Abstract
Reichardia Roth is a small Mediterranean genus comprising ten homogeneous species with basic chromosome numbers of 7, 8, and 9. To assess the plastid genome evolution and differentiation of Reichardia species, we assembled the complete plastome sequences of seven Reichardia and two Launaea species and conducted various phylogenomic analyses comparatively with nuclear ribosomal DNA ITS sequences. Reichardia and Launaea plastomes were highly conserved in gene content and order, containing 130 genes. Plastid phylogenomic reconstruction strongly suggested that Reichardia was a sister to Launaea, and its common ancestor initially diverged into two major lineages: the first containing species with n = 8 chromosomes exclusively, and the other with n = 9, 8, and 7 chromosomes. Although the ancestral Reichardia karyotype was suggested to most likely be n = 9 from ancestral chromosome number reconstruction, the pattern of descending dysploidy indicated by the phylogenetic trees based on nuclear ribosomal DNA ITS was less evident in the trees based on the plastome. Possible reasons for these findings are discussed.
Subject terms: Evolution, Plant sciences
Introduction
Reichardia is a small genus in the subtribe Hyoseridinae Less. (formerly known as Sonchinae) from the tribe Cichorieae Lam. & DC. (Asteraceae). It comprises ten herbaceous species, of which seven are perennials with woody bases [R. dichotoma (DC.) Freyn, R. macrophylla (Vis. & Pančić) Pančić, R. albanica F. Conti & D. Lakušić, R. picroides (L.) Roth, R. crystallina (Sch. Bip.) Bramwell, R. famarae Bramwell & G. Kunkel, and R. ligulata (Vent.) G. Kunkel and Sunding]; a biennial-perennial cycle [R. gaditana (Willk.) Samp.]; and two are annuals [R. tingitana (L.) Roth and R. intermedia (Sch. Bip.) Samp.]. Although all of these are native to the Mediterranean and/or Macaronesia (North Atlantic), those with wider distribution, namely R. tingitana and R. picroides, also colonize Middle East Asia. In addition, R. tingitana reaches Pakistan, northwestern India, and some areas of tropical and subtropical eastern Africa, showing a range roughly coinciding with the paleo-geographical limits of the Mediterranean or ‘Madrean-Tethyan’ region1,2. These two widely distributed species were introduced in Hawaii and Australia3–6. R. intermedia is also a circum-Mediterranean taxon but is quite common in the western area. R. gaditana is limited to the western Mediterranean region, whereas R. dichotoma is exclusively present in the eastern Mediterranean region. Five species show endemic distributions: the close relatives R. macrophylla and R. albanica, which are restricted to the Balkan Peninsula7; and R. crystallina, R. famarae, and R. ligulata, which are endemic to the Canary Islands. R. famarae occurs exclusively in five localities in Lanzarote and Fuerteventura8,9. The genus Reichardia is quite homogeneous in morphology and is easily recognizable (Fig. 1), mainly by the flower heads, which are often large and typically broadly conical when fruiting, and heteromorphic fruits. Outer and medium phyllaries show typical scarious margins and a subterminal mucro. Although florets are generally yellow, in two species (R. tingitana and R. gaditana), the base of the ligule is deep purple. Achenes are four-ribbed, outer transversely rugose, while compressed inner ones are paler and variably smooth. The pappus consists of numerous fine scabrid bristles.
Figure 1.
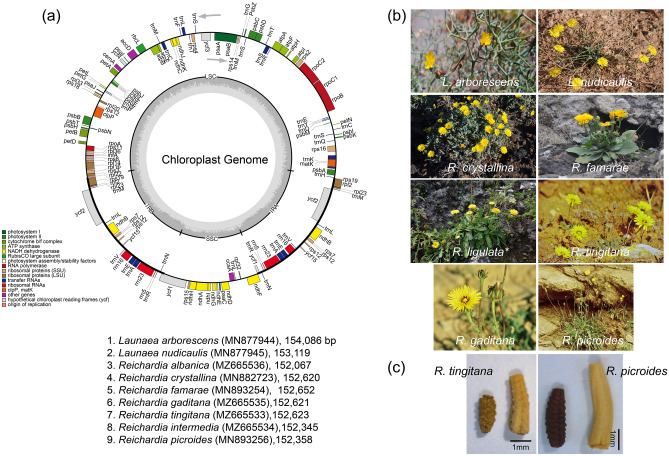
(a) Gene map of the chloroplast genomes of seven Reichardia and two Launaea species sequenced in this study. The genes inside and outside of the circle are transcribed in the clockwise and counterclockwise directions, respectively. Genes belonging to different functional groups are shown in different colors. The thick lines indicate the extent of the inverted repeats that separate the genomes into small single copy (SSC) and large single copy (LSC) regions. (b) Photos of Reichardia and Launaea species. (c) Photos of outer (left) and inner (right) heteromorphic achenes in R. tingitana and R. picroides. Photo credit: José A. Mejías (L. nudicaulis, R. gaditana, R. tingitana, R. picroides, and fruits), Myong-Suk Cho (L. arborescens), Arnoldo Santos-Guerra (R. crystallina), and Seung-Chul Kim (R. famarae and R. ligulata).
Despite its low species diversity, Reichardia presents three basic chromosome number groups of n = 7, 8, and 93,10–16. All the species are diploid, and no cases of polyploidy have been documented. The chromosome numbers n = 9 and 2n = 18 have been reported for the Western Asiatic R. dichotoma and the Balkan R. macrophylla, which can be considered vicarious species7 and perhaps tertiary relicts17. The recently described Albanian endemic R. albanica also showed a chromosome number of 2n = 18. Several chromosomes count in the widespread species R. tingitana, R. gaditana, and R. intermedia have shown n = 8 and 2n = 16, which are also the numbers found in the three species endemic to the Canary Islands, namely R. crystallina, R. famarae, and R. ligulata. R. picroides showed chromosome numbers of n = 7 and 2n = 14. Although some authors have reported a somatic number of 2n = 14 for R. intermedia11,18,19 in samples from the Iberian Peninsula, we chose to apply the somatic numbers from a monographic study of the genus by Gallego13 based on their subsequent reconfirmation from the Iberian materials15.
The diversity observed in chromosome numbers in the genus is presumed to be the result of dysploidy during karyotypic changes in the basic chromosome number19–22, as no polyploidy numbers have ever been reported for them, despite it being a widespread phenomenon reported in Asteraceae23. Previous studies have shown a correlation between this diversity and morphological characteristics6, chemical compounds24, and phylogenetic lineages19, claiming both ascending and descending patterns of dysploidy in Reichardia. Exceptional chromosomal diversity in Reichardia has been the subject of evolutionary studies as a good model for the analysis of genome evolution and differentiation. For example, Lӧve and Kjellqvist11 suggested that the most primitive number was n = 7, and the variation from primitive to higher chromosome numbers (for example, n = 8) was derived by means of the appearance of B chromosomes. However, Gallego13 proposed an alternative hypothesis that the primitive basic chromosome number was n = 9, and the secondary basic numbers of n = 8 and n = 7 arose from the loss of chromosomes or fragmentation. The analysis of phenolic compounds in R. tingitana and R. picroides did not support any specific pattern, neither ascending nor descending, but found that chemo-taxonomic differences related to morphological and karyological characteristics were in agreement with the separation of the Reichardia species into two major groups24. Recently, the chromosomal and genomic evolution of Reichardia species has been investigated within a molecular phylogenetic framework. Siljak-Yakovlev et al.19 plotted the karyological, cytogenetic, and pollen traits of Reichardia species onto the nuclear ribosomal DNA (nrDNA) internal transcribed spacer (ITS) phylogeny and suggested that the descending dysploidy in Reichardia chromosomal numbers was accompanied by changes in heterochromatin patterns and modifications of the location and organization of ribosomal genes. They also reported smaller pollen sizes in R. picroides and R. intermedia, which correlated with their smaller genome sizes and the reduced chromosome number within the genus (n = 7 for both species); however, in our view, these reports for R. intermedia may be erroneous. Moreover, these cytological studies did not include appropriate closest relatives or progenitors of the genus. Without robust phylogenetic relationships between Reichardia and other closely related genera and precise species relationships within the genus, it may be unsuitable to explore genome evolution and differentiation.
The phylogenetic position of Reichardia was previously inferred using nrDNA ITS and chloroplast DNA (cpDNA) psbA-trnH intergenic spacer and matK gene sequences25–28. In all phylogenies, the genera Launaea and Reichardia represented early diverging lineages within the subtribe Hyoseridinae, and Reichardia was monophyletic, which is concordant with the first comprehensive molecular analyses of the tribe Cichorieae29. The ITS tree strongly supported the monophyly of Reichardia25,26,28, which was sister to the clade containing the genus Launaea and the remaining genera of the subtribe Hyoseridinae, that is, the currently circumscribed Sonchus sensu lato (s.l.). The cpDNA trees based on either the psbA-trnH intergenic spacer27 or matK28 gene regions also supported the monophyly of Reichardia and the basal position of Reichardia and Launaea. However, cpDNA phylogenies in previous studies27,28 were too limited to determine the phylogenetic position of Reichardia and robust species relationships due to insufficient molecular markers and species representation. The relationships between species and genera were either highly unresolved or weakly supported, specifically in the psbA-trnH tree27. Unlike the nrDNA ITS-based phylogeny, Reichardia and Launaea shared the most recent common ancestor, and these two genera were sister to the remaining genera of the subtribe Hyoseridinae, which was also weakly supported (< 50% in the psbA-trnH tree)27 or moderately supported (86% in the matK tree)28. These conclusions are consistent with subsequent studies based on ITS datasets19,30.
With the advent of high-throughput sequencing technologies for next-generation sequencing (NGS), massive amounts of data have now become available based on the considerable genome-wide variation of entire plastid genomes through plastome sequencing. The benefits of genome-wide variation could increase phylogenetic resolution tremendously and significantly enhance our understanding of plant evolution and diversity in the field of plastid genetics and genomics31. Comparative genomic analysis using whole plastome sequences is now an efficient option to improve phylogenetic resolution at lower taxonomic levels that are currently hindered by limited sequence variation due to recent divergence, rapid radiation, and conservative genome evolution of plastomes32.
In this study, we sequenced and assembled the whole plastid genomes of seven Reichardia species in addition to R. ligulata, sequenced in our previous study33. The plastomes of two additional species from the closely related genus Launaea were also sequenced25–28. Based on these complete plastome sequences, we first tested the previous phylogenetic hypotheses proposed by nrDNA ITS and cpDNA markers; plastome-based phylogeny was reconstructed using genome-wide variation and compared with the nrDNA ITS phylogeny, newly reconstructed in this study. Second, through the ancestral karyotype reconstruction, we inferred the karyotype evolution in the dysploidy process of Reichardia species in the framework of the plastome and nrDNA ITS-based phylogenies. We also performed several comparative plastome analyses to determine the structure, gene content, and rearrangements of the plastid genomes of Reichardia and two Launaea species. In addition, we identified highly variable plastid regions that could be utilized as useful markers for further population genetic or phylogeographic studies of Reichardia and closely related genera.
Results
Genome features, content, order, and organization of Reichardia plastomes
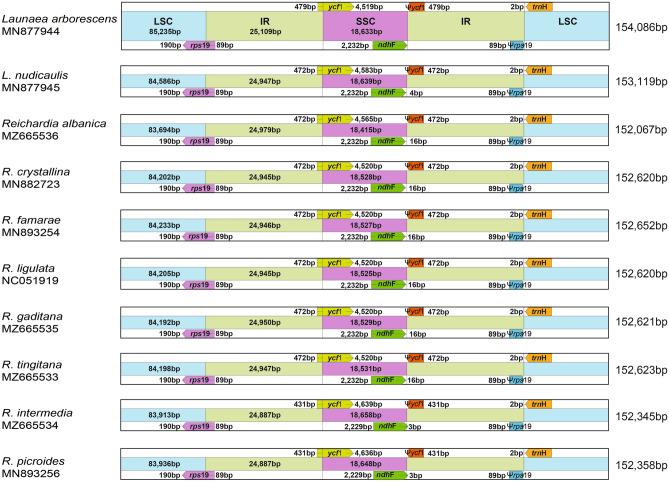
The eight plastomes of Reichardia species (R. albanica, R. crystallina, R. famarae, R. gaditana, R. intermedia, R. ligulata, R. picroides, and R. tingitana) and two species of Launaea (L. arborescens and L. nudicaulis) were highly conserved in terms of gene content and arrangement, with 98.6% pairwise sequence similarity (99.2% among eight Reichardia species only) (Table 1). The total length of eight Reichardia plastomes ranged from 152,067 (R. albanica) to 152,652 (R. famarae) base pairs (bp) with an overall guanine-cytosine (GC) content of 37.6%, whereas two plastomes of Launaea were slightly longer than Reichardia plastomes, that is, 153,119 bp for L. nudicaulis and 154,086 bp for L. arborescens with an overall GC content of 37.4%. The plastomes of Reichardia and Launaea species consisted of four typical regions: LSC, SSC, and a pair of inverted repeat regions (IRs), sharing exactly the same genes and similar gene contents at all boundaries among the four regions, with slight changes in the length of intergenic regions. They all contained the functional protein-coding gene ycf1 at SSC/IR with its pseudogene copy, ycf1Ψ at IR/SSC, and functional rps19 at LSC/IR with pseudogene copy rps19Ψ at IR/LSC endpoints (Fig. 2).
Table 1.
Summary of the genomic characteristics of two Launaea and eight Reichardia chloroplast genomes used for comparative genomic analyses in this study.
| Species GenBank accession no |
Voucher (Herbarium) | Total size (bp) GC content (%) |
LSC (bp) GC content (%) |
IR (bp) GC content (%) |
SSC (bp) GC content (%) | No. of total genes | No. of protein coding genes | No. of tRNA genes | No. of rRNA genes |
|---|---|---|---|---|---|---|---|---|---|
| Genus Launaea | |||||||||
|
L. arborescens |
Kim et al. 1040 (OS) |
154,086 37.4% |
85,235 35.5% |
25,109 43.0% |
18,633 31.2% |
130 | 87 | 37 | 6 |
|
L. nudicaulis |
Kim et al. 1053 (OS) |
153,119 37.5% |
84,586 35.7% |
24,947 43.1% |
18,639 31.1% |
130 | 87 | 37 | 6 |
| Genus Reichardia | |||||||||
|
R. albanica |
APP 56321 (APP) |
152,067 37.6% |
83,694 35.8% |
24,979 43.0% |
18,415 31.2% |
130 | 87 | 37 | 6 |
|
R. crystallina |
Santos-Guera & Kim 210 (OS) |
152,620 37.6% |
84,202 35.7% |
24,945 43.1% |
18,528 31.1% |
130 | 87 | 37 | 6 |
|
R. famarae |
Santos-Guera & Kim 211 (OS) |
152,652 37.6% |
84,233 35.7% |
24,946 43.1% |
18,527 31.1% |
130 | 87 | 37 | 6 |
|
R. ligulataa NC051919 |
Kim et al. 1044 (OS) |
152,620 37.6% |
84,205 35.7% |
24,945 43.1% |
18,525 31.1% |
130 | 87 | 37 | 6 |
|
R. gaditana |
Mejías JA SEV289342 (SEV) |
152,621 37.6% |
84,192 35.7% |
24,950 43.1% |
18,529 31.1% |
130 | 87 | 37 | 6 |
|
R. tingitana |
Mejías JA SEV289341 (SEV) |
152,623 37.6% |
84,198 35.7% |
24,947 43.1% |
18,531 31.1% |
130 | 87 | 37 | 6 |
|
R. intermedia |
Mejías JA SEV289340 (SEV) |
152,345 37.6% |
83,913 35.8% |
24,887 43.1% |
18,658 31.2% |
130 | 87 | 37 | 6 |
|
R. picroides |
Jansen 2873 (TEX) |
152,358 37.6% |
83,936 35.8% |
24,887 43.1% |
18,648 31.1% |
130 | 87 | 37 | 6 |
Herbarium code: APP: Parco Nazionale del Gran Sacco e Mont della Laga-Universita di Camerino, Italy, OS: Ohio State University herbarium, SEV: Herbario de la Universidad de Sevilla, TEX: University of Texas Herbarium.
aSequenced from a previous study33. The remaining nine cp genomes were sequenced for this study.
Figure 2.
Comparison of the border positions of the large single copy (LSC), small single copy (SSC), and inverted repeat (IR) regions among eight Reichardia and two Launaea plastomes.
Each of the eight Reichardia and two Launaea cp genomes contained 130 genes, including 87 protein-coding genes (excluding pseudogenes), six rRNA genes, and 37 tRNA genes (Tables 1 and 2). Twenty-four genes contained introns, including nine tRNA genes. Three genes, clpP, rps12, and ycf3, had two introns. The trnK-UUU tRNA gene harbors the largest intron, which contains the matK gene. In total, 17 genes were duplicated in the IR regions, including seven tRNAs, three rRNAs, and seven protein coding genes. The trans-splicing gene rps12, consisting of three exons, was located in the LSC region of exon 1, whereas exons 2 and 3 of the gene were embedded in the IR regions. Parts of ycf1 and rps19 duplicated in the IR region were considered pseudogenes in all cp genomes sequenced in this study.
Table 2.
Genes encoded by two Launaea and eight Reichardia chloroplast genomes.
| Category | Group | Genes |
|---|---|---|
| Photosynthesis | Subunits_of_photosystem_I | psaA, psaB, psaC, psaI, and psaJ |
| Subunits_of_photosystem_II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, and psbZ | |
| Subunits_of_NADH_dehydrogenase | ndhA*, ndhB(2)*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, and ndhK | |
| Subunits_of_cytochrome_b/f_complex | petA, petB*, petD*, petG, petL, and petN | |
| Subunits_of_ATP_synthase | atpA, atpB, atpE, atpF*, atpH, and atpI | |
| Large_subunit_of_Rubisco | rbcL | |
| Self-replication | Large_subunits_of_ribosome | rpl2(2)*, rpl14, rpl16*, rpl20, rpl22, rpl23(2), rpl32, rpl33, and rpl36 |
| Small_subunits_of_ribosome | rps2, rps3, rps4, rps7(2), rps8, rps11, rps12 (2)**, rps14, rps15, rps16*, rps18, and rps19 | |
| DNA-dependent_RNA_polymerase | rpoA, rpoB, rpoC1*, and rpoC2 | |
| translation initiation factor | infA | |
| Ribosomal_RNAs | rrn5(2), rrn16(2), and rrn23(2) | |
| Transfer_RNAs | trnA-UGC(2)*, trnC-GCA, trnD-GUC, trnE-UUC(3)*, trnF-GAA, trnG-GCC, trnH-GUG, trnK-UUU*, trnL-CAA(2)*, trnL-UAA, trnL-UAG, trnM-CAU(4), trnN-GUU(2), trnP-UGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-CGA, trnS-GCU*, trnS-GGA, trnS-UGA, trnS-GGU, trnT-UGU, trnV-GAC(2), trnV-GCA, trnW-CCA, and trnY-GUA | |
| Other genes | Maturase | matK |
| Protease | clpP** | |
| Envelope_membrane_protein | cemA | |
| Acetyl-CoA_carboxylase | accD | |
| C-type_cytochrome_synthesis_gene | ccsA | |
| Genes of unknown function | Proteins_of_unknown_function | ycf1, ycf2(2), ycf3**, ycf4, ycf15(2) |
(×N) indicates the genes that have N copies. * and ** indicate genes containing one and two introns, respectively.
Comparative plastome analyses of Reichardia and Launaea
The frequency of codon usage in the eight Reichardia and two Launaea plastomes was calculated based on protein coding gene sequences (Fig. 3 and Supplementary Table S1). The codon usage numbers in the ten Reichardia and Launaea plastomes ranged from 22,765 to 22,791, and the patterns of frequently used codons were also consistent among them except for slight variations in codon usage numbers. Within Reichardia, species with chromosome numbers of n = 8, except R. intermedia, showed lower codon usage (22,765) than species with chromosome numbers of n = 9 (R. albanica: 22,781) or n = 7 (R. picroides: 22,790), and R. intermedia (22,791). Codon usage values are described by relative synonymous codon usage (RSCU), which reflects how often a particular codon is used relative to the expected number of times that codon would be used in the absence of codon usage bias. All the RSCU values for each amino acid considered in this study were similar among the ten plastomes. The highest RSCU value was indicated in the usage of the UUA codon for leucine (1.9–1.92), followed by AGA for arginine (1.84–1.86), while the lowest were indicated in the usages of the AGC codon for serine (0.33–0.36) and CUG for leucine (0.35–0.38).
Figure 3.
The relative synonymous codon usage (RSCU) of the protein-coding genes in chloroplast genomes of eight Reichardia and two Launaea species. The codon usages of amino acids are plotted along the x-axis, while the stacked RSCU values in each bar column are plotted along the y-axis respectively. Each amino acid contains ten clustered bar columns representing ten species; 1st column through 10th column for R. albanica, R. crystallina, R. famarae, R. ligulata, R. gaditana, R. tingitana, R. intermedia, R. picroides, L. arborescens, and L. nudicaulis.
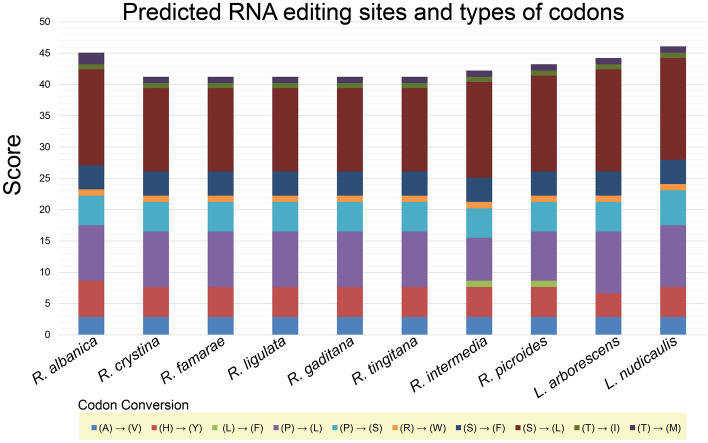
The possible RNA editing sites predicted among the Reichardia and Launaea plastomes ranged from 43 to 48 in the 19 genes of the 35 protein-coding genes (Fig. 4 and Supplementary Table S2). We found that the RNA editing patterns across the ten Reichardia and Launaea plastomes were similar in gene location and codon conversion type of the predicted RNA editing sites, and only slight changes were observed in the number of editing sites for several codon conversions. Within Reichardia, all species with chromosome numbers of n = 8 showed lower numbers of predicted editing sites (43 for all species, with the exception of 44 for R. intermedia) than species with chromosome numbers of n = 7 (45 for R. picroides) and n = 9 (R. albanica), with 47 sites showing the highest number in the genus. Genes with predicted sites included photosynthesis-related genes (atpA, ndhA, ndhB, ndhD, ndhF, ndhG, petB, psbF, and psbL), self-replication genes (rpl20, rpoA, rpoB, rpoC1, rpoC2, rps2, and rps14), and others (accD, ccsA, and matK). We detected no RNA editing sites in atpB, atpF, atpI, clpP, petD, petG, petL, psaB, psaI, psbB, psbE, rpl2, rpl23, rps8, rps16, and ycf3. The highest numbers of potential editing sites were found in the NADH dehydrogenase genes, which was consistent with previous findings in tobacco, maize, rice, and other plants34–37; the ndhB gene was the highest at 9–10 sites, followed by the ndhD gene at 4–5 sites. The highest conversions in the editing frequencies of codons associated with the corresponding amino acid changes were represented by the changes from serine (S) to leucine (L) (average confidence score of 14.52), followed by proline (P) to leucine (L) (average confidence score of 8.76).
Figure 4.
Amino acid changes in predicted RNA editing sites in ten chloroplast genomes of eight Reichardia and two Launaea species. The scores (the proportion of sites that have the same amino acid at that position) of each edit site are stacked in each bar column. Color bricks indicate RNA editing effect: Alanine to Valine, A → V; Histidine to Tyrosine, H → Y; Leucine to Phenylalanine, L → F; Proline to Phenylalanine, P → F; Proline to Leucine, P → L; Proline to Serine, P → S; Arginine to Tryptophan, R → W; Serine to Phenylalanine, S → F; Serine to Leucine, S → L; Threonine to Iso-leucine, T → I; and Threonine to Methionine, T → M.
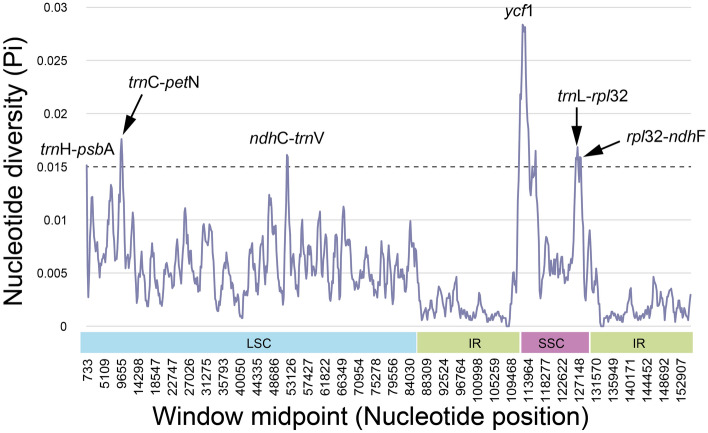
The divergence level of nucleotide diversity between the Reichardia and Launaea plastomes was compared using DnaSP38 and visualized by plotting with mVISTA39. The results showed a high degree of synteny and gene order conservation in the mVISTA graph (Fig. 5). The overall nucleotide diversity (Pi) of the ten plastomes was 0.00520, with 2586 polymorphic sites, ranging from 0 to 0.02836, which was higher than that of Sonchus species (average Pi value of 0.00283, ranging from 0 to 0.01593) belonging to the same subtribe Hyoseridinae33. Genetic polymorphisms in different regions of the chloroplast genome varied substantially. The SSC region, where the most variable gene, ycf1, was located, was the most divergent (average Pi, 0.0102), whereas the two IR regions were highly conserved (average Pi, 0.00173). Six divergence hotspots among the Reichardia and Launaea plastomes are suggested as potential chloroplast markers: five intergenic regions (trnH-psbA, trnC-petN, ndhC-trnV, trnL-rpl32, and rpl32-ndhF) and one protein-coding gene (ycf1) (Fig. 6).
Figure 5.
Comparison of the chloroplast genomes of eight Reichardia and two Launaea species against L. arborescens using mVISTA. Grey arrows indicate genes with their orientation and position. Genome regions are color-coded, as indicated.
Figure 6.
Six mutation hotspot regions were observed in eight Reichardia plastomes and two Launaea plastomes.
Selective pressure in genes or genomic regions is inferred by the proportion of amino acid substitutions, and the ratio (ω = dN/dS) of nonsynonymous substitution (dN) and synonymous substitution rates (dS) has been widely used as a genomic signature of selective pressure acting on a protein-coding gene, that is, ω = 1 indicates neutral mutations, ω < 1 indicates purifying selection, and ω > 1 indicates diversifying positive selection40. We identified that seven genes potentially evolved under positive selection in ten Reichardia and Launaea plastomes by calculating the dN/dS ratio using various site-specific substitution models implemented in EasyCodeML (Table 3 and Supplementary Table S3)41,42. These genes included NADH-dehydrogenase subunit genes (ndhB and ndhG), subunit genes of photosystem I (psaI) and II (psbH), and large subunit genes of ribosomal protein (rpl20), ycf2, and ycf15. Positively selected sites were suggested based on the posterior probability calculated using the Bayes empirical Bayes (BEB) method43 with a cutoff > 0.95 and > 0.99 indicated with asterisks (* and **, respectively) in Table 3. Despite the critical importance of the genes that a plastome carries and its high conservativeness, the latest empirical evidence revealed that the variable genes potentially evolve under positive selection in the plastomes of a few other plant groups; three genes (rps2, rbcL, and ndhG) have been identified in Paulownia44, five (rbcL, clpP, atpF, ycf1, and ycf2) in Panax45, three (clpP, ycf1, and ycf2) in the tribe Sileneae46, and six (accD, rbcL, rps3, ndhB, ndhD, and ndhF) in Rosaceae37.
Table 3.
Positively selected sites with dN/dS values > 1 detected in two Launaea and eight Reichardia chloroplast genomes.
| Gene name | Site models | np | ln L | Model compared | LRT p-value | Positively selected sites |
|---|---|---|---|---|---|---|
| ndhB | M8 | 23 | − 2072.828684 | M7 vs. M8 | 0.250784820 | 87 F 0.990*, 205 S 0.990*, 210 E 0.990*, 424 G 0.990*, 428 G 0.990*, and 431 F 0.990* |
| M7 | 21 | − 2074.211844 | ||||
| ndhG | M8 | 23 | − 748.902533 | M7 vs. M8 | 0.000639245 | 18 G 0.952* and 90 F 0.991** |
| M7 | 21 | − 756.257756 | ||||
| psaI | M8 | 23 | − 142.875382 | M7 vs. M8 | 0.521237233 | 4 L 0.962* and 23 M 0.962* |
| M7 | 21 | − 143.526932 | ||||
| psbH | M8 | 23 | − 298.305462 | M7 vs. M8 | 0.057472483 | 11 R 0.960* and 21 D 0.989* |
| M7 | 21 | − 301.161911 | ||||
| rpl20 | M8 | 23 | − 523.911808 | M7 vs. M8 | 0.384904126 | 25 R 0.984*, 85 L 0.984*, 110 M 0.984*, and 122 K 0.984* |
| M7 | 21 | − 524.866569 | ||||
| ycf2 | M8 | 23 | − 9554.618258 | M7 vs. M8 | 0.005774440 | 1778 I 0.984* |
| M7 | 21 | − 9559.772572 | ||||
| ycf15 | M8 | 23 | − 264.859366 | M7 vs. M8 | 0.408814407 | 23 A 0.962* and 33 R 0.962* |
| M7 | 21 | − 265.753860 |
np: number of parameters in the ω distribution; ln L: log-likelihood values; LRT p-value: likelihood ratio test p-value; positive selection sites are inferred by significant posterior probability value with * > 0.95 or ** > 0.99.
Phylogenomic analyses and ancestral state reconstructions
Phylogenetic trees were constructed using maximum likelihood (ML) and Bayesian inference (BI) methods based on the plastome and nrDNA ITS sequences to infer the phylogenetic relationships among Reichardia and the closely related species in the subtribe Hyoseridinae, with Lactuca sativa as an outgroup. The topological structures of ML phylogenetic trees constructed by IQ-TREE47 and BI trees by MrBayes 3.2.7a48 were consistent, and BI trees showed higher support values than ML trees (Fig. 7 and Supplementary Fig. S1). The best-fit evolutionary models in ML analyses were selected as “TVM + F + I” for plastome trees and “TIM3e + G4” for the nrDNA ITS tree using ModelFinder49 implemented in IQ-TREE. The plastome phylogenies were reconstructed using both whole plastome sequences (Fig. 7a) and 80 chloroplast protein-coding genes (Supplementary Fig. S1) of 18 representative Hyoseridinae species. The phylogenies based on plastome datasets shared the same topology and provided much greater resolution with strong bootstrap (BS) values for inter- and intra-generic relationships compared to previous studies27,28. The genus Reichardia was resolved as monophyletic and shared the most recent common ancestor with the genus Launaea, placing the clade of Reichardia and Launaea sister to the genus Sonchus s.l. Within Reichardia, R. albanica (n = 9) shared the most recent common ancestor with R. picroides and R. intermedia with chromosomal numbers of n = 7 and 8 robustly (99% or 97% BS support value in ML trees and posterior probability, PP, 1 in BI trees). The remaining five species (R. ligulata, R. famarae, R. crystallina, R. tingitana, and R. gaditana) with chromosome numbers of n = 8 formed a clade with full support (100% BS for ML and PP 1 for BI phylogenies), which was sister to the clade of n = 7, 8, and 9. On the other hand, the nrDNA ITS phylogeny showed incongruence in inter- and intra-generic relationships compared with the plastome phylogeny (Fig. 7b). Unlike plastome phylogenies, Launaea shared the most recent common ancestor with Sonchus s.l., instead of Reichardia; this relationship, however, was weakly supported (68% BS in ML and 0.7867 PP in BI trees). Reichardia was monophyletic in both nrDNA ITS ML and BI phylogenies (100% BS and 1 PP, respectively), but differed in intra-generic relationships from the plastome trees; R. intermedia (n = 8), and R. picroides (n = 7) displayed a closer affinity with the other species with a chromosome number of n = 8 (R. ligulata, R. famarae, R. crystallina, R. tingitana, and R. gaditana) than species with a chromosome number of n = 9 (R. albanica, R. macrophylla, and R. dichotoma) (98% BS in ML and 1 PP in BI trees).
Figure 7.
Maximum likelihood (ML) tree of Reichardia and closely related Launaea and Sonchus species based on (a) the complete plastome and (b) nrDNA ITS sequences. Support values are provided above and below branches (ML bootstrap value with 1000 replicates on the left and Bayesian Inference posterior probabilities on the right). “//” indicates that the branch lengths between outgroup and ingroup taxa are shortened to improve readability of trees. Newly sequenced nine chloroplast plastid genomes in this study are marked with an asterisk (*). Within Reichardia, the species with same chromosome numbers are colored in red for n = 9, navy blue for n = 8, and aqua blue for n = 7 chromosomes.
The ancestral chromosome numbers optimized on the plastome and nrDNA ITS ML phylogenies also reflected the differences between both phylogenies. The ancestral chromosome number of the most recent common ancestor (MRCA) shared by three genera, Reichardia, Launaea, and Sonchus, was inferred to be equivocal with 2n = 14, 16, 18, and 32, and 2n = 36 for both plastome (node P21) and nrDNA ITS (node 29) datasets (Fig. 8). Within Reichardia, ancestral state reconstruction using the nrDNA ITS tree suggested that the MRCA of Reichardia most likely had 2n = 18 (56.5% probability at node 40), suggesting a decrease in dysploidy (Fig. 8b and Supplementary Table S4). Unlike the nrDNA ITS ancestral state reconstruction, the plastome reconstruction proposed that the MRCA (node P31) with 2n = 18 (27.4%, the highest probability) evolved into two lineages: one with 2n = 16 species exclusively (R. tingitana, R. gaditana, and species endemic to the Canary Islands), and the other with 2n = 14 (R. picroides), 16 (R. intermedia), and 18 (R. albanica) species (Fig. 8a and Supplementary Table S4).
Figure 8.
The ancestral states and shared-derived karyotypes reconstructed using MrBayes Ancestral States with R (MBASR) mapped on the ML trees based on (a) plastome and (b) nrDNA ITS sequences. The tree plots overlay state marginal likelihood pie charts at each lineage split for the probabilities of each basic chromosome numbers.
Discussion
This study provides, for the first time, greater resolution with high support for the plastome-based inter- and intra-generic relationships among Reichardia and closely related genera. All three genera, Reichardia, Launaea, and Sonchus, were confirmed as monophyletic with strong support values (BS 100% each) in both phylogenies of full plastome sequences and concatenated sequences of plastid protein-coding genes (Fig. 7 and Supplementary Fig. S1). However, with regard to inter-generic relationships, some topological incongruences were found between cp genomes and nrDNA ITS sequences, which were in agreement with the earlier results on nrDNA ITS25,26,28 and cpDNA marker27,28 phylogenies. In both plastome based phylogenies, Reichardia was sister to Launaea, and the clade of Reichardia and Launaea was sister to Sonchus (Fig. 7a), while Reichardia was sister to the clade containing Launaea and Sonchus in nrDNA ITS phylogeny (Fig. 7b). It is likely difficult to decisively determine the inter-generic relationships in this study based on the current sampling, which included only two Launaea species. Given its species diversity (approximately 54 species and 10 subspecies), life form variability (perennial and annual herbs, subshrubs, cushion-forming rosette shrubs, and spinescent shrubs), and chromosome variation (n = 9, 8, 7, 6, and 5)50, the phylogenetic position of the genus Launaea remains to be clarified in future studies.
Species relationships within Reichardia also demonstrated incongruences between the cp genome and nrDNA ITS phylogenies. One novel finding of the current plastid phylogenomic study was that species with different chromosome numbers of n = 7 (R. picroides), n = 8 (R. intermedia), and n = 9 (R. albanica) were resolved in the same clade. This clade was sister to another clade with n = 8 species exclusively. In the nrDNA ITS phylogeny reconstructed previously19 and in this study (Fig. 7b), a clade composed of species with n = 9 chromosomes (R. albanica, R. macrophylla, and R. dichotoma) was resolved as sister to a clade of the species with chromosome numbers of n = 8 and n = 7, clearly supporting the descending dysploidy process for chromosomal evolution in Reichardia. One of the derived groups included two species with n = 7 and 8 (R. picroides and R. intermedia, respectively), whereas the other was exclusively comprised of five n = 8 species (R. ligulata, R. famarae, R. crystallina, R. tingitana, and R. gaditana). However, the current study strongly suggested a close relationship among n = 7, n = 9, and a representative with n = 8 (R. intermedia) species on both plastome-based phylogenies (Fig. 7a and Supplementary Fig. S1). According to our results, two major lineages may have diverged early from the common ancestor of the Reichardia species, that is, one lineage with n = 8 exclusively and the other lineage containing species with n = 9, 8, and 7. These relationships were not in agreement with a simple sequential descending dysploidy pattern from n = 9 to n = 7, as suggested by the nrDNA ITS phylogeny. The ancestral Reichardia karyotype was suggested to be equivocal, but n = 9 was most likely (56.5% probability at node 40; Fig. 8b and Supplementary Table S4) from the reconstruction mapped onto the nrDNA ITS ML tree. Even from the plastome tree, it was considered equivocal, but with the highest probability for n = 9 (27.4% at node P31), followed by n = 8 (24.6%), and n = 7 (17.3%). Based on these results, we can hypothesize that the common ancestor of Reichardia (with the most likely chromosome number of n = 9) initially diverged into two lineages: one with n = 8 species and the other including n = 9, n = 8, and n = 7 species. The hypothesis that n = 9 is the ancestral chromosome number in Reichardia coincides with the closely related genera Launaea and Sonchus, where this number is the most common and widely distributed13,50,51. Furthermore, it has been assumed as the ancestral basic chromosome number of the tribe Cichorieae52–54.
Given their current geographical distribution ranges, the affinity of n = 9 species with R. intermedia (n = 8) and R. picroides (n = 7) based on plastome sequences is an unexpected finding. The last two species show a wide distribution in large circum-Mediterranean regions in Africa, Asia, and Europe which hardly overlapped with those of n = 9 species and only partly with R. dichotoma in certain regions of West Asia (Turkey and Lebanon-Syria)3–5. In contrast to these two species, n = 9 species are currently found in very limited and isolated areas with a disjunctive geographical distribution pattern; Albanian endemic R. albanica is found only in Mount Çika in southern Albania at altitudes between 1000 and 1700 m above sea level (a.s.l.), Balkan endemic R. macrophylla in the central part of the Balkan Peninsula (East Dinarides) at altitudes between 500 and 1750 m a.s.l., and Western Asiatic R. dichotoma in Turkey, Lebanon-Syria, Iran, and North and Trans-Caucasus7. Considering the putative ancestral Reichardia karyotype (n = 9) and relic species restricted to the eastern Mediterranean17, it is highly plausible that Reichardia originated in the eastern Mediterranean. The mean divergence time based on the nrDNA ITS sequences for the two closely related genera Launaea and Sonchus was estimated to be 6.4 million years (myr)55. The estimated divergence time of Reichardia from its close relatives could be earlier than 6.4 myr based on the ITS tree topology (i.e., Reichardia is sister to the clade of Launaea and Sonchus) or younger than 6.4 myr on the plastome tree (i.e., Sonchus is sister to the clade of Launaea and Reichardia). Thus, according to the proposed divergence estimation, we can hypothesize that Reichardia originated around the Messinian salinity crisis in the Mediterranean basin56. It is plausible that after its origin in the eastern region, Reichardia rapidly expanded its range and diversified during dramatic climatic changes in the Mediterranean, forming n = 8 (R. intermedia) and n = 7 (R. picroides) lineages. In addition, the rapid speciation of n = 8 species in the western Mediterranean region, including species endemic to Macaronesian Islands (R. famarae, R. crystallina, and R. ligulata), most likely occurred after initial splitting from the n = 9 lineages. The Macaronesian Islands are well known for adaptive radiation and the diversification of explosive insular species of numerous plant lineages, including Aeonium57, Argyranthemum58,59, Echium60,61, Tolpis62 and woody Sonchus alliance25–28. The uncertain phylogenetic position of Reichardia relative to Launaea and Sonchus and n = 9 likely ancestral may also suggest that the common ancestor of Reichardia existed widely in the circum-Mediterranean region in the late Miocene and subsequently diversified during climatic turmoil, forming a relictual n = 9 lineage in the eastern Mediterranean and speciating in the western part of the Mediterranean. Furthermore, we cannot ignore the role of Quaternary climatic oscillations in shaping distribution areas, particularly in current relicts. The wide distribution range of R. tingitana, which roughly coincides with the paleo-geographical limits of the Mediterranean, seems to indicate, at least partially, that the expansion and diversification of this lineage may not have been recent and must have taken place prior to the present geologic configuration of the Mediterranean region. Given the phylogenetic position of R. tingitana (i.e., positioned in a recently derived clade), we cannot completely rule out the possibility of its recent rapid range expansion during climatic oscillation in the region.
The phylogenetic relationships among Reichardia species based on plastome sequences were also supported by morphological and chemotaxonomic data. Achene heteromorphy was less marked in the n = 9 species than in the other groups. R. intermedia and R. picroides showed plainly smooth inner achenes with differentiated shapes (Fig. 1c). In addition, species with purple base ligules were exclusively present in R. tingitana, R. gaditana, and the Canary Islands endemic clade. Recio et al.24 divided Reichardia species into two groups by scanning electron microscopy, in support of the findings of the present study. In group I, which contained R. intermedia and R. picroides, the outer achene surface was made up of isodiametric papillated cells, while the cylindrical/conic-truncated inner cells showed long and more or less rectangular cells without papillae. They also included R. macrophylla in group I, although the inner achenes did not show the same level of distinctiveness. Contrarily, group II contained n = 8 species; R. ligulata, R. famarae, R. crystallina, R. tingitana, and R. gaditana showed similar microscopic surface traits on outer and inner achenes, that is, more or less isodiametric cells, which were clearly distinguishable in shape, surface, and color (Fig. 1c; fruits of R. tingitana). The separation of Reichardia species into two major groups seems to be corroborated further by differences in chemical compositions between R. picroides (group I; n = 7, 8, and 9 species) and R. tingitana (group II; n = 8 species)24.
Lastly, the phylogenetic incongruence between the plastome and nrDNA ITS phylogenies could be the result of past gene flow occurring in Reichardia species. Hybridization and introgression often appear to be responsible for the phylogenetic incongruence between organellar and nuclear markers, as observed in many groups of plants; however, other factors, such as phylogenetic sorting or the retention of ancestral polymorphisms, could also play a role in generating incongruences63,64. The R. intermedia and R. picroides group shared the most recent common ancestor with n = 9 species in plastome-based phylogenies with strong support values (BS 99% or 97% in ML and PP 1 in BI trees) (Fig. 7a and Supplementary Fig. S1), whereas they were more closely related to the n = 8 group of species in the nrDNA ITS phylogeny (BS 98% in ML and PP 1 in BI trees) (Fig. 7b). Considering the wide distribution range of both groups in large circum-Mediterranean regions in Africa, Asia, and Europe, they may easily overlap in terms of distribution. Therefore, it is plausible that R. intermedia and R. picroides captured the chloroplast genome from the ancient n = 9 group with a much broader distribution, resulting in a phylogenetic incongruence between the cpDNA and ITS phylogeny. Several cases of hybridization among Reichardia species have been documented, further supporting the likelihood of this hypothesis, including R. × baetica Gallego & Talavera (R. tingitana × R. intermedia), R. x canariensis Gallego & Talavera (R. tingitana × R. ligulata), and R. × sventenia Gallego & Talavera (R. tingitana × R. famarae)6. While the R. × sventenia and R. × canariensis cases are examples of hybridization between widely distributed R. tingitana and the locally restricted species endemic to the Canary Islands, the hybrid origin of R. × baetica occurred between two divergent n = 8 lineages with a much wider geographical distribution.
In conclusion, we found highly conserved plastomes, including gene order and content, in Reichardia and Launaea. We achieved the first full resolution regarding inter and intra-generic phylogenetic relationships based on the complete plastome sequences of most Reichardia species with variable chromosome numbers (n = 7, 8, and 9). The plastid phylogenomics strongly suggested the early divergence of two major lineages of Reichardia, one with a group of species with exclusively n = 8 chromosome numbers and the other with n = 9, 8, and 7 species together. Although the plastome-based relationships were not in full agreement with the stepwise descending dysploid hypothesis in Reichardia, the ancestral Reichardia karyotype was highly likely to be n = 9 species. It is necessary to utilize whole nuclear genome data and a thorough synteny analysis to elucidate convincingly the species relationships and chromosomal evolution within the genus Reichardia. Based on thorough characterization and comparative analyses of the plastomes, we discovered several informative mutation hotspots, which will increase the efficiency and feasibility of phylogenetic reconstruction among Reichardia and closely related genera.
Methods
Plant sampling, DNA isolation, and plastome sequencing/annotation
Plant materials of Reichardia and Launaea species were collected in the field (see plant sources in25,26,28), except for R. albanica, which was obtained from herbarium specimens (voucher number 56321, APP). Total genomic DNA was isolated using CTAB65 and a DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA, USA) following the manufacturer’s protocol. An Illumina paired-end (PE) genomic library was constructed and sequenced using the Illumina HiSeq platform (Illumina, Inc., San Diego, CA, USA) at Macrogen Corporation (Seoul, Korea). Sequence reads of the chloroplast genomes were assembled using the de novo genomic assembler Velvet 1.2.1066. Annotation was performed using Geneious R10 (Biomatters, Auckland, New Zealand)67 and ARAGORN v1.2.3668. The annotated plastome sequences were deposited in GenBank under the accession numbers listed in Table 1: MZ665536 for R. albanica, MN882723 for R. crystallina, MN893254 for R. famarae, MZ665535 for R. gaditana, MZ665534 for R. intermedia, MN893256 for R. picroides, MZ665533 for R. tingitana, MN877944 for L. arborescens, and MN877945 for L. nudicaulis. R. ligulata (GenBank accession number NC051919) was obtained from our previous study33. The annotated GenBank (NCBI, Bethesda, MD, USA) format sequence file was used to draw a circular plastid genome map (Fig. 1a) using the OGDRAW software v1.2 (CHLOROBOX)69.
Comparative plastome analyses
We performed several comparative plastome analyses of the eight Reichardia, representing variable chromosomal numbers. The analyses also included two related Launaea plastomes (L. arborescens and L. nudicaulis). The codon usage frequency was calculated using MEGA770 with the relative synonymous codon usage (RSCU) value, which is the relative frequency of occurrence of the synonymous codon for a specific amino acid. The online program predictive RNA editor for plants (PREP) suite71 was used to predict the potential RNA editing sites for annotated protein-coding genes with 35 reference genes available with known edit sites, based on a cutoff value of 0.8 (suggested as optimal for PREP-Cp). The overall sequence divergence was estimated using the Lagan alignment mode72 in mVISTA39. The nucleotide diversity (Pi) was calculated using sliding window analysis (window length = 1000 bp and step size = 200 bp, excluding sites with alignment gaps) to detect the most divergent regions (i.e., mutation hotspots) in DnaSP38. To evaluate the natural selection pressure in the protein-coding genes of the ten plastomes, site-specific models implemented in EasyCodeML41 were used in the preset running mode based on CodeML algorithms42. Seven codon substitution models with heterogeneous ω values across sites were investigated and compared to detect positively selected sites based on likelihood ratio tests (M0, M1a, M2a, M3, M7, M8, and M8a).
Phylogenetic analyses and ancestral state reconstructions
The phylogenetic positions of the newly sequenced plastomes of Reichardia and Launaea assembled in this study were investigated in the context of their relationships with closely related species in the subtribe Hyoseridinae, including the outgroup species Lactuca sativa from the same tribe (Cichorieae). We further compared the plastome phylogeny with the nrDNA ITS phylogeny to explore phylogenetic incongruence and gain insights into chromosome evolution. To reconstruct the plastome phylogenies, we analyzed 19 plastomes of major genera of the subtribe Hyoseridinae and one outgroup based on whole plastome sequences, as well as concatenated sequences of 80 common protein-coding genes, including only nine newly sequenced species and downloaded plastomes from GenBank (Table 1 and Supplementary Table S5). The nrDNA ITS phylogeny was also reconstructed using the retrieved sequences of 26 accessions from GenBank as specified in Supplementary Table S5, which were selected similarly as the taxa in the plastome phylogeny for the species belonging to the genera Reichardia, Launaea, and Sonchus s.l. in the subtribe Hyoseridinae. The plastome and nrDNA ITS sequences were aligned using MAFFT v. 773, and phylogenetic trees were constructed using IQ-TREE v. 1.4.2, with 1000 bootstrap replicates47 for ML and MrBayes v. 3.2.7a48 for BI. The best-fit evolutionary models in ML trees were scored according to Bayesian information criterion (BIC) scores and weights by testing 88 DNA models of ModelFinder49 implemented in IQ-TREE v. 1.4.2. The evolutionary model of GTR + I + Г (General Time Reversible substitution model with a proportion of invariable sites and a gamma-distributed rate across sites) was selected for Bayesian inference. The Markov chain Monte Carlo simulation (MCMC) had a length of 4,000,000 generations, sampling every 100 generations, for both plastome and nrDNA ITS phylogeny. The average standard deviation of the split frequencies reached below 0.01, which indicates that two runs reached stationarity. The first 25% of samples were discarded as burn-in, and the remaining samples were retained for the construction of a 50% majority-rule consensus tree with clade frequencies.
The reconstructed ML trees for the plastome and nrDNA ITS sequences were used as phylogenetic hypotheses to perform ancestral state reconstruction of chromosome numbers using the R toolkit74 and MrBayes ancestral states with R (MBASR)75. The coded trait scores of chromosome numbers of each taxon were mapped onto both phylogenies, and the ancestral states and shared-derived karyotypes were inferred using MrBayes 3.2.7a48. The parameters in the analyses were set to “character.type = unordered” and “n.samples = 500”.
Supplementary Information
Acknowledgements
We thank Fabio Conti for providing the leaf samples of R. albanica. We also thank Arnoldo Santos-Guerra for his assistance in fieldwork and for providing a photo of R. crystallina. This research was funded by the Basic Science Research Program through the National Research Foundation of Korea (NRF) (Grant Number 2019R1A2C2009841), and the APC was funded by 2019R1A2C2009841.
Author contributions
M.-S.C., J.Y.Y., and S.-C.K. conceived and designed the study; M.-S.C. and J.Y.Y. conducted the experiment; J.A.M. and S.-C.K. provided experimental supports; M.-S.C. prepared the original draft; M.-S.C., J.A.M., and S.-C.K. revised and finalized the manuscript. All authors read and approved the final manuscript.
Data availability
The plastome datasets sequenced and analyzed in the current study are available from GenBank under the accession numbers specified in Table 1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Myong-Suk Cho and JiYoung Yang.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-15235-1.
References
- 1.Axelrod DI. Evolution and biogeography of Madrean-Tethyan sclerophyll vegetation. Ann. Mo. Bot. Gard. 1975;62:280–334. doi: 10.2307/2395199. [DOI] [Google Scholar]
- 2.Takhtajan, A. Floristic regions of the world. 522 (University California Press, 1986).
- 3.Kilian, N., Hand, R. & Raab-Straube, E. von (general editors) (continuously updated). Cichorieae Systematics Portal. http://cichorieae.e-taxonomy.net/portal/, accessed (Accessed on 2 Sept 2021) (2009).
- 4.POWO. Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. http://www.plantsoftheworldonline.org/taxon/urn:lsid:ipni.org:names:10818-1 (Accessed 26 Aug 2021).
- 5.GBIF. Global Biodiversity Information Facility. Reichardia Roth. https://www.gbif.org/es/species/2965909 (Accessed 24 Dec 2021).
- 6.Gallego MJ, Talavera S, Silvestre S. Revision del género Reichardia Roth (Compositae) Lagascalia. 1980;9:159–217. [Google Scholar]
- 7.Conti F, Niketić M, Vukojičić S, Siljak-Yakovlev S, Barina Z, Lakušić D. A new species of Reichardia (Asteraceae, Cichorieae) from Albania and re-evaluation of R. macrophylla. Phytotaxa. 2015;236:121–134. doi: 10.11646/phytotaxa.236.2.2. [DOI] [Google Scholar]
- 8.Bramwell D, Bramwell Z. Wild flowers of the Canary Islands. 2. Editorial Rueda; 2001. pp. 384–386. [Google Scholar]
- 9.Moreno, J. C. Lista Roja de la flora vascular española. Actualización con los datos de la Adenda 2010 al Atlas y Libro Rojo de la Flora Vascular Amenazada, Dirección General de Conservación de la Naturaleza y Sociedad Española de Biología de la Conservación de Plantas: Madrid, Spain, 1–46 (2011).
- 10.Index to Chromosome numbers in Asteraceae. http://www.lib.kobe-u.ac.jp/infolib/meta_pub/sresult (Accessed 9 Sept 2021).
- 11.Löve Á, Kjellqvist E. Cytotaxonomy of Spanish plants. IV. Dicotyledons: Ceslpiniaceae-Asteraceae. Lagascalia. 1974;4:153–211. [Google Scholar]
- 12.Van Loon JC. A cytological investigation of flowering plants from the Canary Islands. Acta. Bot. Neerl. 1974;23:113–124. doi: 10.1111/j.1438-8677.1974.tb00926.x. [DOI] [Google Scholar]
- 13.Gallego MJ. Estudio cariológico de las especies españolas del género Reichardia Roth (Compositae) Lagascalia. 1980;9:149–158. [Google Scholar]
- 14.Siljak-Yakovlev S. Analyse comparative des caryotypes de deux especes du genre Reichardia Roth (R. macrophylla Vis. & Pancic et R. picroides (L.) Roth) et leur relation taxonomique. Caryologia. 1981;34:267–274. doi: 10.1080/00087114.1981.10796891. [DOI] [Google Scholar]
- 15.Kamari G, Felber F, Garbari F. Mediterranean chromosome number reports-8, Reports (936–940) by José A, Mejías. Fl. Medit. 1998;8:245–251. [Google Scholar]
- 16.Kamari G, Blanché C, Siljak-Yakovlev S. Mediterranean chromosome number reports-22. Fl. Medit. 2012;22:211–232. doi: 10.7320/FlMedit22.211. [DOI] [Google Scholar]
- 17.Ritter-Studnička H. Reliktgesellschaften auf Dolomitböden in Bosnien und der Hercegovina. Plant Ecol. 1967;1967(15):190–212. doi: 10.1007/BF01963748. [DOI] [Google Scholar]
- 18.Fernandes, A. & Queirós, M. Contribution à la connaissance cytotaxonomyque des Spermatophytes du Portugal. II. Compositae. Soc. Brot. Bol. 5–121 (1971).
- 19.Siljak-Yakovlev S, Godelle B, Zoldos V, Vallès J, Garnatje T, Hidalgo O. Evolutionary implications of hetero-chromatin and rDNA in chromosome number and genome size changes during dysploidy: A case study in Reichardia genus. PLoS ONE. 2017;12:e0182318. doi: 10.1371/journal.pone.0182318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 2001;16:351–358. doi: 10.1016/S0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- 21.Levin DA. The Role of Chromosomal Change in Plant Evolution. Oxford University Press; 2002. [Google Scholar]
- 22.Schubert I. Chromosome evolution. Curr. Opin. Pl. Evol. 2007;10:109–115. doi: 10.1016/j.pbi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Vallès J, et al. Genome size variation and evolution in the family Asteraceae. Caryologia. 2013;66:221–235. doi: 10.1080/00087114.2013.829690. [DOI] [Google Scholar]
- 24.Recio MC, et al. Phenolics of Reichardia and their taxonomic implications. Biochem. Syst. Ecol. 1992;20:449–452. doi: 10.1016/0305-1978(92)90085-R. [DOI] [Google Scholar]
- 25.Kim S-C, Crawford DJ, Jansen RK. Phylogenetic relationships among the genera of the subtribe Sonchinae (Asteraceae): Evidence from ITS sequences. Syst. Bot. 1996;21:417–432. doi: 10.2307/2419668. [DOI] [Google Scholar]
- 26.Kim S-C, Crawford DJ, Francisco-Ortega J, Santos-Guerra A. A common origin for woody Sonchus and five related genera in the Macaronesian islands: Molecular evidence for extensive radiation. P. Natl. Acad. Sci. USA. 1996;93:7743–7748. doi: 10.1073/pnas.93.15.7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S-C, Crawford DJ, Jansen RK, Santos-Guerra A. The use of a non-coding region of chloroplast DNA in phylogenetic studies of the subtribe Sonchinae (Asteraceae: Lactuceae) Plant Syst. Evol. 1999;215:85–99. doi: 10.1007/BF00984649. [DOI] [Google Scholar]
- 28.Kim SC, Lee C, Mejías JA. Phylogenetic analysis of chloroplast DNA matK gene and ITS of nrDNA sequences reveals polyphyly of the genus Sonchus and new relationships among the subtribe Sonchinae (Asteraceae: Cichorieae) Mol. Phylogenet. Evol. 2007;44:578–597. doi: 10.1016/j.ympev.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Whitton J, Wallace RS, Jansen RK. Phylogenetic relationships and patterns of character change in the tribe Lactuceae (Asteraceae) based on chloroplast DNA restriction site variation. Can. J. Bot. 1995;73:1058–1073. doi: 10.1139/b95-115. [DOI] [Google Scholar]
- 30.Kilian N, Gemeinholzer B, Lack HW. Cichorieae. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, editors. Systematics, evolution, and biogeography of Compositae. I.A.P.T; 2019. pp. 343–383. [Google Scholar]
- 31.Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parks M, Cronn R, Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009;7:84. doi: 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho M-S, Kim SH, Yang J, Crawford DJ, Stuessy TF, López-Sepúlveda P, Kim S-C. Plastid phylogenomics of Dendroseris (Cichorieae, Asteraceae): Insights into structural organization and molecular evolution of an endemic lineage from the Juan Fernández Islands. Front. Plant Sci. 2020;11:594272. doi: 10.3389/fpls.2020.594272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corneille S, Lutz K, Maliga P. Conservation of RNA editing between rice and maize plastids: Are most editing events dispensable? Mol. Gen. Genet. 2000;264:419–424. doi: 10.1007/s004380000295. [DOI] [PubMed] [Google Scholar]
- 35.Kim S-H, Yang JY, Park JS, Yamada T, Maki M, Kim S-C. Comparison of whole plastome sequences between thermogenic skunk cabbage Symplocarpus renifolius and nonthermogenic S. nipponicus (Orontioideae, Araceae) in East Asia. Int. J. Mol. Sci. 2019;20:4678. doi: 10.3390/ijms20194678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsudzuki T, Wakasugi T, Sugiura M. Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol. 2001;53:327–332. doi: 10.1007/s002390010222. [DOI] [PubMed] [Google Scholar]
- 37.Yang JY, Kang GH, Pak JH, Kim S-C. Characterization and comparison of two complete Plastomes of Rosaceae species (Potentilla dickinsii var. glabrata and Spiraea insularis) endemic to Ulleung Island, Korea. Int. J. Mol. Sci. 2020;21:4933. doi: 10.3390/ijms21144933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 39.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z, Nielsen R, Goldman N, Pedersen AMK. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao F, Chen C, Arab DA, Du Z, He Y, Ho SYW. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evolut. 2019;9:3891–3898. doi: 10.1002/ece3.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Bioinformatics. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 43.Yang Z, Wong WS, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 44.Li P, Lou G, Cai X, Zhang B, Cheng Y, Wang H. Comparison of the complete plastomes and the phylogenetic analysis of Paulownia species. Sci. Rep. 2020;10:2225. doi: 10.1038/s41598-020-59204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang P, Shi FX, Li MR, Liu B, Wen J, Xiao HX, Li LF. Positive selection driving cytoplasmic genome evolution of the medicinally important ginseng plant genus Panax. Front. Plant Sci. 2018;9:359. doi: 10.3389/fpls.2018.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sloan DB, Triant DA, Forrester NJ, Bergner LM, Wu M, Taylor DR. A recurring syndrome of accelerated plastid genome evolution in the angiosperm tribe Sileneae (Caryophyllaceae) Mol. Phylogenet. Evol. 2014;72:82–89. doi: 10.1016/j.ympev.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ronquist F, et al. MRBAYES 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kilian N. Revision of Launaea Cass. (Compositae, Lactuceae, Sonchinae) Englera. 1997;17:3–478. doi: 10.2307/3776755. [DOI] [Google Scholar]
- 51.Mejías JA, Andrés C. Karyological studies in Iberian Sonchus (Asteraceae: Lactuceae): S. oleraceus, S. microcephalus and S. asper and a general discussion. Folia Geobot. 2004;39:275–291. doi: 10.1007/BF02804782. [DOI] [Google Scholar]
- 52.Stebbins GL, Jenkins JA, Walters MS. Chromosomes and phylogeny in the Compositae, tribe Cichorieae. Univ. Calif. Publ. bot. 1953;26:401–430. [Google Scholar]
- 53.Tomb AS. Lactuceae-systematic review. In: Heywood VH, Harborne JB, Turner BL, editors. The biology and chemistry of the Compositae. Academic Press; 2013. pp. 1067–1079. [Google Scholar]
- 54.Tomb AS, Chambers KL, Kyhos DW, Powell AM, Raven PH. Chromosome numbers in the Compositae. XIV. Lactuceae. Am. J. Bot. 1978;65:717–721. doi: 10.1002/j.1537-2197.1978.tb06129.x. [DOI] [Google Scholar]
- 55.Tremetsberger K, Gemeinholzer B, Zetzsche H, Blackmore S, Kilian N, Talavera S. Divergence time estimation in Cichorieae (Asteraceae) using a fossil-calibrated relaxed molecular clock. Org. Divers. Evol. 2013;13:1–13. doi: 10.1007/s13127-012-0094-2. [DOI] [Google Scholar]
- 56.Krijgsman W, Hilgen FJ, Raffi I, Sierro FJ, Wilson DS. Chronology, causes and progression of the Messinian salinity crisis. Nature. 1999;400:652–655. doi: 10.1038/23231. [DOI] [Google Scholar]
- 57.Jorgensen TH, Olesen JM. Adaptive radiation of island plants: Evidence from Aeonium (Crassulaceae) of the Canary Islands. Perspect. Plant Ecol. Evol. Syst. 2001;4:29–42. doi: 10.1078/1433-8319-00013. [DOI] [Google Scholar]
- 58.Francisco-Ortega J, Jansen RK, Santos-Guerra A. Chloroplast DNA evidence of colonization, adaptive radiation, and hybridization in the evolution of the Macaronesian flora. Proc. Natl. Acad. Sci. USA. 1996;93:4085–4090. doi: 10.1073/pnas.93.9.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Imamura R, Santos-Guerra A, Kondo K. A molecular phylogenetic relationship of certain species of Argyranthemum found in the Canary Islands of Spain on the basis of the internal transcribed spacer (ITS) Chromosome Bot. 2015;10:75–83. doi: 10.3199/iscb.10.75. [DOI] [Google Scholar]
- 60.Böhle UR, Hilger HH, Martin WF. Island colonization and evolution of the insular woody habit in Echium L. (Boraginaceae) Proc. Natl. Acad. Sci. USA. 1996;93:11740–11745. doi: 10.1073/pnas.93.21.11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia-Maroto F, Manas-Fernández A, Garrido-Cárdenas JA, López Alonso D, Guil-Guerrero JL, Guzmán B, Vargas P. D6-Desaturase sequence evidence for explosive Pliocene radiations within the adaptive radiation of Macaronesian Echium (Boraginaceae) Mol. Phylogenet. Evol. 2009;52:563–574. doi: 10.1016/j.ympev.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 62.Gruenstaeudl M, Santos-Guerra A, Jansen RK. Phylogenetic analyses of Tolpis Adans. (Asteraceae) reveal patterns of adaptive radiation, multiple colonization and interspecific hybridization. Cladistics. 2013;29:416–434. doi: 10.1111/cla.12005. [DOI] [PubMed] [Google Scholar]
- 63.Wendel JF, Doyle JJ. Phylogenetic incongruence: window into genome history and molecular evolution. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants II DNA Sequencing. Springer; 1998. pp. 265–296. [Google Scholar]
- 64.Rieseberg LH, Whitton J, Linder CR. Molecular marker incongruence in plant hybrid zones and phylogenetic trees. Acta Bot. Neerl. 1996;45:243–262. doi: 10.1111/j.1438-8677.1996.tb00515.x. [DOI] [Google Scholar]
- 65.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 66.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lohse M, Drechsel O, Bock R. Organellar genome DRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2009;25:1451–1452. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 70.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis v7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mower JP. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009;37:W253–W259. doi: 10.1093/nar/gkp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. NISC comparative sequencing program. LAGAN and multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.R Development Core Team. R: A language and environment for statistical computing. http://www.R-project.org/ (Accessed 21 Oct 2021) (R Foundation for Statistical Computing, 2005).
- 75.Heritage S. MBASR: Workflow-simplified ancestral state reconstruction of discrete traits with MrBayes in the R environment. bioRxiv. 2021 doi: 10.1101/2021.01.10.426107. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The plastome datasets sequenced and analyzed in the current study are available from GenBank under the accession numbers specified in Table 1.