Abstract
We studied genetic variability of 100 isolates of Claviceps purpurea by using randomly amplified polymorphic DNA (RAPD), an EcoRI restriction site polymorphism in the 5.8S ribosomal DNA (rDNA), the alkaloids produced, and conidial morphology. We identified three groups: (i) group G1 from fields and open meadows (57 isolates), (ii) group G2 from shady or wet habitats (41 isolates), and (iii) group G3 from Spartina anglica from salt marshes (2 isolates). The sclerotia of G1 isolates contained ergotamines and ergotoxines; G2 isolates produced ergosine and ergocristine along with small amounts of ergocryptine; and G3 isolates produced ergocristine and ergocryptine. The conidia of G1 isolates were 5 to 8 μm long, the conidia of G2 isolates were 7 to 10 μm long, and the conidia of G3 isolates were 10 to 12 μm long. Sclerotia of the G2 and G3 isolates floated on water. In the 5.8S rDNA analysis, an EcoRI site was found in G1 and G3 isolates but not in G2 isolates. The host preferences of the groups were not absolute, and there were host genera that were common to both G1 and G2; the presence of members of different groups in the same locality was rare. Without the use of RAPD or rDNA polymorphism, it was not possible to distinguish the three groups solely on the basis of phenotype, host, or habitat. In general, populations of C. purpurea are not host specialized, as previously assumed, but they are habitat specialized, and collecting strategies and toxin risk assessments should be changed to reflect this paradigm shift.
Claviceps purpurea is an ergot fungus with a wide host range that includes the entire subfamily Pooideae, many members of the Arundinoideae, and some species belonging to the chloridoid and panicoid groups (4, 15). Its distribution is basically Holarctic, but it has been recorded in Arctic regions (14) and also occurs in southern temperate and subtropical regions. Due to movement with cereal and grass seeds by migrants, the center of origin of this species is not known.
C. purpurea is morphologically quite variable. Sclerotial length ranges from 2 to 50 mm, and the color of the stromata varies over a wide range of red shades from wine to purple (25) and even to orange (31). Conidial size and shape also are polymorphic; the conidia range from oval spores that are 5 μm long to cylindric or elongated spores that are up to 13 μm long (15, 25, 31). The sclerotia contain peptide alkaloids that belong to three basic groups, the ergotamines (with alanine as the first amino acid entering the cyclopeptide moiety), the ergotoxines (with valine), and the rarely found ergoxines (with 2-aminoisobutyric acid) (for reviews see references 7 and 33).
For the last 100 years, researchers have tried to use this variation to establish varieties, special forms, or races (1), and the primary focus has been on detection of host-specific groups. Stäger introduced four special forms: secalis, lolii (later joined with secalis) (2), milii (on Milium and Brachypodium only), and glyceriae (suspected to be Claviceps wilsonii) for C. purpurea sensu Tulasne (excluding Claviceps microcephala [Wallr.] Tul.) (26–28, 30). However, biological barriers to host specificity have not been confirmed (2, 3, 5, 16).
Stäger found (29) that sclerotia formed on grasses from wet habitats could float on water but that sclerotia formed on Secale sp., Lolium sp., Brachypodium sylvaticum, Sesleria coerulea, Arrhenatherum elatius, Agropyron repens (now Elytrigia repens), Alopecurus myosuroides, and other land grasses sank in water. On Dactylis glomerata, Calamagrostis epigeios, and some Holcus and Poa spp., sclerotia of both types were observed. For the floating isolates, the new taxon f. sp. Phalaridis arundinaceae natans was defined (29).
Jungehülsing and Tudzynski (10) established two main groups by using randomly amplified polymorphic DNA (RAPD) typing; one group consisted primarily of English isolates from Molinia, Holcus, and Dactylis species, and the other group contained the isolates from land grasses.
Loveless (15) found that the conidia of isolates from grasses from wet and shady habitats were longer (6.5 to 8.5 μm) than those from isolates found on land grasses (5 to 6 μm). Conidial shape and size remained unchanged, even when the isolates from different hosts were inoculated onto wheat or cultivated on agar plates (16), indicating that this trait is characteristic of an isolate and not of the substrate upon which it is raised. However, spores from laboratory cultures vary more than those from a natural host vary. Another group of C. purpurea isolates was found on Spartina spp. populating Atlantic salt marshes in the Americas. This group was characterized (as analyzed by thin-layer chromatography [TLC]) by predominant production of ergocryptine, ergocryptinine, and lysergylvalylmethylester (6). Spartina stands in the British Isles have been colonized by C. purpurea only since 1960 (9, 21). The isolates obtained from these stands have the longest conidia (8.4 μm) of all the British samples studied (15).
Kobel and Sanglier (11) identified 10 chemoraces in sclerotia collected in Europe and North America, with the most common combinations being ergocornine and ergocryptine (23% of the samples), ergocristine and ergosine (20%), and ergotamine (13%). The composition of the alkaloid mixture produced is hereditary and independent of the host (12).
Our objectives in this study were (i) to identify the population structure of C. purpurea and (ii) to characterize the groups and isolates by host or habitat preferences, by phenotypic traits used in previous studies (conidial morphology, alkaloid type, properties of sclerotia) and by DNA polymorphisms. Our goal was to harmonize and relate the ambiguous, even contradictory, groupings made by previous researchers often on the basis of only one or two characters, in a single system.
MATERIALS AND METHODS
Isolates.
The isolates used, their origins, and other properties are summarized in Table 1. Representative cultures have been deposited in the Culture Collection of Fungi, Charles University, Prague, Czech Republic (CCF), under accession numbers CCF 3145 to 3149.
TABLE 1.
Origins of C. purpurea isolates
| Isolate(s) | Host | Designationa | Localityb | Origin |
|---|---|---|---|---|
| G1 strains | ||||
| 134 | Phalaris tuberosa | Station | Victoria, Australia | |
| 150, 151c | Secale cereale | F | Opava, Komárov, Czech Republic | |
| 153 | Unknown | Pepty 695/S | ? | Central Europe |
| 169 | Poa sp. | F | Washington | |
| 170 | Lolium multiflorum | ? | South Africa | |
| 192 | Poa pratensis | Station | Munich, Germany | |
| 193 | P. pratensis | Station | Unknown | |
| 194 | S. cereale | Bay 10 | F | Donaumoos, Germany |
| 198 | Elytrigia repens | W3 | ? | Budleigh, Devon, United Kingdom |
| 199 | Lolium perenne | W10 | ? | Exeter, United Kingdom |
| 200 | Festuca arundinacea | W8 | ? | Weston Mouth, Devon, United Kingdom |
| 201 | Festuca sp. | W15 | ? | Mount Fuji, Japan |
| 202 | Carex sp. | Dla | ? | Hejsager, Denmark |
| 205 | F. arundinacea | CCF 3145 | F | Lauderdale, Ala. |
| 207, 208c | Triticum aestivum | F | Kansas | |
| 215 219c | Elymus caninus | CCF 3146 | Station | Markvartice, Czech Republic |
| 219 | E. caninus | Station | Markvartice, Czech Republic | |
| 220 | E. repens | Station | Markvartice, Czech Republic | |
| 222 | Lolium sp. | Station | Markvartice, Czech Republic | |
| 238 | Phleum sp. | Station | Vyskeř, Czech Republic | |
| 243 | Lolium sp. | M | Vyskeř, Czech Republic | |
| 254 | Dactylis sp. | M | Prague, Czech Republic | |
| 255 | Festuca ovina | Station | Markvartice, Czech Republic | |
| 258 | Spartina fusiformis | 37-1 | ? | Argentina |
| 311 | L. perenne | Station | Markvartice, Czech Republic | |
| 338 | E. repens | R | Freeman, Wash. | |
| 343 | F. arundinacea | M | Pullman, Wash. | |
| 347 | P. pratensis | M | Post Falls, Idaho | |
| 354 | Bromus inermis | M | Freeman, Wash. | |
| 358 | P. pratensis | M | Post Falls, Idaho | |
| 363 | B. inermis | R | Pullman, Wash. | |
| 368 | Alopecurus pratensis | R | Freeman, Wash. | |
| 374 | F. arundinacea | M | Altamont, Ala. | |
| 383 | Lolium sp. | Park | Prague, Czech Republic | |
| 387 | Helictotrichon pubescens | M | Levý Hradec, Czech Republic | |
| 396 | S. cereale | F | Soláň, Czech Republic | |
| 417 | Sesleria tatrae | Station | Zubří, Czech Republic | |
| 426 | P. pratensis | Station | Větrov, Czech Republic | |
| 428 | S. cereale | T5 | F | Hohenheim, Germany |
| 436 | Dactylis sp. | M | Trutnov, Czech Republic | |
| 439 | Dactylis sp. | M | Trutnov, Czech Republic | |
| 442 | F. arundinacea | M | Trutnov, Czech Republic | |
| 445 | E. repens | M | Trutnov, Czech Republic | |
| 447 | Lolium sp. | R | Židněves, Czech Republic | |
| 450 | E. repens | R | Židněves, Czech Republic | |
| 451 | Arrhenatherum elatior | R | Židněves, Czech Republic | |
| 455 | S. cereale | F | Warthe, Usedom, Germany | |
| 477 | Dactylis glomerata | M | Březno, Czech Republic | |
| 478 | D. glomerata | M | Březno, Czech Republic | |
| 479 | S. cereale | F | Dobrá, Šumava, Czech Republic | |
| 483 | Bromus hybrid | M | Saskatoon, Canada | |
| 504 | Ammophila arenaria | C | Zeebrugge, Belgium | |
| 514 | F. arundinacea | M | Rome, Italy | |
| 515 | F. arundinacea | M | Rome, Italy | |
| 516, 517c | F. arundinacea | M | Rome, Italy | |
| G2 strains | ||||
| 162, 163, 164c | Poa annua | Station | Skalička, Czech Republic | |
| 195 | D. glomerata | W9 | ? | Exeter, United Kingdom |
| 196 | Molinia sp. | W12 | ? | Exeter, United Kingdom |
| 197 | Agrostis sp. | T8 | ? | Hohes Venn, Belgium |
| 210, 211, 213c | Phleum sp. | Station | Markvartice, Czech Republic | |
| 231 | P. annua | Station | Lešná, Czech Republic | |
| 232 | P. annua | CCF 3148 | Station | Lešná, Czech Republic |
| 236 | Molinia coerulea | CCF 3147 | M | Vlčí Pole u Bousova, Czech Republic |
| 244 | Festuca rubra | Station | Markvartice, Czech Republic | |
| 247, 250c | D. glomerata | M | Vyskeř, Czech Republic | |
| 256 | Calamagrostis sp. | B | Volyně, Czech Republic | |
| 259 | F. rubra | Station | Markvartice, Czech Republic | |
| 299 | Alopecurus sp. | Station | Zubří, Czech Republic | |
| 379 | A. pratensis | Station | Zubří, Czech Republic | |
| 386 | A. elatior | B | Roztoky, Czech Republic | |
| 389 | P. pratensis | Station | Steinach, Germany | |
| 406 | P. pratensis | Station | Zubří, Czech Republic | |
| 407 | F. rubra | Station | Větrov, Czech Republic | |
| 410 | F. ovina | Station | Větrov, Czech Republic | |
| 413 | P. pratensis | Station | Chyšky, Czech Republic | |
| 415 | Phalaroides arundinacea | B | Bezděkov pond, Czech Republic | |
| 419, 420c | P. pratensis | Station | Zubří, Czech Republic | |
| 421 | F. rubra | Station | Větrov, Czech Republic | |
| 431 | P. arundinacea | MW | Phillipsreuth, Germany | |
| 434 | Dactylis sp. | MW | Phillipsreuth, Germany | |
| 473, 474, 475c | Phragmites communis | B | Sobotka, Czech Republic | |
| 480 | M. coerulea | Station | Zubří, Czech Republic | |
| 503 | A. arenaria | C | Zeebrugge, Belgium | |
| 505 | Dactylis sp. | MW | Phillipsreuth, Germany | |
| 507 | Milium effusum | MW | Phillipsreuth, Germany | |
| 518 | P. arundinacea | MW | Szklarska Poremba, Poland | |
| 521 | P. arundinacea | MW | Szklarska Poremba, Poland | |
| 523 | Calamagrostis villosa | MW | Szklarska Poremba, Poland | |
| G3 strains | ||||
| 481 | Spartina anglica | Salt marsh | Lincegrove Marsh, Hampshire, United Kingdom | |
| 510 | S. anglica | CCF 3149 | Salt marsh | Skeffling, Yorkshire, United Kingdom |
CCF (Prague) collection numbers of representative isolates are given. Strain Pepty 695/S (kindly donated by D. Gröger, Halle/Saale, Germany) produces clavines, ergocornine, and ergocryptine (17). Isolates Bay10 (= 194), W9 (= 195), W12 (= 196), T8 (= 197), W3 (= 198), W10 (= 199), W8 (= 200), W15 (= 201), Dla (= 202), 37-l (= 258), and T5 (= 428) used by Jungehülsing and Tudzynski (10) were kindly supplied by P. Tudzynski (Westphalische Wilhelms-Universität, Münster, Germany).
C, sea coast; F, field; M, meadow or garden; MW, mountain wood; R, along the road; B, river or pond bank; Station, grassland station; ?, unknown.
Multiple isolate entries with identical characteristics originated from sclerotia from the same plant.
Cultivation of fungi.
Sclerotia were surface sterilized for 2 to 5 min (depending on their size) in 1.3% sodium hypochlorite and rinsed three times for 3 to 5 min in distilled water. Sterilized sclerotia were placed on T2 agar plates (18) supplemented with 100 μg of ampicillin per ml. Isolates were maintained on T2 agar slants at 4°C and were subcultured every 6 months.
Preparation of genomic DNA.
Spore or mycelium suspensions were plated on cellophane discs laid on T2 plates and were grown for 1 to 2 weeks at 24°C. Mycelium was scraped into a mortar and ground in liquid nitrogen. From ca. 0.5 g of the powdered mycelium, DNA was extracted (18) with an additional purification step. To the DNA solution, 0.5 volume of 7.5 M ammonium acetate was added. After incubation for 10 min at room temperature, proteinaceous impurities were pelleted by centrifugation for 10 min at 10,000 × g. DNA was precipitated from the supernatant by adding 0.7 volume of isopropanol, incubating the preparation for 2 h at 4°C, and then centrifuging it for 15 min at 10,000 × g.
Analyses of ITS rDNA.
The region containing internal transcribed spacer 1 (ITS1), ITS2, and 5.8S ribosomal DNA (rDNA) was amplified by using ITS1 and ITS4 primers (34). Each mixture (25 μl) contained 50 ng of genomic DNA, 20 pmol of each primer, each deoxynucleoside triphosphate (Promega, Madison, Wis.) at a concentration of 200 μM, and 1 U of DynaZyme along with the buffer supplied by the manufacturer (Finnzymes, Oy, Finland). The reaction mixtures were subjected to 32 cycles in a GeneE thermal cycler (Techne, Cambridge, United Kingdom), as follows: (i) one cycle consisting of 95°C for 3 min, 55°C for 30 s, and 72°C for 1 min; (ii) 30 cycles consisting of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and (iii) one cycle consisting of 95°C for 30 s, 55°C for 30 s, and 72°C for 10 min. The amplicons in 3 to 5 μl of reaction mixture were, without further purification, digested with 10 U of EcoRI for 1 to 2 h in a total volume of 20 μl. Custom sequencing was done with an ABI 373A sequencer (Institute of Biotechnology, Technical University, Graz, Austria) by using dye terminators (18).
RAPD analysis.
Primers 1CR (GCCTTGCGGACGGCAATATC), 1CF (TCCTTGATGCATTCGCAACC), 206 (TCAACAATGTCGGCCTCCGT), 257 (CGTGATGTCAGTGATGC), RP2, and RP4-2 (13), as well as primers OPA01, OPA03, OPA04, OPA08, OPA02, OPE01, OPE04, OPE14, and OPE17 (Operon Technologies, Alameda, Calif.), were tested. Each 20-μl reaction mixture contained each deoxynucleoside triphosphate at a concentration of 200 μM, 20 pmol of primer, DynaZyme reaction buffer, DynaZyme polymerase (1 U), and 1.75 mM MgCl2. DNA (50 to 100 ng) was added, and samples were placed on the cycler plate after the temperature reached 80°C. The following cycling conditions were used: (i) one cycle consisting of 94°C for 3 min, 38°C for 1 min, and 72°C for 20 s; (ii) 33 cycles consisting of 93°C for 20 s, 38°C for 1 min, and 72°C for 20 s; and (iii) one cycle consisting of 93°C for 20 s, 38°C for 1 min, and 72°C for 6 min. The resulting bands were separated on a 1% agarose gel (SeaKem LE agarose; FMC BioProducts, Rockland, Maine) at 4 V/cm in 1× TBE (8.9 mM Tris-borate, 8.9 mM boric acid, 2 mM EDTA) for 2 to 4 h. The same patterns were observed when different DNA preparations from the same isolate were used and also when samples were run on a Mastercycler gradient (Eppendorf AG, Cologne, Germany) instead of GeneE.
Sequencing of amplicons.
Amplicons A and C were purified from agarose gels with a Qiaex II gel extraction kit (catalog no. 20021; Qiagen GmbH, Hilden, Germany) and were cloned into SmaI-digested pBluescript/SK (Stratagene, La Jolla, Calif.). The sequences were determined by using T3 (AATTAACCCTCACTAAAGGG) and T7 (GTAATACGACTCACTATAGGGC) universal primers as described above.
Alkaloid extraction.
Sclerotia (50 to 500 mg) were powdered and extracted with 6 ml of extraction mixture (80% methanol with 1 ml of NH4OH per liter). The mixture was stirred for 2 h and extracted further without stirring for 12 h in the dark before paper filtering. The filtrate was vacuum evaporated to dryness at room temperature, and the residue was dissolved in 50 to 200 μl of chloroform.
TLC.
Ten microliters of alkaloid extract was loaded on a TLC plate (Silica Gel 60; Merck, Darmstadt, Germany). The plate was exposed to ammonia vapor and developed with a chloroform-acetone (3:1, vol/vol) mobile phase. Alkaloids were detected as blue spots by using Ehrlich reagent spray (5 g of 4-dimethylaminobenzaldehyde, 75 ml of ethanol, 25 ml of concentrated hydrochloric acid).
HPLC.
To quantify the compositions of the alkaloid mixtures, reverse-phase high-performance liquid chromatography (HPLC) was used. Separations were carried out on an RP C18 column (150 by 3.3 mm [inside diameter]; Tessek, Prague, Czech Republic) by using gradient elution with the following mobile phases: methanol-water-concentrated ammonium hydroxide (9:1:0.0004) (solvent A) and methanol-water-concentrated ammonium hydroxide (0.5:9.5:0.0004) (solvent B). Gradient elution started with 50% solvent A, which was linearly increased to 95% solvent A within 65 min at a flow rate of 0.5 ml/min. The alkaloids were detected at a wavelength of 310 nm. Chromatographic data were processed with the Millennium 2.15 software (Waters Czech Republic, Prague, Czech Republic). Ergot alkaloid standards were isolated and their identities were confirmed in our laboratory.
Mathematical analyses.
RAPD gel photos were scanned with an HP Scanjet 4P, and the images were converted into binary matrices by the Cross Checker fingerprint analysis program (version 2.9; J. B. Buntjer, Wageningen University and Research Centre, Wageningen, The Netherlands). A distance matrix was calculated by using Jaccard's coefficient (23). An alkaloid-based Euclidean distance matrix for 25 isolates was compared with a RAPD-based distance matrix by using the Mantel test (24), as implemented by Mantel (version 2.0; A. Liedloff, Queensland University of Technology, Brisbane, Australia). This program uses two methods which determine significance with a standard normal variate (g) and 1,000 random permutations of the first matrix to determine the possible variation within the data. The values for Z (Mantel coefficient), g, and r (correlation coefficient) from both matrices specified were calculated. The null hypothesis tested was the hypothesis that there was no relationship between the matrices.
Microscopy.
Conidial measurements were made at a magnification of ×1,125 by using 30 spores washed from sclerotia. For cultures from other laboratories, conidia from plates were measured. The standard deviations ranged from 15 to 20% of the mean.
Floating test.
Sclerotia were placed on the surface of distilled water in a 200-ml beaker. If the sclerotia were still floating after 1 h, the water was swirled with a glass rod. Sclerotia that floated for at least 4 h were considered floaters.
RESULTS
RAPD.
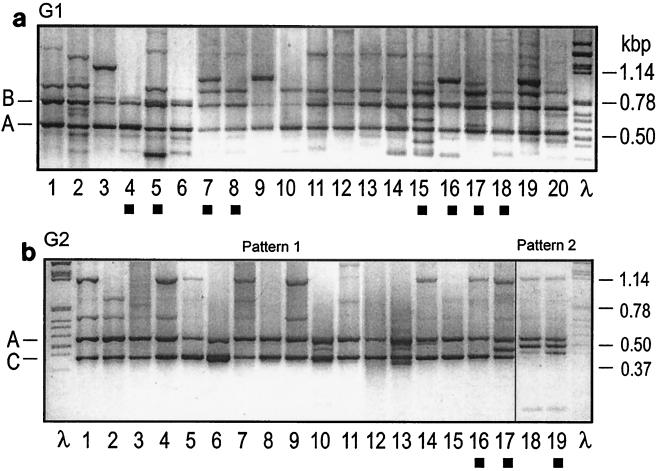
We tested 15 primers that all resulted in band patterns that distinguished three groups. Primer 257 was chosen for routine typing because the patterns obtained with it were the most distinct (Fig. 1). All isolates shared one band, the species-specific amplicon A band derived from a multicopy sequence (19). The first group, group G1 (57 isolates), was characterized by band B; however, other bands were also observed. Forty-one isolates belonging to the second group, group G2, produced the same pattern with most primers. However, with primer 257, 36 of these isolates produced a pattern with a prominent band C, whereas a second pattern appeared only with isolates T8, 236, 473, 474, and 475.
FIG. 1.
Examples of RAPD profiles of G1 and G2 isolates obtained with primer 257. Amplicon A is species specific, amplicon B is specific for G1 isolates, and amplicon C is specific for the majority of G2 isolates. (a) G1 strains. Lane 1, 417; lane 2, 343; lane 3, 374; lane 4, Dla; lane 5, 37-1; lane 6, 134; lane 7, Bay10; lane 8, T5; lane 9, 383; lane 10, 254; lane 11, 478; lane 12, Pepty 695/S; lane 13, 192; lane 14, 150; lane 15, W3; lane 16, W8; lane 17, W10; lane 18, W15; lane 19, 363; lane 20, 317. (b) G2 strains. Lane 1, 231; lane 2, 163; lane 3, 419; lane 4, 420; lane 5, 379; lane 6, 299; lane 7, 413; lane 8, 407; lane 9, 410; lane 10, 421; lane 11, 256; lane 12, 415; lane 13, 386; lane 14, 211; lane 15, 213; lane 16, W9; lane 17, W12; lane 18, 236; lane 19, T8. The solid squares indicate isolates from the study of Jungehülsing and Tudzynski (10).
We included isolates from the two main groups described by Jungehülsing and Tudzynski (10). We found that eight of these isolates (W3, W10, W8, W15, 37-1, Dla, Bay10, and T5), which belong to the Secale-Agropyron clade, grouped with G1 and that three isolates (W9, W12, and T8) from the Molinia-Dactylis clade grouped with G2.
The third group, group G3, contained two British isolates from Spartina anglica growing in salt marshes. These isolates produced the same RAPD pattern that lacked amplicon B and was typical of G1 and also differed from the pattern typical of G2. Isolate 37-1, which originated from Spartina fusiformis, belonged to G1, and its RAPD pattern was therefore not similar to that of isolates from S. anglica.
Intraspecific EcoRI polymorphism.
The 5.8S rDNA of C. purpurea G1 isolates Pepty 695/S (17) and 134 (EMBL nucleotide sequence database accession no. AJ000069 and AJ000070) contains a conserved EcoRI site (22). The sequence of isolate 162 (group G2) has a transversion at this site (GAATTC to GACTTC) and is the same as the sequence (GenBank accession no. U57669) of a C. purpurea isolate collected from Dactylis glomerata in Athens, Ga. (8). All of the G1 and G3 isolates were EcoRI positive, and the G2 isolates were EcoRI negative. The representative isolates from the Secale-Agropyron group of Jungehülsing and Tudzynski (10) were EcoRI positive, whereas representative isolates from the Molinia-Dactylis group were EcoRI negative.
Habitats and host plants.
G1 isolates originated from fields, from open meadows, and from grasses along roads, i.e., from open, sunny localities that often are dry. We found that Alopecurus pratensis, Ammophila arenaria, Arrhenatherum elatior, Dactylis sp., Festuca ovina, Festuca rubra, Phleum sp., and Poa pratensis could be colonized naturally by both G1 and G2 isolates.
G2 isolates were more commonly recovered from hosts growing in shady or wet habitats, and Calamagrostis, Phalaroides, Phragmites, and Molinia were the most frequent host genera. Common habitats included pond and river banks, ditches, forests, and even mountain woods.
Among the sclerotial samples obtained from Markvartice and Zubří grassland stations in the Czech Republic, isolates belonging to both G1 and G2 were recovered from the same agricultural field. There were two natural locations from which we recovered both types of isolates. One of them was the meadow at Vyskeř in the Czech Republic, where samples collected from Lolium and Phleum yielded only G1 isolates but samples collected from Dactylis yielded both G1 and G2 isolates. The other location was at the sand dunes on the sea coast in Zeebrugge (Belgium), where G1 and G2 sclerotia were found on the same plant of A. arenaria.
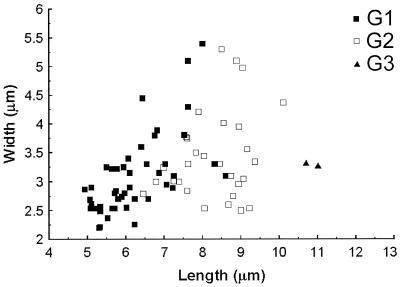
Conidial size.
G3 isolates have the longest conidia (length, more than 10 μm), but the number of isolates that have been obtained so far is limited. Conidia shorter than 6.5 μm were always from G1 strains. Conidia between 8.5 and 10 μm long were always from G2 strains (Fig. 2). In the 6.5- to 8.5-μm range, G1 and G2 strains overlapped. Therefore, conidial size is only of secondary importance in distinguishing the three groups.
FIG. 2.
Average conidial sizes of isolates belonging to the three C. purpurea groups. The standard deviation of spore size was 15 to 20%.
Floating sclerotia.
All of the sclerotia of G2 strains could float, whereas the sclerotia of G1 strains all sank within the first 30 min in the water. The best floating was observed with the sclerotia of G3 strains, which were difficult even to wet.
Alkaloid analyses.
Sclerotia of G1 strains contained one or more of seven different alkaloids (Table 2). Sclerotia of G2 strains contained only ergosine, ergocristine, and small amounts of ergocryptine. Sclerotia of G3 strains contained mixtures of ergocristine and ergocryptine. Thus, G2 and G3 constitute stable chemoraces.
TABLE 2.
Alkaloids from sclerotia of C. purpurea group strains
| Isolate(s) | Amt of alkaloids (mg/g [dry wt])a | % of total area of HPLC peaks
|
||||||
|---|---|---|---|---|---|---|---|---|
| Ergotamine | Ergosine | Ergocryptine | Ergocristine | Ergocornine | Ergine | Ergometrine | ||
| G1 strains | ||||||||
| 136 | 0.4 | 8.9 | 24 | 24 | 14 | 16 | 0 | 13 |
| 141 | 0.3 | 7.2 | 28 | 18 | 22 | 11 | 14 | 0 |
| 150 | 2.3 | 0.8 | 2.2 | 8 | 78 | 0 | 0 | 11 |
| 151 | 1.1 | 0 | 2 | 85 | 0 | 0 | 0 | 13 |
| 169 | 0.2 | 18 | 7.3 | 9.7 | 65 | 0 | 0 | 0 |
| 170 | 0.9 | 19 | 14 | 32 | 0 | 19 | 0 | 16 |
| 205 | 1.4 | 18 | 30 | 14 | 38 | 0 | 0 | 0 |
| 207, 208b | NDc | 45 | 31 | 0 | 17 | 0 | 6.6 | 0 |
| 215, 219b | 1.9 | 0 | 18 | 40 | 0 | 40 | 1.7 | 0 |
| 220 | 0.5 | 0 | 15 | 41 | 2.1 | 42 | 0 | 0 |
| 222 | 0.9 | 0 | 21 | 20 | 27 | 32 | 0 | 0 |
| 243 | 1.6 | 0 | 21 | 26 | 26 | 27 | 0 | 0 |
| 338 | 1.1 | 30 | 0 | 5.2 | 42 | 0 | 0 | 23 |
| 354 | 1.6 | 15 | 0 | 0 | 62 | 0 | 0 | 23 |
| 358 | 1.2 | 14 | 0 | 25 | 38 | 23 | 0 | 0 |
| 368 | 1.4 | 31 | 0 | 0 | 55 | 0 | 0 | 14 |
| 396 | 0.2 | 82 | 5.1 | 0 | 13 | 0 | 0 | 0 |
| 442 | 1.4 | 0 | 20 | 38 | 5.2 | 37 | 0 | 0 |
| 445 | 1.6 | 16 | 0 | 30 | 25 | 29 | 0 | 0 |
| 479 | 0.6 | 0 | 4.8 | 70 | 11 | 11 | 0 | 3.6 |
| 483 | 1.0 | 20 | 0 | 13 | 45 | 0 | 0 | 22 |
| 516, 517b | 1.6 | 19 | 33 | 0 | 48 | 0 | 0 | 0 |
| G2 strains | ||||||||
| 162, 163, 164b | 1.6 | 0 | 55 | 0 | 45 | 0 | 0 | 0 |
| 210, 211, 213b | 1.8 | 0 | 44 | 35 | 20 | 0 | 0 | 0 |
| 231, 232b | 0.2 | 0 | 60 | 6.5 | 32 | 0 | 0 | 0 |
| 236 | 0.63 | 0 | 49 | 8.3 | 42 | 0 | 0 | 0 |
| 247 | 0.4 | 0 | 44 | 3.2 | 53 | 0 | 0 | 0 |
| 259 | 2.6 | 0 | 61 | 4.9 | 34 | 0 | 0 | 0 |
| 299 | 0.6 | 0 | 44 | 7.1 | 49 | 0 | 0 | 0 |
| 389 | 1.5 | 0 | 61 | 4.6 | 34 | 0 | 0 | 0 |
| 406 | 0.4 | 0 | 44 | 12 | 44 | 0 | 0 | 0 |
| 407 | 0.5 | 0 | 59 | 6.9 | 34 | 0 | 0 | 0 |
| 413 | 0.6 | 0 | 52 | 5.1 | 43 | 0 | 0 | 0 |
| 415 | 0.5 | 0 | 56 | 2.4 | 41 | 0 | 0 | 0 |
| 431 | 0.7 | 0 | 55 | 2.1 | 43 | 0 | 0 | 0 |
| 473, 474, 475b | 1.3 | 0 | 46 | 12 | 42 | 0 | 0 | 0 |
| 480 | 2.7 | 0 | 71 | 1.0 | 28 | 0 | 0 | 0 |
| 505 | 1.0 | 0 | 60 | 2.6 | 36 | 0 | 0 | 0 |
| 507 | 0.8 | 0 | 42 | 2.1 | 55 | 0 | 0 | 0 |
| 518 | 2.5 | 0 | 30 | 60 | 9.7 | 0 | 0 | 0 |
| 521 | 0.7 | 0 | 83 | 0 | 17 | 0 | 0 | 0 |
| G3 strains | ||||||||
| 481 | 3.9 | 0 | 0 | 54 | 39 | 0 | 0 | 0 |
| 510 | 3.0 | 0 | 0 | 44 | 54 | 0 | 0 | 0 |
Measurements were obtained twice, and the standard errors were less than 5%.
Multiple isolates with identical alkaloid contents were obtained from the same sclerotial sample.
ND, not determined.
We also attempted to find a correlation between RAPD profiles and the alkaloids produced for 25 G1 isolates for which both types of data were available. We compared a RAPD-based distance matrix with a Euclidean matrix based on percentages of the seven alkaloids but found no evidence for a correlation.
DISCUSSION
Our data suggest that most G1 C. purpurea isolates contain alkaloid mixtures of variable composition and usually occur in meadows, fields and other open, dry localities. Despite considerable internal variation, no subgroups were distinguished on the basis of host plant, locality, or the types of alkaloids produced. In previous studies, this group was probably described as C. purpurea f. sp. secalis (26) or, on the basis of hosts and conidial size, C. purpurea var. purpurea (with conidia that were 5.8 by 3.0 μm) and C. purpurea var. agropyri (with conidia that were 6.7 by 2.4 μm) (31, 32).
G2 isolates differ from G1 isolates in a single base pair in the 5.8S rDNA, are more uniform in terms of the alkaloid production pattern, and produce predominantly elongated conidia and floating sclerotia. They have been found in Belgium and England (10), in the Czech Republic, Poland, and Germany (this study), and in the United States (8), and based on conidial morphology, they probably have been found in Japan as C. purpurea var. alopecuri and C. purpurea var. phalaridis (31, 32). Alkaloid production profiles suggest that about 20% of European and North American isolates are G2 isolates (11). The equivocal nature of host and conidial morphology phenotypes resulted in reduction of specific epithets such as C. wilsonii and C. microcephala to synonyms of C. purpurea (20). As G2 is clearly distinguishable from G1, we suggest that G2 should be called by the original name, C. purpurea f. sp. Phalaridis arundinaceae natans (29). G2 and G3 also are distinct chemoraces.
Regardless of the group, the composition of the alkaloid mixture produced by a given isolate is genetically stable (3, 12). The ratio of ergocornine to ergocryptine can be changed by feeding valine, leucine, and isoleucine to cultures and plants inoculated with an ergocornine and ergocryptine-producing C. purpurea strain. However, feeding phenylalanine did not result in ergocristine production (11). Synthesis of cyclopeptides catalyzed in vitro by the multifunctional enzyme LPS1 from a strain producing high levels of ergotamine yields all three groups of peptide ergot alkaloids if their amino acid precursors are added (33), which implies that this enzyme does not regulate the type of alkaloid produced. Regulation studies are hindered by the fact that natural isolates as a rule do not produce alkaloids in culture and producing mutants may have altered regulatory mechanisms.
Our results suggest that from a large, variable, basal population (group G1) that may correspond to former C. purpurea f. sp. secalis, occasionally smaller homogeneous populations develop that can be differentiated as ecological subspecies or varieties and that specialize to fill adjoining ecological niches. DNA and alkaloid analyses now enable us to clearly define populations, which was hitherto not possible by using only host and conidial data.
ACKNOWLEDGMENTS
This project was supported by Czech Grant Agency grants 206/97/0611 and 522/99/0517 and by Ministry of Education grant project COST 835.30.
We thank D. Sztachová and M. Pavlíček for technical assistance.
REFERENCES
- 1.Barger G. Ergot and ergotism. London, United Kingdom: Gurney and Jackson; 1931. [Google Scholar]
- 2.Békésy N. Ein Beitrag zur Biologie des Mutterkorns. Phytopathol. 1956;26:49–56. [Google Scholar]
- 3.Brejcha V, Kybal J. Faktory ovlivňující obsah alkaloidů ve sklerociích Claviceps purpurea. Preslia (Prague) 1956;28:161–168. [Google Scholar]
- 4.Brewer J, Loveless A R. Ergot of Pennisetum macrourum in South Africa. Kirkia. 1977;10:589–600. [Google Scholar]
- 5.Campbell W P. Studies of ergot infection in gramineous hosts. Can J Bot. 1957;35:315–320. [Google Scholar]
- 6.Eleuterius L N, Meyers C P. Claviceps purpurea on Spartina in coastal marshes. Mycologia. 1974;66:978–986. [Google Scholar]
- 7.Flieger M, Wurst M, Shelby R. Ergot alkaloids—sources, structures and analytical methods. Folia Microbiol. 1997;42:3–30. doi: 10.1007/BF02898641. [DOI] [PubMed] [Google Scholar]
- 8.Glenn A E, Bacon C W, Price R, Hanlin R T. Molecular phylogeny of Acremonium and its taxonomic implications. Mycologia. 1996;88:369–383. [Google Scholar]
- 9.Hubbard J C E. Effect of cutting and seed production in Spartina anglica. J Ecol. 1970;58:329–334. [Google Scholar]
- 10.Jungehülsing U, Tudzynski P. Analysis of genetic diversity in Claviceps purpurea by RAPD markers. Mycol Res. 1997;101:1–6. [Google Scholar]
- 11.Kobel H, Sanglier J J. Formation of ergotoxine alkaloids by fermentation and attempts to control their biosynthesis. FEMS Symp. 1978;5:233–242. [Google Scholar]
- 12.Kybal J, Brejcha V. Problematik der Rassen und Stämmen des Mutterkorns Claviceps purpurea Tul. Pharmazie. 1955;10:752–755. [PubMed] [Google Scholar]
- 13.Lehmann P F, Lin D, Lasker B A. Genotypic identification and characterization of species and strains within the genus Candida using random amplified polymorphic DNA. J Clin Microbiol. 1992;30:3249–3254. doi: 10.1128/jcm.30.12.3249-3254.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linder D H. Fungi. Botany of the Canadian Eastern Arctic. Part II. Thallophyta and Bryophyta. Bull Natl Mus Can. 1948;97:234–297. [Google Scholar]
- 15.Loveless A R. Conidial evidence for host restriction in C. purpurea. Trans Br Mycol Soc. 1971;56:419–434. [Google Scholar]
- 16.Loveless A R, Peach J M. Evidence for the genotypic control of spore size in C. purpurea. Trans Br Mycol Soc. 1974;63:612–615. [Google Scholar]
- 17.Maier W, Erge D, Gröger D. Über Aktivierungsreaktionen bei Claviceps. Biochem Physiol Pflanz. 1972;163:432–462. [Google Scholar]
- 18.Pažoutová S, Fučíkovský L, Leyva-Mir S G, Flieger M. Claviceps citrina sp. nov., a parasite of the halophytic grass Distichlis spicata from Mexico. Mycol Res. 1998;102:850–854. [Google Scholar]
- 19.Pažoutová S, Tudzynski P. Claviceps sp. PRL. 1980, ATCC 26245, 59 and Pepty 695/ch-I: their true story. Mycol Res. 1999;103:1044–1048. [Google Scholar]
- 20.Petch T. More about Claviceps. Naturalist (Hull) 1937;961:25–28. [Google Scholar]
- 21.Raybould A F, Gray A J, Clarke R T. The long-term epidemic of Claviceps purpurea on Spartina anglica in Poole Harbour: pattern of infection, effects on seed production and the role of Fusarium heterosporum. New Phytol. 1998;138:497–505. [Google Scholar]
- 22.Schardl C L, Liu J-S, White J F, Finkel R A, An Z, Siegel M R. Molecular phylogenetic relationships of nonpathogenic grass mycosymbionts and clavicipitaceous plant pathogens. Plant Syst Evol. 1991;178:27–41. [Google Scholar]
- 23.Sneath P H A, Sokal R R. Numerical taxonomy. W. H. San Francisco, Calif: Freeman; 1973. [Google Scholar]
- 24.Sokal R R, Rohlf F J. Biometry. 3rd ed. New York, N.Y: W. H. Freeman and Company; 1995. p. 815. [Google Scholar]
- 25.Sprague R. Diseases of cereals and grasses in North America. New York, N.Y: Ronald Press; 1950. pp. 59–67. [Google Scholar]
- 26.Stäger R. Infectionsversuche mit Gramineen-bewohnenden Claviceps-Arten. Bot Ztg. 1903;61:111–118. [Google Scholar]
- 27.Stäger R. Weitere Beiträge zur Biologie des Mutterkorns. Zentbl Bakteriol Parasitenkd Infektkrankh Hyg Abt II. 1905;14:25–32. [Google Scholar]
- 28.Stäger R. Zur Biologie des Mutterkorns. Zentbl Bakteriol Parasitenkd Infektkrankh Hyg Abt II. 1908;20:272–279. [Google Scholar]
- 29.Stäger R. Beiträge zur Verbreitungsbiologie der Claviceps-Sklerotien. Zentbl Bakteriol Parasitenkd Infektkrankh Hyg Abt II. 1922;56:329–339. [Google Scholar]
- 30.Stäger R. Impfversuchen mit dem Mutterkorn des Weizens. Mitt Naturforsch Ges Bern. 1923;1922:11–20. [Google Scholar]
- 31.Tanda S. Mycological studies on ergot in Japan. Part 9. Distinct variety of C. purpurea Tul. on Phalaris arundinacea L. and P. arundinacea var. picta L. J Agric Sci (Tokyo) 1979;24:79–95. [Google Scholar]
- 32.Tanda S. Mycological studies on the ergot in Japan. Part 14. Ergots on Agropyron spp. J Agric Sci (Tokyo) 1981;1981:85–114. [Google Scholar]
- 33.Walzel B, Riederer B, Keller U. Mechanism of alkaloid cyclopeptide synthesis in the ergot fungus Claviceps purpurea. Chem Biol. 1997;4:223–230. doi: 10.1016/s1074-5521(97)90292-1. [DOI] [PubMed] [Google Scholar]
- 34.White T J, Bruns T, Lee S, Taylor J W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 315–322. [Google Scholar]