Abstract
Pseudomonas syringae pv. tomato DC3000 (PstDC3000) is an important plant pathogen that infects tomatoes and Arabidopsis. Thiamine and its derivative thiamine pyrophosphate (TPP) are cofactors that play an important role in the growth and survival of many bacterial microorganisms. However, the role of thiamine-related genes has not been determined in PstDC3000. Hence, to investigate the role of TPP in growth, resistance to stresses, and virulence of PstDC3000, double and quadruple mutants of thiamine biosynthesis-related genes (thiD/E, thiS/G, and thiD/E/S/G deletion mutants) as well as a single mutant of a lipoprotein-related gene (apbE) were constructed. Our results showed that growth of the thiD/E, thiS/G, and thiD/E/S/G mutants in the mannitol-glutamate (MG) medium was significantly lower than that of the wild type (WT) and their growth could be restored to the WT level with the addition of exogenous thiamine, whereas mutation of the apbE gene did not affect its growth in vitro. While tolerance to acid, osmotic, and oxidative stresses for the double mutants was similar to the WT, tolerance to stresses for the apbE mutant was reduced as compared to the WT. In addition, all four mutants exhibited reduced virulence and growth in tomatoes. However, when the double and quadruple mutants were inoculated with exogenous thiamine, the virulence and growth rate of these mutants were restored to the WT level. These results indicated that the thiD/E, thiS/G, and thiD/E/S/G mutants exhibiting growth deficiency in planta are probably due to a lack of thiamine biosynthesis, thus reducing colonization in tomatoes. On the other hand, it is possible that the apbE mutant exhibited reduced stress tolerances, thus resulting in reduced colonization. Overall, our findings suggest that the thiamine biosynthetic (TBS) pathway plays an important role in the colonization and infection of PstDC3000. Therefore, the thiamine biosynthetic pathway could be used as the target to develop new control measures for a bacterial spot in tomatoes.
Keywords: Pseudomonas syringae pv. tomato DC3000, thiamine, cell growth, stress tolerance, virulence
Introduction
Pseudomonas syringae pv. tomato DC3000 (PstDC3000) is a model pathogen for studies in molecular mechanisms of bacterial pathogenesis and in plant–microbe interactions (Xin and He, 2013). PstDC3000 causes bacterial speck disease on tomato and Arabidopsis thaliana by injecting effectors into plant cells through the type III secretion system (T3SS) and producing the phytotoxin coronatine (COR) (Zhao et al., 2003, 2006; Xie et al., 2019). Besides T3SS and phytotoxin coronatine (COR), extracellular protease, siderophore, and alginate all contribute to the virulence of PstDC3000 (Brooks et al., 2004; Swingle et al., 2008; Vargas et al., 2013; Ishiga et al., 2018). Previous studies showed that guanosine tetra/pentaphosphate [(p)ppGpp] and RsmA affect virulence via regulation of T3SS, motility, siderophore, syringafactin, and alginate production in PstDC3000 (Chatnaparat et al., 2015; Ge et al., 2019; Liu et al., 2020, 2021). In addition, mutations of (p)ppGpp-biosynthetic genes and rsmA significantly affected the expression of thiamine biosynthesis-related genes (Liu et al., 2020, 2021).
Vitamins of the B group are key precursors for the biosynthesis of essential enzyme cofactors that drive many metabolic processes in all the forms of life (Rodionov et al., 2017). The active form of vitamin B1 (thiamine), thiamine pyrophosphate (TPP), is an important cofactor for several anabolic and catabolic reactions in central metabolism (Begley and Ealick, 2010). Thiamine pyrophosphate (TPP)-dependent enzymes, including transketolase, pyruvate dehydrogenase, 2-oxoglutarate dehydrogenase, alpha-ketoglutarate dehydrogenase, and 1-deoxy-D-xylulose-5-phosphate synthase, catalyze essential cellular processes in bacteria, from central metabolism to biosynthesis of amino acids, cofactors, and lipids (Rapala-Kozik et al., 2012; Bunik et al., 2013; Osiezagha et al., 2013; Hwang et al., 2017; Chen et al., 2019). The thiamine biosynthetic (TBS) pathway is well-understood in bacteria, and several enzymes of the pathway have been identified (Rodionov et al., 2002; Jurgenson et al., 2009). Thiamine monophosphate (TMP) is formed by the coupling of two moieties, i.e., 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate (HMP-PP) and 5-(2-hydroxyethyl)-4-methylthiazole phosphate (HET-P) by the thiamine-phosphate pyrophosphorylase (ThiE) (Tylicki et al., 2018). The aminoimidazole ribotide (AIR) derived from the purine pathway is catalyzed by hydroxymethyl pyrimidine synthase (ThiC) and lipoprotein (ApbE) to form hydroxymethyl pyrimidine phosphate (HMP-P), which is phosphorylated to form HMP-PP by the bifunctional HMP kinase/HMP-P kinase (ThiD) (Beck and Downs, 1998; Palmer and Downs, 2013; Rodionov et al., 2017). On the other hand, the thiazole moiety of thiamine HET-P is derived from tyrosine, cysteine, and 1-deoxy-D-xylulose phosphate and catalyzed by the products of the thiF, thiS, thiG, thiH, and thiI genes (Jurgenson et al., 2009; Rodionov et al., 2017). Lastly, TMP is phosphorylated by thiamine monophosphate kinase (ThiL) to form thiamine pyrophosphate (TPP) (Figure 1) (Begley et al., 1999; Melnick et al., 2004; Hwang et al., 2017). In addition to the TPP biosynthetic pathway, bacteria, such as Escherichia coli (E. coli), are able to salvage exogenous sources of thiamine to generate TPP by first converting thiamine to TMP thiamine kinase (Thik) and then subsequently generating TPP by adding an additional phosphate to TMP via ThiL (Rodionov et al., 2002; Melnick et al., 2004). Therefore, TPP is indispensable for bacterial survival, and enzymes involved in bacterial TPP biosynthesis might be important targets for disease control and prevention.
Figure 1.
Diagram showing a putative thiamine biosynthesis pathway. Black arrows indicate thiamine biosynthetic (TBS) pathway. HMP-P, hydroxymethyl pyrimidine phosphate; HMP-PP, 4-amino-5-hydroxymethyl-2-methylpyrimidine diphosphate; HET-P, 5-(2-hydroxyethyl)-4-methylthiazole phosphate; TMP, thiamine monophosphate; AIR, aminoimidazole ribotide; TPP, thiamine pyrophosphate.
It has been previously reported that several enzymes of the TBS pathway are important for host infection. The protein encoded by thiG acts as an essential enzyme in the thiamine biosynthetic pathway, which plays a crucial role in tolerance to oxidative and heat stresses in Fusarium oxysporum (F. oxysporum) (Ruiz-Roldán et al., 2008) and contributes for full virulence in Verticillium dahliae and Xanthomonas oryzae (X. oryzae) pv. oryzae (Hoppenau et al., 2014; Yu et al., 2015). In Listeria monocytogenes (L. monocytogenes), mutation of the thiD gene significantly reduced bacterial replication in epithelial cells and inhibited infection (Schauer et al., 2009). In Xanthomonas oryzae pv. oryzicola, the thiE gene was reported to influence bacterial virulence (Zou et al., 2011). In addition, the apbE gene encoding a lipoprotein is involved in the conversion of AIR to the HMP moiety of TPP in Salmonella typhimurium (S. typhimurium) (Beck and Downs, 1998).
In this study, the open reading frames (ORFs) of ThiD (thiD, pspto_4798), ThiE (thiE, pspto_4799), ThiS (thiS, pspto_0433), ThiG (thiG, pspto_0434), and ApbE (apbE, pspto_2105) in the genome of PstDC3000 (gene bank accession #: AE016853.1) were identified by performing BLAST with homologous enzymes from other organisms. In PstDC3000, both the thiDE and thiSG genes are located adjacent to each other in the genome. However, the role of thiamine biosynthesis-related genes on the virulence of pathogens remains unknown. Thus, in order to determine whether thiamine biosynthesis-related genes and apbE gene affect the virulence of PstDC3000, we constructed the thiD/E, thiS/G, thiD/E/S/G, and apbE mutants using splice overlap extension mutagenesis (Supplementary Figure S1) and their corresponding complementation strains in PstDC3000 and assessed their growth in vitro and in vivo, stress tolerance, and virulence on tomato. Our findings would enhance our understanding of thiamine in the virulence of PstDC3000.
Materials and Methods
Bacterial Strains and Culture Conditions
Bacterial strains and vectors used in this study are given in Supplementary Table S1. The WT PstDC3000 and its mutants were routinely cultured on King's medium B (KB) or mannitol-glutamate (MG) medium (Park et al., 2010) with appropriate antibiotics at 28°C with shaking at 250 rpm. All the E. coli strains were grown in Luria-Bertani (LB) broth or agar medium with appropriate antibiotics at 37°C with shaking at 250 rpm. Bacterial growth was monitored by measuring the absorbance of cell suspensions at 600 nm. Antibiotics were supplied at the following final concentrations: 100 μg ml−1 rifampicin, 50 μg ml−1 kanamycin, 100 μg/ml ampicillin, 15 μg/ml tetracycline, and 100 μg/ml spectinomycin.
Construction of Deletion Mutants
Deletion mutations of thiD/E, thiS/G, thiD/E/S/G, and apbE were generated using splice overlap extension mutagenesis (Supplementary Figure S1), as described previously (Ge et al., 2019). Briefly, approximately, 1 kb upstream and downstream fragments of thiD/E, thiS/G, and apbE in PstDC3000 were amplified using primers unique to these regions (Supplementary Table S2). At the same time, to facilitate the screening of mutants, these primers also contained an extension sequence complementary to a kanamycin resistance cassette flanked with flippase (FLP) recombination target (FRT) sites from pKD13. The two PCR products contain ends overlapping with sequences at both ends of the kanamycin resistance cassette (Datsenko and Wanner, 2000). The FRT-Km-FRT forward and reverse primers were used amplified FRT-flanked kanamycin cassette. The two fragments were then joined with the kanamycin resistance cassette using overlap extension PCR amplified into a single fragment. The final fragment was cloned into the pTok2 suicide vector digested with SmaI, resulting in pTok2::ΔthiD/E, pTok2::ΔthiS/G, and pTok2::ΔapbE, respectively. Each of these plasmids was electroporated into PstDC3000. Subsequently, transformants were plated on KB plates containing rifampicin and kanamycin. Those kanamycin-resistant, tetracycline-sensitive mutants were selected. Gene disruption was confirmed using PCR, with primers specific to the sequences flanking each of the genes. To generate markerless mutants, plasmid pFLP2-omega expressing FLP recombinase was electroporated into the mutant strains. Transformants were plated on KB plates containing spectinomycin to allow FLP recombinase-mediated recombination between the FRT sites, resulting in the loss of the kanamycin resistance cassette. The pFLP2-omega plasmid was cured of the mutants by plating on KB plates containing 10% sucrose. Markerless deletion strains were confirmed by PCR. In order to generate a thiD/E/S/G quadruple deletion mutant, plasmid pTok2::ΔthiD/E was transferred to the thiS/G markerless mutant strain via conjugation.
Complementation of Mutants
For complementation of the thiD/E, thiS/G, and apbE mutants, about 2 kb fragment containing the native promoter and the thiD/E, thiS/G, and apbE genes was amplified by PCR and cloned into the pUCP18 vector to yield plasmids pThiD/E, pThiS/G, and pApbE, respectively. These plasmids were sequenced at the University of Illinois at Urbana-Champaign core sequencing facility. The final plasmids were introduced into the corresponding deletion mutants and PstDC3000 by electroporation. Transformants were selected on KB agar plates amended with 100 μg/ml ampicillin. Finally, the complementation of mutants was confirmed by PCR and used for further study. All the primer sequences used in this study are given in Supplementary Table S2 (see Supplementary Materials).
Growth Curve Assay
Pseudomonas syringae pv. tomato DC3000 WT strain, mutants, and corresponding complementation strains were cultured in KB broth at 28°C for 18 h with shaking at 250 rpm. All the strains were harvested and resuspended to OD600 = 0.01 in fresh KB. Bacterial strains were grown at 28°C and aliquots of the culture were taken after 2, 4, 6, 8, 10, and 24 h. Furthermore, all the strains were harvested and resuspended to OD600 = 0.01 in MG medium with 100 μg/ml thiamine (Fisher Scientific, Waltham, MA, USA) or without thiamine at 28°C and aliquots of the culture were taken after 24, 48, and 72 h. These experiments were performed in triplicate and repeated three times.
Stress Tolerance (Acid, Oxidative, and Osmotic) Assays
Growth assays were performed using a previously described procedure to detect stress response (Ge et al., 2018). Briefly, overnight bacterial cells were harvested by centrifugation and washed twice using phosphate-buffered saline (PBS). After the final wash, the pellet was resuspended in PBS and adjusted to a concentration of OD600 = 0.1. Bacterial cells of overnight cultures in KB were washed and resuspended to OD600 = 0.01 in KB liquid medium with pH 5.5, 0.5 M NaCl, and 0.5 mM hydrogen peroxide (H2O2), respectively. Cells were incubated at 28°C for 24 h and bacterial growth was monitored by measuring OD600. Meanwhile, bacterial cells cultured in a normal KB medium were used for normalization. These experiments were performed in triplicate and repeated three times.
Spot Dilution Plate Assay
Overnight bacterial cultures were harvested by centrifugation and cells were washed two times using PBS. Bacterial cells were resuspended in PBS and were adjusted to an initial concentration of OD600 = 0.1 for assays on KB plates. Ten-fold serial dilution of the bacterial suspension was made using PBS. For each dilution, 5 μl was spotted on the KB plates with or without acid, oxidative, and osmotic stresses, and was incubated at 28°C for 48 h. Growth on normal KB plates was used as a control to show similar growth of the WT and the mutants, whereas the growth of mutants was compared with that of the WT on KB plates with acid, oxidative, and osmotic stresses. If the growth of the mutant was observed at least one dilution lower or higher than that of the WT, this indicated that the mutant was more resistant or sensitive to acid, oxidative, and osmotic stresses and vice versa. The experiments were performed in triplicate and were repeated at least three times.
Virulence Assay and Bacterial Growth in Tomato
Virulence of PstDC3000 and its mutants on tomatoes was evaluated using an injection of tomato leaves, as described previously (Ge et al., 2019). For injection of tomato leaves, overnight cultures of PstDC3000, mutants, and complementation strains were harvested by centrifugation and adjusted to a concentration of about 5 × 104 colony-forming unit (CFU)/ml [diluted from original suspension at OD600 = 0.1 (~108 CFU per milliliter)] with PBS. Each strain was inoculated into 10 tomato leaves (Tomato, Solanum lycopersicum “Big Daddy Hybrid” plants were grown in a greenhouse) using a needleless syringe, which were then observed for disease symptoms at 7 days postinoculation (dpi). For bacterial growth on tomato leaves, bacteria were recovered from plants by taking three samples from three leaves at the site of infiltration using a disk punch (three disks per strain) at 0, 1, 3, and 5 dpi. Leaf disks were homogenized by mechanical disruption using pestles in PBS. Serial 10-time dilutions of the homogenates were plated on KB plates containing rifampicin antibiotic and the number of CFUs per disk (cm2) was calculated. The experiment was repeated three times.
Statistical Analyses
SPSS statistics version 26 was used to perform a one-way ANOVA and least significant difference (LSD) test to determine the significant difference in the growth curve, stress tolerance (acid, oxidative, and osmotic) assays, and bacterial colonization. Differences with P < 0.05 were considered statistically significant.
Results
Effect of Thiamine on Bacterial Growth in vitro
Thiamine (vitamin B1) is an essential cofactor for carbohydrate and branched-chain amino acid metabolic enzymes in all the organisms (Settembre et al., 2003; Du et al., 2011). To investigate whether thiD/E, thiS/G, and apbE had an effect on in vitro growth, we examined the growth of the thiD/E, thiS/G, thiD/E/S/G, and apbE deletion mutants and their complementation strains in the rich medium (KB) and limited nutrition medium (MG). In KB, the growth of the thiD/E, thiS/G, and thiD/E/S/G deletion mutants and their complementation strains were similar to that of PstDC3000 (Figures 2A,B). However, growth of the thiD/E, thiS/G, and thiD/E/S/G mutants was significantly (P < 0.05) reduced as compared with PstDC3000 in MG medium without exogenous thiamine (Figure 2C). All the complementation strains, except the thiD/E/S/G mutant complementation (P < 0.05) with either the thiD/E or thiS/G genes, restored growth levels similar to these of the PstDC3000 in MG medium without exogenous thiamine (Figure 2D). When supplemented with exogenous thiamine in MG medium, wild type, thiD/E, thiS/G, and thiD/E/S/G mutants and its complementation strains exhibited similar growth as PstDC3000 (Figures 2E,F), suggesting that growth arrest of the mutant strains was caused by thiamine starvation. In contrast, the growth of the apbE deletion mutant and its complementation strain was similar to PstDC3000 in both the KB and MG media (Figure 2). These results suggest that the thiD/E and thiS/G genes are required for the growth of PstDC3000 in an MG medium.
Figure 2.
Thiamine-related genes are required for the growth of Pseudomonas syringae pv. tomato DC3000 (PstDC3000) in minimum medium. (A) Growth of PstDC3000 wild type (WT), the thiD/E, thiS/G, thiD/E/S/G, and apbE mutant strains in King's B medium (KB). (B) Growth of complementation strains in KB. (C) Growth of all the mutant strains in mannitol-glutamate (MG) medium. (D) Growth of complementation strains in MG medium. (E) Growth of all the mutant strains in MG medium with exogenous 100 μg/ml thiamine. (F) Growth of complementation strains in MG medium with exogenous 100 μg/ml thiamine. The solid line represents mutant strains; the dashed line represents complementation strains; and the vertical bars represent SDs. “+thi:” added exogenous 100 μg/ml thiamine. Experiments were performed in triplicate and repeated three times. Statistical tests were performed at 24, 48, and 72 h. Statistically significant differences were determined by one-way ANOVA and P-values were < 0.05 [an asterisk (*) indicates P < 0.05, mutant and complementation strains vs. the WT PstDC3000].
Role of Thiamine in Acid, Osmotic, and Oxidative Tolerances
To test whether thiamine plays an important role in stress tolerance, including acid, osmotic, and oxidative responses, we performed growth assays in KB liquid medium. Growth of the thiD/E and thiS/G mutants was essentially unaffected after being incubated in KB liquid medium at pH 5.5, with 0.5 M NaCl or 0.5 mM H2O2, and growth of the thiD/E/S/G mutant showed slightly increase, whereas growth of the apbE mutant significantly (P < 0.05) reduced as compared with that of PstDC3000 (Figures 3A–C). The growth of all the complementation strains was similar with PstDC3000 in KB liquid medium at pH 5.5, with 0.5 M NaCl or 0.5 mM H2O2 (Figures 3D–F). Spot dilution assays on KB plates further illustrated the role of apbE in acid, osmotic, and oxidative tolerances. On KB plates at pH = 7, with no NaCl or H2O2, respectively, growth of the apbE mutant was equivalent to those of the WT strain (Figure 4). When plated on KB at pH = 5.5, with 0.5 M NaCl or 0.5 mM H2O2, the growth of the apbE mutant was reduced at least 10-fold as compared with the WT (Figure 4). These results indicate that the apbE gene plays a role in stress responses in PstDC3000.
Figure 3.
Thiamine-related genes affected tolerance to acid, oxidative, and osmotic stresses of PstDC3000. (A) PstDC3000 wild type, the thiD/E, thiS/G, thiD/E/S/G, and apbE mutant cells of overnight cultures in KB were washed and resuspended to OD600 = 0.01 in KB liquid medium with pH 5.5. The phenotype was observed after incubation at 28°C for 24 h and bacterial growth was monitored by measuring OD600. (B) All the mutant cells were grown in KB liquid medium with 0.5 M NaCl and incubated at 28°C for 24 h. (C) All the mutant cells were grown in KB liquid medium with 0.5 mM hydrogen peroxide (H2O2) and incubated at 28°C for 24 h. (D) All the complementation strains were grown in KB liquid medium with pH 5.5. (E) All the complementation strains were grown in KB liquid medium with 0.5 M NaCl. (F) All the complementation strains were grown in KB liquid medium with 0.5 mM H2O2. The solid line represents mutant strains; the dashed line represents complementation strains; and the vertical bars represent SDs. Experiments were performed in triplicate and repeated three times. Statistical tests were performed and statistically significant differences were determined by one-way ANOVA and P-values were < 0.05 [an asterisk (*) indicates P < 0.05, mutant and complementation strains vs. the WT PstDC3000].
Figure 4.
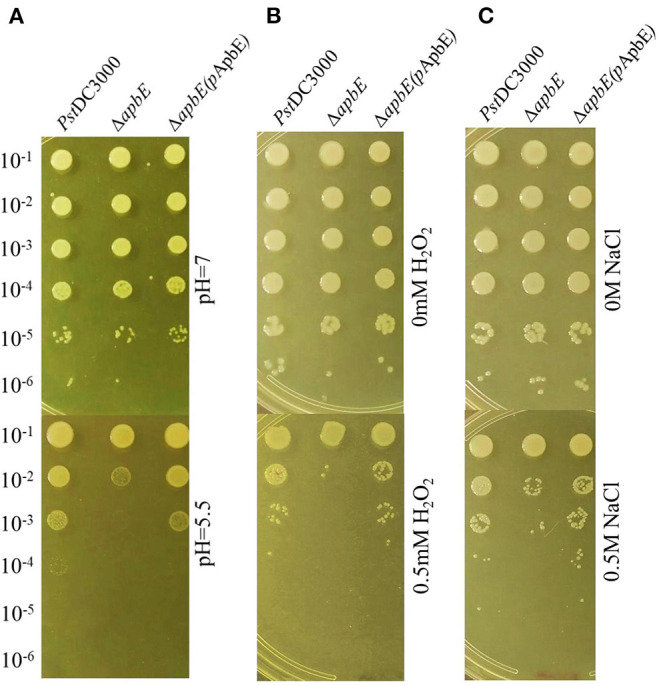
ApbE plays an important role in stress responses in PstDC3000. (A) Acid stress. (B) Oxidative stress. (C) Osmotic stress. Serial 10-fold dilutions were made from optical density at 600 nm (OD600) = 0.1 and 5 μl for each dilution was added to plates containing acid, oxidative, and osmotic stresses, respectively. Photographs were taken 48 h postincubation. Experiments were performed in triplicate and repeated three times.
Role of Thiamine in PstDC3000 Virulence
To determine the effect of thiamine in virulence of PstDC3000 on tomatoes, PstDC3000 wild type, the thiD/E, thiS/G, thiD/E/S/G, and apbE mutants were infiltrated into tomato leaves at a concentration of 5 × 104 CFU/ml. The PstDC3000 WT strain exhibited typical necrotic spots with chlorosis on leaves of tomato plants at 7 dpi, whereas the thiD/E, thiS/G, thiD/E/S/G, and apbE mutant strains all exhibited reduced virulence (Figure 5A). All the complementation strains, except for the thiD/E/S/G mutant complementation with either the thiD/E or thiS/G genes, restored virulence similar to these of the PstDC3000 (Figure 5A). On the other hand, to determine whether thiamine could restore the ability of the mutants to cause disease, we inoculated plants with the mutants and complementation strains by supplementing exogenous thiamine at 100 μg/ml. As shown in Figure 5B, these mutants and complementation strains, except the apbE mutant, all exhibited similar virulence as that of PstDC3000 (Figure 5B). These results indicate that thiamine is required for full virulence of PstDC3000.
Figure 5.

Thiamine-related genes contribute to PstDC3000 virulence. (A) Tomato leaves inoculated by infiltration with PstDC3000 wild type and the thiD/E, thiS/G, thiD/E/S/G, apbE mutants, and complementation strains. Phosphate-buffered saline (PBS) was used as a negative control. (B) PstDC3000 wild type, the thiD/E, thiS/G, thiD/E/S/G mutants, and their corresponding complementation strains with exogenous 100 μg/ml thiamine inoculated into tomato leaves. Pictures were taken at 7 days postinoculation (dpi). “+thi:” added exogenous 100 μg/ml thiamine. Each strain was inoculated onto 10 tomato leaves with a needleless syringe in each experiment. Experiments were repeated three times.
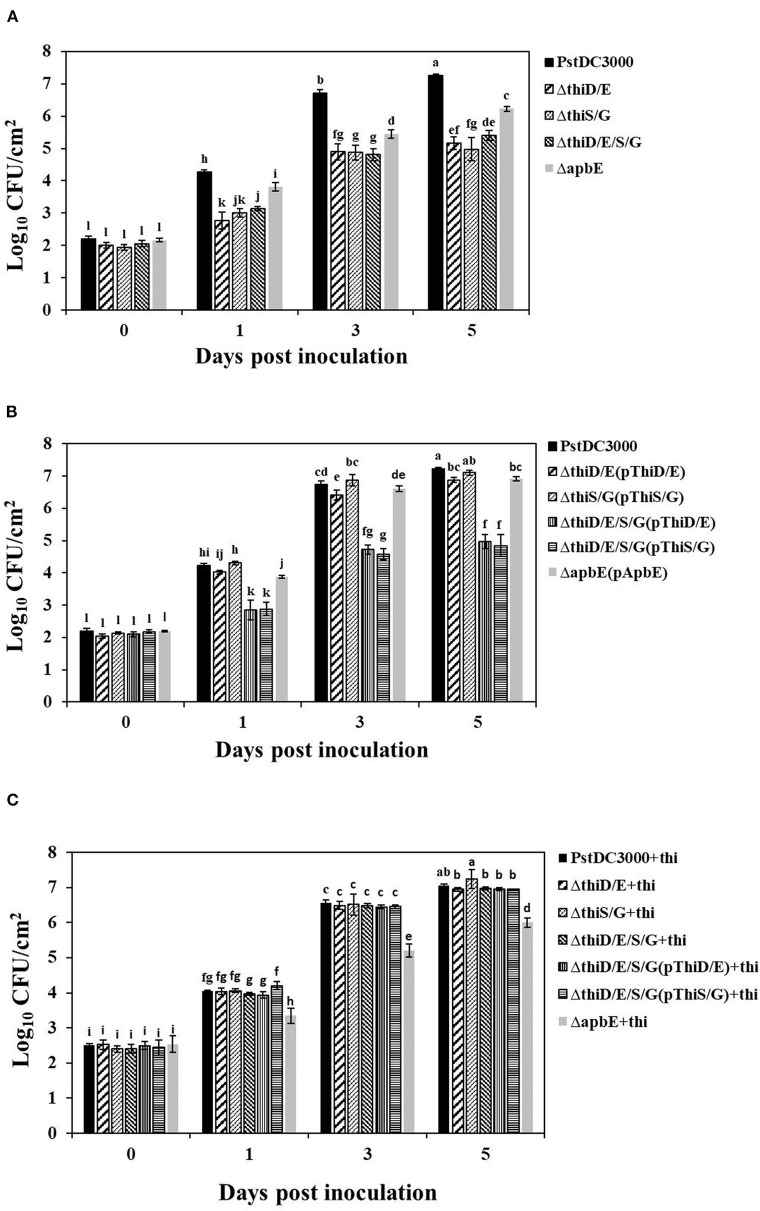
Deletion of the thiD/E, thiS/G, and apbE Genes in PstDC3000 Reduced Bacterial Growth on Tomato Leaves
To determine whether reduced virulence by the mutants was correlated with bacterial growth on tomato leaves, bacterial growth was monitored in planta at 0, 1, 3, and 5 dpi. Bacterial growth of the thiD/E, thiS/G, thiD/E/S/G, and apbE mutants was about 100-fold lower than (P < 0.05) those of the PstDC3000 at 3 and 5 dpi (Figure 6A), whereas growth of the apbE mutant was about 10-fold lower than (P < 0.05) those of the PstDC3000 at 3 and 5 dpi (Figure 6A). The reduced bacterial growth of the mutants could be completely rescued in the complementation strains, except for the thiD/E/S/G mutant complementation with either the thiD/E or thiS/G genes (P < 0.05) (Figure 6B). Similarly, when the double mutants and the thiD/E/S/G mutant complementation, with either the thiD/E or thiS/G genes, were infiltrated with supplemented exogenous thiamine, the growth of these strains was similar to PstDC3000, whereas the growth of the apbE mutant was lower than (P < 0.05) those of the PstDC3000 (Figure 6C). These results suggested that thiamine starvation may lead to reduced growth of the thiD/E, thiS/G, and thiD/E/S/G mutants in plants, whereas reduced ability for stress tolerance in the apbE mutant might be the cause of its reduced growth in tomatoes.
Figure 6.
Thiamine is required for bacterial growth of PstDC3000 in tomato leaves. (A) PstDC3000 wild type and the thiD/E, thiS/G, thiD/E/S/G, and apbE mutant strains. (B) PstDC3000 wild type and all the complementation strains. (C) PstDC3000 wild type and the thiD/E, thiS/G, thiD/E/S/G, and apbE mutants with exogenous 100 μg/ml thiamine. Bacterial growth was monitored at 0, 1, 3, and 5 dpi. “+thi:” added exogenous 100 μg/ml thiamine. Each strain was inoculated onto 10 tomato leaves with a needleless syringe in each experiment. Experiments were repeated three times. Data represent the means of three replicates ± SDs (error bars). Different lowercase letters indicate a significant difference between the strains. Statistically significant differences were determined by one-way ANOVA and P-values were < 0.05.
Discussion
The TBS pathway is essential for the proper function of animals, plants, and microorganisms, as it plays a crucial role in cellular progress, including carbohydrate, lipid, amino acid, and nucleic acid metabolisms (Hwang et al., 2017; Chen et al., 2019). The TBS pathway in E. coli (Rodionov et al., 2002; Melnick et al., 2004) and Salmonella typhimurium (Webb et al., 1998) has been extensively studied through biochemical analysis and comparative genomes. In previous studies, 12 genes have been identified for thiamine biosynthesis in prokaryotes. These include thiF, thiS, thiG, thiH, thiI, and dxs, which are required for thiazole biosynthesis, thiC, which is involved in pyrimidine biosynthesis, thiE, which is required for linking of thiazole and pyrimidine, and thiD, thiM, thiL, and thiK, which are kinase genes (Jurgenson et al., 2009; Hwang et al., 2017). However, there are no reports about the role of thiamine biosynthesis during the interaction between bacteria and host plants in PstDC3000. In present study, our results showed that growth of the thiD/E, thiS/G, and thiD/E/S/G mutants in MG medium was significantly decreased in MG medium, whereas mutation of the apbE gene did not affect its growth in vitro, while tolerance to acid, osmotic, and oxidative stresses for the apbE mutant was reduced as compared to the WT. In addition, all the mutants exhibited reduced virulence and growth in tomatoes. Therefore, our results suggest that the thiamine biosynthetic (TBS) pathway plays an important role in the colonization and infection of PstDC3000.
Thiamine (vitamin B1) is an essential cofactor for carbohydrate metabolism (Hwang et al., 2017; Chen et al., 2019) and biosynthesis and degradation of the branched-chain amino acids, including isoleucine, leucine, and valine (Schauer et al., 2009). Therefore, the availability of thiamine has a key effect on the growth of many organisms, e.g., X. oryzae pv. oryzae (Yu et al., 2015), Listeria monocytogenes (Madeo et al., 2012), and F. oxysporum (Ruiz-Roldán et al., 2008). Consistent with other organisms, thiD/E, thiS/G, and thiD/E/S/G play an important role in the growth of a low nutrition environment, suggesting that thiamine biosynthesis is required for maintaining the growth of PstDC3000. On the other hand, the growth of the apbE mutant was not affected, which is inconsistent with an earlier report on S. typhimurium (Beck and Downs, 1998). This result indicates that the abpE gene may not be directly involved in the biosynthesis of thiamine in PstDC3000.
The stress response of microorganisms is indispensable since the living environment of microorganisms is constantly changing (Vorob'eva, 2003; Jordan et al., 2008; Swiecilo and Zych-Wezyk, 2013). Therefore, microorganisms must continuously sense and respond to environmental stimuli, e.g., osmotic, oxidative, and acid stresses (Moat et al., 2003; Bremer and Krämer, 2019). Many studies have shown that stress resistance mechanisms play an important role in virulence. As an example, OxyR of PstDC3000 plays an important role in virulence by regulating the expression of genes related to oxidative stress (Ishiga and Ichinose, 2016). The DnaK/DnaJ of S. typhimurium is essential for stress resistance and invasion of epithelial cells (Takaya et al., 2004). Moreover, the thiT deletion mutant is necessary for acid tolerance in L. monocytogenes and thiamine starved cells are more acid-sensitive than the wild type (Madeo et al., 2012). In contrast, the thiD/E and thiS/G double mutants exhibited similar stress tolerance as that of PstDC3000, indicating that thiamine is not a key factor in stress tolerance in PstDC3000. However, the thiD/E/S/G quadruple mutant showed increased acid, osmotic, and oxidative tolerances, suggesting either a synergistic effect between them or some unknown functions of the intermediates in the pathways they are involved in. On the other hand, the apbE mutant exhibited significantly reduced acid, osmotic, and oxidative tolerances as compared to PstDC3000, indicating that ApbE might play an important role in protecting the bacteria from stress responses.
It has been reported that thiamine-related genes play a critical role in virulence. The thiD mutation of L. monocytogenes exhibited significantly reduced bacterial replication in epithelial cells and infection (Schauer et al., 2009). In X. oryzae pv. oryzicola, the thiE gene was reported to influence bacterial virulence (Zou et al., 2011). The protein encoded by thiG acts as an essential enzyme in the thiamine biosynthetic pathway, which plays a crucial role in the virulence of Verticillium dahliae (Hoppenau et al., 2014). In X. oryzae pv. oryzae, the thiG mutant showed reduced virulence, while bacterial growth in planta was similar to the WT (Yu et al., 2015). In this study, the thiD/E, thiS/G, and thiD/E/S/G mutants exhibited significantly reduced virulence and bacterial growth in planta, which could be restored by supplementing with thiamine in an extracellular environment. These results demonstrated that lack of thiamine biosynthesis leads to growth deficiency, thus disease symptoms in planta. In contrast, the apbE mutant was normal growth in vitro, but still exhibited reduced virulence and bacterial growth in planta, suggesting that the loss of virulence of the apbE mutant may be due to reduced stress tolerance in tomatoes.
In summary, we provide evidence that the TBS pathway and ApbE play an important role in growth, colonization, and disease development in PstDC3000. These results provide new insight into the role of the thiamine-related genes in PstDC3000 virulence. In the future, we will investigate whether stress tolerance-related genes are directly or indirectly regulated by ApbE. Taken together, the results of this study demonstrate that thiamine plays an important role in the infection and survival of PstDC3000 and provides a new target for the development of new drugs to prevent and control tomato bacterial spots.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
Author Contributions
JL, YZ, and HW designed the study. JL, XZ, and SD performed the study and analyzed the data. JL wrote the manuscript. YZ and HW revised the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This study was financially supported by the National Key Research and Development Program of China (2019YFD1002000), the Key Technology Research and Demonstration Project of the Hubei Agricultural Science and Technology Innovation Center (2020-620-000-002-07) and Postdoctoral Innovative Research Positions in Hubei Province.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.903258/full#supplementary-material
References
- Beck B. J., Downs D. M. (1998). The apbE gene encodes a lipoprotein involved in thiamine synthesis in Salmonella typhimurium. J. Bacteriol. 180, 885–891. 10.1128/JB.180.4.885-891.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley T. P., Downs D. M., Ealick S. E., McLafferty F. W., Van Loon A. P., Taylor S., et al. (1999). Thiamin biosynthesis in prokaryotes. Arch. Microbiol. 171, 293–300. 10.1007/s002030050713 [DOI] [PubMed] [Google Scholar]
- Begley T. P., Ealick S. E. (2010). “Thiamin biosynthesis,” in Comprehensive Natural Products II: Chemistry and Biology, Vol. 7, eds L. N. Mander and H. Liu (Kidlington: Elsevier Ltd.), 547–559. [Google Scholar]
- Bremer E., Krämer R. (2019). Responses of microorganisms to osmotic stress. Annu. Rev. Microbiol. 73, 313–334. 10.1146/annurev-micro-020518-115504 [DOI] [PubMed] [Google Scholar]
- Brooks D. M., Hernández-Guzmán G., Kloek A. P., Alarcón-Chaidez F., Sreedharan A., Rangaswamy V., et al. (2004). Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato strain DC3000. Mol. Plant Microbe. Interact. 17, 162–174. 10.1094/MPMI.2004.17.2.162 [DOI] [PubMed] [Google Scholar]
- Bunik V. I., Tylicki A., Lukashev N. V. (2013). Thiamin diphosphate-dependent enzymes: from enzymology to metabolic regulation, drug design and disease models. FEBS J. 280, 6412–6442. 10.1111/febs.12512 [DOI] [PubMed] [Google Scholar]
- Chatnaparat T., Li Z., Korban S. S., Zhao Y. F. (2015). The stringent response mediated by (p) ppGpp is required for virulence of Pseudomonas syringae pv. tomato and its survival on tomato. Mol. Plant Microbe. Interact. 28, 776–789. 10.1094/MPMI-11-14-0378-R [DOI] [PubMed] [Google Scholar]
- Chen Y. Y., Wang L. L., Shang F., Liu W., Lan J., Chen J. L., et al. (2019). Structural insight of the 5-(Hydroxyethyl)-methylthiazole kinase ThiM involving vitamin B1 biosynthetic pathway from the Klebsiella pneumoniae. Biochem. Biophys. Res. Commun. 518, 513–518. 10.1016/j.bbrc.2019.08.086 [DOI] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q., Wang H., Xie J. (2011). Thiamin (vitamin B1) biosynthesis and regulation: a rich source of antimicrobial drug targets? Int. J. Biol. Sci. 7, 41–52. 10.7150/ijbs.7.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y. X., Lee J. H., Hu B. S., Zhao Y. F. (2018). Loss-of-function mutations in the Dpp and Opp permeases render Erwinia amylovora resistant to kasugamycin and blasticidin S. Mol. Plant Microbe Interact. 31, 823–832. 10.1094/mpmi-01-18-0007-r [DOI] [PubMed] [Google Scholar]
- Ge Y. X., Lee J. H., Liu J., Yang H. W., Tian Y. L., Hu B. S., et al. (2019). Homologues of the RNA binding protein RsmA in Pseudomonas syringae pv. tomato DC3000 exhibit distinct binding affinities with non-coding small RNAs and have distinct roles in virulence. Mol. Plant Pathol. 20, 1217–1236. 10.1111/mpp.12823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppenau C. E., Tran V. T., Kusch H., Aßhauer K. P., Landesfeind M., Meinicke P., et al. (2014). Verticillium dahlia VdTHI4, involved in thiazole biosynthesis, stress response and DNA repair functions, is required for vascular disease induction in tomato. Environ. Exp. Bot. 108, 14–22. 10.1016/j.envexpbot.2013.12.015 [DOI] [Google Scholar]
- Hwang S., Cordova B., Abdo M., Pfeiffer F., Maupin-Furlow J. A. (2017). ThiN as a versatile domain of transcriptional repressors and catalytic enzymes of thiamine biosynthesis. J. Bacteriol. 199, e00810–e00816. 10.1128/JB.00810-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga T., Ishiga Y., Betsuyaku S., Nomura N. (2018). AlgU contributes to the virulence of Pseudomonas syringae pv. tomato DC3000 by regulating production of the phytotoxin coronatine. J. Gen. Plant Pathol. 84, 189–201. 10.1007/s10327-018-0775-6 [DOI] [Google Scholar]
- Ishiga Y., Ichinose Y. (2016). Pseudomonas syringae pv. tomato OxyR is required for virulence in tomato and Arabidopsis. Mol. Plant Microbe. Interact. 29, 119–131. 10.1094/MPMI-09-15-0204-R [DOI] [PubMed] [Google Scholar]
- Jordan S., Hutchings M. I., Mascher T. (2008). Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32, 107–146. 10.1111/j.1574-6976.2007.00091.x [DOI] [PubMed] [Google Scholar]
- Jurgenson C. T., Begley T. P., Ealick S. E. (2009). The structural and biochemical foundations of thiamin biosynthesis. Annu. Rev. Biochem. 78, 569–603. 10.1146/annurev.biochem.78.072407.102340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitten T., Willis D. K. (1996). Suppression of a sensor kinase dependent phenotype in Pseudomonas syringae by ribosomal proteins L35 and L20. J. Bacteriol. 178, 1548–1555. 10.1111/j.1365-2672.1996.tb03229.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yu M. G., Chatnaparat T., Lee J. H., Tian Y. L., Hu B. S., et al. (2020). Comparative transcriptomic analysis of global gene expression mediated by (p)ppGpp reveals common regulatory networks in Pseudomonas syringae. BMC Genom. 21, 296. 10.1186/s12864-020-6701-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yu M. G., Ge Y. X., Tian Y. L., Hu B. S., Zhao Y. F. (2021). The RsmA RNA-binding proteins in Pseudomonas syringae exhibit distinct and overlapping roles in modulating virulence and survival under different nutritional conditions. Front. Plant Sci. 12, 637595. 10.3389/fpls.2021.637595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo M., O'Riordan N., Fuchs T. M., Utratna M., Karatzas K. A., O'Byrne C. P. (2012). Thiamine plays a critical role in the acid tolerance of Listeria monocytogenes. FEMS Microbiol. Lett. 326, 137–143. 10.1111/j.1574-6968.2011.02442.x [DOI] [PubMed] [Google Scholar]
- Melnick J., Lis E., Park J. H., Kinsland C., Mori H., Baba T., et al. (2004). Identification of the two missing bacterial genes involved in thiamine salvage: thiamine pyrophosphokinase and thiamine kinase. J. Bacteriol. 186, 3660–3662. 10.1128/JB.186.11.3660-3662.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moat A. G., Foster J. W., Spector M. P. (2003). “Microbial stress responses,” in Microbial Physiology, eds A. G. Moat, J. W. Foster, and M. P. Spector (New York, NY: Wiley; ), 582–611. [Google Scholar]
- Norrander J., Kempe T., Messing J. (1983). Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26, 101–106. 10.1016/0378-1119(83)90040-9 [DOI] [PubMed] [Google Scholar]
- Osiezagha K., Ali S., Freeman C., Barker N. C., Jabeen S., Maitra S., et al. (2013). Thiamine deficiency and delirium. Innov. Clin. Neurosci. 10, 26–32. [PMC free article] [PubMed] [Google Scholar]
- Palmer L. D., Downs D. M. (2013). The thiamine biosynthetic enzyme ThiC catalyzes multiple turnovers and is inhibited by S-adenosylmethionine (AdoMet) metabolites. J. Biol. Chem. 288, 30693–30699. 10.1074/jbc.M113.500280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D. H., Mirabella R., Bronstein P. A., Preston G. M., Haring M. A., Lim C. K., et al. (2010). Mutations in γ-aminobutyric acid (GABA) transaminase genes in plants or Pseudomonas syringae reduce bacterial virulence. Plant J. 64, 318–330. 10.1111/j.1365-313X.2010.04327.x [DOI] [PubMed] [Google Scholar]
- Rapala-Kozik M., Wolak N., Kujda M., Banas A. K. (2012). The upregulation of thiamine (vitamin B1) biosynthesis in Arabidopsis thaliana seedlings under salt and osmotic stress conditions is mediated by abscisic acid at the early stages of this stress response. BMC Plant Biol. 12, 2. 10.1186/1471-2229-12-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov D. A., Leyn S. A., Li X., Rodionova I. A. (2017). A novel transcriptional regulator related to thiamine phosphate synthase controls thiamine metabolism genes in Archaea. J. Bacteriol. 199, e00743–e00716. 10.1128/JB.00743-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov D. A., Vitreschak A. G., Mironov A. A., Gelfand M. S. (2002). Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J. Biol. Chem. 277, 48949–48959. 10.1074/jbc.M208965200 [DOI] [PubMed] [Google Scholar]
- Ruiz-Roldán C., Puerto-Galán L., Roa J., Castro A., Di Pietro A., Roncero M. I. G., et al. (2008). The Fusarium oxysporum sti35 gene functions in thiamine biosynthesis and oxidative stress response. Fungal Genet. Biol. 45, 6–16. 10.1016/j.fgb.2007.09.003 [DOI] [PubMed] [Google Scholar]
- Schauer K., Stolz J., Scherer S., Fuchs T. M. (2009). Both thiamine uptake and biosynthesis of thiamine precursors are required for intracellular replication of Listeria monocytogenes. J. Bacteriol. 191, 2218–2227. 10.1128/jb.01636-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre E., Begley T. P., Ealick S. E. (2003). Structural biology of enzymes of the thiamin biosynthesis pathway. Curr. Opin. Struct. Biol. 13, 739–747. 10.1016/j.sbi.2003.10.006 [DOI] [PubMed] [Google Scholar]
- Swiecilo A., Zych-Wezyk I. (2013). Bacterial stress response as an adaptation to life in a soil environment. Pol. J. Environ. Stud. 22, 1577–1587. 10.2166/wh.2013.242 [DOI] [PubMed] [Google Scholar]
- Swingle B., Thete D., Moll M., Myers C. R., Schneider D. J., Cartinhour S. (2008). Characterization of the PvdS-regulated promoter motif in Pseudomonas syringae pv. tomato DC3000 reveals regulon members and insights regarding PvdS function in other Pseudomonads. Mol. Microbiol. 68, 871–889. 10.1111/j.1365-2958.2008.06209.x [DOI] [PubMed] [Google Scholar]
- Takaya A., Tomoyasu T., Matsui H., Yamamoto T. (2004). The DnaK/DnaJ chaperone machinery of Salmonella enterica serovar Typhimurium is essential for invasion of epithelial cells and survival within macrophages, leading to systemic infection. Infect. Immun. 72, 1364–1373. 10.1128/IAI.72.3.1364-1373.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylicki A., Lotowski Z., Siemieniuk M., Ratkiewicz A. (2018). Thiamine and selected thiamine antivitamins-biological activity and methods of synthesis. Biosci. Rep. 38, BSR20171148. 10.1042/BSR20171148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas P., Farias G. A., Nogales J., Prada H., Carvajal V., Barón M., et al. (2013). Plant flavonoids target Pseudomonas syringae pv. tomato DC3000 flagella and type III secretion system. Environ. Microbiol. Rep. 5, 841–850. 10.1111/1758-2229.12086 [DOI] [PubMed] [Google Scholar]
- Vorob'eva L. I. (2003). Stressors, stress reactions, and survival of bacteria: a review. Appl. Biochem. Microbiol. 40, 217–224. 10.1023/B:ABIM.0000025941.11643.19 [DOI] [PubMed] [Google Scholar]
- Webb E., Claas K., Downs D. (1998). thiBPQ encodes an ABC transporter required for transport of thiamine and thiamine pyrophosphate in Salmonella typhimurium. J. Biol. Chem. 273, 8946–8950. 10.1074/jbc.273.15.8946 [DOI] [PubMed] [Google Scholar]
- Xie Y., Shao X., Deng X. (2019). Regulation of type III secretion system in Pseudomonas syringae. Environ. Microbiol. 21, 4465–4477. 10.1111/1462-2920.14779 [DOI] [PubMed] [Google Scholar]
- Xin X. F., He S. Y. (2013). Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51, 473–498. 10.1146/annurev-phyto-082712-102321 [DOI] [PubMed] [Google Scholar]
- Yu X., Liang X., Liu K., Dong W., Wang J., Zhou M. (2015). The thiG gene is required for full virulence of Xanthomonas oryzae pv. oryzae by preventing cell aggregation. PLoS ONE 10, e0134237. 10.1371/journal.pone.0134237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Thilmony R., Bender C. L., Schaller A., He S. Y., Howe G. A. (2003). Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 36, 485–499. 10.1046/j.1365-313X.2003.01895.x [DOI] [PubMed] [Google Scholar]
- Zhao Y, He S. Y., Sundin G.W. (2006). The Erwinia amylovora avrRpt2EA gene contributes to virulence on pear and AvrRpt2EA is recognized by Arabidopsis RPS2 when expressed in Pseudomonas syringae. Mol. Plant Microbe Interact. 19, 644–654. 10.1094/MPMI-19-0644 [DOI] [PubMed] [Google Scholar]
- Zou H. S., Yuan L., Guo W., Li Y. R., Che Y. Z., Zou L. F., et al. (2011). Construction of a Tn5-tagged mutant library of Xanthomonas oryzae pv. oryzicola as an invaluable resource for functional genomics. Curr. Microbiol. 62, 908–916. 10.1007/s00284-010-9804-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.