Abstract
Background
Biliary atresia (BA) is the most common cholestatic liver disease in neonates. Herein, we aimed at characterizing the gut microbiota and fecal bile acid profiles of BA patients, defining the correlations between them, and evaluating the relationship between the clinical pathogenesis and changes in the gut microbiota and bile acid profiles.
Methods
A total of 84 fecal samples from BA patients (n = 46) and matched healthy controls (HCs, n = 38) were subjected to sequencing by 16S rRNA gene amplification, and fecal bile acid were analyzed by targeted metabolomics.
Findings
Compared with the controls, a structural separation of the intestinal flora of BA patients was uncovered, which was accompanied by changes in the composition of fecal bile acids. In the BA group, Actinobacillus, Monoglobus, and Agathobacter were enriched in patients without cholangitis (p < 0.05). Selenomonadaceae and Megamonas were more abundant in patients without recurrent cholangitis episodes (p < 0.05), while Lachnospiraceae and Ruminococcaceae were enriched in patients with multiple recurrences of cholangitis (p < 0.05). Postoperative jaundice clearance was associated with Campylobacter and Rikenellaceae (p < 0.05), and tauroursodeoxycholic acid was associated with jaundice clearance (p < 0.001).
Conclusion
BA patients are characterized by different compositions of gut microbiota and bile acids, and their interaction is involved in the process of liver damage in BA, which may be closely related to the occurrence of postoperative cholangitis and jaundice clearance.
Keywords: biliary atresia, gut microbiome, bile acid spectrum, cholangitis, jaundice clearance
Introduction
Biliary atresia (BA) is a severe neonatal disease of the hepatobiliary system, in which the intrahepatic and extrahepatic bile ducts are occluded, resulting in cholestasis and progressive liver fibrosis (Kasahara et al., 2017; Lane and Murray, 2017). Even after successful Kasai surgery (portoenterostomy), the majority of children progress to end-stage cirrhosis (Asai et al., 2015). A study showed that the native liver survival rates at 10 and 20 years after Kasai was 70.7% and 61.5% respectively (Chung et al., 2021).
The enterohepatic axis is an anatomical and physiological bridge that connects the intestine and liver (Long et al., 2017). Intestinal microflora balance is the key to maintaining immune homeostasis of the gut-liver axis. Changes in intestinal permeability and gut bacteria profiles are related to many diseases, and dysbacteriosis is an important cause of liver diseases (Feenstra et al., 1985; Hiippala et al., 2018; Tripathi et al., 2018). Gut microbiota disorders have been found in a variety of liver diseases, such as alcoholic hepatitis, cirrhosis, ischemic liver injury, rejection in liver transplantation, and cholestatic liver diseases (Sun et al., 2017; Song et al., 2021; Song et al., 2021; Liu et al., 2021). Previous study has reported that disturbed gut microbiota structures can be observed during the early and late stages of BA (Song et al., 2021). Liver damage in BA is directly or indirectly exacerbated by the interplay between Klebsiella, Veronella, and Enterococcus enrichment and impaired tryptophan and bile acid metabolism (Song et al., 2021). Several studies have found the dysbiosis of the gut microbiome to be associated with BA, as well as an increase in the abundances of Proteus, Enterococcus, Streptococcus, and other opportunistic pathogens, while the abundances of some butyrate-producing bacteria were found to be reduced (Wang et al., 2019; Wang et al., 2020; Song et al., 2021).
Bile acids are produced by the liver from cholesterol through classical or alternative pathways, and primary bile acids are dehydroxylated by the gut microbiome to form secondary bile acids in the intestine. Individual bile acids have recently been found to be useful for the differentiation and assessment of liver injury (Fu et al., 2020). Plasma levels of selected bile acids were higher in BA patients than that in cholestatic non-BA patients; more specifically, plasma levels of taurochenodeoxycholic acid (TCDCA) and chenodeoxycholic acid (CDCA) were increased and decreased, respectively. Fecal bile acid levels were significantly lower in BA patients due to the disruption of normal biliary drainage, with a significant difference between BA and healthy control (HC) groups (Golden et al., 2018).
Previous studies have provided a preliminary description of the compositions of the gut microbiota and bile acids in patients with BA, but whether a correlation exists between the two is still unclear. In particular, there is a lack of knowledge on their association with important disease features of BA, such as post-Kasai cholangitis, jaundice clearance, and prognosis. Thus, a comprehensive analysis of the gut microbiota in a large BA cohort and a thorough exploration of its relationship with the bile acid profile are needed. Herein, we aimed at characterizing the gut microbiota and fecal bile acid profiles of BA patients compared with those of HCs, unraveling possible correlations between them and evaluating the relationship between the clinical pathogenesis of BA and changes in gut microbiota and bile acid profiles.
Methods
Study Group
Patients with BA (n = 46) and age-matched healthy controls (HC, n = 38) were recruited from Beijing Children’s Hospital, Capital Medical University, from October 2019 to June 2021. Forty-six patients with BA had their diagnosis based on intraoperative cholangiography and liver biopsy. The patients were enrolled with the following criteria: age less than 6 months; no remaining gastrointestinal diseases; no history of antibiotic and probiotic use within 2 weeks prior to enrollment. HC consists of children in the same age group as BA with non-digestive diseases, e.g. children with polydactylism, precocious heart disease, inguinal hernia, etc. Stool samples were freshly collected as soon as possible after admission preoperatively, and frozen them at -80°C within one hour. Follow-up and long-term sequelae were collected from outpatient and inpatient records, as well as via clinical and telephone communication. All patients with BA received antibiotics, ursodeoxycholic acid, steroids and probiotics post-operatively. This study was approved by the Ethics Committee of Beijing Children’s Hospital (2019-k-386), and informed consent was obtained from the guardians of each participant before sample collection.
Gene Amplicon Sequencing of 16S rRNAs and Data Processing
DNA was extracted using the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) according to the manufacturer’s instructions. The hypervariable V3-V4 regions of the bacterial 16S rRNA gene were amplified with 338F/806R primers using an ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, USA). Purified amplicons were pooled in equimolar amounts and paired-end sequenced on an Illumina MiSeq PE300 platform (Illumina, San Diego, USA) according to the standard protocols employed by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Raw 16S rRNA gene sequencing reads were demultiplexed, quality-filtered using fastp version 0.20.0 (Chen et al., 2018), and merged using FLASH version 1.2.7 (Magoč and Salzberg, 2011). Operational taxonomic units (OTUs) with a 97% similarity cutoff were clustered using UPARSE version 7.1 (Edgar, 2013), and chimeric sequences were identified and removed. Taxonomic classification of the sequences of each sample was carried out individually using RDP Classifier version 2.2 (Wang et al., 2007) according to the RDP database.
Quantitative Analysis of Bile Acids
The bile acids from the human fecal samples were quantitatively conducted by Metabo-Profile Inc. (Shanghai, China) according to their previously published methods (Xie et al., 2015; Lan et al., 2016; Zheng et al., 2017). All of the bile acid standards were purchased from Steraloids Inc. (Newport, RI, USA) and TRC Chemicals (Toronto, ON, Canada) or synthesized from the MPB laboratory, and 6 stable isotope-labeled standards were obtained from C/D/N Isotopes Inc. (Quebec, Canada) and Steraloids Inc. (Newport, RI, USA). Ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS, ACQUITY UPLC-Xevo TQ-S, Waters Corp., Milford, MA, USA) was used to quantify for the bile acids in human fecal samples. Fecal samples (10 mg) were accurately weighed and homogenized with 20 µL of deionized water and 180 µL of acetonitrile/methanol (v/v=8:2) containing 6 internal standards for 3 min (BB24, Next Advance, Inc, Averill Park, NY, USA). After centrifugation at 13,500 rpm for 20 min at 4°C (Microfuge 20R, Beckman Coulter, Inc., Indianapolis, IN, USA). The supernatant (20 µL) was transferred to a 96-well plate and lyophilized in a freeze dryer (Labconco, Kansas City, MO, USA), and then diluted with 1:1 solvent mixture of acetonitrile/methanol (80/20) and ultrapure water, mixed, and centrifuged. 5 µL aliquot of the supernatant of each sample was used for UPLC-MS/MS analysis. The raw data files generated by UPLC-MS/MS analysis were processed using MassLynx software (v4.1, Waters, Milford, MA, USA), upon which they were integrated, standard curves were created, and all bile acids were quantified. The bile acids detected in this study as follows: 3-DHCA: 3-dehydrocholic acid; 6-ketoLCA: 6-ketolithocholic acid; 7-ketoDCA: 7-ketodeoxycholic acid; 12-ketoLCA: 12-ketolithocholic acid; βUDCA: 3β-ursodeoxycholic acid; C4: 7a-Hydroxy-cholestene-3-one; CA: cholic acid; CDCA: chenodeoxycholic acid; CDCA-3Gln: chenodeoxycholic acid-3-β-D-glucuronide; DCA: deoxycholic acid; GCA: glycocholic acid; GCDCA: glycochenodeoxycholate; GDCA: glycodeoxycholic acid; GHCA: glycohyocholate; GLCA: glycolithocholate; GUDCA: glycoursodeoxycholic acid; HCA: γ-muricholic acid/hyocholic acid; LCA: lithocholic acid; LCA-3S: lithocholic acid 3 sulfate; NorCA: Nor Cholic acid; TCA: taurocholic acid; TCDCA: taurochenodeoxycholate; TDCA: taurodeoxycholate; TUDCA: tauroursodeoxycholic acid; UDCA: ursodeoxycholic acid.
Statistical Analysis
In this study, SPSS Statistics 26 software was used for statistical analysis of the data. Wilcoxon rank-sum test and Fisher’s exact test were used to identify statistical differences in continuity variables and categorical variables, respectively, between the two groups. p < 0.05 was defined as statistically significant. Redundancy analysis (RDA) was conducted to determine the effect of environmental factors on the structure of the intestinal flora. Principal coordinates analysis (PCoA) between species was used to evaluate the β diversity of the gut microbial composition. Linear discriminant analysis effect size (LEfSe) analysis using linear discriminant analysis (LDA) was used to determine the taxonomic differences between BA and HC groups. The community heatmap is a color gradient illustrating community composition and abundance information at the genus level. Unidimensional statistical analysis was used to screen for differential bile acids between BA and HC groups. The non-parametric Wilcoxon test was performed to analyze statistically significant differences at different taxonomic levels between the different cohorts. Spearman correlation coefficients were calculated to investigate the relationship between clinical parameters, microbial composition, and bile acids.
Results
Characteristics of Gut Microbiota Profiles in the Study Population
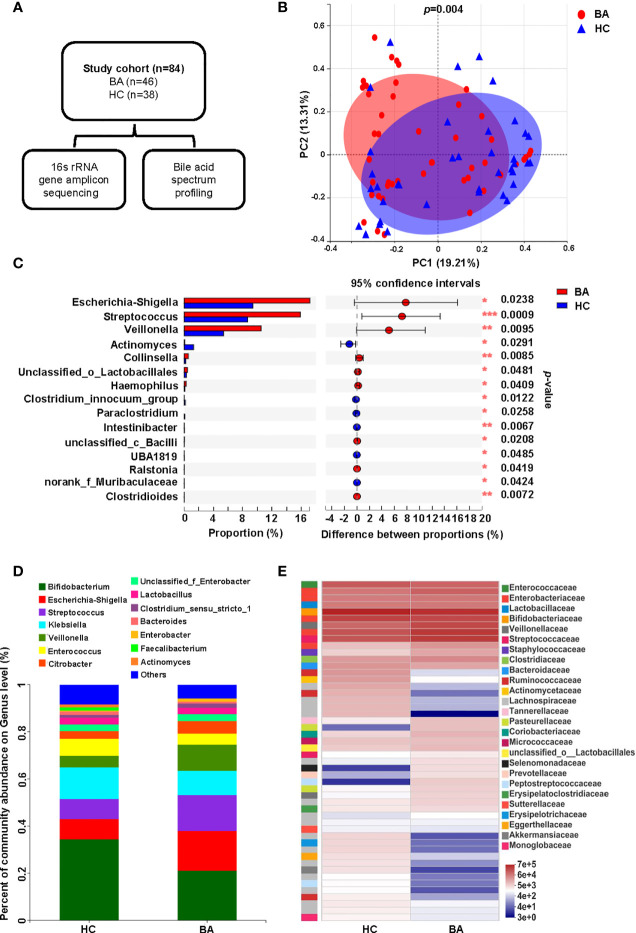
In this study, fecal samples from 84 individuals were sequenced after 16S rRNA gene amplification to assess the composition of the gut microbiome, and a targeted metabolomics approach was used to detect the bile acid composition in each fecal sample ( Figure 1A ). Common demographic data and clinical characteristics are shown in Table 1 . A further RDA analysis was conducted to determine the effect of age, sex, mode of delivery, gestational age and feeding pattern on the structure of the intestinal flora ( Supplementary Figure 1 ). The permutation test ( Supplementary Table 1 ) showed that none of the above environmental factors had a significant effect (all p > 0.05) on the composition of the fecal microbiota, so we did not consider these confounding effects in the subsequent analysis. To investigate the differences in gut microbiota between patients with BA and HC individuals, we determined the changes in the gut microbiota of the two groups and evaluated the differences in community composition using PCoA based on Bray-Curtis differences, which showed that the overall microbial composition in the feces of the BA group deviated from that of the HC group ( Figure 1B ). LDA scores of the gut microbiota are shown in Supplementary Figure 2A , and the correlation between fecal microbiota composition and samples is shown in Supplementary Figures 2B, C . At the genus level, the BA and HC groups were enriched with 77 and 45 differential species, respectively. A specific list of differential species is shown in Supplementary Table 2 . Figures 1C , 1D show the composition of the fecal microbiota in BA and HC groups, indicating significant differences between the two groups at the genus level. Among them, Bifidobacterium, Escherichia-Shigella, Klebsiella, Streptococcus, and Veillonella were the dominant species in both the BA and HC groups. Escherichia-Shigella, Streptococccus, Veillonella, and Citrobacter were more abundant in the individuals of the BA group than in HC individuals, whereas Bifidobacterium, Klebsiella, and Enterobacter were more abundant in the case of the latter ( Figure 1D ). As shown in Figure 1C , Escherichia-Shigella, Streptococccus, and Veillonella were significantly enriched in the BA group, whereas Actinomyces was significantly less abundant in BA patients. These differences were determined to be statistically significant (all p < 0.05). Figure 1E shows the differences in microbial abundance between BA and HC groups, with Faecalibacterium, Actinomyces, Agathobacter, Blautia, Eggerthella, as well as other genera being of lower abundance in the BA group than in the HC group. In conclusion, the overall structure of the gut microbiota of BA patients was significantly different from that of the HC group.
Figure 1.
Fecal microbiome variations and composition in BA and HC groups. (A) Overview of the study design. (B) Beta diversity (principal coordinates analysis based on the Bray-Curtis distance of genus abundance) between the two groups. (C) Microbiota with significantly different abundances between the two groups at the genus level (*p < 0.05, **p < 0.01, and ***p < 0.001). (D) Bar diagram of the fecal microbial composition in the two groups. (E) Community heatmap presenting information on the genus-level composition and abundance of the community between the two groups, with color changes reflecting the similarities and differences in community composition. BA, biliary atresia; HC, health control; PCoA, principal coordinated analysis.
Table 1.
Clinical characteristics of the BA and HC patients.
| Clinical characteristics | BA (n = 46) | HC (n = 38) | p-value |
|---|---|---|---|
| Female, n (%) | 22 (47.8) | 24 (63.2) | 0.273 |
| Age, days, median (IQR, Q1-Q3) | 55.0 (40.8-75.0) | 58.0 (46.5-115.5) | 0.100 |
| AST, U/L, median (IQR, Q1-Q3) | 242.5 (137.4-338.0) | 43.2 (37.1-67.9) | < 0.001 |
| ALT, U/L, median (IQR, Q1-Q3) | 131.0 (75.0-237.3) | 24.6 (19.2-41.1) | < 0.001 |
| GGT, U/L, median (IQR, Q1-Q3) | 336.5 (185.3-723.1) | 35.9 (18.1-103.6) | 0.001 |
| TBIL, μmol/L, median (IQR, Q1-Q3) | 168.1 (131.0-202.2) | 22.3 (9.9-71.0) | < 0.001 |
| DBIL, μmol/L, median (IQR, Q1-Q3) | 114.5 (83.1-146.1) | 4.1 (1.9-13.8) | < 0.001 |
| IBIL, μmol/L, median (IQR, Q1-Q3) | 49.6 (42.3-67.2) | 17.8 (7.8-34.1) | < 0.001 |
| TBA, μmol/L, median (IQR, Q1-Q3) | 114.3 (90.8-150.3) | 7.1 (3.5-11.9) | < 0.001 |
ALT, alanine aminotransferase; AST, aspartate aminotransferase; BA, biliary atresia; DBIL, direct bilirubin; GGT, gamma-glutaryl transferase; HC, health control; IBIL, indirect bilirubin; TBA, total bile acid; TBIL, total bilirubin.
Characteristics of Bile Acid Profiles in the Study Population
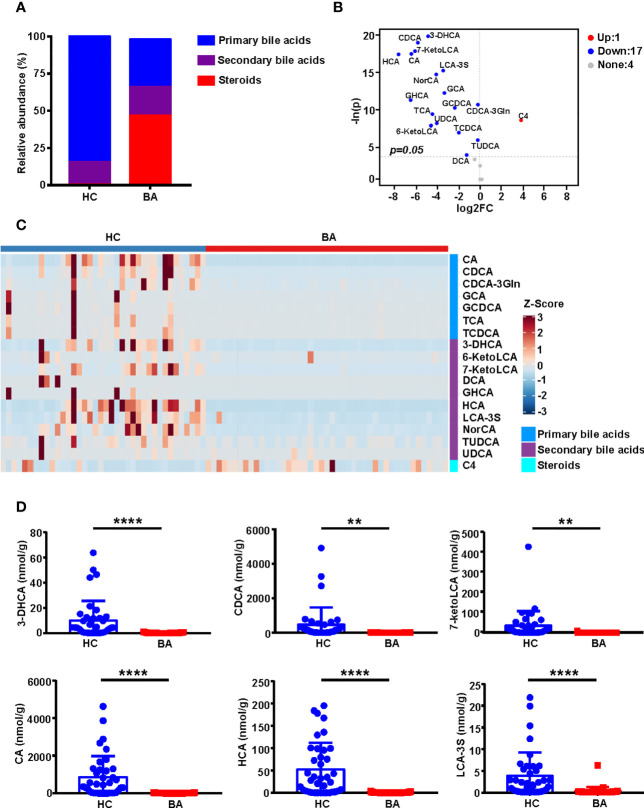
Figures 2A and 2C show statistically significant differences in the composition of primary bile acids, secondary bile acids, and steroids between BA and HC groups. The specific bile acid composition of each sample is shown in Supplementary Figure 3A . Compared to the HC group, the BA group was characterized by a lower abundance of primary bile acids and comparable levels of secondary bile acids but a higher abundance of steroids ( Figure 2A ). A total of 18 differential metabolites were screened based on unidimensional statistical analysis (all p < 0.05, Figure 2C ). As shown in the volcano plot presented in Figure 2B , compared to the HC group, the metabolite C4, highlighted in the upper right corner, was elevated in BA patients, whereas 17 bile acids, including CDCA, CA, HCA, GCA, and TCA, which are presented in the upper left corner, were low in the BA group. Box plots of the top 6 differential metabolites ranked by unidimensional statistical analysis are shown in Figure 2D .
Figure 2.
Bile acid profile composition in BA and HC groups. (A) Stacked histograms of the relative abundances of median values of various bile acids in the two groups of samples. (B) Volcano plot showing the differential metabolites screened based on a unidimensional statistical analysis. The thresholds in the plot were set as follows: p < 0.05 and between-group variation multiplier, absolute value of log2 fold change (FC) ≥ 0. A total of 18 differential metabolites were obtained from the unidimensional analysis according to the screening criteria. Bile acids elevated or decreased are highlighted in red and blue, respectively. (C) The heatmap highlights the difference in abundance of bile acid composition between the two groups, with the changes in color reflecting the similarities and differences between the different groups. (D) Box plots of the top 6 differential metabolites for unidimensional statistical analysis of p values (**p < 0.01 and ****p < 0.0001). 3-DHCA, 3-dehydrocholic acid; 6-ketoLCA, 6-ketolithocholic acid; 7-ketoLCA, 7-ketolithocholic acid; BA, biliary atresia; C4, 7a-Hydroxy-cholestene-3-one; CA, cholic acid; CDCA, chenodeoxycholic acid; CDCA-3Gln, chenodeoxycholic acid-3-β-D-glucuronide; DCA, deoxycholic acid; FC, Fold Change; GCA, glycocholic acid; GCDCA, glycochenodeoxycholate; GHCA, glycohyocholate; HC, health control; HCA, γ-muricholic acid/hyocholic acid; LCA-3S, lithocholic acid 3 sulfate; NorCA, nor cholic acid; TCA, taurocholic acid; TCDCA, taurochenodeoxycholate; TUDCA, tauroursodeoxycholic acid; UDCA, ursodeoxycholic acid.
Association Between Disease-Related Fecal Microbiota, Fecal Bile Acid Profiles, and Preoperative Serum Biochemical Indices
To evaluate the correlations between indicators of liver function and fecal microbiome profiles in BA, we calculated Spearman correlation coefficients ( Figure 3A ). Some intestinal microbiota genera were highly correlated with clinical indicators. More specifically, Eggerthella, Eubacterium_halli_group, Fusicatenibacter, Bacteriodes, and Faecalibacterium were found to be significantly negatively correlated with indicators of liver function (all p < 0.05); therefore, the above-mentioned bacteria with reduced abundance in BA patients were considered potentially beneficial genera. In addition, Dialister, Megasphaera, and an unclassified Villonellaceae strain showed significant positive correlations with total bile acids (TBA, p < 0.05), and these species might be potentially pathogenic. Furthermore, most of the fecal bile acids were negatively correlated with indicators of liver function ( Supplementary Figure 3B ).
Figure 3.
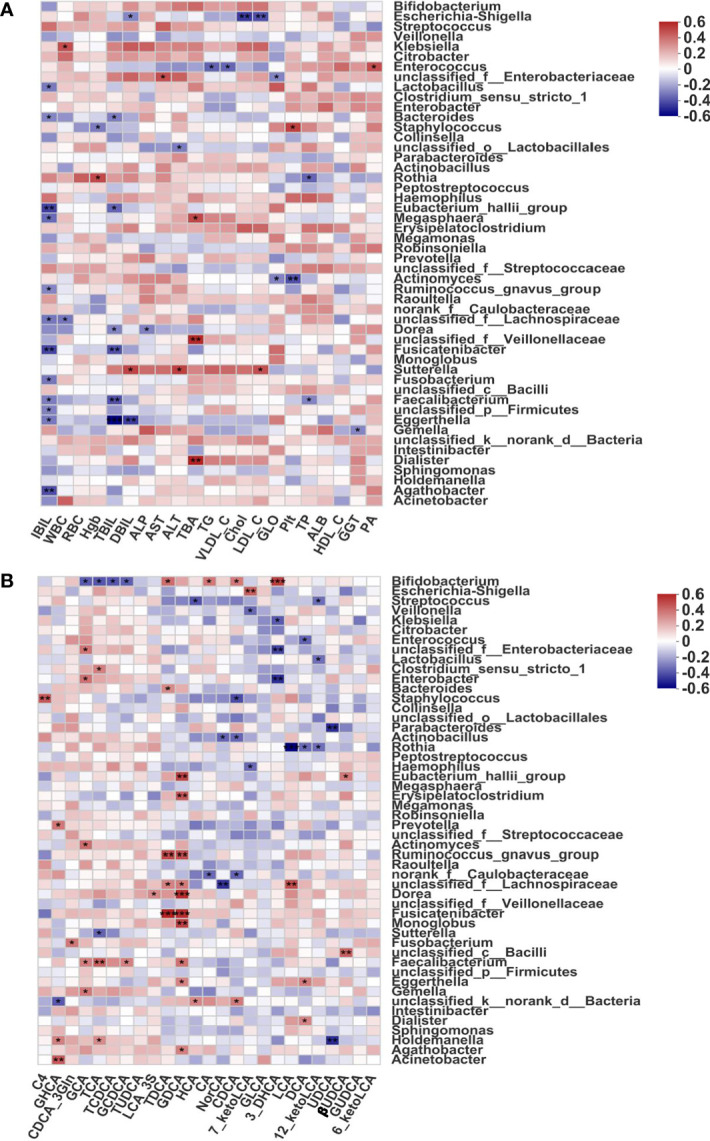
Associations of disease-linked microbiota with clinical indicators and bile acids. (A) Heatmap of Spearman correlation coefficients between the fecal microbiome and indicators of liver function. (B) Heatmap of Spearman correlation coefficients between the fecal microbiome and bile acid profiles in BA. Blue and red denote negative and positive correlations, respectively. The depth of the color indicates the correlation between the genus and environmental factors. *p < 0.05, **p < 0.01, ***p < 0.001. ALB, albumin; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; Chol, cholesterol; DBIL, direct bilirubin; GGT, gamma-glutaryl transferase; GLO, globulin; HDL_C, high-density lipoprotein cholesterol; Hgb, hemoglobin; IBIL, indirect bilirubin; LDL_C, low-density lipoprotein cholesterol; PA, prealbumin; Plt, platelet; RBC, red blood cell; TBA, total bile acid; TBIL, total bilirubin; TG, triglyceride; TP, total protein; VLDL_C, very low-density lipoprotein cholesterol; WBC, white blood cell; 3-DHCA, 3-dehydrocholic acid; 6-ketoLCA, 6-ketolithocholic acid; 7-ketoLCA, 7-ketolithocholic acid; 12-ketoLCA, 12-ketolithocholic acid; βUDCA, 3β-ursodeoxycholic acid; C4, 7a-hydroxy-cholestene-3-one; CA, cholic acid; CDCA, chenodeoxycholic acid; CDCA-3Gln, chenodeoxycholic acid-3-β-D-glucuronide; DCA, deoxycholic acid; GCA, glycocholic acid; GCDCA, glycochenodeoxycholate; GDCA, glycodeoxycholic acid; GHCA, glycohyocholate; GLCA, glycolithocholate; GUDCA, glycoursodeoxycholic acid; HCA, γ-muricholic acid/hyocholic acid; LCA, lithocholic acid; LCA-3S, lithocholic acid 3 sulfate; NorCA, nor cholic acid; TCA, taurocholic acid; TCDCA, taurochenodeoxycholate; TDCA, taurodeoxycholate; TUDCA, tauroursodeoxycholic acid; UDCA, ursodeoxycholic acid.
In addition, one of the typical characteristics of BA is a decrease in fecal bile acids (Wang et al., 2020), which are synthesized in the liver from cholesterol, converted into primary bile acids, and metabolized in the intestine via the enterohepatic circulation. Normal biliary drainage was shown to be disrupted in BA, and we further found that the abundance of fecal bile acids was significantly decreased. Thus, we hypothesized that decreased biliary drainage could be one of the most important causes of gut microbiome dysbiosis among patients with BA. To investigate potential correlations between the gut microbiome and bile acids in BA, we calculated Spearman correlation coefficients and selected the 50 most abundant microorganisms for analysis. As shown in Figure 3B , Eubacterium_halli_group, Erysipelatoclostridium, Ruminococcus_gnavus_group, Fusicatenibacter, Dorea, and Monoglobus were all highly positively correlated with GDCA (all p < 0.01), a predictive marker for liver damage (Drzymała-Czyż et al., 2021). Ruminococcus_gnavus_group and Fusicatenibacter also showed a highly significant positive correlation with TDCA (all p < 0.01), Bifidobacterium showed a significant negative correlation with conjugated bile acids (GCA, TCA, TCDCA, and GCDCA; all p < 0.05), and Rothia was negatively correlated with secondary bile acids (DCA and LCA; both p < 0.05).
Gut Microbiota Composition Correlates With Cholangitis After Kasai Surgery in BA Patients
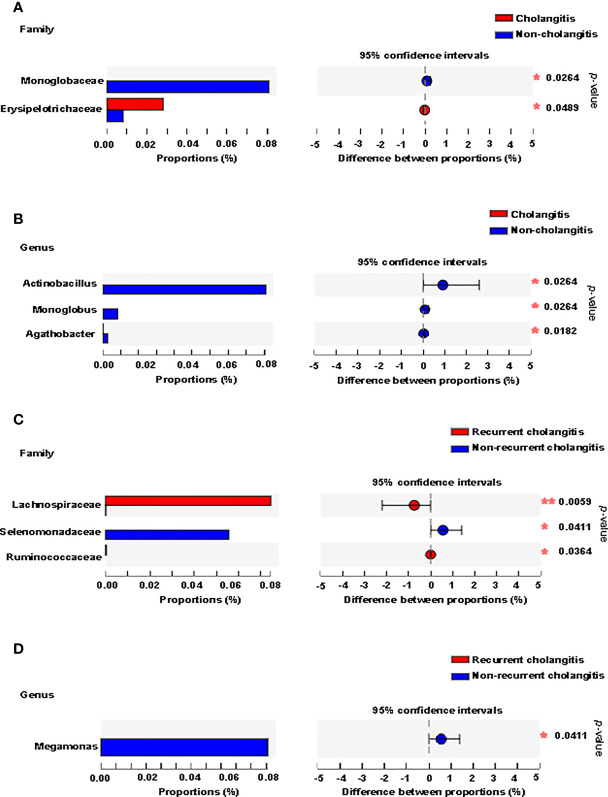
Clinical signs (unexplained fever, recurrent jaundice, and acholic stools) and laboratory tests (raised bilirubin levels, elevated indicators of inflammation) are used to make the diagnosis of cholangitis. To further understand whether gut microbiota composition was associated with postoperative cholangitis after Kasai surgery among patients with BA, we divided the patients into two groups according to the occurrence of cholangitis. We analyzed the microbiota distribution at the family and genus levels and compared the differences between the groups. At the family level, Erysipelotrichaceae was enriched in the cholangitis group, whereas Monoglobaceae was enriched in the non-cholangitis group ( Figure 4A ). At the genus level, Actinobacillus, Monoglobus, and Agathobacter were enriched in the non-cholangitis group ( Figure 4B ).
Figure 4.
Fecal microbiota distribution of BA patients with cholangitis after Kasai surgery. Microbiota with significantly different abundances between the cholangitis and non-cholangitis groups at the family (A) and genus (B) levels are shown. Microbiota with significantly different abundances between recurrent cholangitis and non-recurrent cholangitis groups at the family (C) and genus (D) levels are presented. *p < 0.05, **p < 0.01.
Previous studies have suggested that the greater the number of recurrences of cholangitis, the more severe the patient’s liver damage (Chen et al., 2018). Thus, we divided patients with postoperative episodes of cholangitis into two groups based on cholangitis recurrence. Analysis of the gut community composition was also performed at the family and genus levels. At the family level, Lachnospiraceae and Ruminococcaceae were enriched in patients with multiple recurrences of postoperative cholangitis, while Selenomonadaceae was enriched in the group with non-recurrent cholangitis ( Figure 4C ). At the genus level, although Megamonas was characterized by an abundance score below 0.01 in both groups, the difference in relative abundance of Megamonas between the two groups was statistically significant (p = 0.041, Figure 4D ).
Gut Microbiota and Bile Acid Profiles Are Associated With Jaundice Clearance in BA
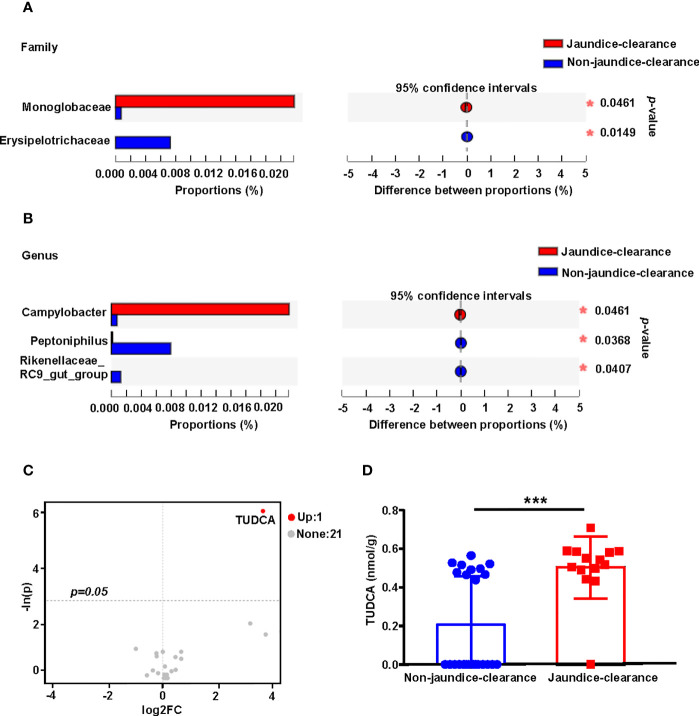
Previous studies have confirmed that jaundice clearance after Kasai surgery is an important predictor of BA prognosis (Nakajima et al., 2018). In order to explore the relationship between the gut microbiota and bile acid profiles and jaundice clearance at 6 months after Kasai surgery (total bilirubin < 1.5 mg/dL), the BA patients were divided into jaundice-clearance and non-jaundice-clearance groups. The distribution of microbiota in the two groups was compared at the family and genus levels. At the family level, Campylobacteraceae was enriched in the jaundice-clearance group, while Rikenellaceae was enriched in the non-jaundice-clearance group ( Figure 5A ). At the genus level, Campylobacter was enriched in the jaundice-clearance group, whereas Peptoniphilus and Rikenellaceae_RC9_gut_group were enriched in the non-jaundice-clearance group ( Figure 5B ).
Figure 5.
Fecal microbiota distribution and bile acid profile for patients with jaundice clearance at 6 months after Kasai surgery. Microbiota with significantly different abundances at the family (A) and genus (B) levels between jaundice-clearance and non-jaundice-clearance groups. (C) The volcano plot shows the differential metabolites screened based on a unidimensional statistical analysis. The thresholds in the plot were set as follows: p < 0.05 and between-group variation multiplier, absolute value of FC ≥ 0. (D) Box diagram of different levels of TUDCA between jaundice-clearance and non-jaundice-clearance groups. *p < 0.05, ***p < 0.001. FC, Fold Change; TUDCA, tauroursodeoxycholic acid.
To understand the relationship between fecal bile acids and postoperative jaundice clearance in patients with BA, we screened for differential bile acids between the two groups based on unidimensional statistical analysis. As shown in the volcano plot in Figure 5C , only one differential bile acid was identified, with TUDCA highlighted in red in the upper right corner of the jaundice-clearance group compared to the non-jaundice-clearance group. Figure 5D also shows the increased level of TUDCA in the non-jaundice-clearance group.
Discussion
In this study, we performed a comprehensive multi-omics analysis of the gut microbiota and bile acid compositions in BA. There is growing evidence in support of gut barrier dysfunction and dysbiosis constituting important causes of diseases affecting the liver as well as other organs (Tang et al., 2018; Wei et al., 2020). Bile acids, important substances associated with liver injury and cholestatic disease, are currently being investigated for their potential to serve as identifiers of cholestatic disease (Zhou et al., 2012). In this study, we explored the relationship between gut microbiota and bile acid composition in BA and its impact on BA pathogenesis.
Gut microbiota entering bile through the gut-liver axis or changes in the composition of the gut microbiota can lead to the activation of mucosal immune responses, which in turn leads to bacterial translocation and immune cell migration to the liver, thereby damaging the biliary tract and hepatocytes, consequently being associated with inflammation-mediated liver injury (Tajeddin et al., 2016; Yang et al., 2020). Compared to HC individuals, the fecal microbial compositions of BA patients were significantly distinct, suggesting that a disruption of the gut microbiota occurs in BA. In terms of specific genus composition, the abundances of Escherichia-Shigella, Streptococccus, and Veillonella were significantly increased in the BA group. Escherichia-Shigella has been shown to be increased in abundance in the feces of patients with ulcerative colitis (UC) (Kojima et al., 2012). Streptococcus and Escherichia-Shigella have been proven to be potential risk factors for UC deterioration. The severity of UC has been shown to be positively correlated with the abundance of Escherichia-Shigella in the inflamed mucosa (Kojima et al., 2012). Furthermore, increased abundance of Escherichia-Shigella has also been demonstrated in animal models of UC and was previously shown to be positively correlated with serum IL-6 levels (Jialing et al., 2020). indicating that Escherichia-Shigella is involved in the development of inflammatory diseases. Streptococccus is a major human pathogen that mainly causes localized infections of the skin and mucous membranes, which can adsorb high molecular weight kininogen in plasma, activate contact factors, and induce bradykinin release (Köhler et al., 2020). Veillonella is a bile acid-sensitive bacterium that is enriched when bile acids are inhibited. It has been reported in previous studies that bile acids are inhibited in the intestine of patients with cirrhosis and that Veillonella is enriched in the intestinal flora of patients with cirrhosis (Chen et al., 2016; Oh et al., 2020; Loomba et al., 2021; Song et al., 2021). Therefore, we suggest that the above-mentioned flora may be potentially pathogenic in BA. In contrast, Actinomyces is a normal bacterium colonizing the gastrointestinal tract, which cannot release exotoxins but produces formate, acetate, succinate, and lactate (Li et al., 2018). The metabolic potential of Actinomyces may be to break down and recycle organic matter in the human gastrointestinal system (Hanning and Diaz-Sanchez, 2015). In the present study, Actinomyces was significantly less abundant in BA patients; however, there is a lack of relevant studies on the biological functions of Actinomyces in the gastrointestinal system.
The intestinal microbiota is closely linked to the bile acid profile. Bile acids are synthesized in the liver and secreted into the small intestine as free or conjugated forms, where the microbiota deconjugates bile acids and converts primary bile acids into secondary bile acids (Camilleri and Gores, 2015). The depletion of bile acids in the gut and liver fibrosis may be major factors inducing dysbiosis of the intestinal flora. Zhou et al. found significant differences in various bile acids and TCDCA/CDCA ratios in serum samples from patients suffering from BA or neonatal hepatitis syndrome (Zhou et al., 2015). Another study showed that the abundance of bile acids were lower in the feces of patients with cholestasis than in that of HC individuals, with the greatest reduction in bile acid abundance being in BA patients (Zhou et al., 2015). Our data showed that the abundance of primary bile acids in feces was generally lower in patients with BA than in the HC individuals, whereas the abundance of steroids followed the reverse trend.
Patients with BA have clinically significant hyperbilirubinemia due to biliary occlusion, resulting in cholestasis and hepatic fibrosis. In our study, some intestinal genera were highly correlated with clinical indicators of liver disease. Specifically, Eggerthella, Eubaterium_halli_group, Fusicatenibacter, Bacteriodes, and Faecalibacterium were negatively correlated with indicators of liver function. Previous studies have shown that Eggerthella (Cho et al., 2016) and Bacteriodes (Wexler, 2007) play critical roles in liver metabolism and detoxification processes. Faecalibacterium is an important component of the gut microbiota and the most important butyric acid-producing bacterium in the human colon. Moreover, drugs and nutrients increase the levels of healthy flora in the human gut, including Faecalibacterium ( Spanogiannopoulos et al., 2016 ). A decrease in Faecalibacterium is currently thought to exacerbate inflammatory processes, and a negative association between Faecalibacterium and intestinal inflammation has been reported in some studies (Miquel et al., 2013). Therefore, we hypothesize that the above-mentioned genera exert potentially beneficial effects in BA. Conversely, Dialister and Megasphaera are both genera associated with inflammatory diseases, such as oral infections, pneumonia, and vaginitis (Rôças and Siqueira, 2006; Ravel et al., 2021). According to our data, Dialister and Megasphaera showed a significant positive association with TBA, thus representing potential pathogens in BA.
Cholangitis is the most common complication in patients with BA after Kasai surgery, with an incidence of 30%-70% (Gunadi et al., 2018). Several studies have suggested that cholangitis may be a risk factor for poor prognosis in patients with post-Kasai BA (Jiang et al., 2018). Cholangitis can lead to increased cholestasis and accelerated liver injury. An increasing number of studies have reported a potential role for Erysipelotrichaceae in human physiology and disease processes, such as enrichment after antibiotic treatment (Dinh et al., 2015). In this study, Erysipelotrichaceae was found to be significantly enriched in the feces of patients who had developed cholangitis after surgery. This indicated that a high abundance of Erysipelotrichaceae might increase the occurrence of cholangitis. Monoglobus, a pectin-degrading bacterium in the human colon (Kim et al., 2019), was enriched in non-cholangitis patients, along with Actinobacillus. Previous studies have suggested that the greater the number of cholangitis recurrences, the more severe will the patient’s liver damage be (Chen et al., 2018), The etiology of cholangitis is unknown, but based on the available studies, it is thought that intestinal bacterial migration and inflammatory responses may be dominant factors. In our study, Lachnospiraceae was observed to be enriched in the feces of patients with multiple postoperative recurrences of cholangitis. Lachnospiraceae is a major producer of short-chain fatty acids, but different taxa have also been associated with different intestinal and extraintestinal diseases, and the physiological effects on hosts are often inconsistent across studies (Vacca et al., 2020). In liver disease, enrichment of Lachnospiraceae may lead to mucosal immune dysregulation, promoting lymphocyte activation and increasing intestinal permeability. Lachnospiraceae was significantly increased in patients with primary sclerosing cholangitis-inflammatory bowel disease compared to HCs (Zhu et al., 2013), In addition, Selenomonadaceae was enriched in the intestines of patients with a single episode of cholangitis. Megamonas was not the dominant flora in patients with postoperative cholangitis after Kasai but was significantly enriched in the feces of patients with non-multiple, recurrent cholangitis. A higher abundance of Megamonas in control individuals has now been demonstrated by animal studies of intestinal inflammation (Maldonado-Contreras et al., 2020). Based on these findings, further research into Megamonas and its potentially beneficial effects are warranted in the future.
Jaundice-free survival is an important influencing factor for native liver survival after a Kasai procedure (Hukkinen et al., 2018; Pakarinen et al., 2018), which was defined as a total bilirubin (TBIL) < 1.5 mg/dL (Bezerra et al., 2014). Rikenellaceae abundance, previously reported to be significantly negatively correlated with fecal bile acids (Ikeda et al., 2020), was also confirmed in the present study; Rikenellaceae was found to be significantly enriched in the non-jaundice-clearance group. Campylobacter was enriched in the jaundice-clearance group, but the specific mechanism involved was not determined. The TUDCA level was higher in the jaundice-clearance group. Bear bile is thought to have anti-inflammatory, anti-bacterial, and gallstone-dissolving effects, and the bile acids in bear bile are in the form of taurine conjugates, of which the beneficial components are probably TUDCA and UDCA. Both have been shown to be potent cholestatic agents in animal studies (Cho et al., 2014; Li et al., 2016). TUDCA, the specific bile acid in bear bile, is a hydrophilic bile acid that is synthesized in the UDCA conjugation pathway and functions as a secretagogue and immunomodulator. The results of this study also support a beneficial effect of UDCA treatment after Kasai procedures in patients with BA.
There are several limitations to this study that have to be addressed. Firstly, this was a single-center study with a relatively small sample size, which may have influenced the results. Future multi-center studies are needed to validate the data presented. Secondly, the use of 16S rRNA amplification and sequencing in this study may have caused limitations with respect to functional prediction analysis. To compensate for this, we analyzed the fecal bile acid profiles. Thirdly, although our data showed an association between microbial communities, bile acid profiles, and clinical indicators, it did not yield a defined causal relationship. Further dissection of the potential mechanisms underlying the interaction is necessary. Finally, collect stool sample again after Kasai during the follow-up and looked at the bile acid and microbiome again was necessary in the future.
Our study describes alterations in the gut microbiome and bile acids in BA, and for the first time, we describe alterations in patients with postoperative cholangitis and jaundice clearance. In addition, we identified unique microbial-bile acid interactions in feces of BA patients, providing a possible disease-specific mechanism for future studies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Beijing Children’s Hospital. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
JSH, TY and SY conceptualized and supervised the study; TY, SY, YZ, JML, SSL, KYH, YCG and DDW managed the resources. JSH, TY and SY developed the methodology. TY, SY, JWZ, PZW, SQL, ZZL, YYJ, XYZ and YNZ performed the investigation. TY and SY wrote the manuscript. JSH reviewed and edited the manuscript. All authors contributed the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China grants (#81660092).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We appreciate all the subjects who participated in the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.914247/full#supplementary-material
RDA analysis of the effect of environmental factors on the fecal microbiota structure. BA, biliary atresia; HC, health control; RDA, redundancy analysis.
Histogram of LDA scores of the gut microbiota and the correlations between fecal microbiota structure and samples. (A) The composition of the gut microbiota between the HC and BA groups was analyzed by LEfSe. Histograms of the LDA scores calculated from the different abundances of microbes between the two groups. The critical point of the LDA score was 2.0, indicating a significant difference. Correlations between fecal microbiota structure and samples form BA (B) and HC (C) groups. BA, biliary atresia; HC, health control; LEfSe, linear discriminant analysis effect size; LDA, linear discriminant analysis.
Bile acid composition of each sample and correlation of bile acids in feces with clinical indicators. (A) Stacked histograms of relative abundances of various bile acids in each sample. (B) Heatmap of the Spearman correlation coefficients between fecal bile acid compositions and indicators of liver function. ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BA, biliary atresia; Chol, cholesterol; DBIL, direct bilirubin; GGT, gamma-glutaryl transferase; HDL_C, high-density lipoprotein cholesterol; HC, health control; IBIL, indirect bilirubin; LDL_C, low-density lipoprotein cholesterol; PA, prealbumin; TBA, total bile acid; TBIL, total bilirubin; TG, triglyceride; TP, total protein; VLDL_C, very low-density lipoprotein cholesterol; 3-DHCA, 3-dehydrocholic acid; 6-ketoLCA, 6-ketolithocholic acid; 7-ketoLCA, 7-ketolithocholic acid; 12-ketoLCA, 12-ketolithocholic acid; βUDCA, 3β-ursodeoxycholic acid; C4, 7a-hydroxy-cholestene-3-one; CA, cholic acid; CDCA, chenodeoxycholic acid; CDCA-3Gln, chenodeoxycholic acid-3-β-D-glucuronide; DCA, deoxycholic acid; GCA, glycocholic acid; GCDCA, glycochenodeoxycholate; GHCA, glycohyocholate; GUDCA, glycoursodeoxycholic acid; HCA, γ-muricholic acid/hyocholic acid; LCA, lithocholic acid; LCA-3S, lithocholic acid 3 sulfate; NorCA, nor cholic acid; TCA, taurocholic acid; TCDCA, taurochenodeoxycholate; TUDCA, tauroursodeoxycholic acid; UDCA, ursodeoxycholic acid.
References
- Asai A., Miethke A., Bezerra J. A. (2015). Pathogenesis of Biliary Atresia: Defining Biology to Understand Clinical Phenotypes. Nat. Rev. Gastroenterol. Hepatol. 12, 342–352. doi: 10.1038/nrgastro.2015.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra J. A., Spino C., Magee J. C., Shneider B. L., Rosenthal P., Wang K. S., et al. (2014). Use of Corticosteroids After Hepatoportoenterostomy for Bile Drainage in Infants With Biliary Atresia: The START Randomized Clinical Trial. JAMA 311, 1750–1759. doi: 10.1001/jama.2014.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri M., Gores G. J. (2015). Therapeutic Targeting of Bile Acids. Am. J. Physiol. Gastrointestinal liver Physiol. 309, G209–G215. doi: 10.1152/ajpgi.00121.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Ji F., Guo J., Shi D., Fang D., Li L. (2016). Dysbiosis of Small Intestinal Microbiota in Liver Cirrhosis and its Association With Etiology. Sci. Rep. 6, 34055. doi: 10.1038/srep34055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.-Y., Lin C.-C., Tsan Y.-T., Chan W.-C., Wang J.-D., Chou Y.-J., et al. (2018). Number of Cholangitis Episodes as a Prognostic Marker to Predict Timing of Liver Transplantation in Biliary Atresia Patients After Kasai Portoenterostomy. BMC Pediatr. 18, 119. doi: 10.1186/s12887-018-1074-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. (2018). Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 34, i884–ii90. doi: 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho G.-S., Ritzmann F., Eckstein M., Huch M., Briviba K., Behsnilian D., et al. (2016). Quantification of Slackia and Eggerthella Spp. In Human Feces and Adhesion of Representatives Strains to Caco-2 Cells. Front. Microbiol. 7, 658. doi: 10.3389/fmicb.2016.00658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E.-J., Yoon J.-H., Kwak M.-S., Jang E. S., Lee J.-H., Yu S. J., et al. (2014). Tauroursodeoxycholic Acid Attenuates Progression of Steatohepatitis in Mice Fed a Methionine-Choline-Deficient Diet. Digestive Dis. 59, 1461–1474. doi: 10.1007/s10620-014-3217-0 [DOI] [PubMed] [Google Scholar]
- Chung P. H. Y., Chan E. K. W., Yeung F., Chan A. C. Y., Mou J. W. C., Lee K. H., et al. (2021). Life Long Follow Up and Management Strategies of Patients Living With Native Livers After Kasai Portoenterostomy. Sci. Rep. 11, 11207. doi: 10.1038/s41598-021-90860-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh D. M., Volpe G. E., Duffalo C., Bhalchandra S., Tai A. K., Kane A. V., et al. (2015). Intestinal Microbiota, Microbial Translocation, and Systemic Inflammation in Chronic HIV Infection. J. Infect. Dis. 211, 19–27. doi: 10.1093/infdis/jiu409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drzymała-Czyż S., Dziedzic K., Szwengiel A., Krzyżanowska-Jankowska P., Nowak J. K., Nowicka A., et al. (2021). Serum Bile Acids in Cystic Fibrosis Patients-Glycodeoxycholic Acid as a Potential Marker of Liver Disease. Digestive liver Dis 54(1):111–17. doi: 10.1016/j.dld.2021.06.034 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2013). UPARSE: Highly Accurate OTU Sequences From Microbial Amplicon Reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Feenstra H., Miedema F., Gerrits W. B., Veer M., Melief C. J. (1985). Adult T-Cell Leukaemia Lymphoma (ATLL) Associated With Human T-Cell Leukaemia Virus (HTLV). Case Report and Immunological Studies With the Neoplastic T-Cells of a Caribbean Patient. Netherlands J. Med. 28, 285–290. [PubMed] [Google Scholar]
- Fu X., Xiao Y., Golden J., Niu S., Gayer C. P. (2020). Serum Bile Acids Profiling by Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) and its Application on Pediatric Liver and Intestinal Diseases. Clin. Chem. Lab. Med. 58, 787–797. doi: 10.1515/cclm-2019-0354 [DOI] [PubMed] [Google Scholar]
- Golden J., Zagory J. A., Fenlon M., Goodhue C. J., Xiao Y., Fu X., et al. (2018). Liquid Chromatography-Mass Spectroscopy in the Diagnosis of Biliary Atresia in Children With Hyperbilirubinemia. J. Surg. Res. 228, 228–237. doi: 10.1016/j.jss.2018.03.021 [DOI] [PubMed] [Google Scholar]
- Gunadi, Gunawan T. A., Widiyanto G., Yuanita A., Mulyani N. S., Makhmudi A. (2018). Liver Transplant Score for Prediction of Biliary Atresia Patients' Survival Following Kasai Procedure. BMC Res. Notes 11, 381. doi: 10.1186/s13104-018-3498-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanning I., Diaz-Sanchez S. (2015). The Functionality of the Gastrointestinal Microbiome in non-Human Animals. Microbiome 3, 51. doi: 10.1186/s40168-015-0113-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiippala K., Jouhten H., Ronkainen A., Hartikainen A., Kainulainen V., Jalanka J., et al. (2018). The Potential of Gut Commensals in Reinforcing Intestinal Barrier Function and Alleviating Inflammation. Nutrients 10 (8), 988. doi: 10.3390/nu10080988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukkinen M., Kerola A., Lohi J., Heikkilä P., Merras-Salmio L., Jahnukainen T., et al. (2018). Treatment Policy and Liver Histopathology Predict Biliary Atresia Outcomes: Results After National Centralization and Protocol Biopsies. J. Am. Coll. Surgeons 226, 46–57.e1. doi: 10.1016/j.jamcollsurg.2017.09.009 [DOI] [PubMed] [Google Scholar]
- Ikeda T., Aida M., Yoshida Y., Matsumoto S., Tanaka M., Nakayama J., et al. (2020). Alteration in Faecal Bile Acids, Gut Microbial Composition and Diversity After Laparoscopic Sleeve Gastrectomy. Br. J. Surg. 107, 1673–1685. doi: 10.1002/bjs.11654 [DOI] [PubMed] [Google Scholar]
- Jialing L., Yangyang G., Jing Z., Xiaoyi T., Ping W., Liwei S., et al. (2020). Changes in Serum Inflammatory Cytokine Levels and Intestinal Flora in a Self-Healing Dextran Sodium Sulfate-Induced Ulcerative Colitis Murine Model. Life Sci. 263, 118587. doi: 10.1016/j.lfs.2020.118587 [DOI] [PubMed] [Google Scholar]
- Jiang H., Gao P., Chen H., Zhong Z., Shu M., Zhang Z., et al. (2018). The Prognostic Value of CD8+ and CD45RO+ T Cells Infiltration and Beclin1 Expression Levels for Early Postoperative Cholangitis of Biliary Atresia Patients After Kasai Operation. J. Korean Med. Sci. 33, e198. doi: 10.3346/jkms.2018.33.e198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M., Umeshita K., Sakamoto S., Fukuda A., Furukawa H., Uemoto S. (2017). Liver Transplantation for Biliary Atresia: A Systematic Review. Pediatr. Surg. Int. 33, 1289–1295. doi: 10.1007/s00383-017-4173-5 [DOI] [PubMed] [Google Scholar]
- Kim C. C., Healey G. R., Kelly W. J., Patchett M. L., Jordens Z., Tannock G. W., et al. (2019). Genomic Insights From Monoglobus Pectinilyticus: A Pectin-Degrading Specialist Bacterium in the Human Colon. ISME J. 13, 1437–1456. doi: 10.1038/s41396-019-0363-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler J., Maletzki C., Koczan D., Frank M., Springer A., Steffen C., et al. (2020). Kininogen Supports Inflammation and Bacterial Spreading During Streptococccus Pyogenes Sepsis. EBioMedicine 58, 102908. doi: 10.1016/j.ebiom.2020.102908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima A., Nakano K., Wada K., Takahashi H., Katayama K., Yoneda M., et al. (2012). Infection of Specific Strains of Streptococcus Mutans, Oral Bacteria, Confers a Risk of Ulcerative Colitis. Sci. Rep. 2, 332. doi: 10.1038/srep00332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane E., Murray K. F. (2017). Neonatal Cholestasis. Pediatr. Clinics North America 64, 621–639. doi: 10.1016/j.pcl.2017.01.006 [DOI] [PubMed] [Google Scholar]
- Lan K., Su M., Xie G., Ferslew B. C., Brouwer K. L. R., Rajani C., et al. (2016). Key Role for the 12-Hydroxy Group in the Negative Ion Fragmentation of Unconjugated C24 Bile Acids. Analytical Chem. 88, 7041–7048. doi: 10.1021/acs.analchem.6b00573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li Y., Zhou Y., Wang C., Wu B., Wan J. (2018). Actinomyces and Alimentary Tract Diseases: A Review of Its Biological Functions and Pathology. BioMed. Res. Int. 2018, 3820215. doi: 10.1155/2018/3820215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Tan H. Y., Wang N., Hong M., Li L., Cheung F., et al. (2016). Substitutes for Bear Bile for the Treatment of Liver Diseases: Research Progress and Future Perspective. Evidence-Based complementary Altern. Med. 2016, 4305074. doi: 10.1155/2016/4305074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Li B., Li Y., Wei Y., Huang B., Liang J., et al. (2021). Altered Faecal Microbiome and Metabolome in IgG4-Related Sclerosing Cholangitis and Primary Sclerosing Cholangitis. Gut. 71 (5), 899–909. doi: 10.1136/gutjnl-2020-323565 [DOI] [PubMed] [Google Scholar]
- Long S. L., Gahan C. G. M., Joyce S. A. (2017). Interactions Between Gut Bacteria and Bile in Health and Disease. Mol. aspects Med. 56, 54–65. doi: 10.1016/j.mam.2017.06.002 [DOI] [PubMed] [Google Scholar]
- Loomba R., Ling L., Dinh D. M., DePaoli A. M., Lieu H. D., Harrison S. A., et al. (2021). The Commensal Microbe Veillonella as a Marker for Response to an FGF19 Analog in NASH. Hepatology 73, 126–143. doi: 10.1002/hep.31523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T., Salzberg S. L. (2011). FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Contreras A., Ferrer L., Cawley C., Crain S., Bhattarai S., Toscano J., et al. (2020). Dysbiosis in a Canine Model of Human Fistulizing Crohn's Disease. Gut Microbes 12, 1785246. doi: 10.1080/19490976.2020.1785246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel S., Martín R., Rossi O., Bermúdez-Humarán L. G., Chatel J. M., Sokol H., et al. (2013). Faecalibacterium Prausnitzii and Human Intestinal Health. Curr. Opin. Microbiol. 16, 255–261. doi: 10.1016/j.mib.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Nakajima H., Koga H., Okawada M., Nakamura H., Lane G. J., Yamataka A. (2018). Does Time Taken to Achieve Jaundice-Clearance Influence Survival of the Native Liver in Post-Kasai Biliary Atresia? World J. Pediatr. 14, 191–196. doi: 10.1007/s12519-018-0139-5 [DOI] [PubMed] [Google Scholar]
- Oh T. G., Kim S. M., Caussy C., Fu T., Guo J., Bassirian S., et al. (2020). A Universal Gut-Microbiome-Derived Signature Predicts Cirrhosis. Cell Metab. 32, 878–88.e6. doi: 10.1016/j.cmet.2020.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakarinen M. P., Johansen L. S., Svensson J. F., Bjørnland K., Gatzinsky V., Stenström P., et al. (2018). Outcomes of Biliary Atresia in the Nordic Countries - A Multicenter Study of 158 Patients During 2005-2016. J. Pediatr. Surg. 53, 1509–1515. doi: 10.1016/j.jpedsurg.2017.08.048 [DOI] [PubMed] [Google Scholar]
- Ravel J., Moreno I., Simón C. (2021). Bacterial Vaginosis and its Association With Infertility, Endometritis, and Pelvic Inflammatory Disease. Am. J. obstet. gynecol. 224, 251–257. doi: 10.1016/j.ajog.2020.10.019 [DOI] [PubMed] [Google Scholar]
- Rôças I. N., Siqueira J. F. (2006). Characterization of Dialister Species in Infected Root Canals. J. endodontics 32, 1057–1061. doi: 10.1016/j.joen.2006.04.010 [DOI] [PubMed] [Google Scholar]
- Song W., Sun L.-Y., Zhu Z.-J. (2021). Effects of Previous Kasai Surgery on Gut Microbiota and Bile Acid in Biliary Atresia With End-Stage Liver Disease. Front. Med. 8, 704328. doi: 10.3389/fmed.2021.704328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Sun L.-Y., Zhu Z.-J., Wei L., Qu W., Zeng Z.-G., et al. (2021). Association of Gut Microbiota and Metabolites With Disease Progression in Children With Biliary Atresia. Front. Immunol. 12, 698900. doi: 10.3389/fimmu.2021.698900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanogiannopoulos P., Bess E. N., Carmody R. N., Turnbaugh P. J. (2016). The Microbial Pharmacists Within Us: A Metagenomic View of Xenobiotic Metabolism. Nat. Rev. Microbiol. 14, 273–287. doi: 10.1038/nrmicro.2016.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.-Y., Yang Y.-S., Qu W., Zhu Z.-J., Wei L., Ye Z.-S., et al. (2017). Gut Microbiota of Liver Transplantation Recipients. Sci. Rep. 7, 3762. doi: 10.1038/s41598-017-03476-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajeddin E., Sherafat S. J., Majidi M. R. S., Alebouyeh M., Alizadeh A. H. M., Zali M. R. (2016). Association of Diverse Bacterial Communities in Human Bile Samples With Biliary Tract Disorders: A Survey Using Culture and Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis Methods. Eur. J. Clin. Microbiol. 35, 1331–1339. doi: 10.1007/s10096-016-2669-x [DOI] [PubMed] [Google Scholar]
- Tang R., Wei Y., Li Y., Chen W., Chen H., Wang Q., et al. (2018). Gut Microbial Profile is Altered in Primary Biliary Cholangitis and Partially Restored After UDCA Therapy. Gut 67, 534–541. doi: 10.1136/gutjnl-2016-313332 [DOI] [PubMed] [Google Scholar]
- Tripathi A., Debelius J., Brenner D. A., Karin M., Loomba R., Schnabl B., et al. (2018). The Gut-Liver Axis and the Intersection With the Microbiome. Nat. Rev. Gastroenterol. 15, 397–411. doi: 10.1038/s41575-018-0011-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca M., Celano G., Calabrese F. M., Portincasa P., Gobbetti M., Angelis M. D. (2020). The Controversial Role of Human Gut Lachnospiraceae. Microorganisms 8 (4), 573. doi: 10.3390/microorganisms8040573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Gao X., Zhang X., Xiao Y., Huang J., Yu D., et al. (2019). Gut Microbiota Dysbiosis Is Associated With Altered Bile Acid Metabolism in Infantile Cholestasis. mSystems 4 (6), e00463–19. doi: 10.1128/mSystems.00463-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences Into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Qian T., Jiang J., Yang Y., Shen Z., Huang Y., et al. (2020). Gut Microbial Profile in Biliary Atresia: A Case-Control Study. J. Gastroenterol. 35, 334–342. doi: 10.1111/jgh.14777 [DOI] [PubMed] [Google Scholar]
- Wei Y., Li Y., Yan L., Sun C., Miao Q., Wang Q., et al. (2020). Alterations of Gut Microbiome in Autoimmune Hepatitis. Gut 69, 569–577. doi: 10.1136/gutjnl-2018-317836 [DOI] [PubMed] [Google Scholar]
- Wexler H. M. (2007). Bacteroides: The Good, the Bad, and the Nitty-Gritty. Clin. Microbiol. Rev. 20, 593–621. doi: 10.1128/CMR.00008-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie G., Wang Y., Wang X., Zhao A., Chen T., Ni Y., et al. (2015). Profiling of Serum Bile Acids in a Healthy Chinese Population Using UPLC-MS/MS. J. Proteome Res. 14, 850–859. doi: 10.1021/pr500920q [DOI] [PubMed] [Google Scholar]
- Yang X., Lu D., Zhuo J., Lin Z., Yang M., Xu X. (2020). The Gut-Liver Axis in Immune Remodeling: New Insight Into Liver Diseases. Int. J. Biol. Sci. 16, 2357–2366. doi: 10.7150/ijbs.46405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Huang F., Zhao A., Lei S., Zhang Y., Xie G., et al. (2017). Bile Acid is a Significant Host Factor Shaping the Gut Microbiome of Diet-Induced Obese Mice. BMC Biol. 15, 120. doi: 10.1186/s12915-017-0462-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Lin N., Xiao Y., Wang Y., Wen J., Zou G.-M., et al. (2012). Elevated Bile Acids in Newborns With Biliary Atresia (Ba). PloS One 7, e49270. doi: 10.1371/journal.pone.0049270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Wang J., Xie G., Zhou Y., Yan W., Pan W., et al. (2015). Distinct Plasma Bile Acid Profiles of Biliary Atresia and Neonatal Hepatitis Syndrome. J. Proteome Res. 14, 4844–4850. doi: 10.1021/acs.jproteome.5b00676 [DOI] [PubMed] [Google Scholar]
- Zhu L., Baker S. S., Gill C., Liu W., Alkhouri R., Baker R. D., et al. (2013). Characterization of Gut Microbiomes in Nonalcoholic Steatohepatitis (NASH) Patients: A Connection Between Endogenous Alcohol and NASH. Hepatology 57, 601–609. doi: 10.1002/hep.26093 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RDA analysis of the effect of environmental factors on the fecal microbiota structure. BA, biliary atresia; HC, health control; RDA, redundancy analysis.
Histogram of LDA scores of the gut microbiota and the correlations between fecal microbiota structure and samples. (A) The composition of the gut microbiota between the HC and BA groups was analyzed by LEfSe. Histograms of the LDA scores calculated from the different abundances of microbes between the two groups. The critical point of the LDA score was 2.0, indicating a significant difference. Correlations between fecal microbiota structure and samples form BA (B) and HC (C) groups. BA, biliary atresia; HC, health control; LEfSe, linear discriminant analysis effect size; LDA, linear discriminant analysis.
Bile acid composition of each sample and correlation of bile acids in feces with clinical indicators. (A) Stacked histograms of relative abundances of various bile acids in each sample. (B) Heatmap of the Spearman correlation coefficients between fecal bile acid compositions and indicators of liver function. ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BA, biliary atresia; Chol, cholesterol; DBIL, direct bilirubin; GGT, gamma-glutaryl transferase; HDL_C, high-density lipoprotein cholesterol; HC, health control; IBIL, indirect bilirubin; LDL_C, low-density lipoprotein cholesterol; PA, prealbumin; TBA, total bile acid; TBIL, total bilirubin; TG, triglyceride; TP, total protein; VLDL_C, very low-density lipoprotein cholesterol; 3-DHCA, 3-dehydrocholic acid; 6-ketoLCA, 6-ketolithocholic acid; 7-ketoLCA, 7-ketolithocholic acid; 12-ketoLCA, 12-ketolithocholic acid; βUDCA, 3β-ursodeoxycholic acid; C4, 7a-hydroxy-cholestene-3-one; CA, cholic acid; CDCA, chenodeoxycholic acid; CDCA-3Gln, chenodeoxycholic acid-3-β-D-glucuronide; DCA, deoxycholic acid; GCA, glycocholic acid; GCDCA, glycochenodeoxycholate; GHCA, glycohyocholate; GUDCA, glycoursodeoxycholic acid; HCA, γ-muricholic acid/hyocholic acid; LCA, lithocholic acid; LCA-3S, lithocholic acid 3 sulfate; NorCA, nor cholic acid; TCA, taurocholic acid; TCDCA, taurochenodeoxycholate; TUDCA, tauroursodeoxycholic acid; UDCA, ursodeoxycholic acid.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .