Abstract
Harmful cyanobacterial blooms (cyanoHABs) cause recurrent toxic events in global watersheds. Although public health agencies monitor the causal toxins of most cyanoHABs and scientists in the field continue developing precise detection and prediction tools, the potent anticholinesterase neurotoxin, guanitoxin, is not presently environmentally monitored. This is largely due to its incompatibility with widely employed analytical methods and instability in the environment, despite guanitoxin being among the most lethal cyanotoxins. Here, we describe the guanitoxin biosynthesis gene cluster and its rigorously characterized nine-step metabolic pathway from l-arginine in the cyanobacterium Sphaerospermopsis torques-reginae ITEP-024. Through environmental sequencing data sets, guanitoxin (gnt) biosynthetic genes are repeatedly detected and expressed in municipal freshwater bodies that have undergone past toxic events. Knowledge of the genetic basis of guanitoxin biosynthesis now allows for environmental, biosynthetic gene monitoring to establish the global scope of this neurotoxic organophosphate.
Graphical Abstract
INTRODUCTION
Freshwater is essential for drinking and agriculture, yet potable watersheds are increasingly impacted by the undesirable high-density growth of algae and/or cyanobacteria.1–3 Understanding, monitoring, and remediating harmful algal/cyanobacterial blooms (HABs/cyanoHABs) and their associated toxins are essential to reducing their societal impact.4 Recent scientific and technological advances continue to improve environmental cyanoHAB detection and prediction;5–7 however, the vast cyanotoxin structural chemodiversity creates challenges in their comprehensive detection and quantification using standard analytical chemistry assays. In contrast, quantitative molecular biological detection of biosynthetic genes via PCR provides a multiplexable and cost-effective monitoring strategy to identify the toxic potential of blooms independent of active toxin synthesis.8 The biosynthetic gene clusters (BGCs) for important freshwater cyanotoxins like microcystin,9 cylindrospermopsin,10 saxitoxin,11 and anatoxin-a12 have been defined and applied toward detection over the past decades. However, the biosynthetic pathway and genes for guanitoxin (1), the only known natural organophosphate neurotoxin, have yet to be described.
Previously known as anatoxin-a(s),13 guanitoxin is an irreversible inhibitor of acetylcholinesterase,14 sharing an identical mechanism of action with organophosphates like the synthetic chemical warfare agent sarin and the banned pesticide parathion (Figure 1A). Guanitoxin induces acute neurological toxicity that can lead to rapid death, showing comparable lethality (LD50 = 20 μ/kg i.p.)15 to saxitoxin, the most potent known cyanotoxin. Sporadic detection in the Americas,15 Europe,16 and Middle East17 coupled with bloom-related animal deaths consistent with guanitoxin exposure suggests that this toxin could be an under-recognized threat in global watersheds.
Figure 1.
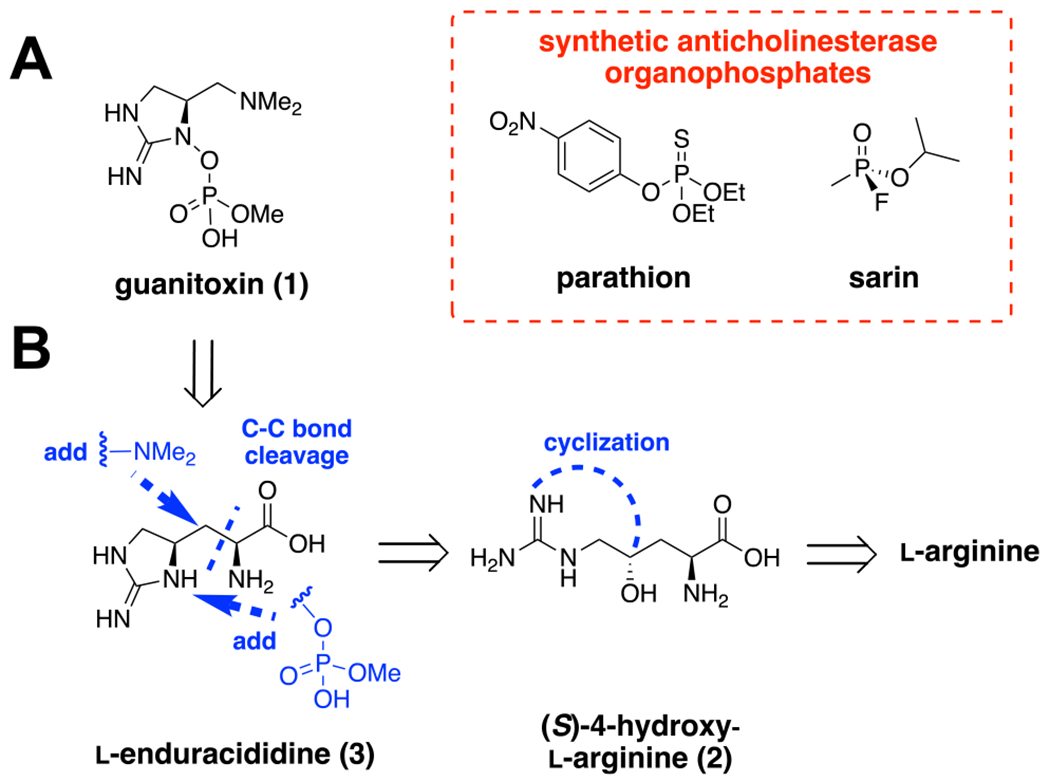
(A) Guanitoxin (1) is a potent cyanobacterial organophosphate neurotoxin with an anticholinesterase mechanism of action comparable to the pesticide parathion and the chemical warfare agent sarin. (B) Retrobiosynthetic proposal to 1 from l-arginine via previously isolated cyanobacterial metabolites 2 and 3.
While its unique pharmacology18 and chemical structure19 have been known for decades, guanitoxin remains largely unmonitored in the environment; this is mainly due to its incompatibility with commonly used analytical detection methods and chemical instability.20 Although previous l-arginine-derived metabolites have been isolated from guanitoxin-producing cyanobacteria and incorporated in vivo via stable isotope labeling experiments (Figure 1B),21,22 a lack of knowledge regarding guanitoxin biosynthesis and accessibility to its stable metabolites as standards has hampered an understanding of its environmental significance. Herein, we describe the complete guanitoxin BGC and pathway, identify its diagnostic intermediates, validate them with synthetic and chemoenzymatic standards, and survey environmental metatranscriptomic and metagenomic data sets to reveal the global prevalence of guanitoxin biosynthetic capability in both rural and populated watersheds. We anticipate that these discoveries will accelerate the understanding of the distribution and impact of this lethal, yet essentially unmonitored toxin.
RESULTS AND DISCUSSION
Discovery of the Guanitoxin Biosynthetic Gene Cluster.
We sequenced the genome of the guanitoxin-producing cyanobacterial strain Sphaerospermopsis torques-reginae ITEP-024 (GenBank accession no. CP080598)28 acquired from a toxic cyanoHAB in the Tapacura Reservoir, Pernambuco, Brazil (Figures S1 and S2).29 Bioinformatic analysis of the resulting 5.24 Mbp genome with prediction tools such as antiSMASH23 identified 11 natural product BGCs (Figure 2A and Table S1). None, however, were consistent with guanitoxin biogenesis involving predicted amino acid and organophosphate biochemistry.
Figure 2.
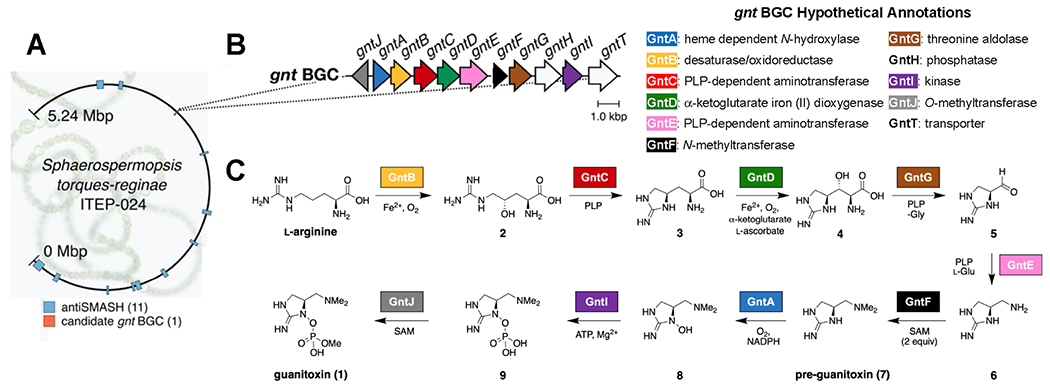
Discovery of a candidate gnt biosynthetic gene cluster (BGC) via sequencing a guanitoxin-producing cyanobacterium. (A) Annotation of the assembled 5.24 Mbp Sphaerospermopsis torques-reginae ITEP-024 genome using antiSMASH v6.023 detected 11 candidate BGCs, while a single candidate guanitoxin BGC was identified via colocalization of relevant candidate enzyme activities. The figure was produced with Circos v0.69-8.24 The background S. torques-reginae image25 is used with permission from Dr. Vera Werner. (B) Organization of the guanitoxin BGC. The gnt locus figure was designed from NCBI accession CP080598.1 (this work) using Clinker v0.0.21.26 gnt BGC deposited under MIBiG27 accession number BGC0002148. (C) Guanitoxin biosynthetic pathway in S. torques-reginae ITEP-024. Individual Gnt enzymes are color-coded after their encoding gnt gene as per (B).
To identify candidate guanitoxin BGCs, we initially focused on genes associated with the arginine metabolites (S)-4-hydroxy-l-arginine (2) and l-enduracididine (3). Both 2 and 3 were previously isolated from guanitoxin-producing Anabaena flos-aquae21,22 and converted in vivo to guanitoxin via stable isotope labeling experiments (Figure 1B).22 We subsequently identified 3 in S. torques-reginae ITEP-024 extracts, further supporting its intermediacy in guanitoxin biosynthesis (Figure S3). As amino acid 3 and its epimers are incorporated in actinobacterial non-ribosomal peptide antibiotics such as mannopeptimycin,30 teixobactin,31 and enduracidin32 and constructed via pyridoxal-5′-phosphate (PLP)-based enzymatic transformations, we queried the S. torques-reginae ITEP-024 genome for homologous genes to known 3-producing enzymes mppP/mppQ/mppR from the mannopeptimycin biosynthetic pathway.33–35 While this strategy did not locate close homologs, it did identify a candidate BGC encoding three PLP-dependent enzymes embedded within a unique 12.5 kb gene cluster consisting of 10 metabolic enzymes (gntA-J) and a putative transporter (gntT) (Figure 2B). This candidate gnt BGC is unique and not found in any reference cyanobacterial genome currently available in the NCBI database nor in the metagenome-assembled genomes (MAGs) of the JGI Earth Microbiome Project.36 While most of the encoded enzymes did not have high sequence similarity to characterized homologs (Figure S4), initial sequence and structural homology analyses identified multiple biosynthetic functions consistent with guanitoxin assembly; these included oxidation (gntA, gntB, gntD), PLP-dependent reactions (gntC, gntE, gntG), S-adenosylmethionine (SAM)-dependent methylations (gntF and gntJ), and phosphate biochemistry (gntI and gntH) (Figures 2B and S4). We therefore constructed a putative guanitoxin biosynthetic pathway based on previously isolated chemical intermediates and gnt bioinformatic annotations (Figure 2C). To experimentally link candidate Gnt enzymes to toxin biosynthesis, we synthesized the gnt genes and then expressed and purified the majority to homogeneity as Escherichia coli codon-optimized N-terminal His6 fusion proteins (Figure S5).
Reconstitution of the Complete Guanitoxin Biosynthetic Pathway.
We first sought to confirm a diagnostic guanitoxin biosynthetic reaction and focused on the presumed N,N-dimethylation of 6 to pre-guanitoxin (7) by predicted N-methyltransferase GntF. The chemical structures of putative intermediates 6 and 7 are novel molecules yet to be observed in other characterized biosynthetic pathways, so their interconversion would strongly implicate the gnt BGC. Intermediates 6 and 7 were chemically synthesized from (S)-Garner’s aldehyde in six and five steps, respectively; upon incubation of synthetic 6 with GntF in the presence of excess SAM, we observed its efficient conversion to dimethylated 7 following hydrophilic interaction liquid chromatography mass spectrometry (HILIC-MS) analyses (Figure 3). We also observed 7 in S. torques-reginae ITEP-024 extracts, lending further physiological relevance to its role in guanitoxin biogenesis (Figure S3). With this encouraging result in hand, we next sought to connect these key biosynthetic intermediates to their primary metabolic precursors and the mature toxin itself.
Figure 3.
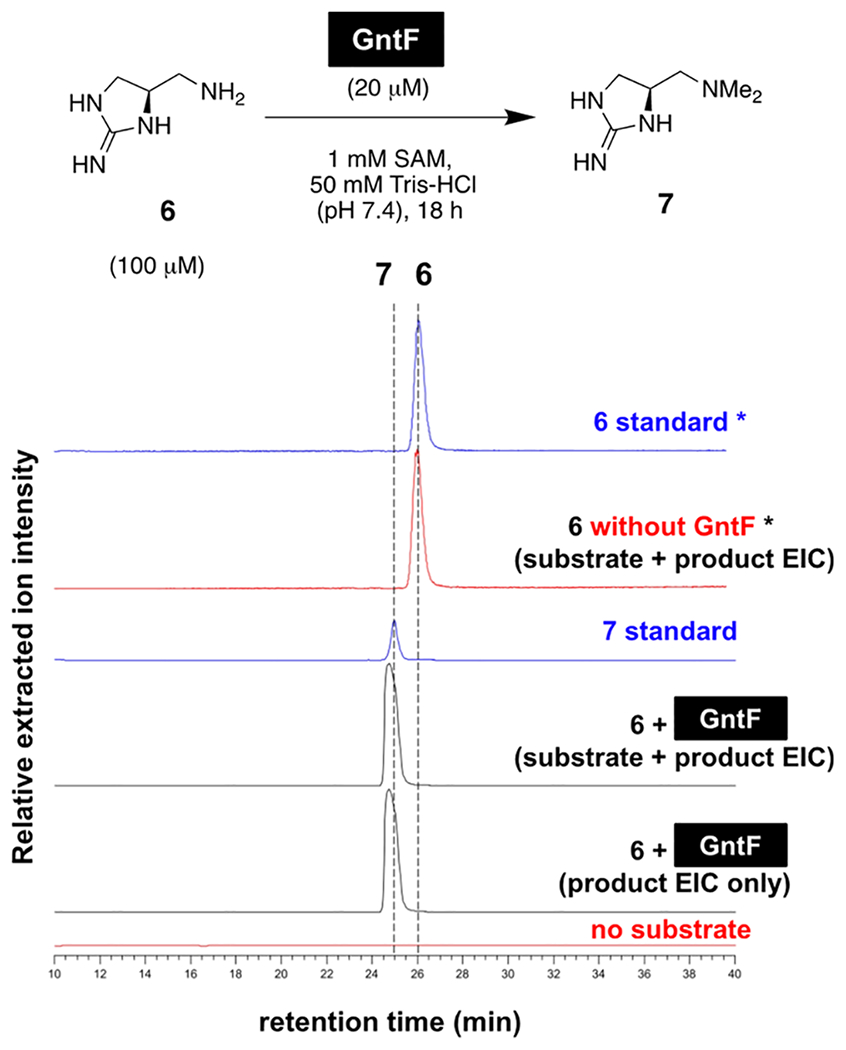
GntF N,N-dimethylates primary amine substrate 6 to form pre-guanitoxin (7) in vitro. Relative intensities of positive mode extracted ion chromatograms were extracted from HILIC-MS traces for both GntF substrate 6 and product 7 masses as appropriate ([M + H]+ (115.0978, 143.1291) ± 0.0100 m/z, respectively). Asterisks indicate that the MS intensities are increased 10-fold relative to other traces for improved visualization of compounds with variable ionization efficiencies.
Multiple characterized biosynthetic pathways employ PLP-dependent enzymes to explore their dehydrogenation, oxidation, and transamination chemistries on l-arginine.37 However, none of the three PLP-dependent enzymes (GntC, GntE, GntG) nor the soluble α-ketoglutarate Fe2+-dioxygenase GntD showed any clear activity toward this amino acid (Figures S6 and S7). We therefore initiated our in vitro enzymology efforts with known biosynthetic intermediates 2 and 3, which were chemically synthesized following adaptation of previously established synthetic procedures.38,39 To our surprise, GntC catalyzed the highly diastereoselective PLP-dependent cyclodehydration of 2 into 3, without any additional enzymes or cosubstrates (Figure S8). The stereochemical differences between the chemically synthesized substrate and product diastereomers were magnified via 1-fluoro-2,4-dinitrophenyl-5-l-alanine amide (l-FDAA, Marfey’s reagent) derivatization and ultra-performance liquid chromatography mass spectrometry (UPLC-MS) analysis, enabling us to determine that GntC was highly diastereoselective for the (S)-hydroxy stereoisomer of 4-hydroxy-l-arginine while exclusively generating 3 over its l-allo-enduracididine epimer (Figure S8). While transmembrane protein GntB proved intractable to soluble protein expression, the in vivo expression of a gntB-gntC-containing plasmid in E. coli successfully produced 3 (Figures S9 and S10), thus corroborating GntB’s initiating role in the construction of 2. Cyanobacterial 3 biosynthesis presents a divergent reaction strategy to known actinobacterial cyclic arginine biosyntheses that either rely on multiple PLP-dependent enzymes33–35 or form six-membered capreomycidine rings from linear β-hydroxylated arginine precursors (Figure S11).40–43
We next established GntD as an enduracididine β-hydroxylase in converting 3 to 4 in the presence of Fe2+, α-ketoglutarate, and l-ascorbate as previously reported for the Streptomyces enzyme MppO (Figure S12).44 To connect 4 with diagnostic intermediate 6, we proposed a two-enzyme cascade with aldolase GntG and transaminase GntE excising glycine and converting the resultant aldehyde to a primary amine, respectively. We validated this GntE/GntG cascade by performing the reaction in reverse to construct 4 from 6, glycine, α-ketoglutarate, PLP, and the two PLP-dependent enzymes (Figure S13). Applying all four biosynthetic enzymes and their requisite cofactors and cosubstrates simultaneously in one pot converted 2 to 6 and the glycine byproduct, with intermediate aldehyde 5 not observed due to its presumed instability and the absence of a primary amine for l-FDAA derivatization (Figures 4 and S14). Exclusion of individual enzymes halted progression along this four-step pathway in a manner consistent with our biosynthetic proposal (Figure S14).
Figure 4.
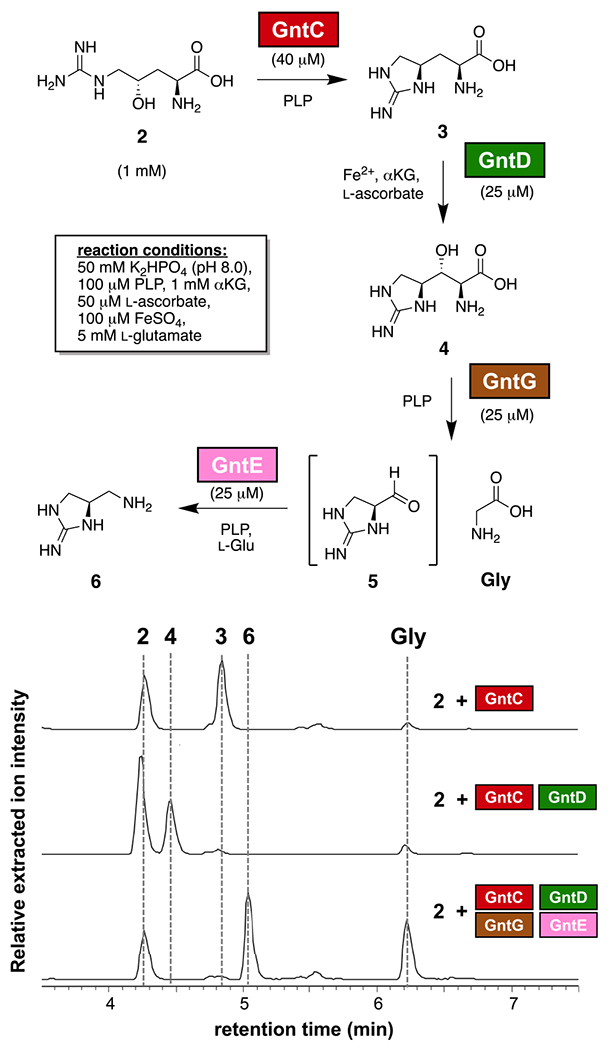
Characterization of GntC/GntD/GntG/GntE biosynthetic enzymes with intermediate 2. Relative intensities of positive mode extracted ion chromatograms were extracted from UPLC-MS traces following 1-fluoro-2,4-dinitrophenyl-5-l-alamne amide (l-FDAA; Marfey’s reagent) derivatization of primary amine-containing intermediates 2, 3, 4, 6, and glycine ([M + H]+ (443.16; 425.15; 441.15; 367.15; 328.09) ± 0.20 m/z, respectively) after incubation of 2 with GntC, GntC/GntD or GntC/GntD/GntG/GntE and all necessary cofactors and cosubstrates.
After assembling the guanitoxin carbon backbone via six biosynthetic transformations, we focused on the enzymatic construction of the N-hydroxylated methyl phosphate that functions as the anticholinesterase pharmacophore (Figure 5A). In the presence of the unusual heme-containing GntA and excess NADPH under ambient aerobic conditions, we observed the selective mono-oxygenation of tertiary amine 7 to product 8 (Figure S15). This reaction is reminiscent of the homologous heme-oxygenase dcsA gene-encoded Nω-hydroxylation of l-arginine that initiates a specialized pathway to d-cycloserine in Streptomyces bacteria.45 Ultrafiltration of the GntA reaction mixture, followed by the addition of kinase GntI, O-methyltransferase GntJ, and their cofactors and cosubstrates, generated phosphorylated intermediate 9 and guanitoxin, with identical HILIC-MS properties to those observed in S. torques-reginae ITEP-024 extracts (Figures 5B and S3, S16, and S17). GntA, GntI, and GntJ in vitro reactions were performed following the stepwise addition of individual enzymes and cosubstrates to the reaction mixture, largely due to the differing ionic strength requirements of N-hydroxylase GntA and kinase GntI. We subsequently investigated recombinant human acetylcholinesterase inhibition with in situ generated 8, 9, and guanitoxin (Figure 5C). Only the mature neurotoxin possessed inhibitory activity, establishing that the GntJ-catalyzed O-methylation is crucial for toxicity.
Figure 5.
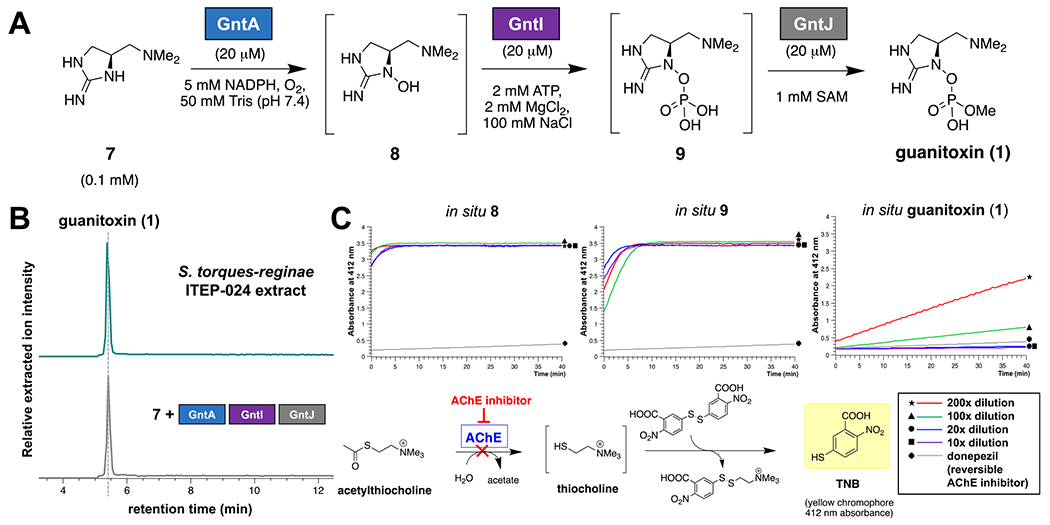
(A) GntA, GntI, and GntJ construct the anticholinesterase organophosphate pharmacophore of guanitoxin. (B) Relative intensities of positive mode extracted ion chromatograms were extracted from HILIC-MS for guanitoxin ([M + H]+ 253.1060 ± 0.0100 m/z) following extraction from S. torques-reginae ITEP-024 and in vitro incubation of intermediate 7 with GntA/GntI/GntJ and all necessary cofactors and cosubstrates. (C) Acetylcholinesterase (AChE) inhibition was assessed via a Bio Vision Acetylcholinesterase Inhibitor Screening Kit (Colorimetric), where AChE inhibition is coupled to the decreased formation of the yellow 5-thio-2-nitrobenzoic acid (TNB) chromophore (412 nm absorbance) via the scheme depicted. In vitro GntA/GntI/GntJ reactions were carried out as previously described, diluted between 10 and 200×, and analyzed for AChE inhibition. In situ generated 8 and 9 showed negligible AChE inhibition at all dilutions tested compared to the reversible inhibitor donepezil positive control (gray trace). However, potent AChE inhibition was observed following the addition of GntJ (in situ guanitoxin) and showed a decreasing inhibitory effect at a higher reaction dilution, highlighting the significance of O-methylation for biological activity.
The details of the nine-step enzymatic biosynthesis of guanitoxin from arginine represent just the second example of an in vitro reconstitution of a cyanotoxin after the recent total enzyme synthesis of aetokthonotoxin from another amino acid, tryptophan.46 Overall, our chemical, biochemical, and metabolomic analyses strongly connect guanitoxin biosynthesis to the gnt BGC and provide a genetic handle to investigate this cyanotoxin environmentally.
Guanitoxin Biosynthetic Genes Are Present in Geographically Diverse Environments.
After validating the guanitoxin biosynthetic genes in S. torques-reginae ITEP-024, we next explored their environmental distribution. As we previously found no gnt BGC homologs in publicly available assembled genomes and metagenomes, we instead queried raw, unassembled environmental sequencing data. Specifically, we searched all available (108,465) NCBI Sequence Read Archive (SRA) metagenomic (metaG) and metatranscriptomic (metaT) data sets using the SearchSRT tool. This search identified 13 data sets with at least one read aligning to the gnt BGC with ≥90% translated-nucleotide to protein sequence identity, even though the computationally efficient SearchSRA tool is sensitivity-limited to detect only highly abundant (metaG) or highly expressed (metaT) genes. Through independent de novo assembly, we confirmed the complete or nearly complete gnt BGC from six of these SRA data sets (Figure 6), including two metaT samples. In the metaT samples, the gnt BGC was discovered as a polycistronic transcript including all genes except gntJ. This polycistronic structure suggested that even limited transcriptomic detection of the gnt genes could be diagnostic for active guanitoxin biosynthesis. Therefore, to explore the feasibility of metaT data for environmental gnt BGC surveys, we comprehensively queried all 2,610 freshwater metatranscriptomic SRA data sets using a custom full-sensitivity SRA search pipeline and ultimately identified 78 gnt BGC hits from Lake Erie, Ohio, USA; Lake Mendota, Wisconsin, USA; the Amazon River, Brazil; the Columbia River, Oregon, USA; and the Delaware River, Delaware, USA (Figure 6 and Table S2). These detections occur in independent sequencing data sets within the same watershed over multiple years, such as in Lake Mendota from 2010 to 2016, indicating the long-term undetected presence of guanitoxin biosynthetic capacity in publicly accessible freshwater sources.
Figure 6.
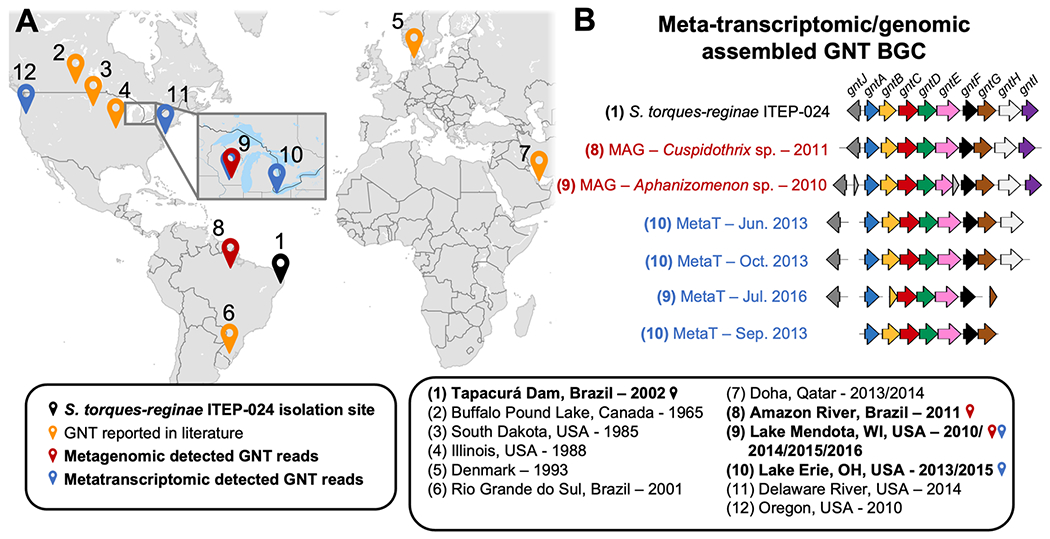
Environmental detection of guanitoxin biosynthetic capability through metagenomic and metatranscriptomic sequencing. (A) Geographic sites with literature reports of guanitoxin or detection of the gnt BGC through environmental sequencing data sets. (B) gnt BGC gene structure from metagenomic and metatranscriptomic de novo assemblies of environmental samples. gnt BGC genes are color-coded as per Figure 1B. The two metagenomic samples were successfully linked to their respective taxon of origin via a metagenome assembled genome (MAG) approach (see Figure S19 for details).
Intriguingly, we were unable to link the known guanitoxin producer genera as the source taxa of the two environmental metaG-derived gnt BGCs. Guanitoxin producer strains have been rarely isolated in the literature, and the taxonomy of guanitoxin producer genera has been unstable, suggesting that the full taxonomic extent of guanitoxin production is not yet known. To explore if other cyanobacterial genera could harbor gnt biosynthetic capacity, we linked the two environmental metagenome-derived gnt BGCs to their best-supported taxonomic origins. Via full MAG assembly, we connected the gnt BGCs from the Amazon River and Lake Mendota data sets to the Cuspidothrix and Aphanizomenon cyanobacterial genera, respectively (Figures S18 and S19). While these genera are known to produce other cyanotoxins such as anatoxin-a, microcystin, and cylindrospermopsin,47 they have not yet been reported to produce guanitoxin. However, their genomic potential warrants future environmental surveying for guanitoxin biosynthesis.
CONCLUSIONS
In conclusion, our detailed characterization of how guanitoxin is biochemically assembled from the standard amino acid l-arginine represents the first biosynthetic report of a natural organophosphate neurotoxin. Significantly, the discovery of the architecturally unique gnt BGC enabled us to identify environmental hotspots in unaware rural and populated areas for the potent yet unmonitored neurotoxin guanitoxin, suggesting that revised cyanoHAB monitoring protocols are warranted in public watersheds vulnerable to toxic cyanobacterial blooms.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge V. R. Werner (Museum of Natural Sciences, Porto Alegre, Brazil) for providing S. torques-reginae ITEP-024 and H.-W. Lee, L. Sanchez, and J. MacMillan (University of California, Santa Cruz) for assistance and maintenance of nuclear magnetic resonance, high-resolution mass spectrometry, and high-performance liquid chromatography machinery, respectively. We dedicate this manuscript to Prof. Shigeki Matsunaga (University of Tokyo) on his retirement for his pioneering work on the isolation and structure elucidation of guanitoxin, originally known as anatoxin-a(s).
Funding
This work was supported by grants and fellowships from the Sao Paulo Research Foundation (FAPESP #2013/50425-8 to M.F.F.; #2017/06869-0 to S.T.L.; #2011/08092-6 to D.O.A.), the National Institute of Environmental Health Sciences (NIEHS, R21-ES032056 to B.S.M.; F32-ES032276 to T.R.F.), the National Council for Scientific and Technological Development (CNPq 306803/2018-6 to M.F.F.), the Simons Foundation Fellowships of the Life Sciences Research Foundation (J.R.C.), the Brazilian Federal Agency for the Support and Evaluation of Graduate Education (E.D.), and the University of California, Santa Cruz, for startup funding and a Faculty Research Grant (S.M.K.M.).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c01424.
General materials and methods, –chemical synthesis, NMR, and compound characterization– (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.2c01424
The authors declare the following competing financial interest(s): A provisional patent application has been filed on the use of gnt enzyme coding sequences in environmental molecular diagnostic monitoring and detection (United States Provisional Patent Application #63/267,862).
Contributor Information
Stella T. Lima, Center for Nuclear Energy in Agriculture, University of São Paulo, Piracicaba, Sao Paulo 13416-000, Brazil; Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California, La Jolla, California 92093, United States; Present Address: Department of Chemistry and Biochemistry, University of North Carolina at Greensboro, NC, 27402, USA.
Timothy R. Fallon, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California, La Jolla, California 92093, United States
Jennifer L. Cordoza, Department of Chemistry and Biochemistry, University of California, Santa Cruz, California 95064, United States
Jonathan R. Chekan, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California, La Jolla, California 92093, United States; Department of Chemistry and Biochemistry, University of North Carolina at Greensboro, Greensboro, North Carolina 27402, United States.
Endrews Delbaje, Center for Nuclear Energy in Agriculture, University of Sao Paulo, Piracicaba, Sao Paulo 13416-000, Brazil.
Austin R. Hopiavuori, Department of Chemistry and Biochemistry, University of California, Santa Cruz, California 95064, United State
Danillo O. Alvarenga, Department of Biology, University of Copenhagen, Copenhagen DK 2100, Denmark.
Steffaney M. Wood, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California, La Jolla, California 92093, United States
Hanna Luhavaya, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California, La Jolla, California 92093, United States.
Jackson T. Baumgartner, Department of Chemistry and Biochemistry, University of California, Santa Cruz, California 95064, United States
Felipe A. Dörr, School of Pharmaceutical Sciences, University of São Paulo, Ribeirao Preto, Sao Paulo 05508-000, Brazil
Augusto Etchegaray, Center for Life Sciences, Graduate Program in Health Sciences, Pontifical Catholic University of Campinas (PUC-Campinas), Campinas, Sao Paulo 13087-571, Brazil.
Ernani Pinto, Center for Nuclear Energy in Agriculture, University of Sao Paulo, Piracicaba, Sao Paulo 13416-000, Brazil.
Shaun M. K. McKinnie, Department of Chemistry and Biochemistry, University of California, Santa Cruz, California 95064, United States.
Marli F. Fiore, Center for Nuclear Energy in Agriculture, University of São Paulo, Piracicaba, Sao Paulo 13416-000, Brazil.
Bradley S. Moore, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California, La Jolla, California 92093, United States; Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, California 92093, United States.
REFERENCES
- (1).Ho JC; Michalak AM; Pahlevan N Widespread global increase in intense lake phytoplankton blooms since the 1980s. Nature 2019, 574, 667–670. [DOI] [PubMed] [Google Scholar]
- (2).Huisman J; Codd GA; Paerl HW; Ibelings BW; Verspagen JMH; Visser PM Cyanobacterial blooms. Nat. Rev. Microbiol 2018, 16, 471–483. [DOI] [PubMed] [Google Scholar]
- (3).Plaas HE; Paerl HW Toxic cyanobacteria: A growing threat to water and air quality. Environ. Sci. Technol 2021, 55, 44–64. [DOI] [PubMed] [Google Scholar]
- (4).Jochimsen EM; Carmichael WW; An J; Cardo DM; Cookson ST; Holmes CEM; Antunes MB; de Melo Filho DA; Lyra TM; Barreto VST; Azevedo SMFO; Jarvis WR Liver failure and death after exposure to microcystins at a hemodialysis center in Brazil. N. Engl. J. Med 1998, 338, 873–878. [DOI] [PubMed] [Google Scholar]
- (5).Mishra DR; Kumar A; Ramaswamy L; Boddula VK; Das MC; Page BP; Weber SJ CyanoTRACKER: A cloud-based integrated multi-platform architecture for global observation of cyanobacterial harmful algal blooms. Harmful Algae 2020, 96, 101828. [DOI] [PubMed] [Google Scholar]
- (6).Rousso BZ; Bertone E; Stewart R; Hamilton DP A systematic literature review of forecasting and predictive models for cyanobacteria blooms in freshwater lakes. Water Res. 2020, 182, 115959. [DOI] [PubMed] [Google Scholar]
- (7).Seegers BN; Werdell PJ; Vandermeulen RA; Salls W; Stumpf RP; Schaeffer BA; Owens TJ; Bailey SW; Scott JP; Loftin KA Satellites for long-term monitoring of inland U.S. lakes: The MERIS time series and application for chlorophyll-a. Remote Sens. Environ 2021, 266, 112685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Baker L; Sendall BC; Gasser RB; Menjivar T; Neilan BA; Jex AR Rapid, multiplex-tandem PCR assay for automated detection and differentiation of toxigenic cyanobacterial blooms. Mol. Cell. Probes 2013, 27, 208–214. [DOI] [PubMed] [Google Scholar]
- (9).Tillett D; Dittmann E; Erhard M; von Döhren H; Börner T; Neilan BA Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: An integrated peptide–polyketide synthetase system. Chem. Biol 2000, 7, 753–764. [DOI] [PubMed] [Google Scholar]
- (10).Mihali TK; Kellmann R; Muenchhoff J; Barrow KD; Neilan BA Characterization of the gene cluster responsible for cylindrospermopsin biosynthesis. Appl. Environ. Microbiol 2008, 74, 716–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kellmann R; Mihali TK; Jeon YJ; Pickford R; Pomati F; Neilan BA Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria. Appl. Environ. Microbiol 2008, 74, 4044–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Méjean A; Mann S; Maldiney T; Vassiliadis G; Lequin O; Ploux O Evidence that biosynthesis of the neurotoxic alkaloids anatoxin-a and homoanatoxin-a in the cyanobacterium Oscillatoria PCC 6506 occurs on a modular polyketide synthase initiated by l-proline. J. Am. Chem. Soc 2009, 131, 7512–7513. [DOI] [PubMed] [Google Scholar]
- (13).Fiore MF; de Lima ST; Carmichael WW; McKinnie SMK; Chekan JR; Moore BS Guanitoxin, re-naming a cyanobacterial organophosphate toxin. Harmful Algae 2020, 92, 101737. [DOI] [PubMed] [Google Scholar]
- (14).Hyde EG; Carmichael WW Anatoxin-a(s), a naturally occurring organophosphate, is an irreversible active site-directed inhibitor of acetylcholinesterase (EC 3.1.1.7). J. Biochem. Toxicol 1991, 6, 195–201. [DOI] [PubMed] [Google Scholar]
- (15).Carmichael WW Health effects of toxin-producing cyanobacteria: “The CyanoHABs.”. Hum. Ecol Risk Assess. Int. J 2001, 7, 1393–1407. [Google Scholar]
- (16).Henriksen P; Carmichael WW; An J; Moestrup Ø Detection of an anatoxin-a(s)-like anticholinesterase in natural blooms and cultures of cyanobacteria/blue-green algae from Danish lakes and in the stomach contents of poisoned birds. Toxicon 1997, 35, 901–913. [DOI] [PubMed] [Google Scholar]
- (17).Chatziefthimiou AD; Richer R; Rowles H; Powell JT; Metcalf JS Cyanotoxins as a potential cause of dog poisonings in desert environments. Vet. Rec 2014, 174, 484–485. [DOI] [PubMed] [Google Scholar]
- (18).Carmichael WW; Gorham PR Anatoxins from clones of Anabaena flos-aquae isolated from lakes of western Canada. Verh. Int. Ver. Theor. Angew. Limnol 1978, 21, 285–295. [Google Scholar]
- (19).Matsunaga S; Moore RE; Niemczura WP; Carmichael WW Anatoxin-a(s), a potent anticholinesterase from Anabaena flos-aquae. J. Am. Chem. Soc 1989, 111, 8021–8023. [Google Scholar]
- (20).Fernandes K; Dörr FA; Pinto E Stability analyses by HPLC-MS of guanitoxin isolated from Sphaerospermopsis torques-reginae. J. Braz. Chem. Soc 2021, 32, 1559–1567. [Google Scholar]
- (21).Moore BS; Ohtani I; de Koning CB; Carmichael WW; Carmichael WW Biosynthesis of anatoxin-a(s). Origin of the carbons. Tetrahedron Lett. 1992, 33, 6595–6598. [Google Scholar]
- (22).Hemscheidt T; Burgoyne DL; Moore RE Biosynthesis of anatoxin-a(s). (2S,4S)-4-Hydroxyarginine as an intermediate. J. Chem. Soc., Chem. Commun 1995, 2, 205–206. [Google Scholar]
- (23).Blin K; Shaw S; Kloosterman AM; Charlop-Powers Z; van Wezel GP; Medema MH; Weber T antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Krzywinski M; Schein J; Birol İ; Connors J; Gascoyne R; Horsman D; Jones SJ; Marra MA Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Werner VR; Laughinghouse HD; Fiore MF; Sant’Anna CL; Hoff C; de Souza Santos KR; Neuhaus EB; Molica RJR; Honda RY; Echenique RO Morphological and molecular studies of Sphaerospermopsis torques-reginae (Cyanobacteria, Nostocales) from South American water blooms. Phycologia 2012, 51, 228–238. [Google Scholar]
- (26).Gilchrist CLM; Chooi Y-H Clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics 2021, 37, 2473–2475. [DOI] [PubMed] [Google Scholar]
- (27).Medema MH; Kottmann R; Yilmaz P; Cummings M; Biggins JB; Blin K; de Bruijn I; Chooi YH; Claesen J; Coates RC; Cruz-Morales P; Duddela S; Düsterhus S; Edwards DJ; Fewer DP; Garg N; Geiger C; Gomez-Escribano JP; Greule A; Hadjithomas M; Haines AS; Helfrich EJN; Hillwig ML; Ishida K; Jones AC; Jones CS; Jungmann K; Kegler C; Kim HU; Kötter P; Krug D; Masschelein J; Melnik AV; Mantovani SM; Monroe EA; Moore M; Moss N; Nützmann H-W; Pan G; Pati A; Petras D; Reen FJ; Rosconi F; Rui Z; Tian Z; Tobias NJ; Tsunematsu Y; Wiemann P; Wyckoff E; Yan X; Yim G; Yu F; Xie Y; Aigle B; Apel AK; Balibar CJ; Balskus EP; Barona-Gómez F; Bechthold A; Bode HB; Borriss R; Brady SF; Brakhage AA; Caffrey P; Cheng Y-Q; Clardy J; Cox RJ; De Mot R; Donadio S; Donia MS; van der Donk WA; Dorrestein PC; Doyle S; Driessen AJM; Ehling-Schulz M; Entian K-D; Fischbach MA; Gerwick L; Gerwick WH; Gross H; Gust B; Hertweck C; Höfte M; Jensen SE; Ju J; Katz L; Kaysser L; Klassen JL; Keller NP; Kormanec J; Kuipers OP; Kuzuyama T; Kyrpides NC; Kwon H-J; Lautru S; Lavigne R; Lee CY; Linquan B; Liu X; Liu W; Luzhetskyy A; Mahmud T; Mast Y; Méndez C; Metsä-Ketelä M; Micklefield J; Mitchell DA; Moore BS; Moreira LM; Müller R; Neilan BA; Nett M; Nielsen J; O’Gara F; Oikawa H; Osbourn A; Osburne MS; Ostash B; Payne SM; Pernodet J-L; Petricek M; Piel J; Ploux O; Raaijmakers JM; Salas JA; Schmitt EK; Scott B; Seipke RF; Shen B; Sherman DH; Sivonen K; Smanski MJ; Sosio M; Stegmann E; Süssmuth RD; Tahlan K; Thomas CM; Tang Y; Truman AW; Viaud M; Walton JD; Walsh CT; Weber T; van Wezel GP; Wilkinson B; Willey JM; Wohlleben W; Wright GD; Ziemert N; Zhang C; Zotchev SB; Breitling R; Takano E; Glöckner FO Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol 2015, 11, 625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lima ST; Alvarenga DO; Etchegaray A; Fewer DP; Jokela J; Varani AM; Sanz M; Dörr FA; Pinto E; Sivonen K; Fiore MF Genetic organization of anabaenopeptin and spumigin biosynthetic gene clusters in the cyanobacterium Sphaerospermopsis torques-reginae ITEP-024. ACS Chem. Biol 2017, 12, 769–778. [DOI] [PubMed] [Google Scholar]
- (29).Molica RJR; Oliveira EJA; Carvalho PVVC; Costa ANSF; Cunha MCC; Melo GL; Azevedo SMFO Occurrence of saxitoxins and an anatoxin-a(s)-like anticholinesterase in a Brazilian drinking water supply. Harmful Algae 2005, 4, 743–753. [Google Scholar]
- (30).He H; Williamson RT; Shen B; Graziani EI; Yang HY; Sakya SM; Petersen PJ; Carter GT Mannopeptimycins, novel antibacterial glycopeptides from Streptomyces hygroscopicus, LL-AC98. J. Am. Chem. Soc 2002, 124, 9729–9736. [DOI] [PubMed] [Google Scholar]
- (31).Ling LL; Schneider T; Peoples AJ; Spoering AL; Engels I; Conlon BP; Mueller A; Schäberle TF; Hughes DE; Epstein S; Jones M; Lazarides L; Steadman VA; Cohen DR; Felix CR; Fetterman KA; Millett WP; Nitti AG; Zullo AM; Chen C; Lewis K A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Asai M; Muroi M; Sugita N; Kawashima H; Mizuno K; Miyake A Enduracidin, a new antibiotic. II Isolation and characterization. J. Antibiot 1968, 21, 138–146. [DOI] [PubMed] [Google Scholar]
- (33).Han L; Schwabacher AW; Moran GR; Silvaggi NR Streptomyces wadayamensis MppP is a pyridoxal 5′-phosphate-dependent l-arginine α-deaminase, γ-hydroxylase in the enduracididine biosynthetic pathway. Biochemistry 2015, 54, 7029–7040. [DOI] [PubMed] [Google Scholar]
- (34).Han L; Vuksanovic N; Oehm SA; Fenske TG; Schwabacher AW; Silvaggi NR Streptomyces wadayamensis MppP is a PLP-dependent oxidase, not an oxygenase. Biochemistry 2018, 57, 3252–3264. [DOI] [PubMed] [Google Scholar]
- (35).Burroughs AM; Hoppe RW; Goebel NC; Sayyed BH; Voegtline TJ; Schwabacher AW; Zabriskie TM; Silvaggi NR Structural and functional characterization of MppR, an enduracididine biosynthetic enzyme from Streptomyces hygroscopicus: Functional diversity in the acetoacetate decarboxylase-like superfamily. Biochemistry 2013, 52, 4492–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Nayfach S; Roux S; Seshadri R; Udwary D; Varghese N; Schulz F; Wu D; Paez-Espino D; Chen I-M; Huntemann M; Palaniappan K; Ladau J; Mukherjee S; Reddy TBK; Nielsen T; Kirton E; Faria JP; Edirisinghe JN; Henry CS; Jungbluth SP; Chivian D; Dehal P; Wood-Charlson EM; Arkin AP; Tringe SG; Visel A; Woyke T; Mouncey NJ; Ivanova NN; Kyrpides NC; Eloe-Fadrosh EA; IMG/M Data Consortium. A genomic catalog of Earth’s microbiomes. Nat. Biotechnol 2021, 39, 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Du Y-L; Ryan KS Pyridoxal phosphate-dependent reactions in the biosynthesis of natural products. Nat. Prod. Rep 2019, 36, 430–457. [DOI] [PubMed] [Google Scholar]
- (38).Rudolph J; Hannig F; Theis H; Wischnat R Highly efficient chiral-pool synthesis of (2S,4R)-4-hydroxyornithine. Org. Lett 2001, 3, 3153–3155. [DOI] [PubMed] [Google Scholar]
- (39).Giltrap AM; Dowman LJ; Nagalingam G; Ochoa JL; Linington RG; Britton WJ; Payne RJ Total synthesis of teixobactin. Org. Lett 2016, 18, 2788–2791. [DOI] [PubMed] [Google Scholar]
- (40).Yin X; Zabriskie TM VioC is a non-heme iron, α-ketoglutarate-dependent oxygenase that catalyzes the formation of 3S-hydroxy-l-arginine during viomycin biosynthesis. ChemBioChem 2004, 5, 1274–1277. [DOI] [PubMed] [Google Scholar]
- (41).Ju J; Ozanick SG; Shen B; Thomas MG Conversion of (2S)-arginine to (2S,3R)-capreomycidine by VioC and VioD from the viomycin biosynthetic pathway of Streptomyces sp. strain ATCC11861. ChemBioChem 2004, 5, 1281–1285. [DOI] [PubMed] [Google Scholar]
- (42).Yin X; McPhail KL; Kim K.-j.; Zabriskie TM Formation of the nonproteinogenic amino acid 2S,3R-capreomycidine by VioD from the viomycin biosynthesis pathway. ChemBioChem 2004, 5, 1278–1281. [DOI] [PubMed] [Google Scholar]
- (43).Chang C-Y; Lyu S-Y; Liu Y-C; Hsu N-S; Wu C-C; Tang C-F; Lin K-H; Ho J-Y; Wu C-J; Tsai M-D; Li T-L Biosynthesis of streptolidine involved two unexpected intermediates produced by a dihydroxylase and a cyclase through unusual mechanisms. Angew. Chem., Int. Ed 2014, 53, 1943–1948. [DOI] [PubMed] [Google Scholar]
- (44).Haltli B; Tan Y; Magarvey NA; Wagenaar M; Yin X; Greenstein M; Hucul JA; Zabriskie TM Investigating β-hydroxyenduracididine formation in the biosynthesis of the mannopeptimycins. Chem. Biol 2005, 12, 1163–1168. [DOI] [PubMed] [Google Scholar]
- (45).Kumagai T; Takagi K; Koyama Y; Matoba Y; Oda K; Noda M; Sugiyama M Heme protein and hydroxyarginase necessary for biosynthesis of d-cycloserine. Antimicrob. Agents Chemother 2012, 56, 3682–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Adak S; Lukowski AL; Schäfer RJB; Moore BS From tryptophan to toxin: Nature’s convergent biosynthetic strategy to aetokthonotoxin. J. Am. Chem. Soc 2022, 144, 2861–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Interstate Technology and Regulatory Council: HCB Team. Strategies for Preventing and Managing Harmful Cyanobacterial Blooms (HCB-1). https://hcb-1.itrcweb.org/introduction/#3_2 (accessed April 22, 2022), Section 3.2 Health, Environment, and Economic Impacts.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


