Abstract
An Escherichia coli strain that accumulated Ni(II) was constructed by introducing the nixA gene (coding for a nickel transport system) from Helicobacter pylori into JM109 cells that expressed a glutathione S-transferase–pea metallothionein fusion protein. The resulting strain accumulated 15 μmol of Ni(II) per g (dry weight) from a 10 μM Ni(II) solution, four times the level taken up by JM109 cells. Ni(II) accumulation did not require an energy source, was inhibited by only 50% by 0.1 M NaCl, and occurred over the pH range from 3 to 9.
Nickel is a toxic heavy metal found in the environment as a result of various natural and industrial activities. Nickel has been implicated as an embryotoxin and teratogen (4). Studies of human cell cultures have indicated that nickel is a possible carcinogen, creating a need for the cleanup of nickel pollution (18). Current methods for nickel removal from water include algal biosorption and chemical precipitation. Both of these methods are sensitive to ambient conditions, namely pH, ionic strength, and the presence of organic or inorganic ligands. Biosorption and precipitation lack specificity in metal binding, and chemical precipitation is effective only at high metal concentrations (1). A highly specific nickel transport system has been identified in Helicobacter pylori (19). The product of the nixA gene is a 37-kDa integral membrane protein consisting of eight transmembrane domains which has a very high affinity for Ni(II), with a Kd of 11.3 nM (9).
The most important properties for biological metal cleanup techniques are flexibility to handle the range of physicochemical parameters in effluents, selectivity to remove only the desired metals, and cost-effectiveness (8). Genetic engineering allows the introduction of desired traits into cells to help meet some of these criteria, and this approach has already been used to construct cells for the bioremediation of mercury (5). In this study, Escherichia coli organisms were genetically engineered to simultaneously express a Ni(II) transport system and overexpress pea metallothionein (PMT) as a carboxyl-terminal fusion to glutathione S-transferase (GST-PMT).
Metallothioneins (MTs) are low-molecular-weight metal binding proteins rich in cysteine residues (14). They are found in most eukaryotic organisms and their induction is an important response to heavy metal poisoning. They are able to bind a variety of heavy metals, including nickel, which is bound at a 1:7 molar ratio [mole of MT/moles of Ni(II)].
MATERIALS AND METHODS
Construction of a Ni(II)-bioaccumulating strain.
Plasmid pUEF202 (19) was digested with the restriction enzymes PstI and KpnI, and the resultant 2.5-kb fragment containing nixA was ligated between the PstI and KpnI sites of pK187 (13) to produce pKNA. E. coli JM109 cells were transformed with either pKNA or pK187 by electroporation (7), and transformed cells were subsequently transformed with either pGPMT (6) or pGEX-2T (23) (Table 1). Transformants were selected on Luria-Bertani (LB) plates containing the appropriate antibiotics (50 μg of ampicillin per ml and/or 25 μg of kanamycin per ml).
TABLE 1.
Plasmids and strains
| Plasmid(s) | Host | Plasmid description |
|---|---|---|
| pUEF202 | SE5000 | pBluescript containing the nixA gene from H. pylori, Ampr19 |
| pK187 | TX1 | p15a replicon, Kanr, lac promoter, medium copy number 13 |
| pKNA | JM109 | 2.5-kb nixA gene from pEUF202 cloned into pK187, Kanr |
| pGEX-2T | JM109 | Codes for GST ColE1 origin Ampr, lac promoter 23 (Pharmacia) |
| pGPMT | JM109 | Codes for GST-PMT ColE1 origin Ampr, lac promoter 6 |
| pGPMT + pKNA | JM109 | Codes for GST-MT fusion protein and Ni(II) transporter, Kanr Ampr |
Ni(II) bioaccumulation.
Cells harboring the above-mentioned plasmids were grown overnight in LB broth containing the appropriate antibiotics, and the following morning they were diluted to an optical density at 600 nm (OD600) of 0.05 with fresh medium and grown at 37°C with vigorous shaking. When the OD600 of a culture reached 0.50 to 0.70, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1.0 mM, and growth was continued for 3 h; then the cells were harvested by centrifugation at 4°C, washed, and resuspended to an OD600 of 1.0 in 10 mM phosphate buffer (pH 7.0) containing 30 μg of chloramphenicol per ml, 50 μg of ampicillin per ml, and 25 μg of kanamycin per ml, and Ni(II) was added to a final concentration of 10 μM. This concentration was chosen because it falls in the range reported for polluted water, sanitary sewage, and industrial wastewater (16). Cells were incubated with Ni(II) at 37°C for 1 h and harvested by centrifugation at 4°C. The pellets were washed twice with cold phosphate buffer, lyophilized, and digested in 70% nitric acid at 42°C for 24 h. Samples were diluted to a nitric acid concentration of 10 to 15% and analyzed for nickel content by flame atomic absorption spectrophotometry on a Perkin-Elmer 2380 atomic absorption spectrophotometer.
RESULTS
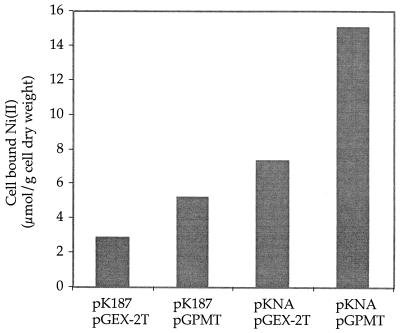
As shown in Fig. 1, E. coli cells without a Ni(II) transport system accumulated the smallest amount of nickel. The accumulation was probably due to the E. coli magnesium transporter (12). Cells expressing the transport system alone had a twofold-higher accumulation, while cells with a transport system and PMT had a fourfold-higher accumulation. Thus, both a transport system and PMT are required to accumulate the highest level of Ni(II). Addition of glucose (data not shown) did not have a significant impact on accumulation, indicating that accumulation does not require an energy source. In more complex media, accumulation was lower. LB broth contains amino acids and organic components that form complexes with metals (3), while M9 medium has a number of salts. Nickel uptake is an energy-dependent process (1, 15). However, addition of the energy poison 2,4-dinitrophenol (2 mM) inhibited Ni(II) bioaccumulation by only 20% (data not shown).
FIG. 1.
Ni(II) bioaccumulation by E. coli cells carrying different plasmids. Induced JM109 cells containing the indicated plasmids were resuspended in phosphate buffer containing 30 μg of chloramphenicol per ml and 10 μM Ni(II) and shaken at 37°C for 1 h.
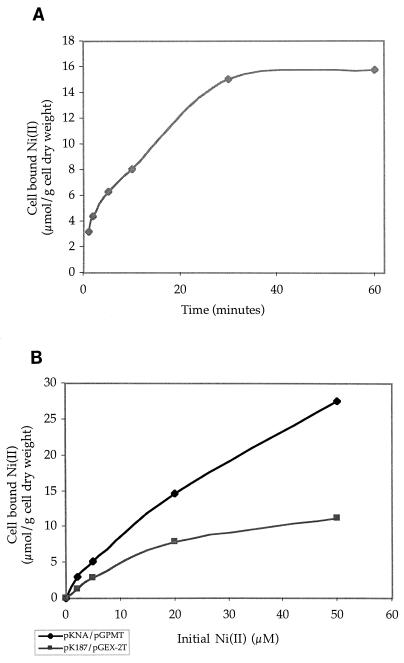
In induced E. coli JM109(pGPMT) cells, PMT accounts for 25% of total protein expression (5). Based on published data on E. coli protein expression (21), the theoretical maximum intracellular Ni(II) binding potential of induced cells is 28 μmol/g of cells (dry weight). To determine the saturation point of cells harboring pKNA and pGPMT, accumulation was measured over a range of initial concentrations and time points. There was a rapid increase in cell-bound Ni(II) in the first 20 min after metal addition (Fig. 2A) but little difference in accumulation between cells incubated for 30 and 60 min. The initial binding (within 1 min) is thought to be due to surface adsorption. Increases in cell-bound Ni(II) beyond that point are due to intracellular bioaccumulation.
FIG. 2.
(A) Time course of Ni(II) bioaccumulation. Induced cells containing pKNA and pGPMT were resuspended in phosphate buffer containing 10 μM Ni(II) and were shaken at 37°C for the indicated amounts of time. (B) Ni(II) bioaccumulation isotherms. Induced cells expressing both a nickel transporter and PMT-GST fusion protein (pKNA/pGPMT) and control cells expressing neither (pK187/pGEX-2T) were resuspended in phosphate buffer at various levels of Ni(II). Cells were shaken at 37°C and harvested after 1 h.
Cells were tested by examining accumulation from solutions containing different levels of Ni(II) (Fig. 2B). Cells expressing both the transporter and PMT accumulated Ni(II) at low concentrations (0 to 5 μM), and approximately 60% of the Ni(II) was removed. At the highest concentration, 28 μmol of Ni(II) was bound per g of cells (dry weight). Though a small amount was due to surface adsorption, the value approached the maximum binding capacity of the intracellular GST-PMT. Control cells had a smaller increase. Accumulation by these cells indicates surface binding and accumulation through the endogenous transporter. Since Ni(II) is toxic to bacteria only at high levels, control cells are able to accumulate a significant amount of Ni(II), peaking at 11 μmol of Ni(II) per g of cells (dry weight).
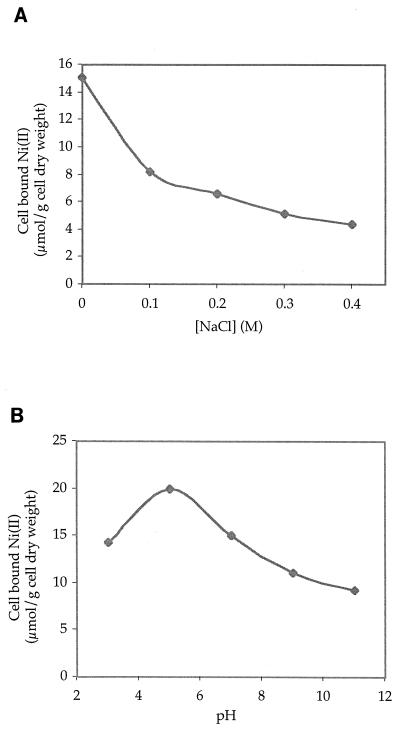
Na(I) and Mg(II) are common in natural waters; average river concentrations have been reported to be 0.26 and 0.18 mM, respectively (11), and in contaminated areas, these values can be much higher. Na(I) levels of 40 mM and Mg(II) levels of 26 mM have been measured in coal mine tailings and acid mine waters (10). Existing techniques, such as those using biosorbents and ion-exchange resins, are very sensitive to ionic strength, since a concentration of 150 mM sodium chloride can diminish the accumulation capability of biosorbents by over 90% (2). Ni(II) accumulation assays were performed with E. coli at increasing concentrations of sodium chloride (Fig. 3A). There was a decrease in Ni(II) accumulation, but not to the extent seen with biosorbents; at 100 mM NaCl the cells retained 55% of their accumulation capability, making them potentially useful as cleanup agents at this NaCl level.
FIG. 3.
(A) Effect of ionic strength on Ni(II) bioaccumulation. Induced cells containing pKNA and pGPMT were resuspended in phosphate buffer containing 10 μM Ni(II) and the indicated concentrations of NaCl. Cells were shaken at 37°C and harvested after 1 h. (B) Effect of pH on Ni(II) bioaccumulation. Induced cells containing pKNA and pGPMT were resuspended in phosphate buffer adjusted to the indicated pH and containing 10 μM Ni(II). Cells were shaken at 37°C and harvested after 1 h.
The E. coli Mg(II) transporter, which takes up Ni(II), is inhibited by Mg(II) (24). Bioaccumulation was tested in phosphate buffer containing different concentrations of MgCl2, and it was observed that the addition of 20 mM MgCl2 dramatically inhibited Ni(II) accumulation (by 80%). However, higher MgCl2 concentrations did not cause much additional inhibition. The high level of inhibition suggests that Mg(II) inhibits uptake not only through the endogenous transporter but also through the high-affinity Ni(II) system.
Dissolved organic matter, synthetic complexing agents, and other chelators are frequently found in contaminated and natural waters. Though they are far less abundant than inorganic ligands, such as Cl− and SO42−, they play an important role in metal uptake. These agents are able to form tight complexes with metals, which may decrease their bioavailability. Metals also adsorb to suspended particulate matter, which may affect their phase partitioning, mobility, and availability and inhibit metal recovery by treatments such as ion exchange and biosorption.
Previous studies have shown that genetically engineered bacteria are able to accumulate Hg(II) even in the presence of chelators (6). Ni(II) was added to induced cells in mixtures containing either 0.5 mM EDTA or 0.5 mM sodium citrate. EDTA completely inhibited accumulation, while sodium citrate inhibited accumulation by 80%. The log of the equilibrium stability constant for the formation of the Ni-EDTA complex is 20.4 (20), which is 13 orders of magnitude greater than that of nickel citrate (20). The differences between the Ni(II)-accumulating strain and the Hg(II)-accumulating strain may be due to the form in which the metal-EDTA complex is transported. The NixA protein has a much higher affinity for Ni(II) than the MerT-MerP system has for mercury (22), but it is inhibited by EDTA. This suggests that transport in the Hg(II)-accumulating strain may involve uptake of the Hg(II)-EDTA complex. Another possibility is that the affinity of MT for Ni(II) is much lower than for Hg(II), since the affinity of MT for Ni(II) has not been reported.
The process of accumulation and uptake of metals by organisms is highly dependent on their bioavailability, which is greatly influenced by pH. At a basic pH stable metal complexes, such as hydroxides, sulfides, and carbonates, form, making the metal less available for removal by ion-exchange resins or biosorbents. A low pH leads to increased mobility of metals in their cationic form. However, acidic conditions can also lead to competition for binding sites and decrease adsorption of heavy metals to biosorbents.
The effect of pH on bioaccumulation by the genetically engineered E. coli cells was tested across a broad range of pH values (between 3 and 9) (Fig. 3B). Induced cells were resuspended in phosphate buffer which had been previously adjusted to the desired pH, Ni(II) was added to a concentration of 10 μM, and the cells were shaken at 37°C for 1 h. Significant bioaccumulation was seen at pH values between 3 and 9, with a peak at pH 5. The transport system encoded by nixA is from H. pylori, a pathogen of the gastrointestinal tract. Given the acidic conditions of the stomach, it is not surprising that the genetically engineered cells were able to accumulate Ni(II) at low pH. These results further suggest that bioaccumulation is indeed an intracellular process, as surface adsorption is likely to be inhibited at low pH.
DISCUSSION
Like the genetically engineered Hg(II)-accumulating E. coli strains, the Ni-accumulating strain was able to accumulate Ni(II) from diluted (<10 μM) solutions, making it especially promising for situations where low levels of Ni(II) need to be decreased further. Neither the Hg(II)- nor the Ni(II)-accumulating strains required a carbon source, nor were they inhibited by dinitrophenol. It is not known for certain why accumulation does not require an energy source while transport does. However, the very high affinity of metallothionein for the metals could mean that facilitative diffusion, not active transport, is what is required for accumulation.
In both strains of E. coli, accumulation was effectively carried out over a wide pH range, unlike what is observed in most current methods. The nickel-accumulating strain was especially effective at low pH, which is significant since a major source of nickel pollution is battery disposal. In addition, Ni(II) uptake was only weakly inhibited by 100 mM NaCl. These results suggest that genetically engineered E. coli strains have some desirable properties for bioremediation not found in other cleanup agents.
However, the Ni(II)-accumulating organisms were not as resistant to ambient conditions as the genetically engineered Hg(II)-accumulating strain of E. coli. In the nickel-accumulating strain, accumulation was inhibited at higher salt concentrations. Accumulation was also inhibited by Mg(II) and agents that form strong metal complexes. This is in contrast to the mercury-accumulating strain, which was resistant to 400 mM NaCl and retained 95% of its Hg(II)-bioaccumulating activity in the presence of metal chelators (6). For both the mercury- and the nickel-accumulating strains, the E. coli hosts and the GST-PMT plasmids were the same, suggesting that the differences are due to the transporters.
REFERENCES
- 1.Baudet C, Sprott G D, Patel G B. Adsorption and nickel uptake in Methanothrix concilii. Arch Microbiol. 1988;150:338–342. [Google Scholar]
- 2.Chang J, Hong J. Biosorption of mercury by the inactivated cells of Pseudomonas aeruginosa PU21 (Rip64) Biotechnol Bioeng. 1994;44:999–1006. doi: 10.1002/bit.260440817. [DOI] [PubMed] [Google Scholar]
- 3.Chang J, Hong J, Ogunseitan O A, Olson B H. Interaction of mercuric ions with the bacterial growth medium and its effects on enzymatic reduction of mercury. Biotechnol Prog. 1993;9:526–532. [Google Scholar]
- 4.Chen C Y, Lin T H. Nickel toxicity to human term placenta: in vitro study on lipid peroxidation. J Toxicol Environ Health Part A. 1998;54:37–47. doi: 10.1080/009841098159015. [DOI] [PubMed] [Google Scholar]
- 5.Chen S, Wilson D B. Construction and characterization of Escherichia coli genetically engineered for bioremediation of Hg2+-contaminated environments. Appl Environ Microbiol. 1997;63:2442–2445. doi: 10.1128/aem.63.6.2442-2445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S, Wilson D B. Genetic engineering of bacteria and their potential for Hg(II) bioremediation. Biodegradation. 1997;8:97–103. doi: 10.1023/a:1008233704719. [DOI] [PubMed] [Google Scholar]
- 7.Dower W J, Miller J F, Van der Lelie D, Baeyens W, Mergeay A. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6146. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eccles H. Removal of heavy metals from effluent streams—why select a biological process? Int Biodeterior Biodegrad. 1995;35:5–16. [Google Scholar]
- 9.Fulkerson J F, Jr, Garner R M, Mobley H L T. Conserved residues and motifs in the NixA protein of Helicobacter pylori are critical for the high affinity transport of nickel ions. J Biol Chem. 1998;273:235–241. doi: 10.1074/jbc.273.1.235. [DOI] [PubMed] [Google Scholar]
- 10.Gang M A, Langmuir D. Proceedings of the 5th Symposium on Coal Mine Drainage Research. Washington, D.C.: National Coal Mine Association; 1974. Controls on heavy metals in surface and ground waters affected by coal mine drainage; pp. 39–69. [Google Scholar]
- 11.Holland H D. The chemistry of the atmosphere and oceans. New York, N.Y: John Wiley & Sons; 1978. [Google Scholar]
- 12.Jasper P, Silver S. Magnesium transport in microorganisms. In: Weinberg E D, editor. Microorganisms and minerals. New York, N.Y: Marcel Dekker; 1977. pp. 7–47. [Google Scholar]
- 13.Jobling M G, Holmes R K. Construction of vectors with the p15a replicon, kanamycin resistance, inducible lacZ alpha and pUC18 or pUC19 multiple cloning sites. Nucleic Acids Res. 1990;18:5315–5316. doi: 10.1093/nar/18.17.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kagi J H. Overview of metallothionein. Methods Enzymol. 1991;205:613–626. doi: 10.1016/0076-6879(91)05145-l. [DOI] [PubMed] [Google Scholar]
- 15.Kaltwasser H, Frings W. Transport and metabolism of nickel in microorganisms. In: Nriagu J O, editor. Nickel in the environment. New York, N.Y: John Wiley & Sons; 1980. pp. 463–491. [Google Scholar]
- 16.Klein L A, Lang M, Nash N, Kirscher S L. Sources of metals in New York City wastewater. J Water Pollut Control Fed. 1974;46:2653–2662. [PubMed] [Google Scholar]
- 17.Kusano T, Ji G, Inoue C, Silver S. Constitutive synthesis of a transport function encoded by the Thiobacillus ferrooxidans merC gene cloned in Escherichia coli. J Bacteriol. 1990;172:2688–2692. doi: 10.1128/jb.172.5.2688-2692.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin K C, Chou I N. Studies on the mechanisms of nickel ion-induced cell injury: effects of nickel ion on microtubules. Toxicol Appl Pharmacol. 1990;106:209–221. doi: 10.1016/0041-008x(90)90241-l. [DOI] [PubMed] [Google Scholar]
- 19.Mobley H L T, Garner R M, Bauerfeind P. Helicobacter pylori nickel-transport gene nixA: synthesis of catalytically active urease in Escherichia coli independent of growth conditions. Mol Microbiol. 1995;16:97–109. doi: 10.1111/j.1365-2958.1995.tb02395.x. [DOI] [PubMed] [Google Scholar]
- 20.Morel F M M. Principles of aquatic chemistry. New York, N.Y: John Wiley & Sons; 1983. [Google Scholar]
- 21.Neidhardt F C, Umbarger H E. Chemical composition of Escherichia coli. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 13–16. [Google Scholar]
- 22.Sahlmen L, Granstroem Skaerfstad E. Mercuric ion binding abilities of merP variants containing only one cysteine. Biochem Biophys Res Commun. 1993;196:583–588. doi: 10.1006/bbrc.1993.2289. [DOI] [PubMed] [Google Scholar]
- 23.Smith D B, Johnson N J. Single step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 24.Webb M. Interrelationships between the utilization of magnesium and the uptake of other bivalent cations by bacteria. Biochim Biophys Acta. 1970;222:428–439. doi: 10.1016/0304-4165(70)90133-9. [DOI] [PubMed] [Google Scholar]