) | 77.3 (87.3) | 71.3 (83.8) |
| Nitrosospira multiformis C71 | 69.5 (76.6) | 71.7 (79.4) | 72.3 (80.1) | 71.1 (80.9) | 77.5 (83.1) | 75.0 (83.7) | 74.6 (84.4) | 77.0 (85.9) | 75.7 (85.2) |
| Nitrosospira tenuis Nv12 | 71.3 (76.6) | 71.5 (78.9) | 71.9 (77.3) | 69.8 (77.5) | 76.6 (81.0) | 73.5 (81.6) | 74.0 (82.4) | 78.4 (85.9) | 74.6 (82.4) |
| Nitrosospira sp. NpAV | 69.3 (78.4) | 69.0 (79.9) | 69.9 (79.1) | 70.2 (81.3) | 78.8 (85.7) | 77.2 (86.4) | 75.5 (87.1) | 78.6 (89.3) | 75.7 (87.2) |
| % amoA (AmoA) sequence similarity
| |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nitrosomonas cryotolerans cluster Nm55 |
Nitrosomonas europaea/Nitrosococcus mobilis cluster
|
Nitrosospira cluster
|
|||||||||
| Nm50 | Nm103 | Nm57 | Nm1 | Nc2 | Nm104 | Nm107 | Nm93 | C128 | C71 | Nv12 | |
| 75 (80.9) | |||||||||||
| 74 (79.6) | 99.7 (99.3) | ||||||||||
| 75 (80.9) | 87 (94.4) | 87.1 (93.5) | |||||||||
| 72.8 (79.4) | 81.9 (95.1) | 81.3 (94.2) | 80.6 (90.8) | ||||||||
| 71.8 (75.2) | 77 (88.7) | 76.5 (87.7) | 76.5 (85.2) | 76.7 (91.5) | |||||||
| 71.8 (75.2) | 77 (88.7) | 76.5 (87.7) | 76.5 (85.2) | 76.7 (91.5) | 99.8 (100) | ||||||
| 71.8 (75.2) | 77 (88.7) | 76.5 (87.7) | 76.5 (85.2) | 76.7 (91.5) | 100 (100) | 99.8 (100) | |||||
| 72.1 (75.2) | 77.2 (88.7) | 76.8 (87.7) | 76.7 (85.2) | 76.5 (91.5) | 99.8 (100) | 99.5 (100) | 99.8 (100) | ||||
| 74.2 (85.9) | 70.5 (78.2) | 69.8 (76.8) | 70.1 (76.8) | 71.7 (76.8) | 65.0 (72.5) | 64.9 (72.5) | 64.9 (72.5) | 64.7 (72.5) | |||
| 75.5 (86.6) | 72.7 (80.9) | 72.3 (79.6) | 71.2 (76.6) | 73.7 (79.4) | 67.6 (75.2) | 67.5 (75.2) | 67.5 (75.2) | 67.8 (75.2) | 83.5 (92.3) | ||
| 75.3 (85.9) | 71.2 (78.2) | 70.5 (77.5) | 69.9 (76.8) | 73.5 (76.8) | 65.6 (71.8) | 65.6 (71.8) | 65.6 (71.8) | 65.8 (71.8) | 85.9 (93.0) | 85.7 (90.8) | |
| 75.9 (89.3) | 70.8 (80.6) | 73.2 (84.2) | 69.0 (77.7) | 72.6 (78.4) | 66.2 (74.1) | 66.2 (74.1) | 66.2 (74.1) | 66.4 (74.1) | 85.3 (93.6) | 85.1 (92.1) | 84.2 (90.7) |
Nucleic acid similarities include the third codon position; the lowest sequence similarity within a cluster is bold.
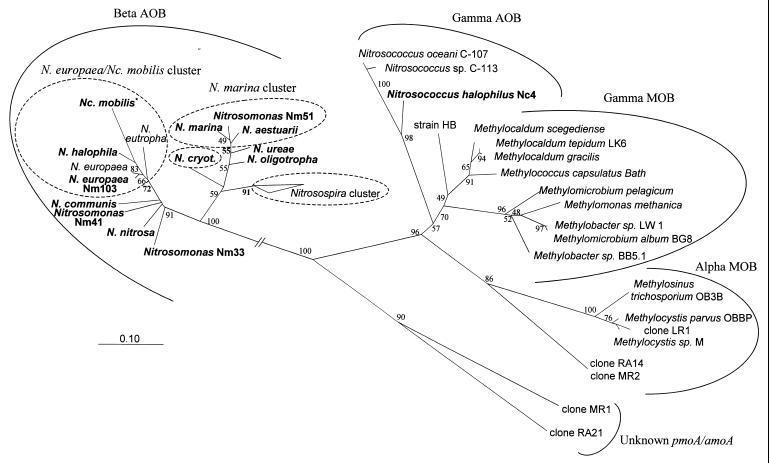
Phylogenetic trees for amoA and AmoA were calculated from the nucleotide and amino acid data sets by distance, parsimony, and maximum-likelihood methods. Overall, highly similar orderings of taxa were found between amoA and AmoA and the 16S rRNA trees described above. For all methods with both DNA (with and without the third codon position) and amino acid amoA and AmoA data sets, Nitrosococcus halophilus grouped together with Nitrosococcus oceani and Nitrosococcus sp. strain C-113 (Fig. 2). The amoA and AmoA sequences of the other AOB investigated clustered together with the beta-subclass AOB Nitrosomonas europaea, Nitrosomonas eutropha, and the members of the Nitrosospira cluster. Three of the five beta-subclass AOB clusters revealed by comparative 16S rRNA analysis were also found in all or most of the amoA and AmoA trees (Fig. 2). The monophyly of the Nitrosospira cluster, the Nitrosomonas marina cluster, and the Nitrosomonas europaea-Nitrococcus mobilis cluster was supported by all methods and data sets. However, comparatively low parsimony bootstrap values were calculated for the latter two clusters (55 and 72%). Furthermore, the topology of the Nitrosomonas europaea and Nitrococcus mobilis cluster differed significantly between the 16S rRNA- and AmoA-based trees, demonstrating the limited phylogenetic resolution provided by these biopolymers for highly related organisms. All methods and data sets suggested a grouping of Nitrosomonas oligotropha and Nitrosomonas ureae with the Nitrosomonas marina cluster. The monophyly of the Nitrosomonas communis cluster was supported by the different treeing methods only if a nucleic acid data set including the third codon position was analyzed. Consistent with the 16S rRNA phylogeny, the phylogenetic position of Nitrosomonas cryotolerans varied within the beta-subclass AOB dependently on the treeing method and data set used. As for the 16S rRNA, comparative amoA and AmoA sequence analysis does not suggest a bifurcation of the beta-subclass AOB into nitrosomonads and nitrosospiras (Fig. 2).
FIG. 2.
Phylogenetic Fitch-Margoliash tree (using global rearrangement and randomized input order [7 jumbles]) reflecting the relationships of AOB and methane-oxidizing bacteria (MOB) based on deduced AmoA and PmoA sequences. Parsimony bootstrap values (100 replicates) for branches are reported. Missing bootstrap values indicate that the branch in question was not recovered in the majority of bootstrap replicates by the parsimony method. The bar indicates 10% estimated sequence divergence. Clones RA14 and RA21 (20) and MR1 and MR2 (23) were retrieved in previous studies from soil. Whether clones RA21 and MR1 represent AOB or MOB has not been clarified yet. ∗, to enhance clarity, AmoA sequences of Nitrosococcus mobilis Nm93 and of the isolates Nm104 and Nm107, which are identical in sequence to the AmoA sequence of Nitrosococcus mobilis Nc2, are not shown in the tree.
Comparison of AOB DNA-DNA, 16S rRNA, and amoA-AmoA similarity.
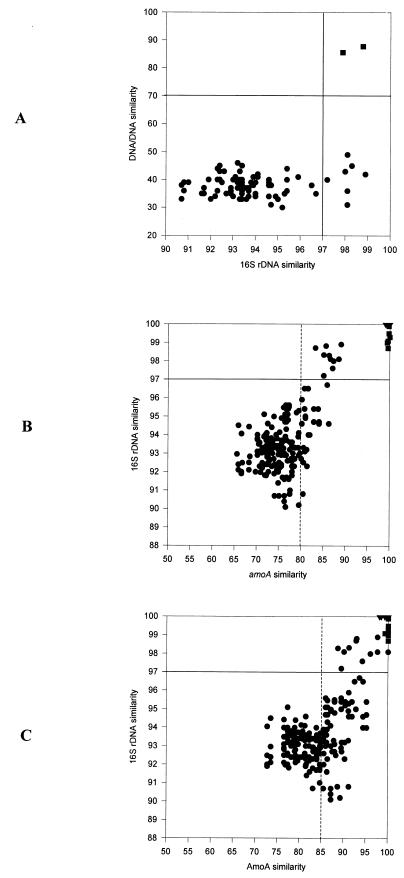
By plotting the 16S rRNA sequence similarity versus the DNA-DNA reassociation values for several bacterial species pairs, Stackebrandt and Goebel demonstrated that at 16S rRNA similarity values below 97%, it is unlikely that two organisms have more than 70% DNA similarity and hence that they are related at no more than the species level (66). We confirmed that the above-mentioned correlation does also apply for beta-subclass AOB species according to published DNA-DNA reassociation values (28, 30, 31, 33, 34, 52) and the 16S rRNA similarities given in Table 3 (Fig. 3A). DNA similarities of AOB species may be as low as 31% at 16S rRNA similarities of 98.1% (Nitrosomonas marina Nm22 and Nitrosomonas aestuarii Nm36), demonstrating again the superior resolution of DNA-DNA hybridization versus comparative 16S rRNA sequencing for closely related microorganisms.
FIG. 3.
Correlation plots of DNA-DNA reassociation, 16S rRNA similarity, and amoA and AmoA similarity values of AOB. (A) Comparison of 16S rRNA similarity and DNA-DNA similarity values. DNA-DNA hybridization data were obtained from studies by Juretschko et al. (28), Koops et al. (34), Koops et al. (30), Koops and Harms (31), and Pommerening-Röser et al. (52). (B) Comparison of amoA similarity and 16S rRNA similarity values. (C) Comparison of AmoA and 16S rRNA similarity values. Sequences of multiple amoA gene copies of Nitrosomonas eutropha and Nitrosospira sp. strain Np39-19 were obtained from GenBank (accession no. AF006692, AF016002, AF042170, U51630, and U72670). Solid lines indicate the DNA and 16S rRNA threshold values for species delineation. Dotted lines indicate the suggested amoA and AmoA threshold values below which environmentally retrieved amoA and AmoA sequences are indicative of novel AOB species. Circle, pair of different AOB species; square, pair of different strains of a single AOB species; triangle, pair of different amoA operons of a single AOB species.
amoA is increasingly used as phylogenetic marker molecule for molecular diversity inventories of AOB in environmental samples (18, 24, 28, 47, 56, 57, 60, 68; see below). These analyses frequently revealed amoA sequences related to but not identical to known AOB species even when the above-presented amoA data set containing all validly described AOB species was used as a framework (see below). However, it is not possible to estimate whether such an environmental amoA sequence represents a different strain of a described species or whether it originates from a novel species. Correlation plots of amoA and AmoA similarity (Table 4) versus 16S rRNA similarity (Table 3) of all possible pairs of beta-subclass AOB species demonstrate that (i) 16S rRNA is more conserved than amoA and (ii) AOB showing below 83.2% amoA nucleic acid similarity (Nitrosospira sp. C128 and Nitrosolobus multiformis) and 89.1% AmoA amino acid similarity (Nitrosomonas communis and Nitrosomonas sp. strain Nm41) do possess less than 97% 16S rRNA similarity (Fig. 3B and C). We consequently suggest that environmental amoA sequences with lower than 80% nucleic acid similarity (85% amino acid similarity) to described AOB species are indicative of previously undiscovered species. An amoA or AmoA sequence with a higher similarity to a described AOB species can represent multiple gene copies, different strains of this species, or a novel AOB species. The latter possibility exists since 16S rRNA similarities between different species can be higher than 97% (the value used to define the amoA threshold, see above) (for an example, see reference 13).
AmoA sequences from wastewater treatment plants.
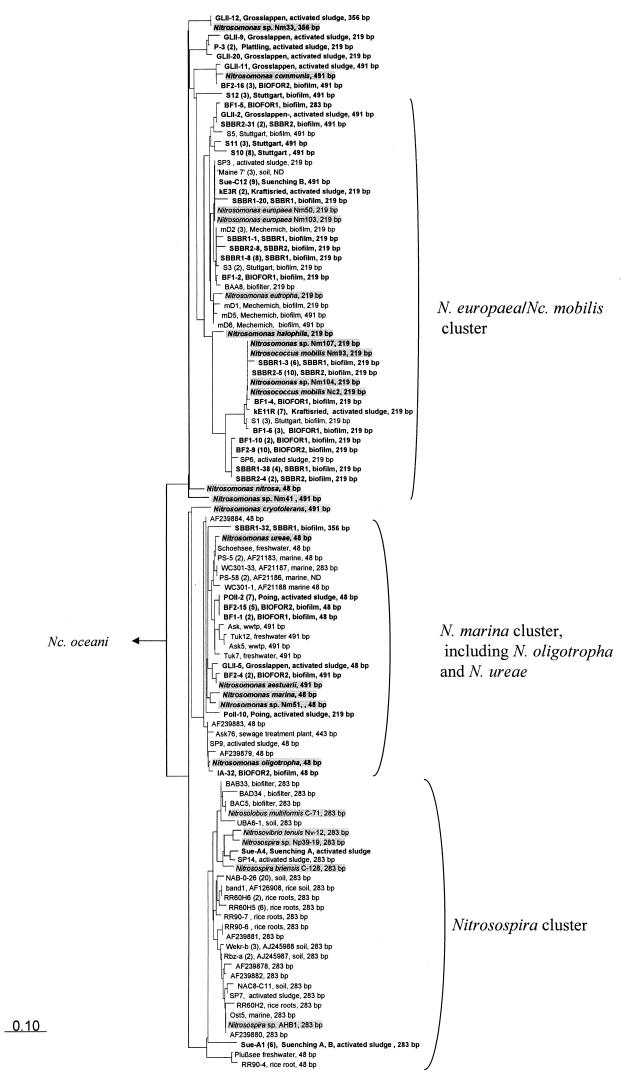
Beta-subclass AOB diversity surveys were performed in 11 nitrifying wastewater treatment samples (Table 2). amoA PCR products (using the primers amoA-1F and amoA-2R) retrieved from the samples were used for the generation of amoA libraries. A total of 122 clones were randomly selected and sequenced. Phylogenetic analysis demonstrated that all clones contained amoA sequences affiliated to the beta-subclass AOB (Fig. 4). Nitrosospira-related sequences could be detected only in the municipal and industrial plant Sünching (the latter plant was inoculated with sludge from the former plant during start-up). However, all 11 plants investigated harbored nitrosomonads. AmoA sequences closely related to those of Nitrosomonas europaea, Nitrosomonas eutropha, Nitrosococcus mobilis, Nitrosomonas communis, Nitrosomonas sp. strain Nm33, Nitrosomonas oligotropha, Nitrosomonas ureae, and the Nitrosomonas marina cluster were detected. No indications for the occurrence of Nitrosomonas sp. strain Nm41, Nitrosomonas cryotolerans, Nitrosomonas halophila, and Nitrosomonas nitrosa in the analyzed wastewater treatment plants could be obtained.
FIG. 4.
Phylogenetic Fitch-Margoliash AmoA dendrogram (using global rearrangement and randomized input order [3 jumbles]) showing the positions of cultured ammonia oxidizers (shaded in gray) in relation to environmental sequences recovered from 11 wastewater treatment plants (bold [this study]) and other previously published environmental sequences (18, 19, 23, 24, 56, 57, 60, 68). The bar indicates 10% estimated sequence divergence. The root was determined by using the AmoA sequences of gamma-subclass AOB. Cloned AmoA sequences with amino acid similarities of >99% which originated from the same sample are represented by a single clone—the number in parentheses indicates the number of amoA clones for each representative. For each clone, the calculated fragment length in the TaqI-based restriction fragment length polymorphism analysis (24) is listed.
DISCUSSION
In general, the phylogenetic analyses of the completed 16S rRNA AOB data set supported the previously published perception of AOB phylogeny (17, 52, 73). As expected from DNA-DNA hybridization data (34), the 16S rRNA sequence of Nitrosococcus halophilus groups together with the gamma-subclass AOB Nitrosococcus oceani (C-107T, C-27) and Nitrosococcus sp. strain C-113, which is most probably a strain of Nitrosococcus oceani. The obtained 16S rRNA tree topology of the beta-subclass AOB is overall consistent with the one reported by Pommerening-Röser et al. (52), who suggested six lines of descent among the beta-subclass nitrosomonads. Based on our analyses, however, we suggest grouping the Nitrosococcus mobilis cluster together with the Nitrosomonas europaea cluster since (i) 16S rRNA similarities between both clusters are comparable to similarities within the other five proposed clusters (Table 3), (ii) both clusters are monophyletic in all treeing analyses, and (iii) no physiological traits separating members of both clusters are known. These facts were considered to be more decisive than the morphological differences between members of both clusters, which obviously evolved relatively recently. We would like to point out again (52, 73) that a taxonomic revision of Nitrosococcus mobilis is required to express its phylogenetic affiliation with the genus Nitrosomonas.
Based on the completed 16S rRNA sequences of the beta-subclass AOB, we reevaluated the specificity of previously published PCR primers and hybridization probes for the direct detection of these organisms in the environment (Table 5). None of the primers and probes intended to target all beta-subclass AOB showed both 100% sensitivity (targeting all beta-subclass AOB) and 100% specificity (excluding all non-beta-subclass AOB). For general beta-subclass AOB diversity surveys in environmental samples using 16S rDNA libraries (7, 69) or fingerprinting techniques (36, 37) we recommend using PCR primer pairs with high sensitivity [e.g., βAMOf and βAMOr (43) accepting unwanted amplification of non-AOB 16S rDNA fragments which subsequently have to be identified by phylogenetic analysis or hybridization with probes with excellent specificity (e.g., Nso1225 [48]). For AOB community composition analysis, using in situ hybridization (e.g., see references 28, 63, and 80), probes with nested specificity (and good sensitivity) should be simultaneously applied (for example, Nso1225, Nsv443, and Nso 156 [48]). However, apparently inconsistent results from simultaneous in situ hybridization experiments with multiple probes can also be indicative of the presence of novel AOB.
TABLE 5.
Specificity and sensitivity of published 16S rDNA/RNA targeting PCR primers and hybridization probes for beta-subclass AOB
| Primer (OPD nomenclature [3])a | Target regionb | Refer ence | Intended specificityc | No. of mishits withd:
|
Sensitivitye
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Nitrosospira cluster
|
Nitrosomonas communis cluster
|
|||||||||||
| 0MM | 1MM | C128 | C71 | Nv12 | Nm2 | Nm33 | Nm41 | Nm90 | ||||
| Nm-75 (S-*-Nsm-0067-a-S-20) | 67–86 | 21 | Terrestrial Nitrosomonas spp., Nitrosococcus mobilis | >10 | 5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 |
| NS-85 (S-G-Nsp-0076-a-S-20) | 76–95 | 21 | Nitrosospira spp. | 6 | >10 | 0 | 0 | 0 | 1I | 0 | 3 | 0 |
| NmII (S-*-Nsm-0120-a-S-20) | 120–139 | 52 | Nitrosomonas communis lineage | 0 | 0 | 3 | 3 | 3 | 0 | 1 | 2 | 0 |
| NitA (S-F-bAOB-0136-a-S-23) | 136–158 | 78 | β-AOB | 0 | 0 | 4 | 2 | 4 | 3 | 3 | 3 | 4 |
| βAMOf (S-F-bAOB-0142-a-S-21) | 142–162 | 43 | β-AOB | 7 | >10 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Nm0 (S-G-Nsm-0148-a-S-18) | 148–165 | 52 | Nitrosomonas spp. | 1 | 5 | 2 | 2 | 2 | 0 | 0 | 0 | 0 |
| Nsm 156 (S-G-Nsm-0155-a-A-19)f | 155–173 | 48 | Nitrosomonas spp., Nitrosococcus mobilis | 2 | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 0 |
| NmV (S-S-Nmob-0174-a-S-18)f | 174–191 | 52 | Nitrosococcus mobilis | 0 | 2 | 4 | 3 | 4 | 3 | 2 | 2 | 3, 1N |
| Nso 190 (S-F-bAOB-0189-a-A-19)f | 189–207 | 48 | β-AOB | 2 | 2 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| CTO189f, A/B-GC (S-F-bAOB-0189-a-S-19) | 189–207 | 37 | β-AOB | 2 | 7 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| CTO189f, C-GC (S-F-bAOB-0189-a-S-19) | 189–207 | 37 | β-AOB | 0 | 3 | 2 | 2 | 2 | 1 | 1 | 1 | 1 |
| NmI (S-*-Nsm-0210-a-S-19) | 210–225 | 52 | Nitrosomonas europaea lineage | 0 | 1 | 3 | 3 | 3 | 5 | 5 | 5 | 4 |
| AAO258 (S-*-bAOB-0258-a-S-19) | 258–277 | 21 | Terrestrial β-AOB | >10 | >100 | 0 | 0 | 1N | 0 | 0 | 1 | 1 |
| NitD (S-S-Nse-0439-a-S-23) | 439–461 | 83 | Nitrosomonas europaea | 0 | 0 | >5 | >5 | >5 | >5 | >5 | >5 | >5 |
| Nsv 443 (S-G-Nsp-0443-a-S-19)f | 443–461 | 48 | Nitrosospira spp. | 1 | 2 | 0 | 0 | 0 | >5 | >5 | >5 | >5 |
| Nsp0 (S-G-Nsp-0452-a-S-18) | 452–469 | 52 | Nitrosospira spp. | 1 | 1 | 0 | 0 | 0 | >5 | >5 | >5 | >5 |
| Nlm 459r (S-*-Nsp-0458-a-A-20) | 458–477 | 16 | Nitrosospira multiformis, Nitrosospira sp. strain C-141 | 1 | 1 | 2 | 0 | 3 | >5 | >5 | >5 | >5 |
| NSM1B (S-*-Nsm-0478-a-A-17) | 478–494 | 25 | Nitrosomonas europaea lineage, Nitrosococcus mobilis | 6 | >10 | 3 | 3 | 3 | 1 | 1 | 1 | 1 |
| TAOrev (S-F-bAOB-0632-a-A-18) | 632–649 | 11 | β-AOB | 2 | 5 | 0 | 0 | 1 | 3 | 3 | 4 | 3 |
| CTO654r (S-F-bAOB-0632-a-A-17) | 632–653 | 37 | β-AOB | 4 | 3 | 0 | 0 | 1 | 3 | 3 | 3 | 3 |
| NITROSO4E (S-F-bAOB-0632-a-A-22) | 638–657 | 25 | β-AOB | 2 | >10 | 0 | 0 | 1 | 3 | 3 | 3 | 3 |
| NEU (S-*-Nsm-0651-a-A-18)f | 651–668 | 80 | Most halophilic and halotolerant Nitrosomonas | 0 | 3 | 1 | 2, 1N | 1 | 4 | 3 | 3 | 3 |
| Amβ (S-F-bAOB-0738-a-S-21) | 738–758 | 77 | β-AOB | 1 | >10 | 0 | 0 | 0 | 1 | 0 | 0 | 3 |
| NitF (S-F-bAOB-0844-a-A-19)g | 844–862 | 83 | β-AOB | 0 | 0 | 2 | 1 | 4 | 3 | 4 | 4 | 3 |
| NitC (S-F-bAOB-0846-a-A-17)g | 846–862 | 78 | β-AOB | 0 | 1 | 3 | 4 | 5 | 3 | 4 | 4 | 3 |
| NmIII (S-*-Nsm-0998-a-S-21) | 998–1018 | 52 | Nitrosomonas marina lineage | 1 | 0 | >5 | >5 | >5 | >5 | >5 | >5 | >5 |
| RNM-1007 (S-*-Nsm-1005-a-A-25) | 1005–1028 | 21 | Terrestrial Nitrosomonas spp. | 0 | 0 | >5 | >5 | >5 | >5 | >5 | >5 | >5 |
| NS-1009 (S-G-Nsp-1007-a-A-25) | 1007–1026 | 21 | Nitrosospira spp. | 1 | 1 | 1 | 1, 1N | 1 | 5 | >5 | >5 | >5 |
| NmIV (S-S-Nsm-1004-a-S-19)fh | 1004–1022 | 52 | Nitrosomonas cryotolerans lineage | 0 | 0 | 5 | 3, 1N | 4 | 5 | >5 | 4 | >5 |
| NitB (S-F-bAOB-1213-a-A-21) | 1213–1233 | 78 | β-AOB | 5 | >10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Nso 1225 (S-F-bAOB-1224-a-A-20)f | 1224–1243 | 48 | β-AOB | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| βAMOr (S-F-bAOB-1295-a-A-20) | 1295–1314 | 43 | β-AOB | >10 | >100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Sensitivitye
| ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Nitrosomonas marina cluster
|
Nitrosomonas oligotropha cluster
|
Nitrosomonas cryotolerans cluster (Nm55) |
Nitrosomonas europaea-Nitrosococcus mobilis cluster
|
|||||||||||
| Nm22 | Nm63 | Nm51 | Nm36 | Nm45 | Nm10 | Nm50 | Nm103 | Nm57 | Nm1 | Nc2 | Nm104 | Nm107 | Nm93 | |
| >5 | >5 | >5 | >5 | >5 | >5 | >5 | 0, IN | 0, 1D | 0 | >5 | >5 | >5 | >5 | >5 |
| >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | 0 | >5 | >5 | >5 | >5 |
| 4 | >5 | 5 | 4 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 3 | 3 | 3 | 3 |
| 4 | 3 | 4 | 3 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 4N | 0 | 0 | 0 | 0 | 0 |
| 3 | 3 | 3 | 4 | 2 | 4 | 2 | 2, 1D | 2, 1D | 3, 1D | 3 | 0 | 0 | 0 | 0 |
| 0 | 1 | 0 | 1 | 2 | 3 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| 0 | 1 | 0 | 1 | 2 | 3 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| 2 | 3 | 2 | 1 | 0 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 |
| 3 | 3 | 3 | 4 | 5 | >5 | 4 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| 1 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 2 |
| >5 | >5 | >5 | >5 | >5 | >5 | >5 | 0 | 0 | 4 | 5 | >5 | >5 | >5 | >5 |
| >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 |
| >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 |
| >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 |
| 1 | 1 | 1 | 0 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | 1 | 1 | 3 | 1 | 1 | 0 | 0, 3D | 1 | 2 | 1 | 1 | 1 | 1 |
| 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 3D | 0 | 2 | 1 | 1 | 1 | 1 |
| 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 3D | 0 | 2 | 1 | 1 | 1 | 1 |
| 1 | 2 | 2 | 1 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| 0 | 0 | 0 | 0 | 3 | 1 | 0 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| 5 | 5 | 5 | 4 | 3 | 4 | 4 | 2 | 2 | 2 | 2 | 4 | 4 | 4 | 4 |
| 3 | 3 | 3 | 2 | 3 | 4 | 2 | 3 | 3 | 3 | 3 | 2 | 2 | 2 | 2 |
| 0 | 0 | 0 | 3 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 |
| >5 | >5 | >5 | >5 | >5 | >5 | >5 | 0 | 1 | 0, 1D, 3N | >5 | 5 | 5 | 5 | 5 |
| >5 | >5 | >5 | >5 | >5 | >5 | 2 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 |
| >5 | >5 | >5 | >5 | >5 | >5 | 1 | >5 | >5 | >5 | >5 | >5 | >5 | >5 | >5 |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 |
OPD, Oligonucleotide Probe Database.
Nucleotide numbers correspond to E. coli numbering (9).
Each specificity was given by the respective authors when the primers were published.
Shown are the numbers of non-AOB targeted with zero mismatches (0MM) or one mismatch (1MM). Environmental 16S rDNA clones were not included in this analysis.
D, deletion; I, insertion; N, undetermined base in the target region.
Probe has been demonstrated to be suitable for in situ hybridization.
Corrected sequences were used (77).
Probe NmIV (52) should be modified as follows: T should be replaced by A at position 1 of the probe sequence to eliminate a mismatch to the target region of Nitrosomonas cryotolerans.
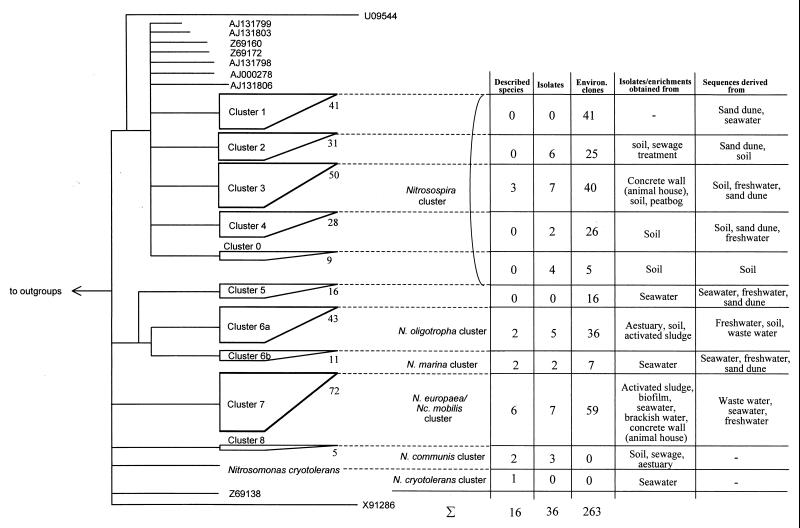
Recently, Stephen et al. (69) suggested a 16S rRNA-based phylogenetic classification scheme for beta-subclass AOB consisting of seven clusters, which has found widespread application (7, 35, 37, 38, 44, 47, 50, 65, 87). We reevaluated this scheme using the completed and newly obtained 16S rRNA AOB sequences of this study by using different treeing methods and data sets. The overall tree topology was determined by exclusively using sequences with more than 1,000 nucleotides. More partial 16S rRNA sequences were subsequently added without changing the overall tree topology (Fig. 5). According to Ludwig et al. (41), this procedure produces more reliable trees than calculating a single tree based on only a few hundred aligned nucleotides (37, 69). This is also exemplified in several obviously incorrect tree topologies obtained in previous studies in which only a few hundred informative positions of the 16S rRNA were analyzed. For example, in the trees constructed by different authors (44, 47, 53, 69, 82, 87), Nitrosococcus mobilis does not belong to cluster 7 but is incorrectly assigned to cluster 6 or to Nitrosomonas cryotolerans.
FIG. 5.
Schematic 16S rRNA-based phylogenetic classification of the beta-subclass AOB. Multifurcations connect branches for which a relative order could not be unambiguously determined by applying different treeing methods. The height of each tetragon represents the number of sequences in the cluster. Due to the presence of many published partial 16S rRNA sequences in the clusters, no meaningful estimate of the sequence diversity within a cluster could be inferred. The cluster designations were adopted from those of Stephen et al. (69). We suggest including two additional clusters in the scheme (Nitrosospira cluster 0; Nitrosomonas cluster 8). Furthermore, cluster 6 should be subdivided into clusters 6a and 6b (see text). In addition to the 16S rRNA sequences determined in this study, 16S rRNA sequences published by Aakra et al. (1, 2), Head et al. (17), Suwa et al. (70), Kowalchuk et al. (35, 37, 38), Logemann et al. (40), McCaig et al. (43), Mendum et al. (47), Phillips et al. (50), Princic et al. (53), Rotthauwe et al. (55), Speksnijder et al. (65), Stehr et al. (67), Stephen et al. (69), Teske et al. (73), Utaker et al. (76), and Whitby et al. (87) as well as unpublished AOB 16S rRNA sequences deposited in GenBank were used to calculate the schematic dendrogram. The composition of each cluster is indicated in the adjacent table. Isolates which have not been analyzed with regard to their species affiliation are as follows: for cluster 2, Nitrosospira sp. strains AHB1 (55), O4 and O13 (2), III7 and B6 (1), and T7 (76); for cluster 3, Nitrosospira sp. strains NpAV and Np22-21 (43) and F3, L115, AF, A4, and A16 (1); for cluster 4, Nitrosospira sp. strains Ka3 and Ka4 (2); for cluster 0, Nitrosospira sp. strains III2, D11, GM4, and 40KI (76); for cluster 6, Nitrosomonas sp. strains Nm80, Nm84, and Nm86 (67) and AL212 and JL21 (70); for cluster 7, Nitrosomonas sp. strains GH22 and HPC101 (71), F5 (1), Koll21 (GenBank accession no. AJ224941), and Nm104 and Nm107 (this study); and for cluster 8, Nitrosomonas sp. strains Nm58 (67) and Nm33 and Nm41 (this study).
Our phylogenetic analyses demonstrated that Nitrosospira clusters 1 to 4 are supported by some but not by all treeing methods. While cluster 1 is recovered with most methods and data sets, clusters 2, 3, and 4 are less stable. It should also be noted that four Nitrosospira isolates (40KI, GM4, D11, and III2 [76, 77]) which form an additional and stable cluster (together with five environmental clones) are not yet included in the current scheme (Fig. 5). Within the nitrosomonads we propose to extend the scheme by the previously excluded Nitrosomonas communis cluster, which thus represents cluster 8. Furthermore, we suggest splitting cluster 6 into clusters 6a and 6b, which are represented by members of the Nitrosomonas oligotropha cluster and the Nitrosomonas marina cluster, respectively (Fig. 5). Most environmental AOB 16S rRNA sequences retrieved so far belong to Nitrosospira clusters 1 and 3 and to the Nitrosomonas europaea-Nitrosococcus mobilis cluster. However, it should be stressed that the relationships inferred from very short 16S rRNA sequences, even using the “combined” treeing method applied here, are still of low confidence.
Despite the discussed limitations, several interesting observations can be made from the hitherto performed AOB diversity studies. First, within the nitrosomonads, only cluster 5 clearly represents a missing species within the AOB culture collection with sequence similarities of <96.5% to previously described AOB species (highest similarity was to a 186-bp 16S rRNA fragment of Nitrosomonas sp. strain Nm84 [67]). In addition, four 340-bp-long molecular wastewater isolates from a reactor with high ammonium level (clones AI-8H, AI-7K, AI-8B1, and AI-9K3 [53]) might represent a new species within cluster 7 (<96% sequence similarity to previously described AOB species). Nitrosospira cluster 1, which does not yet contain a cultured isolate, is nevertheless not demonstrative for the existence of a novel Nitrosospira species since all cluster 1 16S rRNA sequences show more than 97% similarity to available Nitrosospira pure cultures. In addition, some environmentally retrieved partial 16S rDNA sequences (the majority of them related to nitrosospiras) cannot be unambiguously assigned to one of the clusters (Fig. 5). Due to the short sequence lengths, it is difficult to decide whether these sequences represent putative novel AOB species. Second, none of the environmental AOB sequences retrieved so far in the various studies are affiliated with the Nitrosomonas communis cluster (cluster 8), Nitrosomonas halophila, or Nitrosomonas cryotolerans. This might in part be caused by insufficient coverage of these organisms by some of the “AOB-specific” primers used. However, we could detect Nitrosomonas communis and Nitrosomonas sp. strain Nm 33 but not Nitrosomonas halophila and Nitrosomonas cryotolerans in wastewater treatment plants using the amoA approach (see below). Future studies will have to show whether Nitrosomonas halophila and the Nitrosomonas communis and Nitrosomonas cryotolerans clusters are of limited environmental distribution or whether methodological biases cause underestimation of their actual abundance.
The gene encoding the active site subunit of the ammonia monooxygenase (amoA) has increasingly been exploited as a marker molecule for cultivation-independent analyses of ammonia oxidizer diversity. Different sets of PCR primers for the amplification of amoA gene fragments were published (22, 47, 56, 64). In this study, the primers described by Rotthauwe and coworkers (56) were successfully used to amplify the expected amoA fragment from all beta-subclass AOB analyzed, demonstrating the excellent sensitivity of this PCR assay. For amplification of an amoA fragment of the gamma-subclass AOB Nitrosococcus halophilus, a new PCR primer pair was developed. After completion of the amoA database, phylogeny inference based on the nucleic acid and amino acid amoA-AmoA data sets was, both for the beta- and the gamma-subclasses of AOB, overall consistent with the picture described above derived from the 16S rRNA analysis. It is of importance to note that the amoA sequence of Nitrosococcus sp. strain Nm93 reported in this study is, as expected, almost identical to the amoA sequence of Nitrosococcus mobilis Nc2 (99.6% nucleic acid similarity) while we amplified a Nitrosomonas europaea-like amoA sequence from Nitrosococcus sp. strain Nm93 in a previous study (28). Thus, this strain was most likely contaminated at that time with Nitrosomonas europaea. Furthermore, the amoA sequence of Nitrosococcus oceani (C-107, identical with ATCC 19707 and NCIMB 11848) differs significantly in the publications of Holmes et al. (22) and Alzerecca et al. (4) caused by a misidentification of Methylomicrobium pelagicum as Nitrosococcus oceani in the former publication (now corrected by the authors in a recent update of GenBank accession no. U31652). Consequently, gamma-subclass AOB have a lower level of AmoA similarity (<75.5%) to type I methanotrophs than previously considered (22). The separate clustering of gamma-subclass AOB and type I methanotrophs in the AmoA and 16S rRNA trees might reflect their specialization of using either ammonia or methane as preferred substrate. In accordance with this hypothesis, the deduced AmoA sequences of the gamma-subclass AOB do differ in 4 of the 21 signature amino acids of the particulate methane monooxygenase of type I and type II methanotrophs (23). At one (Nitrosococcus oceani; Nitrosococcus sp. strain C-113) or two (Nitrosococcus halophilus) of these signature positions, the gamma-subclass AOB possess amino acids which are absolutely conserved within the ammonia monooxygenases of beta-subclass AOB, which might indicate that these positions are influencing substrate affinity of the respective monooxygenases.
The completed amoA database was also used to perform a specificity check of the primers published by Sinigalliano et al. (64) and Holmes et al. (22). Surprisingly, only Nitrosomonas europaea possesses fully complementary target sequences to the Sinigalliano primers. Most likely, the amoA sequences from Nitrosococcus oceani and Nitrosomonas cryotolerans that were amplified by Sinigalliano et al. (64) originated from a contamination with Nitrosomonas europaea and were thus reported to be identical with the amoA sequence of the latter species. The correct amoA sequences of Nitrosococcus oceani and Nitrosomonas cryotolerans were reported by Alzerecca et al. (4) and in this study, respectively. The Holmes primers do target some beta-subclass AOB and gamma-subclass methanotrophs but possess several mismatches with other beta-subclass AOB and all three gamma-subclass AOB in the database (Table 6). Consequently, conclusions on ecological relevance (19, 20) or diversity of AOB using these primers (57) have to be interpreted with caution.
TABLE 6.
Mismatches of the PCR primers A189 (A-*-MOB-189-a-S-18) and A682 (A-*-MOB-682-a-A-18) (3, 22) with the amoA genes of beta- and gamma-subclass AOB
| Primer | No. of mismatches
|
||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gamma AOB
|
Nitrosospira cluster
|
Nitrosomonas europaea/ Nitrosococcus mobilis cluster
|
Nitrosomonas cryotolerans cluster Nm55 |
Nitrosomonas communis cluster
|
Nitrosomonas oligotropha cluster
|
Nitrosomonas marina cluster
|
|||||||||||||||||||||||||
| C107 | C113 | Nc4 | NpAVa | Np39-19a | Nsp1 | Np22 | AHB1 | L13 | Nv1 | Nv12 | C128 | C71 | Nc2 | Nm93 | Nm104 | Nm107 | Nm1 | Nm50 | Nm103 | Nm57b | Nm41 | Nm2 | Nm33 | Nm90 | Nm45 | Nm10 | Nm51 | Nm22 | Nm36 | ||
| A189 | 0 | 1 | —c | 0 | 0 | 1 | — | 0 | — | — | 1 | 0 | 0 | — | — | — | — | — | 1 | — | 1 | — | — | — | — | — | — | — | — | — | — |
| A682 | 4 | 5 | 2 | 0/1/2 | 3 | — | — | 0 | 1 | 3 | 3 | 4 | 1 | 6 | 6 | 6 | 6 | 4 | 1 | 1 | 3 | 4 | 4 | 3 | 3 | 3 | 1 | 2 | 1 | 1 | 2 |
Contains three amoA operons (29), which differ from Nitrosospira sp. strain NpAV in sequence at the primer A682 target region.
Contains two amoA operons which are identical in sequence at the primer target regions.
—, no sequence information are available.
Comparative sequence analysis of 122 amoA clones obtained from 11 activated-sludge and biofilm samples demonstrated that generally nitrosomonads are responsible for ammonia oxidation in wastewater treatment plants and that nitrosospiras occur only sporadically in these systems. This result is consistent with PCR-independent AOB community structure analysis performed by fluorescent in situ hybridization FISH (28, 81) but disagrees with findings of Hiorns et al. (21), who could detect nitrosospiras but not nitrosomonads in an activated-sludge plant. The latter finding, however, was most likely caused by the very limited coverage of nitrosomonads by probe Nm75 (Table 5). Furthermore, it should be noted that, considering the extended amoA database, the recently developed terminal-restriction fragment length polymorphism (TRFLP) method for identification of major subgroups AOB (24) will not produce meaningful community fingerprint patterns (Fig. 5).
Using the amoA approach, with the exception of Nitrosomonas cryotolerans, Nitrosomonas halophila, and Nitrosomonas nitrosa, sequences related to all recognized Nitrosomonas species were obtained from wastewater treatment plants (Fig. 4). amoA sequences related to Nitrosococcus mobilis were detected in six different wastewater treatment plants, including the industrial plant Kraftisried. In a previous study, Juretschko et al. (28) obtained exclusively Nitrosomonas europaea-like amoA sequences from this plant by using the primers described by Sinigalliano et al. (64) while FISH clearly demonstrated the in situ dominance of Nitrosococcus mobilis. This contradiction was caused by the limited sensitivity of the Sinigalliano primers and was able to be resolved in this study. In different plants, several amoA sequences (for example clones S12 and SBBR1-32) which showed only relatively moderate sequence similarities to known beta-subclass AOB species were recovered. Application of the amoA and AmoA similarity threshold values indicative of novel AOB species (obtained by amoA and AmoA 16S rRNA correlation plots) did not support that these sequences represent previously unrecognized nitrosomonads. However, it is important to clarify that while amoA and AmoA similarities below the suggested threshold values are strongly indicative of the existence of novel species, an amoA and AmoA sequence with a similarity to a described AOB species above the threshold level can originate from either a novel species or the described AOB species. This problem could be solved if the respective threshold values were inferred from correlation plots of amoA and AmoA versus DNA-DNA similarity. However, this analysis has to await the availability of more DNA-DNA hybridization data of cultured AOB.
Different wastewater treatment plants obviously differ significantly in regard to species richness of AOB. While some plants are dominated by a single AOB species (e.g., Nitrosococcus mobilis in the Kraftisried plant), other plants harbor at least four different AOB species (e.g., Munich I-Großlappen). A high AOB diversity could increase the resistance of nitrification against perturbation while the presence of a AOB monoculture in a plant might render its nitrification more susceptible.
In conclusion, a robust phylogenetic framework of AOB was established by comparative sequence analysis of all described AOB species based on the 16S rRNA and the amoA marker molecule. Reevaluation of the specificity of published primers and probes developed for the detection of both biopolymers in environmental samples demonstrated, in many cases, insufficient specificity. High-resolution assignment of all published environmentally retrieved 16S rRNA sequences only provided evidence for the existence of two yet undescribed beta-subclass AOB species, suggesting that available AOB isolates might be more representative of the natural diversity within this physiological group than previously thought. A similar picture emerged from an amoA-based diversity survey of AOB in wastewater treatment plants, which demonstrated that most retrieved molecular isolates were closely related to known nitrosomonads. While almost every amoA or 16S rRNA AOB gene library from environmental samples contains many sequences which are not identical to those of cultured AOB, the degree of divergence is, for most of the sequences obtained up to now, insufficient to unequivocally prove the existence of novel AOB species.
ACKNOWLEDGMENTS
This study was supported by Sonderforschungsbereich 411 from the Deutsche Forschungsgemeinschaft (Project A2 - Research Center for Fundamental Studies of Aerobic Biological Wastewater Treatment).
The excellent technical assistance of Sibylle Schadhauser is acknowledged. We kindly thank Martin Klotz for helpful discussion. We are indebted to Wolfgang Ludwig for providing the 16S rRNA sequence of Nitrosococcus mobilis Nc2.
REFERENCES
- 1.Aakra Å, Utaker J B, Nes I F. RFLP of rRNA genes and sequencing of the 16S–23S rDNA intergenic spacer region of ammonia-oxidizing bacteria: a phylogenetic approach. Int J Syst Bacteriol. 1999;49:123–130. doi: 10.1099/00207713-49-1-123. [DOI] [PubMed] [Google Scholar]
- 2.Aakra Å, Utaker J B, Nes I F, Bakken L R. An evaluated improvement of the extinction dilution method for isolation of ammonia-oxidizing bacteria. J Microbiol Methods. 1999;39:23–31. doi: 10.1016/s0167-7012(99)00094-9. [DOI] [PubMed] [Google Scholar]
- 3.Alm E, Oerther D B, Larsen N, Stahl D A, Raskin L. The Oligonucleotide Probe Database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alzerreca J J, Norton J M, Klotz M G. The amo operon in marine, ammonia-oxidizing gamma-proteobacteria. FEMS Microbiol Lett. 1999;180:21–29. doi: 10.1111/j.1574-6968.1999.tb08773.x. [DOI] [PubMed] [Google Scholar]
- 5.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arthur J W, West C W, Allen K N, Hedke S F. Seasonal toxicity of ammonia to five fish and nine invertebrate species. Bull Environ Contam Toxicol. 1987;38:324–331. doi: 10.1007/BF01606682. [DOI] [PubMed] [Google Scholar]
- 7.Bano N, Hollibaugh J T. Diversity and distribution of DNA sequences with affinity to ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in the Arctic Ocean. Appl Environ Microbiol. 2000;66:1960–1969. doi: 10.1128/aem.66.5.1960-1969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bock E, Atueven R, Mansch R, Vollmer M. Formation and consumption of nitric oxide by nitrifying bacteria. In: Alberghina L, Frontali L, Sensi P, editors. ECB6: Proceedings of the 6th European Congress on Biotechnology. Amsterdam, The Netherlands: Elsevier Science B. V.; 1994. pp. 241–244. [Google Scholar]
- 9.Brosius J, Palmer M L, Kennedy P J, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandler D P, Fredrickson J K, Brockman F J. Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol Ecol. 1997;6:475–482. doi: 10.1046/j.1365-294x.1997.00205.x. [DOI] [PubMed] [Google Scholar]
- 11.Chandler D P, Schreckhise R W, Smith J L, Bolton H., Jr Electroelution to remove humic compounds from soil DNA and RNA extracts. J Microbiol Methods. 1997;28:11–19. [Google Scholar]
- 12.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rDNA identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 14.Frankland P F, Frankland G C. The nitrifying process and its specific ferment. Part I. Phil Trans R Soc B. 1890;181:107–128. [Google Scholar]
- 15.Hall G H. Nitrification in lakes. In: Prosser J I, editor. Nitrification. Vol. 20. Oxford, United Kingdom: IRL Press; 1986. pp. 127–156. [Google Scholar]
- 16.Hastings R C, Ceccherini M T, Miclaus N, Saunders J R, Bazzicalupo M, McCarthy A J. Direct molecular biological analysis of ammonia oxidizing bacteria populations in cultivated soil plots treated with swine manure. FEMS Microbiol Ecol. 1997;23:45–54. [Google Scholar]
- 17.Head I M, Hiorns W D, Embley T M, McCarthy A J, Saunders J R. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J Gen Microbiol. 1993;139:1147–1153. doi: 10.1099/00221287-139-6-1147. [DOI] [PubMed] [Google Scholar]
- 18.Helmer C, Kunst S, Juretschko S, Schmid M C, Schleifer K-H, Wagner M. Nitrogen loss in a nitrifying biofilm system. Water Sci Technol. 1999;39:13–21. [Google Scholar]
- 19.Henckel T, Friedrich M, Conrad R. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl Environ Microbiol. 1999;65:1980–1990. doi: 10.1128/aem.65.5.1980-1990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henckel T, Jackel U, Schnell S, Conrad R. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl Environ Microbiol. 2000;66:1801–1808. doi: 10.1128/aem.66.5.1801-1808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiorns W D, Hastings R C, Head I M, McCarthy A J, Saunders J R, Pickup R W, Hall G H. Amplification of 16S ribosomal RNA genes of autotrophic ammonia-oxidising bacteria. Microbiology. 1995;141:2793–2800. doi: 10.1099/13500872-141-11-2793. [DOI] [PubMed] [Google Scholar]
- 22.Holmes A J, Costello A, Lidstrom M E, Murrell J C. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 23.Holmes A J, Roslev P, McDonald I R, Iversen N, Henriksen K, Murrell J C. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl Environ Microbiol. 1999;65:3312–3318. doi: 10.1128/aem.65.8.3312-3318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horz H P, Rotthauwe J H, Lukow T, Liesack W. Identification of major subgroups of ammonia-oxidizing bacteria in environmental samples by T-RFLP analysis of amoA PCR products. J Microbiol Methods. 2000;39:197–204. doi: 10.1016/s0167-7012(99)00119-0. [DOI] [PubMed] [Google Scholar]
- 25.Hovanec T A, DeLong E F. Comparative analysis of nitrifying bacteria associated with freshwater and marine aquaria. Appl Environ Microbiol. 1996;62:2888–2896. doi: 10.1128/aem.62.8.2888-2896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imhoff K, Imhoff K R. Taschenbuch der Stadtentwässerung. 26th ed. Munich, Germany: Oldenbourg Verlag; 1985. [Google Scholar]
- 27.Jones R D, Morita R Y, Koops H P, Watson S W. A new marine ammonium-oxidizing bacterium, Nitrosomonas cryotolerans sp. nov. Can J Microbiol. 1988;34:1122–1128. [Google Scholar]
- 28.Juretschko S, Timmermann G, Schmid M, Schleifer K H, Pommerening-Röser A, Koops H P, Wagner M. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klotz M G, Norton J M. Sequence of an ammonia monooxygenase subunit A encoding gene from Nitrosospira sp. NpAV. Gene. 1995;163:159–160. doi: 10.1016/0378-1119(95)00392-j. [DOI] [PubMed] [Google Scholar]
- 30.Koops H-P, Böttcher B, Möller U C, Pommerening-Röser A, Stehr G. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. J Gen Microbiol. 1991;137:1689–1699. [Google Scholar]
- 31.Koops H-P, Harms H. Deoxyribonucleic acid homologies among 96 strains of ammonia-oxidizing bacteria. Arch Microbiol. 1985;141:214–218. doi: 10.1007/BF00408061. [DOI] [PubMed] [Google Scholar]
- 32.Koops H-P, Harms H, Wehrmann H. Isolation of a moderate halophilic ammonia-oxidizing bacterium, Nitrosococcus mobilis nov. sp. Arch Microbiol. 1976;107:277–282. doi: 10.1007/BF00425339. [DOI] [PubMed] [Google Scholar]
- 33.Koops H-P, Möller U C. The lithotrophic ammonia-oxidizing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The Prokaryotes. 2nd ed. Vol. 3. New York, N.Y: Springer-Verlag; 1992. pp. 2625–2637. [Google Scholar]
- 34.Koops H-P, Böttcher B, Möller U C, Pommerening-Röser A, Stehr G. Description of a new species of Nitrosococcus. Arch Microbiol. 1990;154:244–248. [Google Scholar]
- 35.Kowalchuk G A, Bodelier P L E, Heilig G H J, Stephen J R, Laanbroek H J. Community analysis of ammonia-oxidising bacteria, in relation to oxygen availability in soils and root-oxygenated sediments, using PCR, DGGE and oligonucleotide probe hybridisation. FEMS Microbiol Ecol. 1998;27:339–350. [Google Scholar]
- 36.Kowalchuk G A, Naoumenko Z S, Derikx P J, Felske A, Stephen J R, Arkhipchenko I A. Molecular analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in compost and composted materials. Appl Environ Microbiol. 1999;65:396–403. doi: 10.1128/aem.65.2.396-403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kowalchuk G A, Stephen J R, De Boer W, Prosser J I, Embley T M, Woldendorp J W. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kowalchuk G A, Stienstra A W, Heilig G H, Stephen J R, Woldendorp J W. Molecular analysis of ammonia-oxidising bacteria in soil of successional grasslands of the Drentsche A (The Netherlands) FEMS Microbiol Ecol. 2000;31:207–215. doi: 10.1111/j.1574-6941.2000.tb00685.x. [DOI] [PubMed] [Google Scholar]
- 39.Larsen N, Olsen G J, Maidak B L, McCaughey M J, Overbeek R, Macke T J, Marsh T L, Woese C R. The ribosomal database project. Nucleic Acids Res. 1993;21:3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Logemann S, Schantl J, Bijvank S, Loosdrecht M v, Kuenen J G, Jetten M. Molecular microbial diversity in a nitrifying reactor system without sludge retention. FEMS Microbiol Ecol. 1998;27:239–249. [Google Scholar]
- 41.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer K-H. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 42.MacDonald R M. Nitrification in soil: an introductory history. In: Prosser J I, editor. Nitrification. Vol. 20. Oxford, United Kingdom: IRL Press; 1986. pp. 1–16. [Google Scholar]
- 43.McCaig A E, Embley T M, Prosser J I. Molecular analysis of enrichment cultures of marine ammonia oxidisers. FEMS Microbiol Lett. 1994;120:363–368. doi: 10.1111/j.1574-6968.1994.tb07059.x. [DOI] [PubMed] [Google Scholar]
- 44.McCaig A E, Phillips C J, Stephen J R, Kowalchuk G A, Harvey S M, Herbert R A, Embley T M, Prosser J I. Nitrogen cycling and community structure of proteobacterial beta-subgroup ammonia-oxidizing bacteria within polluted marine fish farm sediments. Appl Environ Microbiol. 1999;65:213–220. doi: 10.1128/aem.65.1.213-220.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McTavish H, Fuchs J A, Hooper A B. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J Bacteriol. 1993;175:2436–2444. doi: 10.1128/jb.175.8.2436-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meincke M, Krieg E, Bock E. Nitrosovibrio spp., the dominant ammonia-oxidizing bacteria in building sandstone. Appl Environ Microbiol. 1989;55:2108–2110. doi: 10.1128/aem.55.8.2108-2110.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mendum T A, Sockett R E, Hirsch P R. Use of molecular and isotopic techniques to monitor the response of autotrophic ammonia-oxidizing populations of the beta subdivision of the class Proteobacteria in arable soils to nitrogen fertilizer. Appl Environ Microbiol. 1999;65:4155–4162. doi: 10.1128/aem.65.9.4155-4162.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mobarry B K, Wagner M, Urbain V, Rittmann B E, Stahl D A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. . (Erratum, 63:815, 1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Painter H A. Nitrification in the treatment of sewage and waste-waters. In: Prosser J I, editor. Nitrification. Vol. 20. Oxford, United Kingdom: IRL Press; 1986. pp. 185–211. [Google Scholar]
- 50.Phillips C J, Smith Z, Embley T M, Prosser J. Phylogenetic differences between particle-associated and planctonic ammonia-oxidizing bacteria of the β-subdivision of the class Proteobacteria in the northwestern Mediterranean Sea. Appl Environ Microbiol. 1999;65:779–786. doi: 10.1128/aem.65.2.779-786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polz M F, Cavanaugh C M. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pommerening-Röser A, Rath G, Koops H-P. Phylogenetic diversity within the genus Nitrosomonas. Syst Appl Microbiol. 1996;19:344–351. [Google Scholar]
- 53.Princic A, Mahne I, Megusar F, Paul E A, Tiedje J M. Effects of pH and oxygen and ammonium concentrations on the community structure of nitrifying bacteria from wastewater. Appl Environ Microbiol. 1998;64:3584–3590. doi: 10.1128/aem.64.10.3584-3590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reysenbach A L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rotthauwe J H, de Boer W, Liesack W. Comparative analysis of gene sequences encoding ammonia monooxygenase of Nitrosospira sp. AHB1 and Nitrosolobus multiformis C-71. FEMS Microbiol Lett. 1995;133:131–135. doi: 10.1111/j.1574-6968.1995.tb07873.x. [DOI] [PubMed] [Google Scholar]
- 56.Rotthauwe J H, Witzel K P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakano Y, Kerkhof L. Assessment of changes in microbial community structure during operation of an ammonia biofilter with molecular tools. Appl Environ Microbiol. 1998;64:4877–4882. doi: 10.1128/aem.64.12.4877-4882.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 59.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;7:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmid M, Twachtmann U, Klein M, Strous M, Juretschko S, Jetten M, Metzger J W, Schleifer K-H, Wagner M. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst Appl Microbiol. 2000;23:93–106. doi: 10.1016/S0723-2020(00)80050-8. [DOI] [PubMed] [Google Scholar]
- 61.Schramm A, De Beer D, Wagner M, Amann R. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol. 1998;64:3480–3485. doi: 10.1128/aem.64.9.3480-3485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schramm A, Larsen L H, Revsbech N P, Ramsing N B, Amann R, Schleifer K H. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol. 1996;62:4641–4647. doi: 10.1128/aem.62.12.4641-4647.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schramm A, Santegoeds C M, Nielsen H K, Ploug H, Wagner M, Pribyl M, Wanner J, Amann R, de Beer D. On the occurrence of anoxic microniches, denitrification, and sulfate reduction in aerated activated sludge. Appl Environ Microbiol. 1999;65:4189–4196. doi: 10.1128/aem.65.9.4189-4196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sinigalliano C D, Kuhn D, Jones R D. Amplification of the amoA gene from diverse species of ammonium-oxidizing bacteria from an indigenous bacterial population from seawater. Appl Environ Microbiol. 1995;61:2702–2706. doi: 10.1128/aem.61.7.2702-2706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Speksnijder A G, Kowalchuk G A, Roest K, Laanbroek H J. Recovery of a Nitrosomonas-like 16S rDNA sequence group from freshwater habitats. Syst Appl Microbiol. 1998;21:321–330. doi: 10.1016/S0723-2020(98)80040-4. [DOI] [PubMed] [Google Scholar]
- 66.Stackebrandt E, Goebel M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Bacteriol. 1994;44:846–849. [Google Scholar]
- 67.Stehr G B, Böttcher B, Dittberner P, Rath G, Koops H P. The ammonia oxidizing nitrifiying population of the River Elbe estuary. FEMS Microbiol Ecol. 1995;17:177–187. [Google Scholar]
- 68.Stephen J R, Chang Y J, Macnaughton S J, Kowalchuk G A, Leung K T, Flemming C A, White D C. Effect of toxic metals on indigenous soil beta-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl Environ Microbiol. 1999;65:95–101. doi: 10.1128/aem.65.1.95-101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stephen J R, McCaig A E, Smith Z, Prosser J I, Embley T M. Molecular diversity of soil and marine 16S rRNA gene sequences related to beta-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996;62:4147–4154. doi: 10.1128/aem.62.11.4147-4154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suwa Y, Sumino T, Noto K. Phylogentic relationships of activated sludge isolates of ammonia-oxidizers with different sensitivities to ammonium sulfate. J Gen Appl Microbiol. 1997;43:373–379. doi: 10.2323/jgam.43.373. [DOI] [PubMed] [Google Scholar]
- 71.Suzuki M, Rappe M S, Giovannoni S J. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl Environ Microbiol. 1998;64:4522–4529. doi: 10.1128/aem.64.11.4522-4529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teske A, Alm E, Regan J M, Toze S, Rittman B E, Stahl D A. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J Bacteriol. 1994;176:6623–6630. doi: 10.1128/jb.176.21.6623-6630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tortoso A C, Hutchinson G L. Contributions of autotrophic and heterotrophic nitrifiers to soil NO and N2O emission. Appl Environ Microbiol. 1990;56:1799–1805. doi: 10.1128/aem.56.6.1799-1805.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trüper H G, de Clari L. Taxonomic note: necessary correction of specific epithets formed as substantives (nouns) “in apposition.”. Int J Syst Bacteriol. 1997;47:908–909. [Google Scholar]
- 76.Utåker J B, Bakken L, Jiang Q, Nes J. Phylogenetic analysis of seven new isolates of ammonia oxidizing bacteria based on 16S rRNA gene sequences. Syst Appl Microbiol. 1995;18:549–559. [Google Scholar]
- 77.Utåker J B, Nes I F. A qualitative evaluation of the published oligonucleotides specific for the 16S rRNA gene sequences of the ammonia-oxidizing bacteria. Syst Appl Microbiol. 1998;21:72–88. doi: 10.1016/S0723-2020(98)80010-6. [DOI] [PubMed] [Google Scholar]
- 78.Voytek M A, Ward B B. Detection of ammonium-oxidizing bacteria of the beta-subclass of the class Proteobacteria in aquatic samples with the PCR. Appl Environ Microbiol. 1995;61:1444–1450. doi: 10.1128/aem.61.4.1444-1450.1995. . (Erratum, 61:2811.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wagner M, Amann R, Lemmer H, Schleifer K H. Probing activated sludge with proteobacteria-specific oligonucleotides: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wagner M, Rath G, Amann R, Koops H-P, Schleifer K H. In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol. 1995;18:251–264. [Google Scholar]
- 81.Wagner M, Rath G, Koops H-P, Flood J, Amann R. In situ analysis of nitrifiying bacteria in sewage treatment plants. Water Sci Technol. 1996;34:237–244. [Google Scholar]
- 82.Ward B B. Nitrification and denitrification: probing the nitrogen cycle in aquatic environments. Microb Ecol. 1996;32:247–261. doi: 10.1007/BF00183061. [DOI] [PubMed] [Google Scholar]
- 83.Ward B B, Voytek M A, Witzel K F. Phylogenetic diversity of natural populations of ammonia oxidizers investigated by specific PCR amplification. Microb Ecol. 1997;33:87–96. doi: 10.1007/s002489900011. [DOI] [PubMed] [Google Scholar]
- 84.Watson S W. Characteristics of a marine nitrifying bacterium, Nitrosocystis oceanus sp. n. Limnol Oceanogr. 1965;10(Suppl.):R274–R289. [Google Scholar]
- 85.Watson S W, Mandel M. Comparison of the morphology and deoxyribonucleic acid composition of 27 strains of nitrifying bacteria. J Bacteriol. 1971;107:563–569. doi: 10.1128/jb.107.2.563-569.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Watson S W, Valois F W, Waterbury J B. The family Nitrobacteraceae. In: Starr M P, Stolp H, Trüper H G, Balows A, Schlegel H G, editors. The Prokaryotes. Vol. 1. Berlin, Germany: Springer Verlag; 1981. pp. 1005–1022. [Google Scholar]
- 87.Whitby C B, Saunders J R, Rodriguez J, Pickup R W, McCarthy A. Phylogenetic differentiation of two closely related Nitrosomonas spp. that inhabit different sediment environments in an oligotrophic freshwater lake. Appl Environ Microbiol. 1999;65:4855–4862. doi: 10.1128/aem.65.11.4855-4862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Winogradsky S. 2nd memoire. Recherches sur les organismes de la nitrification. Ann Inst Pasteur. 1890;4:257–275. [Google Scholar]
- 89.Winogradsky S. Contribution a la morphologie des organismes de la nitrification. Arch Sci Biol. 1892;1:88–137. [Google Scholar]
- 90.Wintzigerode F, Goebel U B. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 91.Woese C R, Weisburg W G, Hahn C M, Paster B J, Zablen L B, Lewis B J, Macke T J, Ludwig W, Stackebrandt E. The phylogeny of purple bacteria: the gamma subdivision. Syst Appl Microbiol. 1985;6:25–33. [Google Scholar]
- 92.Woese C R, Weisburg W G, Paster B J, Hahn C M, Tanner R S, Krieg N R, Koops H-P, Harms H, Stackebrandt E. The phylogeny of purple bacteria: the beta subdivision. Syst Appl Microbiol. 1984;5:327–336. doi: 10.1016/s0723-2020(84)80034-x. [DOI] [PubMed] [Google Scholar]