Abstract
STUDY QUESTION
How does hormonal contraceptive use and menstrual cycle phase affect the female microbiome across different body sites?
SUMMARY ANSWER
The menstrual cycle phase, but not hormonal contraceptive use, is associated with the vaginal and oral but not the gut microbiome composition in healthy young women.
WHAT IS KNOWN ALREADY
Women with low vaginal levels of Lactobacillus crispatus are at increased risk of pre-term birth, fertility treatment failure, sexually transmitted infections and gynaecological cancers. Little is known about the effect of hormonal fluctuations on other body site’s microbiomes as well as the interplay between them.
STUDY DESIGN, SIZE, DURATION
This study includes a cohort of 160 healthy young Danish women using three different contraceptive regimens: non-hormonal methods (n = 54), combined oral contraceptive (COC, n = 52) or levonorgestrel intrauterine system (LNG-IUS, n = 54). Samples were collected from four body sites during the menstrual cycle (menses, follicular and luteal phases) at Copenhagen University Hospital, Rigshospitalet, Denmark.
PARTICIPANTS/MATERIALS, SETTING, METHODS
The oral, vaginal, rectal and faecal microbiomes were characterized by shotgun sequencing. Microbial diversity and community distance measures were compared between study groups, menstrual phase timepoints and body sites. All participants answered an extensive questionnaire on current health, lifestyle and sex life. Confounding factors such as smoking, BMI and diet were analysed by PERMANOVA. Plasma oestradiol and progesterone levels are correlated with microbiome composition.
MAIN RESULTS AND THE ROLE OF CHANCE
The use of COC and LNG-IUS was not associated with the microbiome composition or diversity. However, increased diversity in the vaginal microbiome was observed during menses, followed by a subsequent expansion of Lactobacillus spp. during the follicular and luteal phases which correlated with measured serum oestradiol levels (r = 0.11, P < 0.001). During menses, 89 women (58%) had a dysbiotic vaginal microbiome with <60% Lactobacillus spp. This declined to 49 (32%) in the follicular phase (P < 0.001) and 44 (29%) in the luteal phase (P < 0.001). During menses, bacterial richness and diversity in saliva reached its lowest point while no differences were observed in the faecal microbiome. The microbiome in different body sites was on average more similar within the same individual than between individuals, despite phase or hormonal treatment. Only the vagina presented a clear cluster structure with dominance of either L. crispatus, Lactobacillus iners, Gardnerella vaginalis or Prevotella spp.
LARGE SCALE DATA
The microbiome samples analysed in this study were submitted to the European Nucleotide Archive under project number PRJEB37731, samples ERS4421369–ERS4422941.
LIMITATIONS, REASONS FOR CAUTION
The cohort is homogenous which limits extrapolation of the effects of ethnicity and socio-economic status on the microbiome. We only present three defined timepoints across the menstrual phase and miss potential important day to day fluctuations.
WIDER IMPLICATIONS OF THE FINDINGS
The use of hormonal contraception did not significantly associate with the microbiome composition in the vagina, faeces, rectum or saliva in healthy young women. This is a welcome finding considering the widespread and prolonged use of these highly efficient contraceptive methods. The menstrual cycle is, however, a major confounding factor for the vaginal microbiome. As such, the time point in the menstrual cycle should be considered when analysing the microbiome of women of reproductive age, since stratifying by vaginal dysbiosis status during menstruation could be misleading. This is the first study to confirm by direct measurements of oestradiol, a correlation with the presence of L. crispatus, adding evidence of a possible hormonal mechanism for the maintenance of this desirable microbe.
STUDY FUNDING/COMPETING INTEREST(S)
This work was partly funded by the Ferring Pharmaceuticals through a research collaboration with The Centre for Translational Microbiome Research (CTMR) at the Karolinska Institutet (L.W.H., E.F., G.E. and I.S.-K.). Ferring Pharmaceuticals also funded the infrastructure to obtain the clinical samples at Copenhagen University Hospital ([#MiHSN01], M.C.K., Z.B., and H.S.N.). This work was also supported by funding from Rigshospitalet’s Research Funds ([#E-22614-01 and #E-22614-02] to M.C.K.) and Oda and Hans Svenningsen’s Foundation ([#F-22614-08] to H.S.N.). M.C.K., L.W.H., E.F., Z.B., G.E., L.E., I.S.-K. and H.S.N., are partially funded by Ferring Pharmaceuticals, which also provided funds for the collection and processing of the samples analysed in this study. H.S.N.’s research is further supported by Freya Biosciences and the BioInnovation Institute. H.S.N. has received honoraria from Ferring Pharmaceuticals, Merck A/S, Astra-Zeneca, Cook Medical and Ibsa Nordic. A.N.A. reports no competing interests.
Keywords: hormonal contraceptives, microbiome, womens reproductive health, shotgun sequencing, menstrual cycle
Introduction
Microbial communities inhabit every inch of the human body and are thought to actively contribute to the homeostasis and health of every individual (Nelson et al., 2010; Lloyd-Price et al., 2017; Young, 2017). The implications of the microbiota in various body sites and their potential interaction and individual importance in maintaining general health are, however, inadequately understood. Most studies focus on one body site at a time and extensive work has been published documenting associations between the digestive tract microbiome and numerous health conditions, from gastrointestinal diseases to mental illnesses (Kostic et al., 2014; Meijnikman et al., 2018; Winter et al., 2018; Figuero et al., 2020; Hernández-Ceballos et al., 2021). The most studied niche in the female reproductive tract is the vaginal microbiome, which has been connected to gynaecological health (Green et al., 2015; Kroon et al., 2018). The gut and oral microbiomes have been shown to have systemic effects and the question remains whether there is an interaction between the microbiota in various body sites and how they could potentially impact women’s reproductive health.
Among the most common colonizers of the vaginal tract in reproductive aged women are Lactobacillus species which have been established as the healthy vaginal microbiome linked to positive health outcomes (Ravel et al., 2011; Romero et al., 2014; Haahr et al., 2016; Kroon et al., 2018; Van Houdt et al., 2018; Brusselaers et al., 2019). Some women of reproductive age lack this Lactobacillus dominance in the vagina but have a diverse composition of other bacteria including anaerobic bacteria. This is similar to the microbial composition in bacterial vaginosis (BV), a condition characterized by thin, greyish vaginal discharge, unpleasant odour and increased vaginal pH. While common, especially among women of African descent (Zhou et al., 2010; Borgdorff et al., 2017; Gosmann et al., 2017), this microbial composition has been connected to susceptibility to sexually transmitted infections (Atashili et al., 2008; Edwards et al., 2019), difficulties achieving pregnancy after fertility treatments (Haahr et al., 2016; Koedooder et al., 2019), pre-term birth (Elovitz et al., 2019; Fettweis et al., 2019), HPV infection and gynaecological cancers (Brusselaers et al., 2019; Coudray and Kiplagat, 2019). This has led many research groups to classify a diverse vaginal microbiome with low abundance of Lactobacillus spp. as dysbiotic, even in the absence of symptoms (Forney et al., 2006; Balle et al., 2020). Interestingly, it is suspected that Lactobacillus dominance in the vagina is oestradiol- and/or progesterone-dependent as it is more common in the reproductive years of life, leaving girls and postmenopausal women with a more diverse microbiome with less abundance of Lactobacillus (Hickey et al., 2015; Muhleisen and Herbst-Kralovetz, 2016; Kaur et al., 2020).
The possible effect of hormonal contraception on the vaginal microbiome is debated. Some studies argue for a beneficial impact of the synthetic oestrogens in the combined oral contraceptive pill (COC) favouring Lactobacillus dominance (Vodstrcil et al., 2013; Brooks et al., 2017), others find no impact on the vaginal microbiome (Achilles and Hillier, 2013; Donders et al., 2017). A more diverse vaginal microbiome in levonorgestrel intrauterine systems (LNG-IUS) users have been reported (Song et al., 2020), although this alteration has also been found to be temporarily after insertion (Donders et al., 2018). Many women, especially younger women, use the monophasic COC and are continually exposed to the synthetic ethinyl oestradiol and a progestin. Both hormones work synergistically to suppress ovulation (De Leo et al., 2016), whereas most women using the progestin-only (LNG-IUS) continue to ovulate but with suppression of endometrial growth, resulting in less or absent menstrual bleeding (Grandi et al., 2018). The possible systemic effect of synthetic hormones on the microbiome composition across the body has not been extensively investigated to date, although we have demonstrated some effects for saliva (Bostanci et al., 2021).
Extensive work by Ravel’s group has categorized specific clusters of microorganisms in the vagina and described the temporal dynamics of the composition in the individual woman (Ravel et al., 2011; Gajer et al., 2012). They found that some women remain stable and others shift between a Lactobacillus dominated microbiome and a more diverse community state type (CST) over time (Gajer et al., 2012). Previous studies of the vaginal microbiome have primarily used 16S rRNA gene sequencing but with time, shotgun metagenomic sequencing techniques have emerged and can provide an extensive mapping of all DNA present in the sample.
A recent study has simultaneously assessed the faecal, vaginal and cervical microbiomes of a cohort of 28 women at a single time-point, but each body site was analysed separately (Ata et al., 2019). No previous studies have investigated the longitudinal effect of the fluctuations of sex hormones during the menstrual cycle and the possible effect of hormonal contraception on the composition of the microbiome across body sites. This study compares the shotgun metagenomic profiles of four different body sites in women during a natural menstrual cycle and in women using COC or LNG-IUS to explore an impact of supressed ovulation, menstrual bleeding and shifts in sex hormones on the overall microbiome composition.
Materials and methods
Participant recruitment
Women were recruited by advertisements in student magazines, university noticeboards and social media and 160 women were included between September 2017 and January 2018 at Copenhagen University Hospital, Rigshospitalet, Denmark. Women were excluded if they were pregnant or planning to become pregnant during the study period of 6 weeks or were taking or had taken antimicrobial medication (antibiotics, antiviral or antifungal medication) 14 days prior to enrolment. If participants had to use any kind of antimicrobial medication or vaginal lactic acid suppositories, they completed the project but were replaced one to one and their data were not included in this study. Women were recruited to fit into three contraception groups: Group 1, women with a regular menstrual cycle and no use of hormonal contraception (NHC), Group 2, women using combined (ethinyl oestradiol and progestin) monophasic oral contraceptive (COC) and Group 3, women using levonorgestrel intrauterine system (LNG-IUS). Women in Groups 2 and 3 had a regular cycle for at least six months prior to commencing their contraceptive regimen. The contraceptive regimens were chosen because they are commonly used in women of reproductive age and provide a unique opportunity to investigate the effects of the fluctuations in sex hormones (NHC group), suppressed ovulation without progesterone rise in the luteal phase (COC group) and sparse/absent menstrual bleeding (LNG-IUS group).
Sample collection
Participants were followed during a 6-week period including three hospital visits (Fig. 1). The participants contacted the study coordinator on the first day of bleeding and were then scheduled for the first hospital visit cycle day (CD) 1–3. If the participant had oligo-/amenorrhoea (Group 3, LNG-IUS, n = 32), she was scheduled at a random day. The second visit was scheduled at CD 8–12 and the third CD 18–22, representing the follicular and luteal phase of the menstrual cycle. For women with oligo-/amenorrhoea, the assignment to cycle phase was performed later, based on serum levels of oestradiol and progesterone.
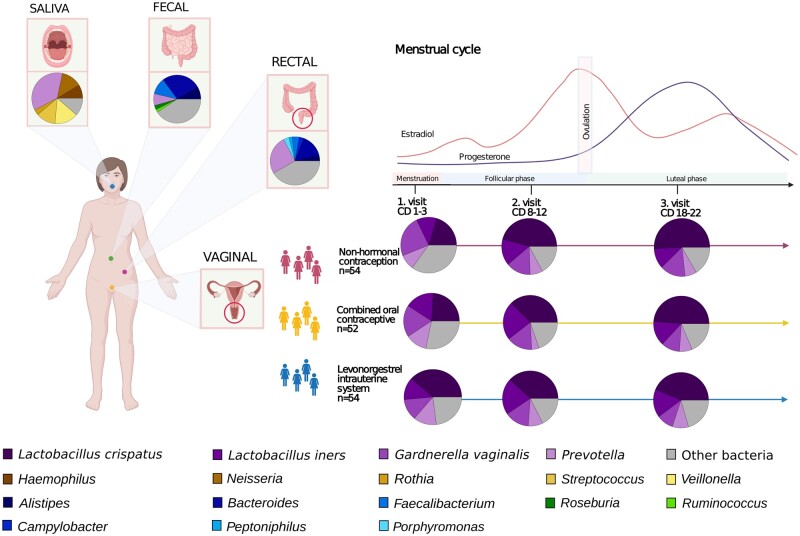
Figure 1.
Schematic representation of the sampling scheme and taxonomic profiles. Over 50 women were recruited using one of three contraceptive regimens: NHC (non-hormonal contraceptives), COC (combined oral contraceptives) and LNG-IUS (levonorgestrel intrauterine system). Included women were sampled at the hospital during the menstrual phase (cycle days 1–3), follicular phase (cycle days 8–12) and luteal phase (cycle days 18–22). Blood samples, saliva and rectal swabs were collected at the hospital; vaginal swabs and faeces were collected by the women at home. Women on LNG-IUS with oligo-/amenorrhoea started their sampling at a random day. The average taxonomic profile observed at each body site is depicted as circle charts. Only the vaginal swabs are divided by contraceptive and sampling point in this figure, since they are the only ones with clear visible differences.
Vaginal swabs, saliva and blood samples were collected at every hospital visit and a rectal sample was collected at the first hospital visit. After each hospital visit, participants collected a stool sample at home within 48 h. The faecal samples were collected with Zymo’s DNA/RNA-shield Fecal Collection Tube and stored at ambient temperature until the next hospital visit.
At every hospital visit, the women underwent a gynaecological examination by medical staff including a transvaginal ultrasound to exclude pathological conditions in the uterus or a pregnancy. A rectal sample was obtained by inserting the swab approximately 1–2 cm in the rectum and the swab was turned three times, touching the walls of the rectum. Vaginal and rectal samples were taken with FLOQSwabs (Copan Flock Technologies, Brescia, Italy) and put directly into FluidX tubes (Brooks Life Sciences, Chelmsford, MA, USA) containing 0.8 ml DNA/RNA-shield (Zymo Research, Irvine, CA, USA).
The patients were fasting 30 min before saliva collection, including no drinking, chewing gum and smoking. Saliva samples were collected in a SalivaGene Collector (STRATEC Molecular GmbH, Germany) containing lyophilized DNA stabilization buffer, according to the instructions of the manufacturer.
Blood samples were collected at the three hospital visits during the menstrual cycle in 9 ml EDTA tubes and plasma was stored in −80°C. Oestradiol and progesterone were measured in plasma using the standard automated system (Cobas® 8000 by Roche Diagnostics). Associations between sex hormones during the menstrual cycle and the microbiome were only calculated for women not using hormonal contraception, as women using hormonal contraceptives are influenced by other synthetic sex hormone derivatives (such as ethinyl oestradiol and progestins), not routinely measured in plasma.
Because of the large prevalence of absent menstrual bleeding in women using LNG-IUS (n = 32), the order of samples was adjusted based on their progesterone levels, with the sample with the highest progesterone level being set as ‘luteal’ and the others distributed accordingly (Supplementary Fig. S1).
Questionnaires
At the first hospital visit, the participants answered a comprehensive online questionnaire about general and reproductive health, lifestyle factors such as diet and exercise, and the use of menstrual hygiene products (Questionnaire 1). After every hospital visit the participants received a 48-h food-recall questionnaire and current gynaecological issues questionnaire (Questionnaire 2). Additionally, they filled out a diary where they registered bleedings (including spotting) and sexual intercourses.
Fibre intake was recorded using a 4-week food recall. Participants reported on a 9-point scale from ‘0 times in the past 4 weeks’ to ‘>3 times/day for the past 4 weeks’ with the following fibre categories: rye bread; whole wheat bread, fruit, vegetable dishes, salad and prepared vegetables. Intake of at least one daily high fibre category was categorized as ‘daily’. Free-sugar intake was derived from a similar scale, as previously described, including the items: chocolate milk, juice, soda with sugar, ice-cream, biscuits and cookies, sweet bread and rolls, dry cake, cake with filling and candy (Bostanci et al., 2021).
Ethical approval and consent to participate
All participants gave oral and written consent to participate. The participants were remunerated by receiving 3000 DKK before taxes when they completed participation. All data were collected and managed using REDCap electronic data capture tools (Harris et al., 2019), hosted at the Capital Region of Denmark. The study is approved by The Regional Committee on Health Research Ethics (H-17017580) and the Data Protection Agency in the Capital Region of Denmark (2012-58-0004).
DNA extraction and sequencing
Samples for microbiome analysis were shipped to CoreBiome (OraSure, Bethlehem, PA, USA) where they were extracted with MO Bio PowerFecal kit (Qiagen, Hilden, Germany) automated for high throughput on QiaCube (Qiagen), with bead-beating in 0.1 mm glass bead plates. Three spaced negative controls and one positive control were included in each extraction. All negative extraction controls had undetectable amounts of DNA, and all positive controls were also approved. The DNA concentration of samples and controls was quantified using Quant-iT Picogreen dsDNA Assay (Invitrogen, ThermoFisher Scientific, Carlsbad, CA, USA).
Libraries were prepared using an adapted Nextera (Illumina Inc, San Diego, CA, USA) procedure and sequenced on an Illumina NextSeq using single-end 150 bp reads with a NextSeq 500/550 High Output v2 kit. Reads were processed with CoreBiome’s BoosterShot shallow shotgun sequencing technology as described previously (Hillmann et al., 2018).
Annotation of metagenomic reads
Because the taxonomic annotation of BoosterShot technology is optimized for the human gut, saliva samples were reannotated using Kraken2 (Wood et al., 2019) and Bracken (Lu et al., 2017) based on the Human Oral Microbiome Database v9.0.3 (Chen et al., 2010). Similarly, vaginal samples were re-annotated with the same tools and the OptiVag database (Hugerth et al., 2020). Due to the uneven amount of annotated bacterial reads generated, each of these samples was filtered to keep species corresponding to at least 0.5% of annotated reads. Samples not attaining 10 000 annotated reads were discarded. Faecal and rectal samples were analysed based on the filtered BoosterShot file. We did not use the annotations described above when comparing strains across body sites, for which raw, unfiltered, BoosterShot annotations were used, as these preserve strain-level information.
Statistical analyses
The descriptive table statistics were performed using Kruskal–Wallis and chi-square with SPSS (v26.0; IBM Corp, Armonk, NY, USA). All other statistical analyses were performed in R v3.5.2. Alpha-diversity was calculated as the observed number of species as well as Simpson’s inverted index. Beta-diversity was calculated on Bray-Curtis distances. Alpha- and beta-diversity were calculated with package Vegan (v2.5-3; CRAN—Package vegan, n.d.) and graphs were generated with packages RColorBrewer (v1.1-2), Vioplot (v0.2) and Pheatmap (v1.0.10).
Explorative analysis of possible confounding factors was conducted with a PERMANOVA based on Bray-Curtis distances of each sample type in turn. Significant factors for each body site were treated as a possible confounder for all other body sites. The following factors were considered: smoking: daily, occasional, not currently; snus (moist non-smoke tobacco): current, not-currently, alcohol: up to 7 weekly units, >7 weekly units; Bristol stool form scale: normal transit (3–4), slow transit (1–4), fast (3–7), varied (both 1–2 and 5–7); BMI: lowest quartile (≤20.6), middle 50% and top quartile (≥23.9); fibre intake: daily versus non-daily; free-sugar consumption: lowest quartile (≤4 occasions/week), middle 50% and top 25% (≥22.7 occasions/week); age: young (18–24) and older (25–40). This has been described in more detail (Bostanci et al., 2021).
For vaginal samples, the role of menstrual hygiene product usage (pads, tampons, cup, panty liner), frequency of sexual intercourse in the preceding 3 months, number of sexual partners in the preceding 3 months and condom usage (protected, unprotected) was assessed as a variation partitioning based on Bray-Curtis together with contraceptive method. Due to limitations in the method, each time-point was considered separately.
Comparisons between alpha-diversity at baseline compared to follicular and luteal phases for the same individual were calculated as paired t-tests, while comparisons across individuals were calculated using Welch’s t-test. For binary outcomes (presence/absence), the chi-squared test was used. For beta-diversity comparisons, because the distribution for vaginal samples was strongly non-normal, Wilcoxon’s rank sum test with continuity correction was used. All tests were performed with a 95% CI and a significance cut-off of P < 0.05. Multiple testing correction was conducted with the Benjamini–Hochberg procedure where applicable.
Associations between taxa and hormones or metadata parameters were calculated in Maaslin2 (Mallick et al., 2021). For the hormonal analysis, serum levels of oestradiol and progesterone were considered as fixed effects, with subject identity as a random effect. For an analysis on the combined effects of contraceptive usage and the menstrual cycle, the following co-variables were included: smoking status, snus usage, alcohol consumption and sugar consumption, with subject identity as a random effect.
Results
Cohort description
Characteristics and health parameters of the participating women in the three groups (NHC, COC and LNG-IUS) are presented in Table I. Some differences were observed between the groups, mostly related to reproductive health, with fewer women having a regular menstrual cycle in the LNG-IUS group (57% vs 100% in the other groups), but also that fewer women in the COC group reported to have a healthy diet (COC: 38%; NHC: 55%; LNG-IUS: 68%) and a larger proportion of women with LNG-IUS reported being in a relationship (78% vs NHC: 46% and COC: 50%).
Table I.
Background characteristics of participants
| No hormonal contraceptives (n = 54) | Combined oral contraceptive (n = 52) | LNG-IUS (n = 54) | P-value | |
|---|---|---|---|---|
| Background | ||||
| Born in Denmark, n (%) | 47 (87.0) | 50 (96.2) | 50 (92.6) | 0.22 |
| Age in years, median (range) | 23.5 (19–40) | 23.0 (18–36) | 23.0 (19–40) | 0.23 |
| Level of education, ongoing or completed university level | 49 (90.7) | 49 (94.2) | 47 (87.0) | 0.47 |
| General health | ||||
| Overweight—BMI >25, n (%) | 12 (22.2) | 6 (11.5) | 4 (7.4) | 0.07 |
| Smoking at least once a week, n (%) | 13 (24.1) | 6 (11.5) | 13 (24.1) | 0.18 |
| Snus at least once a week, n (%) | 4 (7.4) | 6 (11.5) | 4 (7.4) | 0.69 |
| Poor self-rated health, less than good, n (%) | 5 (9.3) | 2 (3.8) | 3 (5.6) | 0.14 |
| Self-rated healthy diet, n (%) | 30 (55.5) | 20 (38.4) | 37 (68.5) | <0.001 |
| High alcohol consumption (>7 units/week) | 12 (22.2) | 19 (36.5) | 13 (24.1) | 0.20 |
| Frequent free sugar consumption (>9 occasions/week), n (%) | 16 (29.6) | 16 (30.8) | 8 (14.8) | 0.10 |
| Daily fibre consumption, n (%) | 46 (90.2) | 44 (84.6) | 50 (92.6) | 0.26 |
| Reproductive health | ||||
| Have children, n (%) | 5 (9.3) | 2 (3.8) | 6 (11.1) | 0.37 |
| Preserved menstrual cycle, n (%) | 54 (100) | 52 (100) | 31 (57.4) | <0.001 |
| Days of bleeding per period, median (range) | 5.5 (2.5–8.5) | 5 (1–10) | 2.75 (0.12) | <0.001 |
| Menstrual cycle length, median (range) | 28 (21–39) | 27 (21–38) | 27 (22–37)# | 0.06 |
| Symptoms of bacterial vaginosis, n (%) § | 11 (20.4) | 10 (19.2) | 9 (16.7) | 0.88 |
| In a relationship, n (%) | 25 (46.3) | 26 (50) | 42 (77.8) | 0.001 |
| Partner | 0.001 | |||
| Male | 22 (40.7) | 26 (50) | 41 (75.9) | |
| Female | 3 (5.5) | 0 | 1 (1.9) | |
| Single | 29 (53.7) | 26 (50) | 12 (22.2) | |
| Number of sexual intercourses during the study period, mean (SD) | 4.4 (4.6) | 3.4 (4.1) | 5.2 (4.3) | 0.049 |
Participants are divided according to contraceptive method: non-hormonal contraceptives, combined oral contraceptives and LNG-IUS.
Only for those with a preserved menstrual cycle.
Self-reports of vaginal discharge in combination with fishy odour at least at one time-point during the study period.
LNG-IUS, levonorgestrel intrauterine system.
The following factors correlated with the overall spread in microbial samples and were adjusted for in following analyses: sugar consumption, smoking status, snus usage and alcohol intake. Age was initially observed to affect the microbiome of vaginal samples only (ANOSIM R = 0.046, P = 0.018). However, since a higher proportion of young women (18–24) were taking COC (39% vs. 20% of women 25+), we also assessed this effect for each contraceptive group separately and found no effect (all R < 0.01, all P > 0.1). Still, age was also adjusted for where appropriate.
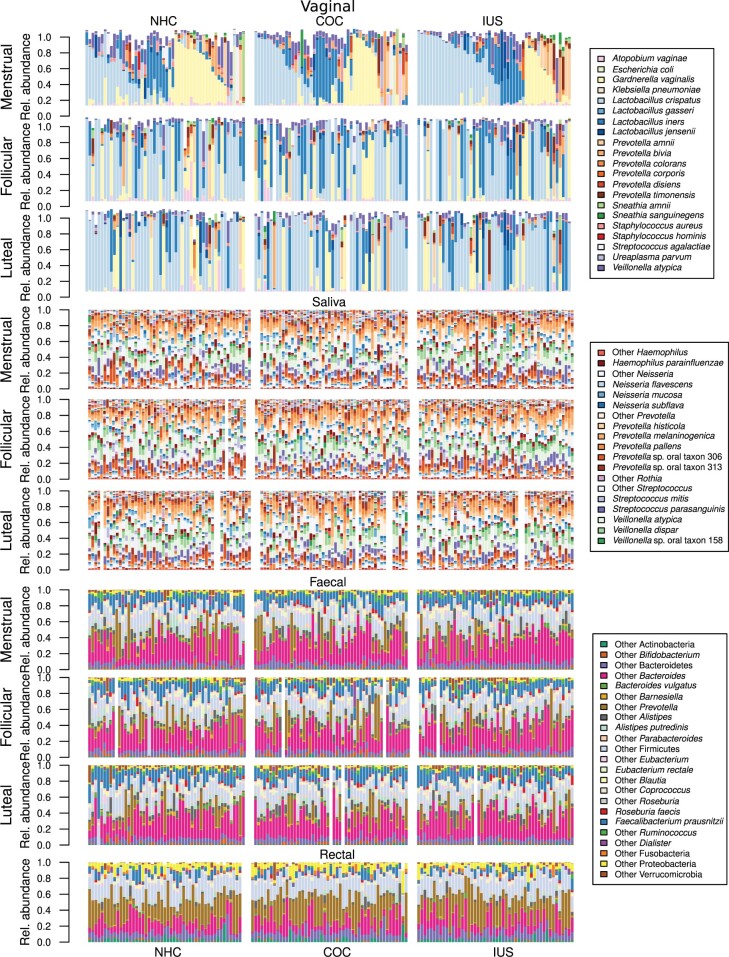
Taxonomic composition across body sites
Vaginal samples were dominated by Lactobacillus and Prevotella species as well as Gardnerella vaginalis (Fig. 2). Saliva samples were dominated by Haemophilus, Neisseria, Prevotella, Streptococcus and Veillonella, regardless of contraceptive method or phase of the menstrual cycle (Fig. 2). Faecal samples were dominated by Bacteroides, Allistipes and Faecalibacterium regardless of contraceptive method or phase of the menstrual cycle (Fig. 2). Rectal samples, which were only collected at the first hospital visit, differed from faecal samples in that they have higher diversity and a larger proportion of unclassified Bacteroidales (Fig. 2). No species were associated with hormonal contraceptive usage or the menstrual cycle in saliva, faecal or rectal samples.
Figure 2.
Taxonomic profiles across body sites, contraceptive groups and phase of the menstrual cycle. Vaginal samples are dominated by Lactobacillus crispatus, Lactobacillus iners and Gardnerella vaginalis, as well as various Prevotella spp. Women are separated by contraceptive and sorted after their dominant group in the menstrual phase and are in the same order in each panel, so each individual’s progression through the menstrual cycle can be followed directly. For some women, the microbiome remains fairly consistent across all three time-points, while other women exhibit sharp changes. Saliva samples are dominated by Veillonella spp., Neisseria spp., Haemophilus spp. and Prevotella spp., while faecal and rectal samples are dominated by Faecalibacterium prausnitzii, Bacteroides spp., Roseburia spp. and Prevotella spp., the latter being more abundant in the rectum than in faeces. Samples are remarkably stable over time. COC, combined oral contraceptives; LNG-IUS, levonorgestrel intrauterine system; NHC, non-hormonal contraceptives.
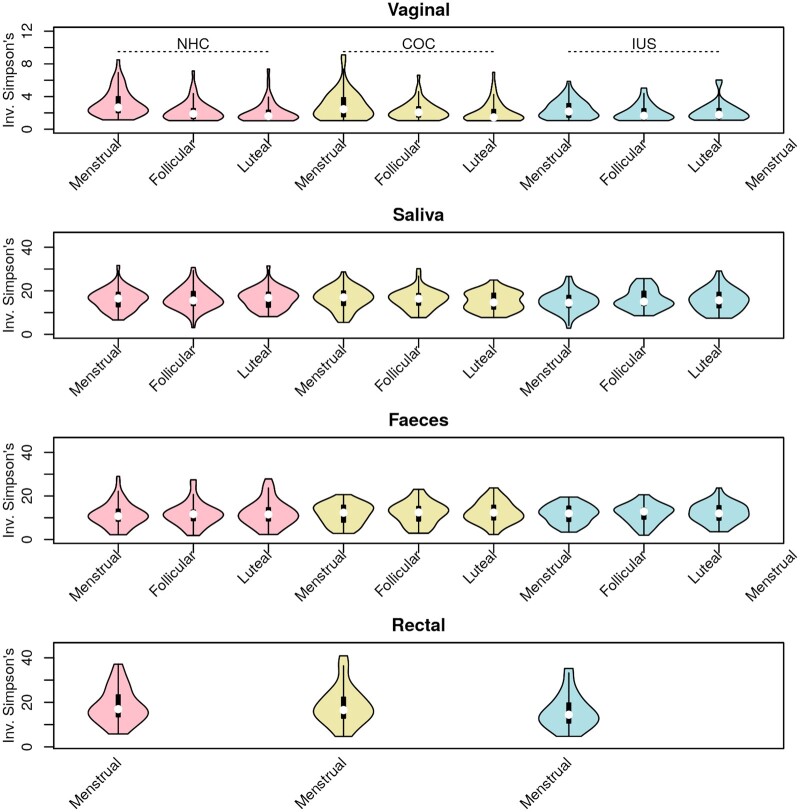
Alpha-diversity and species richness
No significant differences in alpha-diversity (Fig. 3) and species richness (Supplementary Fig. S2) were observed between contraceptive groups. For each individual, rectal samples presented the highest richness, followed by faecal, saliva and vaginal samples (paired t-tests, P < 0.0001) (Supplementary Fig. S2). Faeces were to a larger extent than saliva dominated by a few species, so despite their higher richness, they have slightly lower diversity (paired t-test, P < 0.0001). Vaginal samples have significantly higher diversity during the menstrual phase compared with the follicular or luteal phase (both P < 0.0001).
Figure 3.
Only vaginal swabs have significant differences in diversity across the menstrual cycle. Violin plots of bacterial diversity (Simpson’s inverted index) for vaginal, saliva, faecal and rectal samples. In each panel, samples from women using NHC are depicted in pink, COC in yellow and LNG-IUS in blue. The phase of the menstrual cycle is given in the x-axes. COC, combined oral contraceptives; LNG-IUS, levonorgestrel intrauterine system; NHC, non-hormonal contraceptives.
During the menstrual phase, bacterial richness and diversity in saliva reached their lowest point, while the vaginal microbiome was at its highest diversity (Supplementary Fig. S2); these differences were only significant for vaginal samples (both P < 0.00001). Faecal alpha-diversity was positively correlated to rectal (Pearson’s r = 0.27, P = 0.0009) and vaginal diversity (r = 0.11, P = 0.02), and negatively correlated to oral diversity (r = −0.12, P = 0.01). The other body sites’ diversity indexes were not significantly correlated. The negative correlation between oral and faecal diversity was also supported by an inverse correlation in their richness (r = −0.16, P = 0.0007).
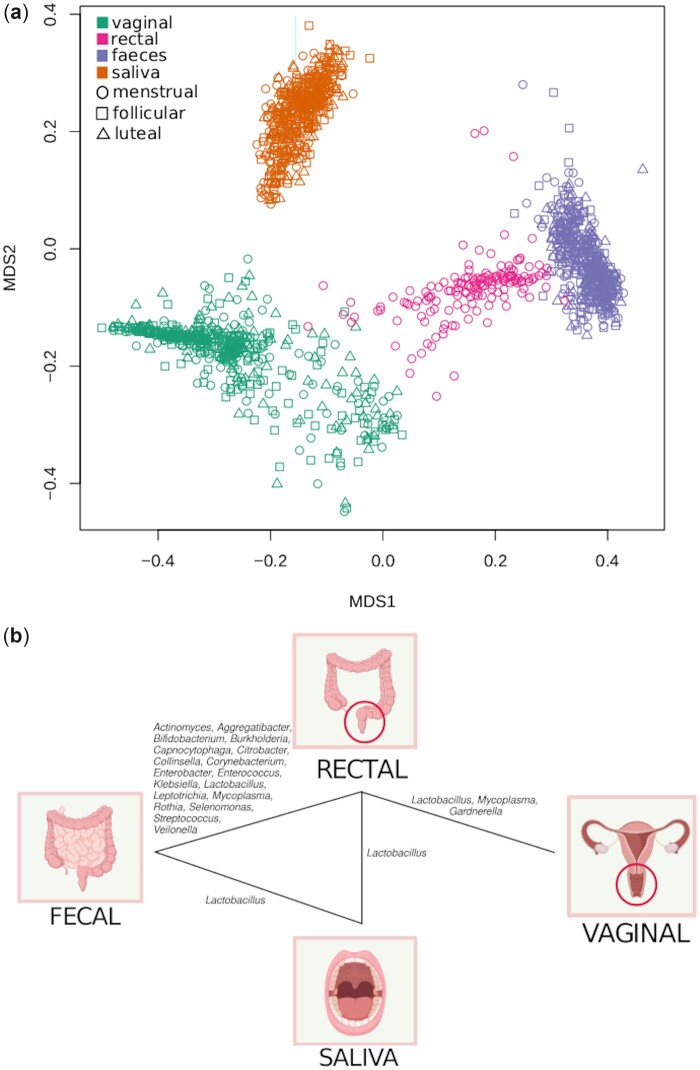
Sample to sample distance (beta-diversity)
Samples from each body site were clearly separated from each other (Fig. 4a), despite certain genera being abundant in two or more sites (Supplementary Fig. S3). Within each body site, the separation between samples was not driven by contraceptive or phase of the menstrual cycle (Fig. 5a). Within a given individual and body site, distances between samples were on average significantly lower than between individuals (P < 0.0001 for all body sites; Supplementary Fig. S4).
Figure 4.
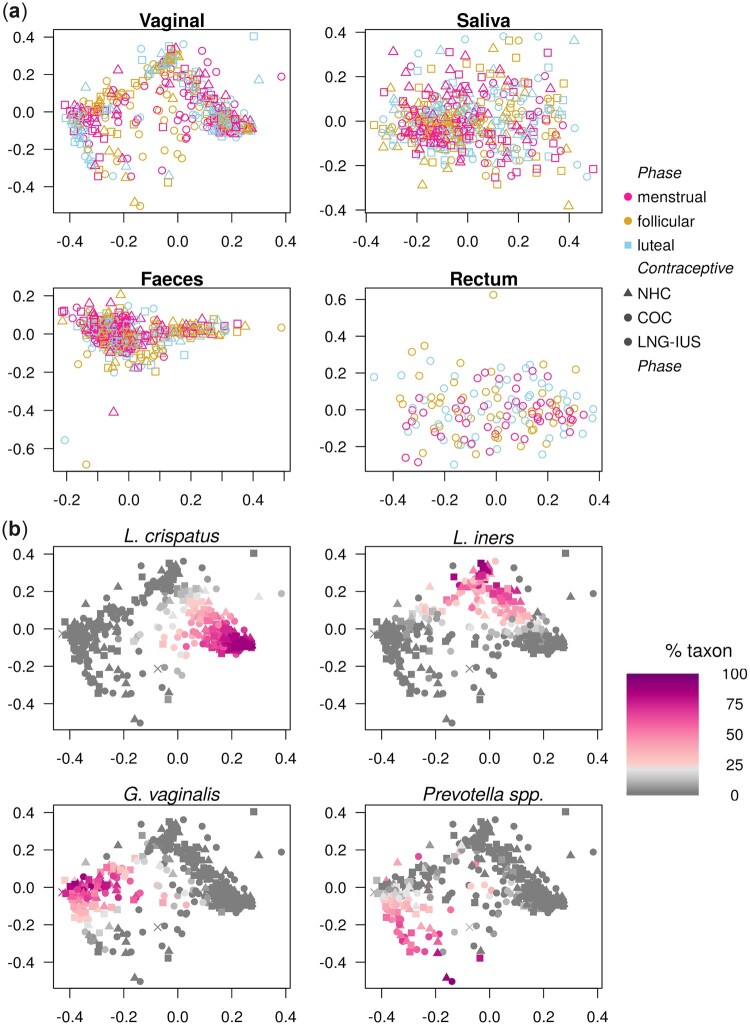
Different body sites have a unique signature, but there are points of contact. (a) Scatter plot of a two-dimensional non-metric multidimensional scaling (NMDS) of distances between samples (Bray-Curtis). Samples separate primarily by sample type, and not by phase of the cycle. Each colour represents a body site, and sampling time-point is shown in the shape of the points. (b) Genera with significant shared strains across body sites. Genera with at least 10 strains in every body site were analysed (n = 63 genera). For each genus, we tested whether separate body sites within the same individual shared strains to a larger extent than body sites from different individuals. Genera for which a few strains are prevalent over the majority of individuals can therefore not be shown to be significantly shared in this analysis.
Figure 5.
Menstrual cycle and hormonal contraceptive usage are not main drivers of sample separation, but vaginal samples present distinct clusters. (a) Scatter plots of two-dimensional non-metric multidimensional scaling of distances between samples for vaginal samples, saliva, faecal and rectal samples. None of the body sites presents a clear clustering structure, nor do samples segregate by phase of the menstrual cycle nor contraceptive. Vaginal samples present a striking triangular separation structure. (b) The same scatter plot of two-dimensional non-metric multidimensional scaling of distances between vaginal samples in Fig. 5a is presented again with a different annotation. In each panel, the relative abundance of one key taxon is depicted in colour scale, from 0 (gray) to 100% (purple): Lactobacillus crispatus, Lactobacillus iners, Gardnerella vaginalis and Prevotella spp. The first three of these taxa are more or less mutually exclusive, while Prevotella spp. can be found in mixtures with other taxa, but most prominently with G. vaginalis. COC, combined oral contraceptives; LNG-IUS, levonorgestrel intrauterine system; NHC, non-hormonal contraceptives.
The number of strains retained over time in each body site for a given individual was significantly higher than the overlap across individuals (P < 0.0001, all sites). There was a significant overlap of shared strains between faecal and rectal samples. Sixty-two genera were found with at least 10 strains in each body site. For these genera, the overlap of strains between body sites was assessed for the first time point (Fig. 4b). Of interest, the Lactobacillus genus had significantly more strains shared between the rectum and all other body sites. While rectal, faecal and saliva samples all shared Lactobacillus strains, the vaginal samples only showed significant overlap with rectal samples. Rectal and faecal samples presented a large overlap of Lactobacillusparacasei, Lactobacilluscasei and Lactobacillusplantarum and rectal samples shared strains of Lactobacillusamylovorus with saliva samples and Lactobacillussalivarius, Lactobacillusacidophilus and Lactobacillusvaginalis with vaginal samples. Saliva samples shared Lactobacillusantri, Lactobacillusfermentum and Lactobacillusbrevis with faecal samples.
For the vaginal samples, a clear cluster structure exists, with samples being dominated by one of Lactobacillus crispatus, Lactobacillus iners, G.vaginalis or Prevotella spp. (Fig. 5). None of the other body sites presented equally clear clustering (Supplementary Fig. S5). The clustering structure observed for the vaginal samples was not clearly reflected on the other body sites (Supplementary Fig. S5b–d). There was a statistically significant but there was a weak correlation of the sample-to-sample distance between individuals of rectal samples compared to vaginal samples (Mantel’s test on Spearman’s correlation. r = 0.058, P = 0.009; rectal against faecal, r = 0.26, P = 0.001; other comparisons not significant; all P adjusted).
Shifts in microbiome composition during the menstrual cycle
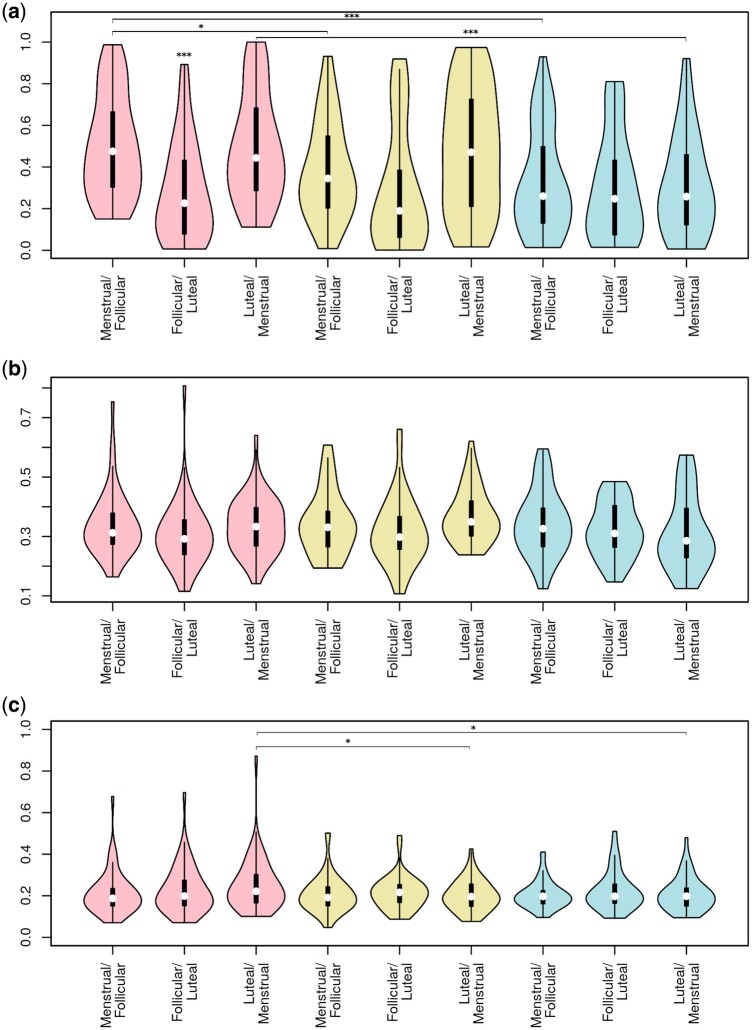
Shifts between consecutive time-points were most subtle for faecal samples, and most extreme for vaginal samples (Fig. 6; P < 0.0001). Shifts in the saliva and vaginal microbiome were smallest in the transition from follicular to luteal phase, and similarly high upon entering and passing out of the menstrual period (P < 10E−5 for vaginal samples; P = 0.04 for saliva). Separating women by their contraceptive group, shifts in the vaginal microbiome were largest for those without hormonal contraception and smallest for LNG-IUS users, even after excluding women without menstrual bleedings (LNG-IUS vs NHC, P = 0.0005; COC vs NHC, P = 0.02). No significant differences between contraceptive groups were observed for saliva or faeces.
Figure 6.
The human microbiome is most variable in the vagina and least stable during the menstrual phase. Violin plots of the within-subject Bray-Curtis sample distance in the transition between each phase of the menstrual cycle for (a) vaginal samples (b) saliva and (c) fecal samples. The phase transition is given in the x-axis. Significant differences within a phase transition are all in relation to non-hormonal contraception, and differences between phase transitions are in relation to the menstrual/follicular transition. *P < 0.05; ***P < 0.001; Pink: non-hormonal contraceptive. Yellow: combined oral contraceptive. Blue: levonorgestrel intrauterine system.
Taxonomic composition of vaginal samples and role of sex hormones
There was no significant difference in the relative abundance of bacterial species between the contraceptive groups. Vaginal samples were dominated by L.crispatus, L.iners, G.vaginalis and Prevotella spp. regardless of contraceptive method (Fig. 2). Genus Lactobacillus and L. crispatus expanded in abundance during the follicular and luteal phases (Welch’s t-test. In the follicular phase P = 0.0002 for Lactobacillus and P = 0.004 for L. crispatus and in the luteal phase, P < 0.0001 for Lactobacillus and P < 0.0001 for L. crispatus). Concomitant to this increase in Lactobacillus spp., there was a decrease in Gardnerella spp. (Figs 1 and 7).
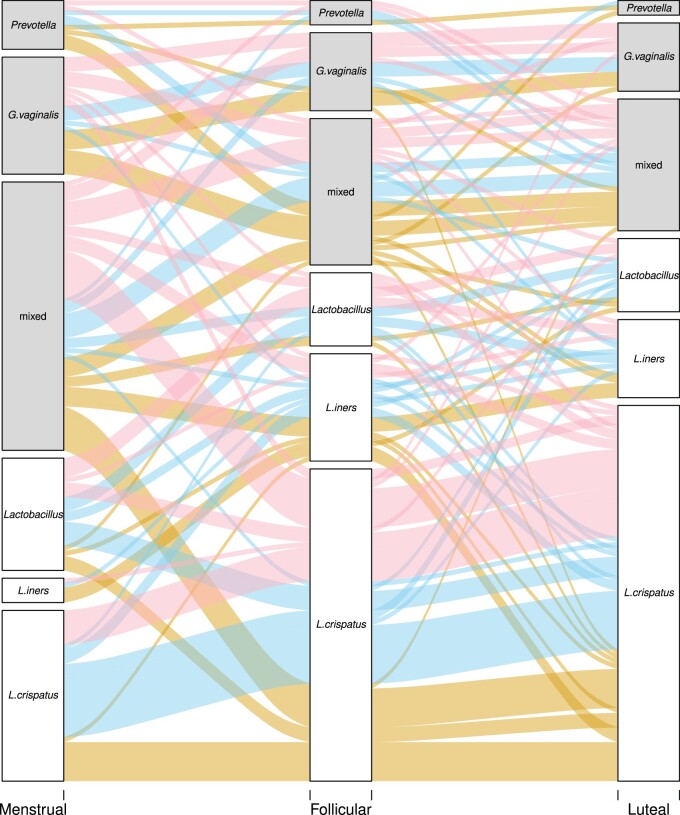
Figure 7.
Over the menstrual cycle, the vaginal microbiome of more women becomes dominated by Lactobacillus crispatus. Alluvial plot showing the fate of each sample across the menstrual cycle. ‘Dominance’ is defined as >60% of reads coming from the same genus or species. The ‘Lactobacillus’ groups comprise samples with a mixture of Lactobacillus species. Samples lacking a dominant group are classified as ‘mixed’. The three groups not dominated by Lactobacillus species are shaded grey. The lines depicting the trajectory of each sample are coloured after the contraceptive used by that individual. Pink: non-hormonal contraceptive. Yellow: combined oral contraceptive. Blue: levonorgestrel intrauterine system.
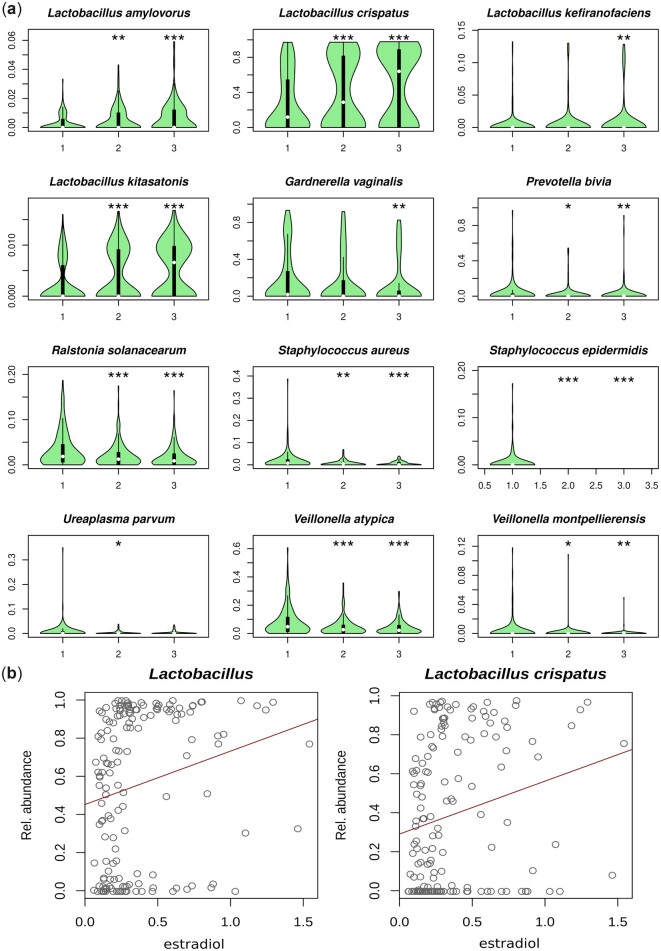
Four Lactobacillus species were increased in vaginal samples in the follicular and luteal phase compared with the menstrual phase, while eight BV-associated species were decreased (Fig. 8a). No vaginal species had significant changes in relationship to other metadata parameters, including age and contraceptive.
Figure 8.
Species whose abundance in the vagina varies over the menstrual cycle and correlate with oestradiol levels. (a) Violin plots showing the relative abundance of species whose abundance is significantly correlated to the phase of the menstrual cycle, adjusted for contraceptive usage and lifestyle variables. Y-axis in each panel is adjusted to show the differences in the data, so scale varies. (1) Menstrual phase, (2) follicular phase, (3) luteal phase. *P < 0.05, **P < 0.01, ***P < 0.001. (b) Both genus Lactobacillus and the species Lactobacillus crispatus are more abundant in the presence of high oestradiol levels, although some women may present low Lactobacillus spp. abundance regardless of their oestradiol level.
Genus Lactobacillus and L. crispatus were positively correlated to serum oestradiol levels (r = 0.11, P < 0.001; Fig. 8b). Another two Lactobacillus species had significant positive correlations with sex hormones (Lactobacilluskitasatonis ∼ oestradiol, r = 0.003, q = 0.041; L. amylovorus ∼ progesterone, r = 8.5E−5, q = 0.013). Women with high levels of L. iners had on average higher serum oestradiol levels (n.s.; Supplementary Fig. S6). No species in the saliva or faecal microbiome were significantly associated to sex hormone levels after adjusting for the individual.
The role of sexual practices and menstrual hygiene on the vaginal microbiome is hard to separate from the effects of contraceptives and the menstrual cycle. We have attempted to study this through partitioning the variation between vaginal samples between the phase of the menstrual cycle, effects of recent sexual and hygienic habits in the past 3 months, and short-term effects, as described in the methods. This technique assigned 5% of the total observed variation to factors prior to the study period, including contraceptive method. Then 1% if the variation was attributed to the phase of the menstrual cycle and 1% could be assigned equally to the phase of the cycle or vaginal bleeding and intercourse in the 24 h before the sample was taken. This leaves about 94% of the variation unaccounted for, of which most (70% of total variation) can be accounted for when individual ID is included as a factor.
Vaginal dysbiosis and sexually transmitted infections
Most women who presented with a dysbiotic vaginal microbiome during menses changed to a Lactobacillus-dominated microbiome in the subsequent follicular and luteal phases (Fig. 7). During menses, 89 women (58%) had a dysbiotic vaginal microbiome with less than 60% Lactobacillus spp. This decreased to 49 (32%) in the follicular phase (P < 0.0001) and 44 (29%) in the luteal phase (P < 0.0001). Most of this shift came from women with a mixed microbiome (no species >60% abundance) becoming more Lactobacillus dominated, but also from a decrease in G.vaginalis. No differences in demographic or health questionnaire data were found in women with Lactobacillus-dominated vaginal microbiomes compared with others and in women dominated by BV-associated bacteria compared with others.
A significant positive correlation was observed between the proportion of human DNA in a sample and the Lactobacillus content (r = 0.54, P < 0.0001). Conversely, typical BV bacteria, such as Gardnerella (r = −0.43, P < 0.0001) and Prevotella (r = −0.53, P < 0.0001) were negatively correlated to the proportion of human DNA in a sample.
Sexually transmitted diseases are identified in two participants. One woman not using hormonal contraception was diagnosed with Chlamydia trachomatis and Neisseria gonorrhoea shortly after providing her samples. These bacteria are also found in relatively low amounts (0.5–1.5% of reads) in all her vaginal samples which also presented with relatively large amounts of Gardnerella (20–30%) and other bacteria such as Prevotella and Veillonella. Another woman, who had completed treatment for C.trachomatis 2 months prior to her enrolment in the study had up to 2.5% of the DNA in her vaginal swab from this species. Her vaginal samples were dominated by L. iners and Lactobacillusjensenii, but also presented about 10–20% of Ralstonia solanacearum and Veillonella atypica.
Discussion
Herein, we present the largest investigation of female microbiomes by shotgun sequencing to date. We investigate four body sites at three points of the menstrual cycle in 160 healthy Caucasian women of reproductive age using three different contraceptive regimens.
The microbiome across body sites, contraceptive groups and phases of the menstrual cycle
In a Danish population of young, healthy women, the type of contraception did not associate with microbiome composition in three body sites: rectum, faeces and saliva, at any of the three time-points in the menstrual cycle (menstrual, follicular and luteal phases). Levels of oestradiol and progesterone also did not associate with microbiome composition in these body sites. This knowledge is novel and important as hormonal contraception is widely used during early reproductive years, often over several years (Lindh et al., 2017; Products—Data Briefs—Number 327—December 2018, n.d.; Hellström et al., 2019), before attempting pregnancy. Indeed, most of the variation in the microbiome could not be explained by the questionnaire data collected, while a significant proportion (70%) was simply attributed to ‘Individual ID’. It is possible that further data, including more detailed food and drug intake profiling, could have covered more of the variation observed.
The hypothesis that different body sites are interconnected and affect each other is here supported by significant sharing of strains across body sites. This connection might be due to other systematic mechanisms (apart from female sex hormones) in the individual, such as genetics, immunological, hormonal and even priority effects. It could also simply be a physical connection, with microorganisms travelling alongside food through the digestive system (oral/faecal/rectal), sexually (oral/vagina/rectal) or due to anatomical proximity (vaginal/rectal/faecal). However, while highly significant, the correlations are small (0.1–0.27) and one body site cannot be sampled as a proxy for another site.
Body site is by far the most prominent feature separating microbiome samples (Costello et al., 2009; Segata et al., 2012; Zhou et al., 2013; Lloyd-Price et al., 2017). We included the rectum, a body site that is not often sampled and observe that it shares many characteristics with faeces (Jones et al., 2018; Fair et al., 2019), but it is also unique. The rectal microbiome has previously been suggested to be a reservoir of bacteria that can readily colonise the vagina (Kostic et al., 2014; Fudaba et al., 2021). This is partially confirmed here in the observation that Lactobacillus strains found in the rectum are significantly more frequent in the vagina of the same individuals, as previously reported (Antonio et al., 2005). Importantly, no such overlap occurs between faecal and vaginal samples, which makes inappropriate wiping routines quite unlikely to explain the shared strains between rectum and vagina. A previous study found a significantly shorter ano-vaginal distance in women diagnosed with BV compared with women without BV, and suggested this as a possible anatomical explanation for the development of dysbiosis (Torky et al., 2021). This strain-level analysis was only possible due to our use of shotgun metagenomics, since it would not have been possible using 16S rRNA or other marker genes.
The composition of the saliva and faecal microbiomes were largely unaffected by menstrual cycle and contraceptives in our study. Accordingly, no specific bacteria in these body sites could be connected to serum levels of oestradiol or progesterone. However, the saliva microbiome seems to be least stable during menses as previously reported (Bostanci et al., 2021).
The vaginal microbiome responds to female sex hormones
Differences according to contraception method were found in the vaginal microbiome as previously reported (Sinha et al., 2019). Women not using hormonal contraceptives (NHC group) had a significantly larger shift in the composition of their vaginal microbiome across the menstrual cycle, compared with the COC and LNG-IUS groups; the latter remained the most stable throughout the menstrual cycle, regardless of whether they had menstrual bleedings or not. Recent work based on transcriptomics has found similar effects of the menstrual cycle in the microbiome of the endometrium, highlighting the regulated crosstalk between female hormones, bacteria and the reproductive tract (Sola-Leyva et al., 2021).
We find an increase of Lactobacillus species and a decrease of eight BV-associated species in the follicular and luteal phases. The cause of the increased diversity in the vaginal microbiome during menstrual bleeding has been debated: are differences observed during menstruation due to hormonal changes or physiological changes with menstrual blood flowing through the vagina leaving iron-sources for Gardnerella spp. (Roberts et al., 2019) or merely the menstrual hygiene products used. Here we found a small effect of menstrual products and sexual practices.
Several previous studies state that low oestradiol levels at the time of menstruation are responsible for low glycogen deposition and thereby lower Lactobacillus presence, resulting in higher pH which allows growth of anaerobic BV-related bacteria (Amabebe and Anumba, 2018). Further evidence of the association between oestradiol and lactobacillus dominance is provided by studies of menopausal women with vaginal dysbiosis returning to a Lactobacillus dominated microbiome after commencing hormone replacement therapy (Ginkel et al., 1993; Heinemann and Reid, 2005; Muhleisen and Herbst-Kralovetz, 2016). Additionally, dysbiosis has been observed in girls before puberty (before oestradiol rise) (Hammerschlag et al., 1978; Gerstner et al., 1982; Hickey et al., 2015) and in transgender men with oestrogen suppression (McPherson et al., 2019), while the opposite is observed during pregnancy, concomitantly with high levels of oestrogen (Romero et al., 2014; MacIntyre et al., 2015). Here, we find a direct correlation between oestradiol levels and Lactobacillus spp. in general and with L. crispatus specifically. While previous studies have argued for this association (Ravel et al., 2011; Song et al., 2020), this is, to our knowledge, the first direct measurement of sex hormones in relation to microbiome composition.
Song et al. (2020) raise the question of whether it could be the decline in progesterone rather than oestradiol before the menstrual bleeding that is responsible for the high diversity during menstruation (Roberts et al., 2019; Song et al., 2020). In this study, we found increased diversity in the vaginal microbiome during menstruation in all three contraceptive groups. They all have a drop in the level of oestradiol/oestrogen synthetics before menstruation, but the COC group does not ovulate and has no large progesterone shifts prior to menstruation, indicating that the dysbiosis is not driven by progesterone dynamics. The direct association between measured oestradiol levels and vaginal microbiome composition suggest that the high diversity observed during menses is mainly due to oestradiol withdrawal before menses. We can, however, not rule out that the bleeding itself could contribute to the high diversity, as LNG-IUS users with no or little menstrual bleeding despite their underlying cycle and ovulation seem to have the most stable vaginal microbiome. This stability could perhaps also be mediated through changes in the composition of the vaginal mucus because of the local effect of the progestin-releasing device.
We found the vagina to be the only body site with a clear clustering with four main clusters: dominated namely by (i) L.crispatus, (ii) L.iners, (iii) G.vaginalis and (iv) Prevotella spp., irrespective of contraception use. This is in contrast with previous findings by Ravel et al. (2011) who identified five specific CSTs in the vaginal microbiome of four ethnic groups of women, including a white north American group, based on 16S rRNA sequencing. Four of these CSTs were dominated by different Lactobacillus species (L. crispatus, Lactobacillusgasseri, L. iners and L. jensenii) leaving the remaining group IV as the diverse group (Ravel et al., 2011), which was later expanded into two clusters (IV-A and IV-B) (Gajer et al., 2012). In our cohort, while L. gasseri and L. jensenii were both detected, these species are not sufficiently abundant to form separate clusters. These two groups were the smallest in Ravel’s pioneering work and were also found in low prevalence in a previous cohort of Nordic women (Cheng et al., 2020).
Unlike most previous work in the vaginal microbiome, here we have used metagenomic shotgun sequencing instead of 16S rRNA gene sequencing. While the large proportion of human reads did not allow us sufficient sequence depth to assess the bacterial functional potential of vaginal swabs, we could nevertheless make a deeper taxonomic analysis. In addition to this, we have measured key sex hormones at each sampling time point. Previous work has used reference values for sex hormones in relation to cycle day (Ravel et al., 2011; Song et al., 2020). While our analysis may present a less clear picture of the complex interaction between hormones and microbes, it is nonetheless a more accurate picture.
Vaginal dysbiosis
Vaginal dysbiosis with a microbiota dominated by BV-associated bacteria, is not associated with contraception use, but is found in 40% of samples in our cohort, most notably during the menstrual phase (58% of samples). A previous study found, in a cohort of healthy women without BV-symptoms, that 27% presented with vaginal dysbiosis (Ravel et al., 2011), similar to what we find in the follicular (32%) and luteal phases (29%). None of our metadata were associated with BV-related species in the vagina. There were no correlations with other related variables (smoking, alcohol, number of intercourses) that could be suspected to be confounding of using snus or related to BV. Recent studies conclude that the presence of Atopobium vaginae together with G.vaginalis provide high predictability of BV (Hardy et al., 2017; Castro et al., 2020). Considering that only few women reported that they used snus (n = 14), this could be a coincident finding. Only two women reported current/recent sexually transmitted disease (C.trachomatis and N.gonorrhoea) which was reflected in the sequencing data. Perhaps a selection bias of non-dysbiotic microbiomes may have been introduced since three women using antimicrobials or lactic acid suppositories during the study were excluded, nevertheless, the cohort can still be considered healthy.
No longitudinal shifts in the cluster structure in the vaginal microbiome were observed during the menstrual cycle regardless of contraceptive use, albeit most women dominated by G.vaginalis or Prevotella during menses shifted towards a Lactobacillus dominated composition throughout the cycle. Other studies have correlated dysbiosis with reproductive complications (Romero et al., 2014; Haahr et al., 2016; Kroon et al., 2018) and future studies need to explore whether those with vaginal dysbiosis throughout the menstrual cycle constitute a different group compared with those with dysbiosis solely during menstruation. It can be speculated that reproductive complications are only associated with women who, despite an increase in oestradiol, maintain a dysbiotic vaginal microbiome throughout the menstrual cycle.
Another interesting finding in this study is the observation that the proportion of human DNA in a vaginal sample was negatively correlated to typical BV-associated bacteria, such as G.vaginalis and Prevotella spp. and, conversely, positively correlated with the Lactobacillus content. This may be due to the shedding of clue cells (Amegashie et al., 2017) with a high bacterium to human DNA ratio, and/or due to the thick, bacteria-dense biofilm formed on top of the epithelial surface by BV-associated bacteria (Chen et al., 2021). This is relevant for future work using shotgun sequencing, since Lactobacillus spp. dominated samples will be sequenced less deeply unless human DNA removal is performed before sequencing.
This is the first large scale shotgun investigation of the microbiota composition across four body sites sampled over a menstrual cycle in a healthy female Caucasian cohort. Another strength is the comparison between a natural menstrual cycle and cycles affected by hormonal contraception use, as well as real-time measurements of endogenous sex hormone levels. The extensive questionnaire data regarding the women’s prior and current health, lifestyle and sex life enabled us to explore and adjust for potential impact on the microbiota compositions across body sites. A limitation is the homogeneity of the cohort which made it impossible to investigate the impact of ethnicity and low socio-economic status. Another limitation is that only three time points in the menstrual cycle were investigated in this study leaving day-to-day variations unexplored.
Conclusions
The type of hormonal contraception does not significantly associate with the microbiome composition in the vagina, faeces, rectum or saliva in healthy young women. This is a welcome finding considering the widespread and prolonged use of these highly efficient contraceptive methods. The menstrual cycle is, however, a major confounding factor for the vaginal microbiome, with large shifts in composition occurring around menstruation. These changes are most pronounced in women with a natural cycle, compared with COC and LNG-IUS users. The time point in the menstrual cycle should be considered when analysing the microbiome of women of reproductive age, since stratifying by vaginal dysbiosis status during menstruation could be misleading. This is the first study to confirm by direct measurements of oestradiol, a correlation of high oestradiol levels with the presence of L.crispatus during a natural cycle, adding evidence of a possible hormonal mechanism for the maintenance of this desirable microbe.
Supplementary data
Supplementary data are available at Human Reproduction online.
Data availability
The sequencing data analysed in this study were submitted to the European Nucleotide Archive under project number PRJEB37731, samples ERS4421369–ERS4422941. Other patient-level data will be shared on reasonable request to the corresponding author.
Supplementary Material
Acknowledgements
We like to thank the nurses employed at the Recurrent Pregnancy Loss Unit in Copenhagen for their huge contribution with including participants: Louise Lunøe, Karen Kirchheiner and Marie Chonovitsch. We thank Marica Hamsten and Fredrik Boulund for invaluable contribution to sample handling logistics.
Authors’ roles
M.C.K. and H.S.N.: planned and organized study cohort, obtained ethics and data protection approval, collected samples, provided data management and wrote the manuscript. Z.B.: included participants and secured informed consent, collected samples, provided data management and wrote the manuscript. A.N.A.: collected samples, analysed data, wrote the manuscript. L.W.H., E.F. and G.E.: analysed data and wrote the manuscript. I.S.-K. and L.E.: planned and organized the study cohort and wrote the manuscript. All authors have read and approved the final manuscript.
Funding
This work was partly funded by the Ferring Pharmaceuticals through a research collaboration with The Centre for Translational Microbiome Research (CTMR) at the Karolinska Institutet (L.W.H., E.F., G.E. and I.S.-K.). Ferring Pharmaceuticals also funded the infrastructure to obtain the clinical samples at Copenhagen University Hospital ([#MiHSN01], M.C.K., Z.B., and H.S.N.). This work was also supported by funding from Rigshospitalet’s Research Funds ([#E-22614-01 and #E-22614-02] to M.C.K.) and Oda and Hans Svenningsen’s Foundation ([#F-22614-08] to H.S.N.).
Conflict of interest
M.C.K., L.W.H., E.F., Z.B., G.E., L.E., I.S.-K. and H.S.N., are partially funded by Ferring Pharmaceuticals, which also provided funds for the collection and processing of the samples analysed in this study. H.S.N.’s research is further supported by Freya Biosciences and the BioInnovation Institute. H.S.N. has received honoraria from Ferring Pharmaceuticals, Merck A/S, Astra-Zeneca, Cook Medical and Ibsa Nordic. A.N.A. reports no competing interests.
Contributor Information
Maria Christine Krog, The Recurrent Pregnancy Loss Unit, The Capital Region, Rigshospitalet and Hvidovre Hospital, Copenhagen University Hospitals, Copenhagen, Denmark; Department of Clinical Immunology, Copenhagen University Hospital, Rigshospitalet, Copenhagen, Denmark.
Luisa W Hugerth, Centre for Translational Microbiome Research, Department of Microbiology, Tumor and Cell Biology (MTC), Karolinska Institutet, Stockholm, Sweden; Science for Life Laboratory, Stockholm, Sweden.
Emma Fransson, Centre for Translational Microbiome Research, Department of Microbiology, Tumor and Cell Biology (MTC), Karolinska Institutet, Stockholm, Sweden.
Zahra Bashir, The Recurrent Pregnancy Loss Unit, The Capital Region, Rigshospitalet and Hvidovre Hospital, Copenhagen University Hospitals, Copenhagen, Denmark; Department of Obstetrics and Gynecology, University Hospital Zealand, Slagelse Hospital, Slagelse, Denmark.
Anders Nyboe Andersen, The Fertility Department Section 4071, Rigshospitalet, Copenhagen University Hospital, Copenhagen, Denmark.
Gabriella Edfeldt, Centre for Translational Microbiome Research, Department of Microbiology, Tumor and Cell Biology (MTC), Karolinska Institutet, Stockholm, Sweden.
Lars Engstrand, Centre for Translational Microbiome Research, Department of Microbiology, Tumor and Cell Biology (MTC), Karolinska Institutet, Stockholm, Sweden; Science for Life Laboratory, Stockholm, Sweden.
Ina Schuppe-Koistinen, Centre for Translational Microbiome Research, Department of Microbiology, Tumor and Cell Biology (MTC), Karolinska Institutet, Stockholm, Sweden; Science for Life Laboratory, Stockholm, Sweden.
Henriette Svarre Nielsen, The Recurrent Pregnancy Loss Unit, The Capital Region, Rigshospitalet and Hvidovre Hospital, Copenhagen University Hospitals, Copenhagen, Denmark; Department of Clinical Medicine, Copenhagen University, Copenhagen, Denmark; Department of Obstetrics and Gynecology, Copenhagen University Hospital, Hvidovre Hospital, Copenhagen, Denmark.
References
- Achilles SL, Hillier SL.. The complexity of contraceptives: understanding their impact on genital immune cells and vaginal microbiota. AIDS 2013;27:S5–S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amabebe E, Anumba DOC.. The vaginal microenvironment: the physiologic role of Lactobacilli. Front Med (Lausanne) 2018;5:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amegashie CP, Gilbert NM, Peipert JF, Allsworth JE, Lewis WG, Lewis AL.. Relationship between nugent score and vaginal epithelial exfoliation. PLoS One 2017;12:e0177797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio MAD, Rabe LK, Hillier SL.. Colonization of the rectum by Lactobacillus species and decreased risk of bacterial vaginosis. J Infect Dis 2005;192:394–398. [DOI] [PubMed] [Google Scholar]
- Ata B, Yildiz S, Turkgeldi E, Brocal VP, Dinleyici EC, Moya A, Urman B.. The endobiota study: comparison of vaginal, cervical and gut microbiota between women with stage 3/4 endometriosis and healthy controls. Sci Rep 2019;9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atashili J, Poole C, Ndumbe PM, Adimora AA, Smith JS.. Bacterial vaginosis and HIV acquisition: a meta-analysis of published studies. AIDS 2008;22:1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balle C, Esra R, Havyarimana E, Jaumdally SZ, Lennard K, Konstantinus IN, Barnabas SL, Happel AU, Gill K, Pidwell T. et al. Relationship between the oral and vaginal microbiota of South African adolescents with high prevalence of bacterial vaginosis. Microorganisms 2020;8:1004–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgdorff H, Van Der Veer C, Van Houdt R, Alberts CJ, De Vries HJ, Bruisten SM, Snijder MB, Prins M, Geerlings SE, Van Der Loeff MFS. et al. The association between ethnicity and vaginal microbiota composition in Amsterdam, the Netherlands. PLoS One 2017;12:e0181135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostanci N, Krog MC, Hugerth LW, Bashir Z, Fransson E, Boulund F, Belibasakis GN, Wannerberger K, Engstrand L, Nielsen HS. et al. Dysbiosis of the human oral microbiome during the menstrual cycle and vulnerability to the external exposures of smoking and dietary sugar. Front Cell Infect Microbiol 2021;11:625229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JP, Edwards DJ, Blithe DL, Fettweis JM, Serrano MG, Sheth NU, Strauss JF, Buck GA, Jefferson KK.. Effects of combined oral contraceptives, depot medroxyprogesterone acetate and the levonorgestrel-releasing intrauterine system on the vaginal microbiome. Contraception 2017;95:405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusselaers N, Shrestha S, van de Wijgert J, Verstraelen H.. Vaginal dysbiosis and the risk of human papillomavirus and cervical cancer: systematic review and meta-analysis. Am J Obstet Gynecol 2019;221:9–18.e8. [DOI] [PubMed] [Google Scholar]
- Castro J, Rosca AS, Cools P, Vaneechoutte M, Cerca N.. Gardnerella vaginalis enhances Atopobium vaginae viability in an in vitro model. Front Cell Infect Microbiol 2020;10:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE.. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010;2010:baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lu Y, Chen T, Li R.. The female vaginal microbiome in health and bacterial vaginosis. Front Cell Infect Microbiol 2021;11:631972–631915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Norenhag J, Hu YOO, Brusselaers N, Fransson E, Ährlund-Richter A, Guðnadóttir U, Angelidou P, Zha Y, Hamsten M. et al. Vaginal microbiota and human papillomavirus infection among young Swedish women. NPJ Biofilms Microbiomes 2020;6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R.. Bacterial community variation in human body habitats across space and time. Science 2009;326:1694–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudray M, Kiplagat S.. Bacterial vaginosis and the risk of human papillomavirus and cervical cancer. Am J Obstet Gynecol 2019;221:171–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRAN—Package vegan, n.d.; Products—Data Briefs—Number 327—December 2018, n.d.
- De Leo V, Musacchio MC, Cappelli V, Piomboni P, Morgante G.. Hormonal contraceptives: Pharmacology tailored to women’s health. Hum Reprod Update 2016;22:634–646. [DOI] [PubMed] [Google Scholar]
- Donders G, Bellen G, Janssens D, Van Bulck B, Hinoul P, Verguts J.. Influence of contraceptive choice on vaginal bacterial and fungal microflora. Eur J Clin Microbiol Infect Dis 2017;36:43–48. [DOI] [PubMed] [Google Scholar]
- Donders GGG, Bellen G, Ruban K, Van Bulck B.. Short- and long-term influence of the levonorgestrel-releasing intrauterine system (Mirena®) on vaginal microbiota and Candida. J Med Microbiol 2018;67:308–313. [DOI] [PubMed] [Google Scholar]
- Edwards VL, Smith SB, McComb EJ, Tamarelle J, Ma B, Humphrys MS, Gajer P, Gwilliam K, Schaefer AM, Lai SK. et al. The cervicovaginal microbiota-host interaction modulates Chlamydia trachomatis infection. mBio 2019;10:e01548-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elovitz MA, Gajer P, Riis V, Brown AG, Humphrys MS, Holm JB, Ravel J.. Cervicovaginal microbiota and local immune response modulate the risk of spontaneous preterm delivery. Nat Commun 2019;10:1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair K, Dunlap DG, Fitch A, Bogdanovich T, Methé B, Morris A, McVerry BJ, Kitsios GD.. Rectal swabs from critically ill patients provide discordant representations of the gut microbiome compared to stool samples. mSphere 2019;4:e00358-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, Parikh HI, Huang B, Arodz TJ, Edupuganti L, Glascock AL. et al. The vaginal microbiome and preterm birth. Nat Med 2019;25:1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figuero E, Han YW, Furuichi Y.. Periodontal diseases and adverse pregnancy outcomes: mechanisms. Periodontol 2000 2020;83:175–188. [DOI] [PubMed] [Google Scholar]
- Forney LJ, Foster JA, Ledger W.. The vaginal flora of healthy women is not always dominated by Lactobacillus species. J Infect Dis 2006;194:1468–1469. [DOI] [PubMed] [Google Scholar]
- Fudaba M, Kamiya T, Tachibana D, Koyama M, Ohtani N.. Bioinformatics analysis of oral, vaginal, and rectal microbial profiles during pregnancy: a pilot study on the bacterial co-residence in pregnant women. Microorganisms 2021;9:1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajer P, Brotman RM, Bai G, Sakamoto J, Schütte UME, Zhong X, Koenig SSK, Fu L, Ma Z, Zhou X. et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med 2012;4:132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner GJ, Grünberger W, Boschitsch E, Rotter M.. Vaginal organisms in prepubertal children with and without vulvovaginitis—a vaginoscopic study. Arch Gynecol 1982;231:247–252. [DOI] [PubMed] [Google Scholar]
- Ginkel PD, Soper DE, Bump RG, Dalton HP.. Vaginal flora in postmenopausal women: the effect of estrogen replacement. Infect Dis Obstet Gynecol 1993;1:94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, Bowman BA, Padavattan N, Desai C, Droit L, Moodley A. et al. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in Young South African Women. Immunity 2017;46:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi G, Farulla A, Sileo FG, Facchinetti F.. Levonorgestrel-releasing intra-uterine systems as female contraceptives. Expert Opin Pharmacother 2018;19:677–686. [DOI] [PubMed] [Google Scholar]
- Green KA, Zarek SM, Catherino WH.. Gynecologic health and disease in relation to the microbiome of the female reproductive tract. Fertil Steril 2015;104:1351–1357. [DOI] [PubMed] [Google Scholar]
- Haahr T, Jensen JS, Thomsen L, Duus L, Rygaard K, Humaidan P.. Abnormal vaginal microbiota may be associated with poor reproductive outcomes: a prospective study in IVF patients. Hum Reprod 2016;31:795–803. [DOI] [PubMed] [Google Scholar]
- Hammerschlag MR, Alpert S, Onderdonk AB, Thurston P, Drude E, McCormack WM, Bartlett JG.. Anaerobic microflora of the vagina in children. Am J Obstet Gynecol 1978;131:853–856. [DOI] [PubMed] [Google Scholar]
- Hardy L, Cerca N, Jespers V, Vaneechoutte M, Crucitti T.. Bacterial biofilms in the vagina. Res Microbiol 2017;168:865–874. [DOI] [PubMed] [Google Scholar]
- Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O'Neal L, McLeod L, Delacqua G, Delacqua F, Kirby J. et al. ; REDCap Consortium. The REDCap consortium: building an international community of software platform partners. J Biomed Inform 2019;95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann C, Reid G.. Vaginal microbial diversity among postmenopausal women with and without hormone replacement therapy. Can J Microbiol 2005;51:777–781. [DOI] [PubMed] [Google Scholar]
- Hellström A, Gemzell Danielsson K, Kopp Kallner H.. Trends in use and attitudes towards contraception in Sweden: results of a nationwide survey. Eur J Contracept Reprod Health Care 2019;24:154–160. [DOI] [PubMed] [Google Scholar]
- Hernández-Ceballos W, Cordova-Gallardo J, Mendez-Sanchez N.. Gut microbiota in metabolic-associated fatty liver disease and in other chronic metabolic diseases. J Clin Transl Hepatol 2021;9:227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey RJ, Zhou X, Settles ML, Erb J, Malone K, Hansmann MA, Shew ML, Van Der Pol B, Dennis Fortenberry J, Forney LJ.. Vaginal microbiota of adolescent girls prior to the onset of menarche resemble those of reproductive-age women. mBio 2015;6:e00097-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmann B, Al-Ghalith GA, Shields-Cutler RR, Zhu Q, Gohl DM, Beckman KB, Knight R, Knights D.. Evaluating the information content of shallow shotgun metagenomics. MSystems 2018;3:e00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugerth LW, Pereira M, Zha Y, Seifert M, Kaldhusdal V, Boulund F, Krog MC, Bashir Z, Hamsten M, Fransson E. et al. Assessment of in vitro and in silico protocols for sequence-based characterization of the human vaginal microbiome. mSphere 2020;5:e00448-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RB, Zhu X, Moan E, Murff HJ, Ness RM, Seidner DL, Sun S, Yu C, Dai Q, Fodor AA. et al. Inter-niche and inter-individual variation in gut microbial community assessment using stool, rectal swab, and mucosal samples. Sci Rep 2018;8:4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Merchant M, Haque MM, Mande SS.. Crosstalk between female gonadal hormones and vaginal microbiota across various phases of women’s gynecological lifecycle. Front Microbiol 2020;11:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koedooder R, Singer M, Schoenmakers S, Savelkoul PHM, Morré SA, De Jonge JD, Poort L, Cuypers WJSS, Beckers NGM, Broekmans FJM. et al. The vaginal microbiome as a predictor for outcome of in vitro fertilization with or without intracytoplasmic sperm injection: a prospective study. Hum Reprod 2019;34:1042–1054. [DOI] [PubMed] [Google Scholar]
- Kostic AD, Xavier RJ, Gevers D.. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 2014;146:1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon SJ, Ravel J, Huston WM.. Cervicovaginal microbiota, women’s health, and reproductive outcomes. Fertil Steril 2018;110:327–336. [DOI] [PubMed] [Google Scholar]
- Lindh I, Skjeldestad FE, Gemzell-Danielsson K, Heikinheimo O, Hognert H, Milsom I, Lidegaard Ø.. Contraceptive use in the Nordic countries. Acta Obstet Gynecol Scand 2017;96:19–28. [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, Brady A, Creasy HH, McCracken C, Giglio MG. et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017;550:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Breitwieser FP, Thielen P, Salzberg SL.. Bracken: estimating species abundance in metagenomics data. PeerJ Computer Science 2017;3:e104. [Google Scholar]
- MacIntyre DA, Chandiramani M, Lee YS, Kindinger L, Smith A, Angelopoulos N, Lehne B, Arulkumaran S, Brown R, Teoh TG. et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep 2015;5:8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick H, Rahnavard A, McIver LJ, Ma S, Zhang Y, Nguyen LH, Tickle TL, Weingart G, Ren B, Schwager EH et al. Multivariable association discovery in population-scale meta-omics studies. PLOS Computat Biol 2021;17:e1009442. https://doi.org/10.1371/journal.pcbi.1009442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson GW, Long T, Salipante SJ, Rongitsch JA, Hoffman NG, Stephens K, Penewit K, Greene DN.. The vaginal microbiome of transgender men. Clin Chem 2019;65:199–207. [DOI] [PubMed] [Google Scholar]
- Meijnikman AS, Gerdes VE, Nieuwdorp M, Herrema H.. Evaluating causality of gut microbiota in obesity and diabetes in humans. Endocr Rev 2018;39:133–153. [DOI] [PubMed] [Google Scholar]
- Muhleisen AL, Herbst-Kralovetz MM.. Menopause and the vaginal microbiome. Maturitas 2016;91:42–50. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, Rusch DB, Mitreva M, Sodergren E, Chinwalla AT et al; Human Microbiome Jumpstart Reference Strains Consortium. A catalog of reference genomes from the human microbiome. Science 2010;328:994–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Products—Data Briefs—Number 327—December 2018. (n.d.). https://www.cdc.gov/nchs/products/databriefs/db327.htm (3 June 2021, date last accessed).
- Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO. et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA 2011;108:4680–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Brabin L, Diallo S, Gies S, Nelson A, Stewart C, Swinkels DW, Geurts-Moespot AJ, Kazienga A, Ouedraogo S. et al. Mucosal lactoferrin response to genital tract infections is associated with iron and nutritional biomarkers in young Burkinabé women. Eur J Clin Nutr 2019;73:1464–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Nikita L, Galuppi M, Lamont RF, Chaemsaithong P, Miranda J. et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2014;2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J.. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol 2012;13:R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha T, Vich Vila A, Garmaeva S, Jankipersadsing SA, Imhann F, Collij V, Bonder MJ, Jiang X, Gurry T, Alm EJ. et al. Analysis of 1135 gut metagenomes identifies sex-specific resistome profiles. Gut Microbes 2019;10:358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola-Leyva A, Andrés-León E, Molina NM, Terron-Camero LC, Plaza-Díaz J, Sáez-Lara MJ, Gonzalvo MC, Sánchez R, Ruíz S, Martínez L. et al. Mapping the entire functionally active endometrial microbiota. Hum Reprod 2021;36:1021–1031. [DOI] [PubMed] [Google Scholar]
- Song SD, Acharya KD, Zhu JE, Deveney CM, Walther-Antonio MRS, Tetel MJ, Chia N.. Daily vaginal microbiota fluctuations associated with natural hormonal cycle, contraceptives, diet, and exercise. MSphere 2020;5:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torky HA, El-Desouky ES, Hussein A, Abo-Louz A, Mohammed A, El-Hamid AA, Galal S, Tawfick MM, Marie H.. Relationship between ano-vaginal distance and bacterial vaginosis (cross-sectional study). Reprod Sci 2021;28:2310–2313. [DOI] [PubMed] [Google Scholar]
- Van Houdt R, Ma B, Bruisten SM, Speksnijder AGCL, Ravel J, De Vries HJC.. Lactobacillus iners-dominated vaginal microbiota is associated with increased susceptibility to Chlamydia trachomatis infection in Dutch women: a case-control study. Sex Transm Infect 2018;94:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodstrcil LA, Hocking JS, Law M, Walker S, Tabrizi SN, Fairley CK, Bradshaw CS.. Hormonal contraception is associated with a reduced risk of bacterial vaginosis: a systematic review and meta-analysis. PLoS One 2013;8:e73055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G, Hart RA, Charlesworth RPG, Sharpley CF.. Gut microbiome and depression: what we know and what we need to know. Rev Neurosci 2018;29:629–643. [DOI] [PubMed] [Google Scholar]
- Wood DE, Lu J, Langmead B.. Improved metagenomic analysis with Kraken 2. Genome Biol 2019;20:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young VB. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ 2017;356:j831. [DOI] [PubMed] [Google Scholar]
- Zhou X, Brotman RM, Gajer P, Abdo Z, Schüette U, Ma S, Ravel J, Forney LJ.. Recent advances in understanding the microbiology of the female reproductive tract and the causes of premature birth. Infect Dis Obstet Gynecol 2010;2010:737425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Gao H, Mihindukulasuriya KA, Rosa PSL, Wylie KM, Vishnivetskaya T, Podar M, Warner B, Tarr PI, Nelson DE. et al. Biogeography of the ecosystems of the healthy human body. Genome Biol 2013;14:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data analysed in this study were submitted to the European Nucleotide Archive under project number PRJEB37731, samples ERS4421369–ERS4422941. Other patient-level data will be shared on reasonable request to the corresponding author.