Abstract
Cannabis is increasingly consumed by women of childbearing age, and the reproductive and epigenetic effects are unknown. The purpose of this study was to evaluate the potential epigenetic implications of cannabis use on the female ovarian follicle. Whole-genome methylation was assessed in granulosa cells from 14 matched case-control patients. Exposure status was determined by liquid chromatography–mass spectrometry (LC-MS/MS) measurements of five cannabis-derived phytocannabinoids in follicular fluid. DNA methylation was measured using the Illumina TruSeq Methyl Capture EPIC kit. Differential methylation, pathway analysis and correlation analysis were performed. We identified 3679 differentially methylated sites, with two-thirds affecting coding genes. A hotspot region on chromosome 9 was associated with two genomic features, a zinc-finger protein (ZFP37) and a long non-coding RNA (FAM225B). There were 2214 differentially methylated genomic features, 19 of which have been previously implicated in cannabis-related epigenetic modifications in other organ systems. Pathway analysis revealed enrichment in G protein-coupled receptor signaling, cellular transport, immune response and proliferation. Applying strict criteria, we identified 71 differentially methylated regions, none of which were previously annotated in this context. Finally, correlation analysis revealed 16 unique genomic features affected by cannabis use in a concentration-dependent manner. Of these, the histone methyltransferases SMYD3 and ZFP37 were hypomethylated, possibly implicating histone modifications as well. Herein, we provide the first DNA methylation profile of human granulosa cells exposed to cannabis. With cannabis increasingly legalized worldwide, further investigation into the heritability and functional consequences of these effects is critical for clinical consultation and for legalization guidelines.
Keywords: cannabis, marijuana, Δ9-THC, DNA methylation, epigenetics, granulosa cells
Introduction
Cannabis, the third most commonly used psychoactive drug by women of childbearing age (Substance Abuse and Mental Health Services Administration (SAMHSA), 2014), has been increasingly legalized worldwide (Smart et al., 2017). This has led, in turn, to increased use across all ages (Rotermann, 2019; Volkow et al., 2019). All cannabis-derived cannabinoids are collectively referred to as phytocannabinoids (PCs). Delta-9-tetrahydrocannabinol (Δ9-THC), the main psychoactive component of cannabis, is used for medicinal and recreational purposes (Sun and Dey, 2012). Other PCs include cannabidiol (CBD), a non-psychotropic PC that counteracts the psychoactive effects of Δ9-THC, and cannabinol (CBN), a byproduct of Δ9-THC degradation, as well as 11-OH-THC and 11-COOH-THC, both byproducts of Δ9-THC metabolism (ElSohly et al., 2017). Phytocannabinoids exert their actions primarily via two G protein-coupled receptors (GPCRs), cannabinoid receptors 1 and 2 (CB1R and CB2R), by acting as agonists, inverse agonists or as antagonists (Howlett, 2002; Fonseca et al., 2013; Yohn et al., 2015; Brents, 2016). Both receptors can be found in the female reproductive system and are essential for folliculogenesis, oocyte maturation and ovulation, among other reproductive functions (Wang et al., 2006; Taylor et al., 2007; Battista et al., 2008; Maccarrone, 2015; Fuchs Weizman et al., 2021).
Several clinical studies involving self-reported PC use, have shown that exposure to PCs can cause ovulatory abnormalities and impact oocyte quality and pregnancy rates amongst patients undergoing IVF (Mueller et al., 1990; Klonoff-Cohen et al., 2006; Jukic et al., 2007). In our previous study, we reported on measuring PCs in human follicular fluid utilizing liquid chromatography–mass spectrometry (LC-MS/MS), thereby paving the way for objective assessments of the impact of cannabis on female fertility (Fuchs Weizman et al., 2021).
Cannabis exposure can also cause epigenetic modifications, like other environmental agents (Yohn et al., 2015; Szutorisz and Hurd, 2016). It is established that Δ9-THC causes genome-wide histone modifications and altered DNA methylation, in brain, sperm, blood cells and, potentially, the ovarian follicle (Yang et al., 2014; Watson et al., 2015; Yohn et al., 2015; Santoroet al., 2017; Murphy et al., 2018; Levin et al., 2019; Osborne et al., 2020; Schrott et al., 2020; Fuchs Weizman et al., 2021). However, there is still a significant gap in the literature regarding the epigenetic effects of cannabis exposure on the somatic cells supporting oocyte growth and maturation. In our previous study, we showed that PCs alter the epigenetic machinery in human granulosa cells, via decreased expression of DNMT3b, a denovo methylating enzyme, which in turn led to decreased global DNA methylation, in vitro (Fuchs Weizman et al., 2021). In the current study, we aimed at (i) establishing methylation profiles of naive human granulosa cells, (ii) exploring the effects of cannabis exposure on these profiles and (iii) comparing the epigenetic consequences of cannabis exposure on human granulosa cells to those observed in other cell types.
Materials and methods
Ethical approval
This study received research ethics board (REB) approval (Veritas #16518). All subjects provided written informed consent for the donation of their biological waste material, which included the collection of follicular fluid (FF) and granulosa cells (GC) as well as de-identified clinical information, including age, BMI, ovarian reserve metrics and treatment regimens.
Sample collection
All follicular fluid (FF) samples were obtained from the CReATe Fertility BioBank (Toronto, Canada) which were biobanked from consenting patients undergoing IVF at CReATe Fertility Center (Toronto, Canada), between January 2018 and July 2019. Patients were treated using a standard antagonist protocol, with initial gonadotropin dosing and subsequent adjustments at the discretion of their treating physician. Ultrasound-guided oocyte retrieval was performed ∼36 h following trigger injection. Participants were of similar ethnic background and socioeconomic status, did not report any polysubstance abuse, and all self-identified as ‘social drinkers’. The study personnel were blinded to all clinical information pertaining to the tested samples prior to the analysis of PC concentrations.
Measurement of phytocannabinoids in follicular fluid
FF samples were assayed for phytocannabinoid levels (Δ9-THC, 11-OH-THC, 11-COOH-THC, CBD and CBN). FF was utilized to determine if the follicle was a privileged site, determine the concentration of PCs that the oocyte and GCs are exposed to, and (unlike measurement of cannabis in urine) to allow for the measurement of not only metabolites of THC (11-COOH-THC), but the active molecule as well (Δ9-THC). Measurements were performed by LC-MS/MS of the FF at the Analytical Facility for Bioactive Molecule (Hospital for Sick Children, Toronto, Canada) as previously described (Fuchs Weizman et al., 2021).
Inclusion criteria
Seven patients whose FF tested positive for one or more of the PCs were assigned to the case group. These case-patients were matched by demographic (age, BMI and ethnicity) and stimulation parameters (anti-Müllerian hormone, LH on trigger, FSH on Day 2/3 of cycle and E2 on trigger) with seven patients whose samples were negative for all measured PCs (control group). To further reduce inter-patient variability, included patients were of similar ethnic background, with the majority of patients of Caucasian/European descent, while two patients were of South Asian descent, and one patient was of Indigenous descent. No patients reported any recreational drug use, tobacco use, polysubstance use or other medication use during the initial patient intake questionnaire. Polycystic ovary syndrome (PCOS) patients were excluded from this study as PCOS has been shown to alter the methylome of cells in the follicular niche (Xu et al., 2016; Sagvekar et al., 2019). All corresponding granulosa cells (GCs) were retrieved from the CReATe Fertility BioBank (Toronto, Canada).
Sample preparation and DNA extraction
Granulosa cells from all large/dominant follicles were thawed rapidly, using a 37°C water bath, and pooled. Cells were washed in DMEM/F12 + 2.5% Fetal Bovine Serum (FBS) to remove cryoprotectants. The resulting cell pellet was resuspended, and cell number and viability were assessed using the Countess automated cell counter (ThermoFisher Scientific, Mississauga, Canada). Genomic DNA was isolated from ∼50 000 cells using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. Briefly, cells were lysed in Buffer AL and homogenized using a Disruptor Genie for 5 min. The genomic DNA was bound to the supplied column and washed using the supplied buffers. Genomic DNA was eluted in 200 µl of Buffer AE. Total genomic DNA concentration was assayed using the Qubit dsDNA HS Assay (ThermoFisher Scientific, Mississauga, Canada).
Bisulfite sequencing
Samples were sequenced at the Princess Margaret Genomics Centre (Toronto, Canada) using the Illumina TruSeq Methyl Capture EPIC kit (Illumina, Canada), according to the manufacturer’s instructions. Briefly, 500 ng of genomic DNA was sheared using a Covaris S220 sonicator (Covaris, MA, USA) to yield fragments of ∼180–220 bp. The fragmented DNA was then end-repaired, poly-A tailed and ligated with uniquely indexed adapters. These indexed libraries were hybridized twice to the EPIC capture oligos to specifically bind regions of interest. The hybridized probes were captured and purified after each hybridization using streptavidin magnetic beads. The enriched library was bisulfite converted, PCR amplified and purified by magnetic beads. Final libraries were quantified by the Qubit dsDNA HS Assay (Thermo Fisher) and quality was assessed using the 2100 Bioanalyzer High Sensitivity DNA chip (Agilent Technologies, USA). Libraries were normalized and sequenced on a NovaSeq 6000 S2 flow cell (Illumina, Canada) (paired-end 2 × 100 bp).
Targeted mRNA sequencing
A custom-designed AmpliSeq targeted panel (97 targets total) (Illumina, Canada) was designed to assess the expression of genes whose DNA methylation was either correlated with PC exposure or previously reported to be impacted by PC exposure in other studies. We also included genes that participate in the signaling pathways of cannabinoids including, cannabinoid receptors, DNMTs, GPCR signaling, Zinc homeostasis and other downstream pathways including MAPK, ERK2 and PKA signaling pathways. The 14 samples used for bisulfite sequencing plus an additional 10 samples (7 cases and 3 controls) were subjected to AmpliSeq library preparation according to manufacturer’s instructions using the AmpliSeq Library PLUS kit (Illumina, Canada). Final libraries were quantified by the Qubit dsDNA HS Assay (Thermo Fisher) and quality was assessed using the 2100 Bioanalyzer High Sensitivity DNA chip (Agilent Technologies, USA). Libraries were normalized and sequenced on a miSeq v2 flow cell (Illumina, Canada) (paired-end 2 × 150 bp).
Bioinformatics: differential methylation analysis
FASTQ files were generated using bcl2fastq2 (v2.17) and read quality was assessed using FASTQC (v0.11.8) (Andrews, 2010). Reads were trimmed using trim_galore (v0.5.0) to remove Illumina adapter sequences and low-quality bases (quality = 20, stringency = 7) (Krueger, 2012). The trimmed reads were aligned to Human Genome Assembly 38 (hg38) using bismark (v0.22.1) and bismark_methylation_extractor was used to extract the methylation call for each cytosine base (Krueger and Andrews, 2011). MethylKit (v1.10.0) was used for differential methylation analysis (Akalin et al., 2012). Bases with coverage of <10 in each of the samples were discarded from further analysis. Samples were clustered based on the similarity of their methylation profiles using hierarchical clustering and principal component analysis. CpG were annotated to genic annotations (enhancer, 1–5 kb upstream of the Transcription Start Site (TSS), promoter, exon/3′UTR, intron and intergenic) using the annotation package (v3.12) (Cavalcante and Sartor, 2017). Differentially methylated sites (DMSs) were defined as having a percent methylation difference >25% and an adjusted P-value (q-value) <0.01. Differentially methylated regions (DMRs) were defined as regions of the genome containing at least three CpG with a concordant (either hypo- or hypermethylated) mean methylation difference >25% between cases and controls, within a 1000-bp interval (Watson et al., 2015).
Pathway analysis
Gene set enrichment analysis (GSEA) was conducted on hypo- and hyper-methylated DMS individually, using g:Profiler-g:GOSt (KEGG, Reactome and Wikipathways) with the g:SCS multiple testing correction method, applying a significance threshold of 0.05 (q-value), to determine the effect that all DMS have on cellular processes and functions (Raudvere et al., 2019). Genes that could not be mapped to any gene-set term were excluded from the comparison. Gene sets with fewer than five genes and a q-value >0.05 were excluded from further analysis.
RNA sequencing analysis
Bioinformatics were conducted using Partek Flow. Briefly, reads were trimmed to remove low-quality bases (Phred score < 25), aligned to hg19 using STAR (v2.5.3a) and quantified to the annotation model RefSeq Transcripts 99 (Dobin et al., 2013). Samples were normalized using trimmed mean of M-values (TMM) and differential expression was conducted using DESeq2 comparing cases to controls (Robinson and Oshlack, 2010; Love et al., 2014). Differentially expressed genes were defined as having a fold change (FC) −2 > FC > 2 and a false discovery rate <0.05.
Data analysis and literature review
We conducted a literature review of the PubMed database for previous studies assessing the effects of cannabis on DNA methylation in any cell type using a variety of methods (i.e. qPCR, microarray, whole-genome/reduced representation sequencing or targeted sequencing), and cross-referenced the findings with significant DMS within DMR in our analysis. The associated features were further explored in depth using the Ovarian Kaleidoscope Database (Leo and Hsueh, 2000) and GeneCards Human Gene database (http://www.genecards.org/) to correlate our bioinformatic findings with hallmark physiological and pathological processes in the ovary.
Statistical analysis
Mean and SE were utilized for continuous variables. Student’s t-test or Fisher’s exact tests were used to assess statistical significance. Correlational analysis was used to assess the relationship between PC measures and methylation. Normality was measured using skew and kurtosis, and Pearson or Spearman correlations were used, as appropriate. To control for multiple comparisons, P values from 0.04 to 0.055 were considered a trend, and values below P = 0.04 were considered significant. Specific tests used are indicated in all figure and table legends.
Results
Fourteen patients were included in this matched case-control study: seven cases and seven controls. Patient demographics and clinical characteristics are presented in Table I. Cases and controls did not differ significantly in the number of sequencing reads, the number of cytosines analyzed or the average methylation percentage. Overall, there were 3679 DMSs in this study, of which 1741 were hypermethylated, (47.3%) and 1938 were hypomethylated (52.7%) in cases compared with controls. (Supplementary Table SI).
Table I.
Patient demographics.
| Case (n = 7) | Control (n = 7) | P-value | |
|---|---|---|---|
| Δ9-THC (nM) | 37.7 ± 9.2 (8.1–84.8) | — | — |
| 11-OH-THC (nM) | 6.3 ± 1.9 (0–12.3) | — | — |
| 11-COOH-THC (nM) | 70.8 ± 16.3 (4.5–143.6) | — | — |
| Age (years) | 31.1 ± 2.0 (23–40) | 29.4 ± 1.7 (23–36) | 0.554 |
| BMI (kg/m2) | 24.2 ± 1.7 (19–32.6) | 26.7 ± 2.2 (20.2–38.4) | 0.413 |
| AMH (pmol/l) | 20.2 ± 2.0 (14.5–29.6) | 23.0 ± 4.1 (11.2–38.6) | 0.601 |
| LH on Trigger (IU/ml) | 3.0 ± 0.7 (0.7–5.4) | 1.9 ± 0.5 (0.3–4.0) | 0.307 |
| FSH on Day 2/3 (mIU/ml) | 6.7 ± 0.3 (5.7–8.3) | 6.0 ± 0.4 (4.7–7.5) | 0.291 |
| E2 on Trigger (pmol/l) | 10 698.4 ± 2295.0 (6106–24 930) | 11 351.0 ± 1350.6 (6279–16 225) | 0.853 |
Data presented as Mean±SEM(Range). THC, Tetrahydrocannabinol; AMH, anti-Müllerian hormone; E2, estradiol; Trigger, day of ovulation trigger.
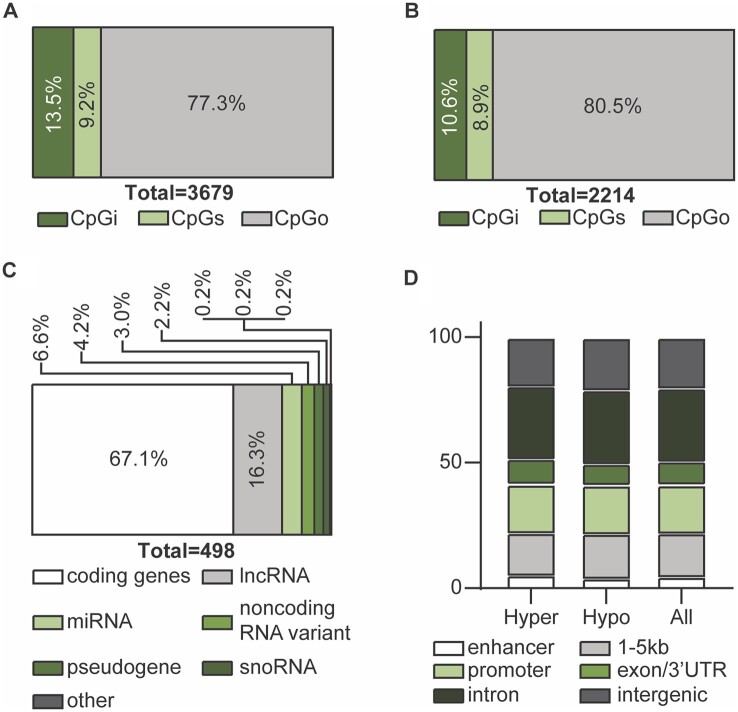
Of all DMS, 497 were in CpG islands (CpGi), 339 were in CpG shores (CpGs) and 2843 were not associated with either CpGi or CpGs (CpG other, CpGo) (Fig. 1A and B) (Supplementary Table SI). The proportion of DMS, in each of the sublocations, is presented in Fig. 1A, and the proportion of unique genomic features, in the same sublocations, is presented in Fig. 1B. Of DMS within ±5kb of the transcription start site, two-thirds of the features were coding genes, whereas a third were non-coding genes, potentially involved in regulatory functions (Fig. 1C). There was no difference in the distribution of genic annotations between the hypo- and the hyper-methylated DMS (Fig. 1D).
Figure 1.
Localization and genomic feature annotation associated with differentially methylated sites (DMS) in case versus control groups. (A) Of the 3679 DMS, 497 were associated with CpG islands (CpGi) (13.5%), 339 were associated with CpG shores (CpGs) (9.2%) and 2843 were not associated with either CpGi or CpGs and deemed CpGother (CpGo) (77.3%). (B) Of the 2214 unique genomic features that the DMS mapped to, 251 were associated with CpGi (10.6%), 210 were associated with CpGs (8.9%) and 1900 were not associated with either and deemed CpGo (80.5%). (C) Stratification of unique genomic feature biotypes of DMS within ±5 kb of the transcription start site. (D) Proportions of DMS in hypo-, hyper-methylated and all DMS associated with specific genic annotation using the ‘annotatr’ package.
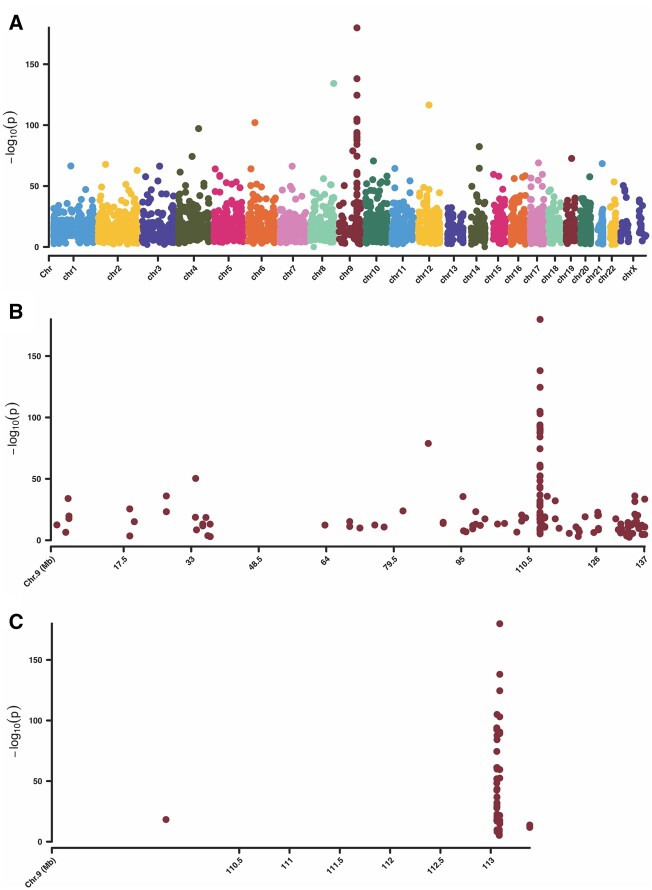
Figure 2A shows a Manhattan plot depicting all CpGs that were identified in this study. A 0.29 Mb region of chromosome 9 (chr9:113059999-113089725) was enriched in DMS (n = 48), driven by two genomic features; ZFP37 and the lncRNA FAM225B (Fig. 2B and C).
Figure 2.
Manhattan plots of the genome-wide differentially methylated sites (DMS) found in the case versus control group. All sites on these plots are significant. Significance is defined as percent methylation difference >25% and an adjusted P-value (q-value) <0.01. The y-axis represents the −log10(adjusted P). (A) Manhattan plot of all autosomes and the X chromosome. (B) Manhattan plot of only chromosome 9, depicting the ‘hotspot’ region on the distal q arm. (C) Manhattan plot of only the chromosome 9 ‘hotspot’ region ranging from chr9:113059999-113089725.
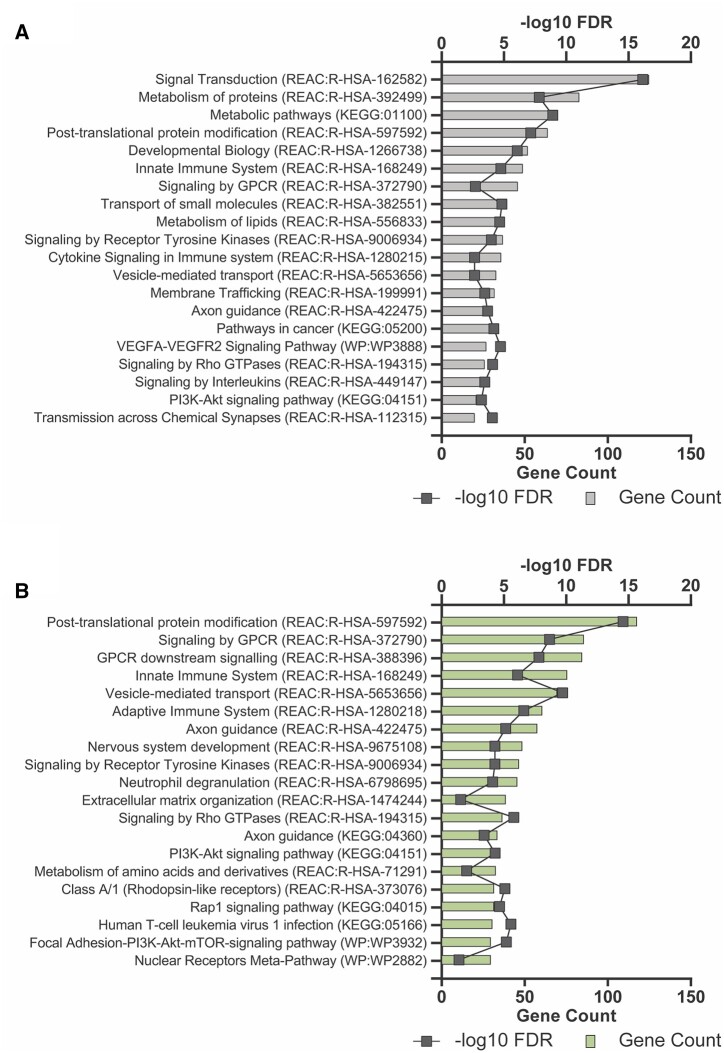
A literature review revealed 19 genomic features (22 DMS) from the current study, which have already been previously annotated in the context of cannabis exposure and epigenetic modifications in human and animal studies exploring its effects in different organ systems (Table II). This study has also identified 2195 differentially methylated features that have not been annotated to date in the context of epigenetic effects attributed to cannabis exposure and are outlined in Supplementary Table SI. Pathway analyses were conducted on the genomic features which were associated with DMS using KEGG, Reactome and Wikipathways using g:Profiler. The hypomethylated DMS (n = 1161) showed enrichment in small molecule transport, innate immune response and developmental biology with the most enriched pathways being involved in signal transduction and metabolism (Fig. 3A, Supplementary Table SII). The hypermethylated DMS (n = 1227) were enriched in similar pathways overall; however, the top pathways were GPCR signaling and immune response (Fig. 3B, Supplementary Table SIII).
Table II.
Differentially methylated sites (DMS) in our study that were also previously reported to be affected by Δ9-THC exposure in other models and organs/systems.
| Current study |
Previous literature |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CpGi |
CpGs |
CpGo |
||||||||||
| Description | Main function | % methylation difference | q value | % methylation difference | q value | % methylation difference | q value | Species | System | Method | Source | |
| TP73 | Tumour protein P73 | Tumour suppression | 31.67 | 1.23E−05 | — | — | — | — | Human | Blood | EPIC array | Osborne et al. (2020) |
| ETV2 | ETS variant transcription factor 2 | Transcription factor | 26.12 | 1.36E−11 | — | — | — | — | Human | Blood | EPIC array | Osborne et al. (2020) |
| KLHL30 | Kelch like family member 30 | Ubiquitin-protein transferase | −45.44 | 5.92E−29 | — | — | — | — | Human and rat | Sperm | RRBS | Murphy et al. (2018) |
| GAS6-AS1 | GAS6 antisense RNA 1 | ncRNA | 25.49 | 1.10E −05 | — | — | — | — | Human and rat | Sperm | RRBS | Murphy et al. (2018) |
| PRKCZ | Protein kinase C zeta | Cellular differentiation and proliferation | 26.9 | 3.22E−05 | 25.78 | 3.31E−11 | — | — | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| PRKCZ | Protein kinase C zeta | Cellular differentiation and proliferation | −38.11 | 4.36E−15 | — | — | — | — | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| MGAT3 | Beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase | Biosynthesis of glycoprotein oligosaccharides | 27.21 | 5.53E−10 | — | — | — | — | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| LSP1 | Lymphocyte specific protein 1 | Regulation of immune cell chemotaxis and adhesion | — | — | −25.08 | 2.13E−05 | 44.55 | 8.48E−30 | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| ABR | ABR activator of RhoGEF and GTPase | Regulation of macrophage motility and function | — | — | −29.83 | 8.36E−10 | — | — | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| KCNMA1 | Potassium calcium-activated channel subfamily M alpha 1 | Regulation of smooth muscle tone and neuronal excitability | — | — | −32.17 | 1.04E−22 | — | — | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| LZTS2 | Leucine zipper tumour suppressor 2 | Tumour suppression | — | — | — | — | −29.95 | 1.85E−12 | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| SMYD3 | SET and MYND domain containing 3 | Histone methyltransferase | — | — | — | — | −50.22 | 4.77E−14 | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| SMYD3 | SET and MYND domain containing 3 | Histone methyltransferase | — | — | — | — | −30.58 | 6.65E−28 | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| SMYD3 | SET and MYND domain containing 3 | Histone methyltransferase | — | — | — | — | −33.76 | 2.07E−13 | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| RFC1 | Replication factor C subunit 1 | Transcription, DNA replication and repair | — | — | — | — | −27.56 | 3.72E−10 | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| ABHD8 | Abhydrolase domain containing 8 | Hydrolase activity | — | — | — | — | −27.48 | 3.31E−17 | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| GRXCR2 | Glutaredoxin and cysteine rich domain containing 2 | Protein S-glutathionylation | — | — | — | — | 30.49 | 1.37E−14 | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| TOX2 | TOX high mobility group box family member 2 | Transcription factor | — | — | — | — | −25.11 | 1.10E−09 | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| MFAP2 | Microfibril-associated protein 2 | Microfibrils component | — | — | — | — | −31.23 | 3.21E−09 | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| EPHA2 | EPH receptor A2 | Regulates apoptosis, chemotactic cell migration, adhesion, proliferation, apoptosis and angiogenesis | — | — | — | — | −26.13 | 4.65E−15 | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| MPPED1 | Metallophosphoesterase domain containing 1 | Metallophosphoesterase | — | — | — | — | 25.09 | 3.56E−17 | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
| AFF3 | AF4/FMR2 family member 3 | Transcription factor | — | — | — | — | 25.49 | 1.31E−07 | Rat | Nucleus accumbens | ERRBS | Watson et al. (2015) |
CpGi, CpG islands; CpGs, CpG shores; CpGo, CpG other (non-island and non-shore); RRBS, reduced-representation bisulfite sequencing; ERRBS, enhanced reduced-representation bisulfite sequencing.
Figure 3.
Gene set enrichment analysis of differentially methylated sites (DMS). Gene set enrichment analysis (GSEA) was conducted using g:Profiler-g:GOSt and interrogating KEGG, Reactome and Wikipathways databases. Genesets were considered significant if they had an adjusted P-value (q-value) <0.05 and five or more genes in the gene set. g:SCS multiple testing correction method was used to determine significance. (A) GSEA of all hypomethylated DMS. (B) GSEA of all hypermethylated DMS. A full list of gene sets can be found in Supplementary Tables SII and SIII, respectively.
In this study, we identified 71 DMR (Supplementary Table SIV). None of the associated features were previously annotated in the context of cannabis exposure and epigenetic modifications. The top 30 DMR are presented in Table III. Of these, 12 are associated with ncRNAs, 6 with enzymes involved in biosynthesis, 5 with regulatory proteins, 4 with adhesion molecules and 3 are associated with genes encoding zinc finger proteins.
Table III.
The top 30 differentially methylated regions (DMR).
| Location | DMR state | Size (bp) | Significant CpG | q value | Mean % methylation difference | Max % methylation difference | Genomic feature | Description | Main function | annotar Genic annotation |
|---|---|---|---|---|---|---|---|---|---|---|
| chr10:1097887-1098690 | Hypo | 803 | 3 | 6.21E−38 | −50.42 | −53.77 | IDI1 | Isopentenyl-diphosphate delta isomerase 1 | Cholesterol synthesis | Intron |
| chr12:32792786-32792901 | Hypo | 115 | 4 | 3.85E−30 | −52.45 | −56.30 | YARS2 | Tyrosyl-TRNA synthetase 2 | Mitochondrial translation | Exon and promoter |
| chr10:133286049-133286495 | Hyper | 446 | 3 | 8.07E−29 | 31.55 | 35.37 | ADAM8 | ADAM metallopeptidase domain 8 | Adhesion | 1 to 5 kb |
| chrX:115888618-115888639 | Hyper | 21 | 5 | 3.20E−26 | 28.49 | 36.23 | DANT1 | DXZ4-associated non-coding transcript 1, proximal | ncRNA | Intron |
| chr19:52596457-52596505 | Hypo | 48 | 3 | 1.26E−23 | −39.64 | −43.79 | ZNF137P | Zinc finger protein 137, pseudogene | ncRNA | Exon |
| chr9:134838539-134839110 | Hypo | 571 | 3 | 2.16E−22 | −38.96 | −43.99 | MIR3689C | MicroRNA 3689c | ncRNA | Intron |
| chr10:80450825-80450885 | Hypo | 60 | 3 | 4.37E−20 | −36.08 | −37.80 | TSPAN14 | Tetraspanin 14 | Innate immunity-MMP trafficking | 1 to 5 kb |
| chr9:113059999-113060198 | Hypo | 199 | 9 | 1.84E−19 | −28.01 | −33.12 | ZFP37 | ZFP37 zinc finger protein | Zinc-finger | 1 to 5 kb |
| chr11:71587803-71588521 | Hyper | 718 | 3 | 3.19E−19 | 34.87 | 37.96 | KRTAP5-11 | Keratin-associated protein 5-11 | Adhesion | 1 to 5 kb |
| chr9:113061674-113061745 | Hypo | 71 | 8 | 1.26E−18 | −33.09 | −41.29 | ZFP37 | ZFP37 zinc finger protein | Zinc-finger | 1 to 5 kb |
| chr18:112336-112400 | Hypo | 64 | 4 | 1.08E−17 | −26.71 | −28.20 | MIR8078 | MicroRNA 8078 | ncRNA | Promoter |
| chr20:61389938-61390521 | Hyper | 583 | 3 | 3.83E−17 | 35.53 | 37.18 | CDH4 | Cadherin 4 | Adhesion | Intron |
| chr20:56987407-56988020 | Hypo | 613 | 3 | 8.94E−17 | −33.66 | −35.22 | BMP7-AS1 | BMP7 antisense RNA 1 | ncRNA | Intergenic |
| chr9:113089352-113089725 | Hypo | 373 | 12 | 1.68E−16 | −28.25 | −33.86 | FAM225B | Family with sequence similarity 225 member B | ncRNA | 1 to 5 kb |
| chr4:2399815-2400293 | Hypo | 478 | 5 | 1.81E−16 | −29.51 | −34.68 | ZFYVE28 | Zinc finger FYVE-type containing 28 | EGFR signaling | Intron |
| chrX:126472512-126472534 | Hypo | 22 | 4 | 2.48E−16 | −41.81 | −44.40 | DCAF12L1 | DDB1 and CUL4-associated factor 12 like 1 | Protein complex formation | Enhancer |
| chr4:521699-522056 | Hyper | 357 | 3 | 5.34E−16 | 31.90 | 33.03 | PIGG | Phosphatidylinositol glycan anchor biosynthesis class G | Post-translational modification | Promoter |
| chr2:130037205-130038141 | Hypo | 936 | 6 | 4.38E−15 | −29.68 | −36.44 | FAR2P1 | Fatty Acyl-CoA reductase 2 pseudogene 1 | ncRNA | 1 to 5 kb |
| chr5:160186362-160186495 | Hyper | 133 | 3 | 5.50E−15 | 40.74 | 46.77 | FABP6 | Fatty acid binding protein 6 | Bile synthesis | Promoter |
| chr2:241666232-241666548 | Hyper | 316 | 3 | 3.57E−14 | 38.93 | 44.52 | DTYMK | Deoxythymidylate kinase | Pyrimidine synthesis | Promoter |
| chr8:134466004-134466775 | Hyper | 771 | 3 | 2.92E−13 | 31.49 | 34.94 | ZFAT-AS1 | ZFAT antisense RNA 1 | ncRNA | Intergenic |
| chr4:1050997-1051032 | Hyper | 35 | 3 | 3.14E−13 | 26.54 | 27.11 | FGFRL1 | Fibroblast growth factor receptor like 1 | Cell proliferation | Intergenic |
| chr6:52995723-52995825 | Hyper | 102 | 12 | 6.59E−13 | 34.20 | 39.77 | RN7SK | RNA component of 7SK nuclear ribonucleoprotein | ncRNA | Promoter |
| chr9:35657904-35657916 | Hyper | 12 | 3 | 1.26E−12 | 28.11 | 29.91 | RMRP | RNA component of mitochondrial RNA processing endoribonuclease | Mitochondrial RNA processing | Promoter |
| chr20:63074755-63075677 | Hypo | 922 | 4 | 1.87E−12 | −37.56 | −42.54 | HAR1A | Highly accelerated region 1A | ncRNA | Intron |
| chr4:536425-537351 | Hyper | 926 | 3 | 2.47E−12 | 33.85 | 44.09 | PIGG | Phosphatidylinositol glycan anchor biosynthesis class G | Post-translational modification | Exon |
| chr17:3981605-3981676 | Hypo | 71 | 4 | 5.95E−12 | −40.27 | −49.37 | LINC01975 | Long intergenic non-protein coding RNA 1975 | ncRNA | Exon |
| chr12:131434953-131435002 | Hyper | 49 | 4 | 1.38E−11 | 35.30 | 36.11 | LINC02370 | Long intergenic non-protein coding RNA 2370 | ncRNA | 1 to 5 kb |
| chr9:113062049-113062461 | Hypo | 412 | 12 | 1.48E−11 | −28.54 | −34.96 | ZFP37 | ZFP37 zinc finger protein | Zinc-finger | 1 to 5 kb |
| chr17:7148150-7148200 | Hypo | 50 | 4 | 2.28E−11 | −35.39 | −41.17 | ASGR1 | Asialoglycoprotein receptor 1 | Glycoprotein homeostasis | Intergenic |
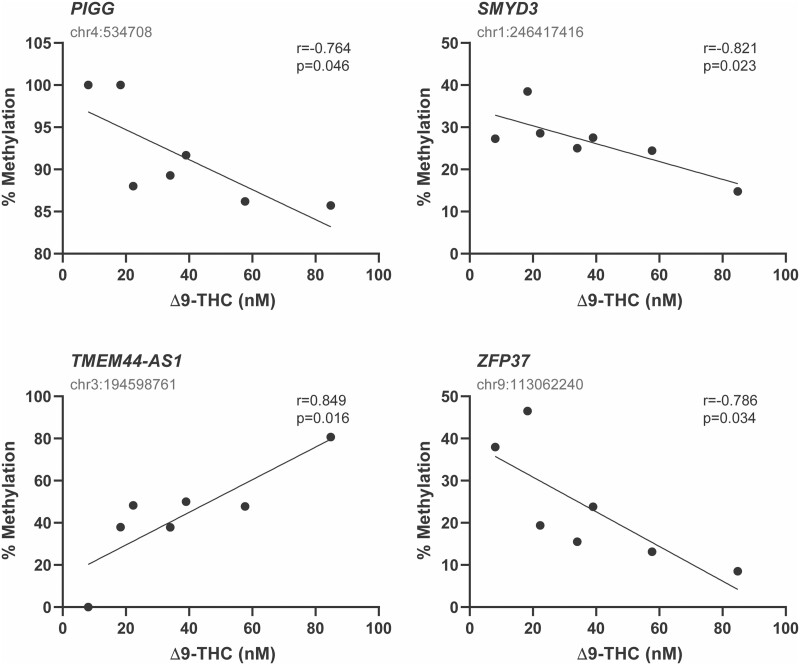
To investigate whether there is a concentration-dependent effect of cannabis, we correlated the concentration of either Δ9-THC or 11-COOH-THC in the case group, with the % methylation of all DMS (n = 421) within DMR identified in this study (n = 71), and with features that were previously reported to be affected by cannabis (n = 19). We identified 25 DMS which correlated with Δ9-THC levels (18 were significant, P < 0.04; 7 were trending, 0.055 > P > 0.04), and 16 DMS that correlated with 11-COOH-THC levels (14 were significant, P < 0.04; 2 were trending, 0.055 > P > 0.04). Together, these DMS correspond to 16 unique genomic features (Table IV), some of which had multiple significantly correlated DMS (FTCD, MLLT10P1 and ZFP37). Figure 4 depicts examples of four genes that were found to be differentially methylated in a concentration-dependent manner.
Table IV.
Correlation with phytocannabinoids (PC) concentration and DNA methylation.
| Location | Feature name | Family | Function | Identified in previous study | THC correlation coefficient | P value | THC-COOH correlation coefficient | P value |
|---|---|---|---|---|---|---|---|---|
| chr17:1028783 | ABR | Rho family of GTP-binding proteins | Regulates neuronal and macrophage activity | Yes | −0.807 | 0.03 | −0.807 | 0.03 |
| chrX:126472512 | DCAF12L1 | WD repeat protein family | Coordinating multi-protein complex assemblies | No | −0.821 | 0.02 | − | − |
| chr1:232142881 | DISC1-IT1 | lncRNA | Unknown | No | −0.818 | 0.03 | − | − |
| chr21:46161446 | FTCD | Transferase enzyme | Folate-dependent enzyme | No | − | − | 0.760 | 0.05 |
| chr21:46161478 | FTCD | Transferase enzyme | Folate-dependent enzyme | No | − | − | 0.794 | 0.03 |
| chr21:46161584 | FTCD | Transferase enzyme | Folate-dependent enzyme | No | 0.793 | 0.03 | 0.803 | 0.03 |
| chr21:46161499 | FTCD | Transferase enzyme | Folate-dependent enzyme | No | 0.881 | 0.01 | 0.847 | 0.02 |
| chr21:46161497 | FTCD | Transferase enzyme | Folate-dependent enzyme | No | 0.858 | 0.01 | 0.902 | 0.01 |
| chr21:46161492 | FTCD | Transferase enzyme | Folate-dependent enzyme | No | 0.870 | 0.01 | 0.930 | 0.002 |
| chr21:46161556 | FTCD | Transferase enzyme | Folate-dependent enzyme | No | 0.903 | 0.01 | 0.961 | 0.001 |
| chr21:46161714 | FTCD | Transferase enzyme | Folate-dependent enzyme | No | 0.648 | 0.01 | − | − |
| chr21:46161540 | FTCD | Transferase enzyme | Folate-dependent enzyme | No | 0.745 | 0.06 | − | − |
| chr21:46161526 | FTCD | Transferase enzyme | Folate-dependent enzyme | No | 0.746 | 0.05 | − | − |
| chr21:46161528 | FTCD | Transferase enzyme | Folate-dependent enzyme | No | 0.781 | 0.04 | − | − |
| chr21:46161115 | FTCD | Transferase enzyme | Folate-dependent enzyme | No | 0.784 | 0.04 | − | − |
| chr10:77640334 | KCNMA1 | Ion channel | Control of smooth muscle tone | Yes | − | − | 0.785 | 0.04 |
| chr12:52317764 | KRT83 | Keratin gene family | Structural molecule | No | −0.859 | 0.01 | −0.903 | 0.01 |
| chr22:50085800 | MLC1 | Predicted integral membrane transporter | Unknown | No | − | − | −0.792 | 0.03 |
| chr20:30585672 | MLLT10P1 | Pseudogene | Unknown | No | −0.753 | 0.05 | −0.881 | 0.03 |
| chr20:30585291 | MLLT10P1 | Pseudogene | Unknown | No | − | − | −0.750 | 0.05 |
| chr20:30586485 | MLLT10P1 | Pseudogene | Unknown | No | −0.809 | 0.03 | − | − |
| chr21:32300477 | MRAP | MRAP family | Signaling in response to corticotropin | No | −0.867 | 0.01 | − | − |
| chr4:534708 | PIGG | Transferase enzyme | post translational modification | No | −0.764 | 0.05 | − | − |
| chr1:246417416 | SMYD3 | Histone methyltransferase | H3K4 di- and tri- methylation | Yes | −0.821 | 0.02 | − | − |
| chr21:46185079 | SPATC1L | Speriolin family | Centrosome component | No | −0.757 | 0.05 | − | − |
| chr2:94871757 | TEKT4 | Tektin family | Structural component of the flagellum | No | − | − | 0.783 | 0.04 |
| chr3:194598761 | TMEM44-AS1 | lncRNA | Unknown | No | 0.849 | 0.02 | 0.894 | 0.01 |
| chr8:134466775 | ZFAT-AS1 | lncRNA | Unknown | No | 0.929 | 0.003 | 0.929 | 0.003 |
| chr9:113062448 | ZFP37 | Zinc finger | Transcriptional regulation | No | −0.788 | 0.04 | − | − |
| chr9:113062240 | ZFP37 | Zinc finger | Transcriptional regulation | No | −0.786 | 0.04 | − | − |
Figure 4.
Correlation analysis of differentially methylated regions (DMR) and cannabis usage. Correlations between follicular fluid Δ9-THC concentration in the cannabis group and granulosa cell DNA methylation levels for DMS identified as differentially methylated, analyzed by Pearson correlation. To control for multiple comparisons, P values from 0.04 to 0.055 were considered a trend, and values below P = 0.04 were considered significant. All DMS displaying a concentration-dependent effect can be found in Table IV.
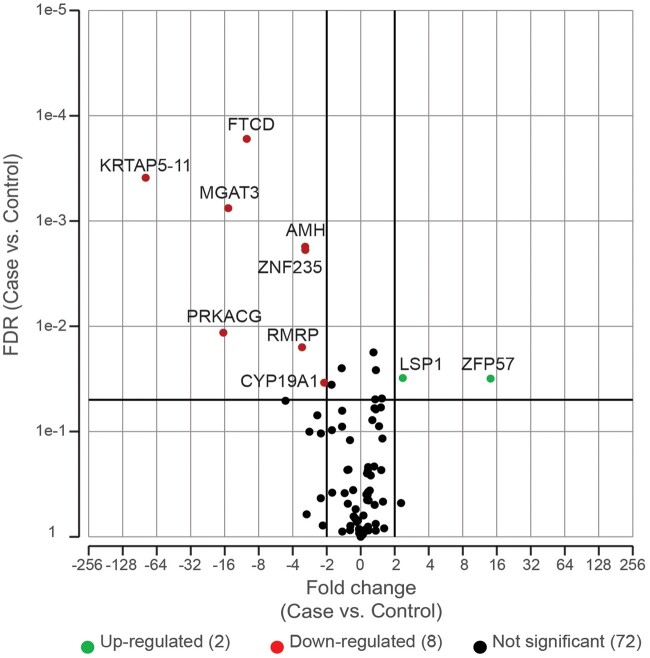
Finally, to determine if there is an effect of cannabis on the transcription of genes differentially methylated in this study, genes previously reported to be affected by cannabis exposure in other tissues, or genes involved in downstream signaling in response to cannabis exposure, we performed targeted RNA sequencing (RNASeq) on 97 genes of interest. Of these, 82 genes were detected, and 10 were differentially expressed between cases and controls (8 downregulated, and 2 upregulated). Of note, the promoters of both upregulated genes (LSP1 and ZFP57) were significantly hypomethylated in the cases, and promoters of four downregulated genes (FTCD, KRTAP5-11, MGAT3 and RMRP) were significantly hypermethylated in the cases. Two genes critical for GC function, AMH and CYP19A1, were significantly downregulated, -3.10 and -2.11, respectively. Neither of these genes were differentially methylated in this study. We did not detect any alterations to the expression of genes involved in GPCR signaling or the downstream ERK1/2 or AKT signaling pathways. PRKACG, a component of the PKA signaling pathway, was significantly downregulated in cases when compared with the control samples, however, this gene was not identified as differentially methylated in this study (Fig. 5 and Supplementary Table SV).
Figure 5.
Volcano plot of targeted RNASeq of cases versus controls. Target RNAseq was conducted on 24 samples (14 cases and 10 controls) using a custom designed ampliseq target panel. Differential expression was conducted using DESeq2. Genes were considered differentially expressed with a fold change (FC) 2 < FC < −2.
Discussion
Cannabis-related research led to the discovery of the endocannabinoid system (Maccarrone, 2015). It is well established that endocannabinoids are important for reproduction-related functions in both males and females (Wang et al., 2006; Taylor et al., 2007; Battista et al., 2015; Maccarrone, 2015). The question of whether the presence of phytocannabinoids (cannabis derivatives) disrupt the reproductive functions of endocannabinoids, remains unanswered. However, it has been shown in non-human models and human studies that phytocannabinoids can exert epigenetic effects (Yang et al., 2014; Watson et al., 2015; Yohn et al., 2015; Santoro et al., 2017; Murphy et al., 2018; Levin et al., 2019; Osborne et al., 2020; Schrott et al., 2020; Fuchs Weizman et al., 2021). With increased legalization of cannabis worldwide, cannabis use is on the rise, as previously reported (Rotermann, 2019; Fuchs Weizman et al., 2021). Hence, this study was conducted to explore the potential epigenetic effects of phytocannabinoids (PCs) on the human follicular environment surrounding the growing oocyte.
Herein, we provide the first comprehensive DNA methylation profile of human granulosa cells from patients who have used cannabis and investigate how phytocannabinoids affect DNA methylation. We measured follicular fluid PC concentrations to better understand the effect that PCs have on the immediate environment of granulosa cells and the resulting changes to the methylome of these cells. This study utilized biobanked samples, and as such, data regarding the actual amount of cannabis used was not available and data regarding concomitant drug or medication exposure was limited. It has been established that the pharmacokinetics of PCs are affected by mode of ingestion, and reflect the time that has passed from ingestion, and we have previously elaborated on this topic (Fuchs Weizman et al., 2021). The presence of Δ9-THC in the FF in most patients indicates recent exposure to cannabis (within 24–48 h) as levels in the plasma drop below the limit of detection following this time (Skopp et al., 2003; Balikova et al., 2014). By measuring FF-PC concentrations, we were able to bypass the above hurdles and highlight the effect this concentration has on the methylome of the immediate environment of the follicular niche.
By virtue of our matched case-control study design, we were able to minimize the biologic variability that could potentially affect the epigenome in question. Furthermore, since PCOS is the only condition that has been shown, to date, to affect the follicular niche methylome we chose to exclude PCOS patients from this dataset to control for the confounding effects this diagnosis may have (Xu et al., 2016; Sagvekar et al., 2019).
Of the differentially methylated sites (DMS) in this study, 47% were hypermethylated and 53% were hypomethylated, similarly to a previous report (Watson et al., 2015). There was no difference in the distribution of genic annotations between hypo- and hyper-methylated DMS, with the largest proportion being either within a confirmed promoter or within a predicted promoter (within 5 kb of the TSS), followed by DMS located in the introns and then intergenic genes. Two-thirds of affected features were coding genes, whereas a third were non-coding regions and potentially involved in regulatory functions.
A striking feature of our bioinformatic analysis was a ‘hotspot’ of differential methylation in a 0.29 Mb region on chromosome 9, which was enriched in 48 DMS. Looking closely, we observed the region to be highly dense with CpGi. This, in turn, affected two genomic features, the zinc finger protein ZFP37, and the long non-coding RNA (lncRNA), FAM225B. ZFP37 is known to be highly expressed in the ovary, second only to its expression in the central nervous system (GTEx Consortium, 2013). In their study, Watson et al. (2015) found eight different ZFPs to be differentially methylated in the rat nucleus accumbens following exposure to Δ9-THC, and ZFPs were also in their top five DMR enrichment analysis. In mice ZFP37 has been identified as a transcript that is enriched in growing/developing follicles and may be a marker of oocyte potential, however its mechanism of action and specific targets have yet to be elucidated (Herrera et al., 2005). Furthermore, the KRAB-ZFP family, of which ZFP37 is a member, has been proposed to mediate histone deacetylation and H3-K9 trimethylation (H3-K9-me3) through the formation of a potent epigenetic silencing complex with KAP1. This complex recruits histone deacetylases and methyltransferases, resulting in the formation of heterochromatin and repression of genes expression. KRAB-ZFPs have been implicated in several biological processes in humans, including protection of imprinting control regions in mice and human embryonic stem cells (hESC) (Juan and Bartolomei, 2019; Takahashi et al., 2019), retroelement silencing in hESC (Rowe et al., 2010; Pontis et al., 2019), and carcinogenesis (Machnik et al., 2019; Cylwa et al., 2020; Sobocinska et al., 2021). Notably, Juknat et al. (2012) suggested that cannabis affects zinc homeostasis which could potentially explain the antioxidant and anti-inflammatory effects of CBD and, to a lesser extent, that of Δ9-THC. Further investigation into ZFP37 and other ZPFs identified in this study is ongoing, specifically with the aim to identify their binding partners and the genomic features they are associated with. This information will be the first step in understanding their mode of action in GCs and how cannabis may be altering their function.
FAM225B belongs to the lncRNA family and, as such, it is difficult to predict by which mechanism it exerts its regulatory function (Yang et al., 2016). However, previous studies have shown it to be involved in Wnt signaling and cell cycle regulation (Li et al., 2020). Our findings point to the need for further exploration of this potentially enhanced sensitivity of ZFPs and lncRNAs to epigenetic changes caused by cannabis derivatives.
Of all differentially methylated genomic features identified in the current study, 19 have been previously annotated in the context of cannabis exposure and epigenetic modifications. Of these, four either activate transcription or support it (AFF3, ETV2, RFC1 and TOX2), four regulate cell proliferation and apoptosis (EPHA2, LZTS2, PRKCZ and TP73), three are involved in post-translational modifications (GRXCR2, KLHL30 and MGAT3), and three are involved in extracellular matrix remodeling (ABHD8, MFAP2 and MPPED) (Table II). Notably, two exert their actions primarily in immune cells (ABR and LSP1) and one is a histone methyltransferase (SMYD3) (Table II). Interestingly, we were also able to demonstrate a concentration-related effect of cannabis on three of the differentially methylated genomic features from our study that were annotated previously in the context of cannabis-associated DNA differential methylation: ABR, known to regulate macrophage actions, was negatively correlated with Δ9-THC concentration; KCNMA1, which encodes for a subunit of potassium channels and plays a key role in controlling excitability in several systems, was positively correlated with Δ9-THC concentration; and SMYD3, which associates with the RNA polymerase complex, was negatively correlated with Δ9-THC concentrations. SMYD3 is a histone methyltransferase, and plays an important role in transcriptional activation. In bovine oocytes, SMYD3 has been shown to regulate transcription during oocyte maturation and early embryonic development (Bai et al., 2016). In fact, the knockdown of SMYD3 expression in GV oocytes causes a significant reduction in NANOG expression resulting in embryos arresting at the 4- to 8-cell stage (Bai et al., 2016). Furthermore, SMYD3 targets not only histones (H3-K4, H4-K5 and H2A.Z.1-K101), but non-histone proteins as well (VEGFR1, MAP3K2, AKT1 and HER2) which, in turn, stimulate angiogenesis and cell growth and survival and are implicated in carcinogenesis (Bottino et al., 2020). Importantly, this is the second gene, along with ZFP37, capable of either directly or indirectly altering histone acetylation/methylation, further implicating cannabis exposure in histone modification alterations in addition to changes in DNA methylation patterns. Further investigation into the impact cannabis has on histone modifications is ongoing.
The current study also identified 2195 novel DMS, in the context of cannabis and epigenetic modifications, which adds significantly to the existing literature and supports future research. Pathway analyses, based on these DMS, portrayed enrichment in cell signaling and metabolism amongst hypomethylated sites, and enriched GPCR signaling and immune response amongst hypermethylated sites. The pathways enriched in DMS are consistent with previous literature investigating the effect cannabis has on the sperm methylome including Hippo signaling, pathways in cancer and MAPK signaling (Murphy et al., 2018).
To elaborate on a key pathway identified, we observed significant hypermethylation of genomic features involved in GPCR signaling. Phytocannabinoids and endocannabinoids exert their effects through GPCRs, namely CB1R and CB2R (Howlett, 2002; Fonseca et al., 2013; Yohn et al., 2015; Brents, 2016). Cecconi et al. (2019) have established that blocking these receptors causes cAMP levels to drop and prevents meiotic resumption in mouse oocytes. Treinen et al. (1993) achieved similar results in a rat model, by competitive inhibition of these receptors with invitro Δ9-THC treatment. To date, there have been two studies assessing the effect of invitro Δ9-THC treatment on bovine oocyte maturation, yielding conflicting results (Lopez-Cardona et al., 2016; Misner et al., 2021). Based on our findings and previous literature, we hypothesize that the hypermethylation of genes involved in GPCR signaling in response to cannabis exposure, and the reduced transcription that may ensue, represent a cellular response mechanism to protect the cell from overstimulation of the GPCR cascade, thereby rescuing meiotic resumption of the oocyte. Hence, further research is needed to determine the effect cannabis has on human oocyte maturation and ART outcomes.
To focus our analysis, and minimize the inclusion of false-positive CpG, we defined a sliding window of 1000-bp containing at least three CpG with a concordant mean methylation difference >25% and a q-value < 0.01, as being a differentially methylated region (DMR), as previously described (Watson et al., 2015). After applying these criteria, we identified 71 DMRs of which, within the top 30, there was representation of ZFP family, as well as genomic features involved in adhesion (ADAM8, KRTAP5-11, CDH4), biosynthesis (IDI1, FABP6, DTYMK, ASGR1), translation and post-translational modifications (YARS2, PIGG, RMRP) (Table III). Once again, there was a significant representation of ncRNAs which may have regulatory functions.
Next, we explored the concentration-related effects of cannabis on DNA methylation. For the purpose of this analysis, we focused on genomic features that are a part of a DMR in the current study, as well as on genomic features which were previously annotated in the context of epigenetic effects of cannabis exposure. Together, there were 16 unique genomic features that exhibited concentration-related differential methylation (Table IV), three of which have been previously annotated as affected by exposure to cannabis (ABR, KCNMA1 and SMYD3). Interestingly, these 16 genomic features participate in diverse cellular processes including GTP hydrolysis (ABR), folate pool regulation (FTCD), voltage-gated potassium channel activity (KCNMA1), structural cellular components (DCAF12L1, KRT83, MLC1, SPATC1L and TEKT4), steroid synthesis (MRAP), chromatin regulation (SMYD3), non-coding RNAs (DISC1-IT1, MLLT10P1, TMEM44-AS1 and ZFAT-AS1) and transcription regulation (PIGG and ZFP37).
Finally, to determine the impact cannabis exposure has on hallmark genes and pathways by changing their methylation, we assessed the transcription of 97 genes of interest which were chosen because they were differentially methylated in this study, previously reported to be affected by cannabis use in other tissues, or involved in downstream signaling in response to cannabis exposure. Of these genes, 10 were differentially expressed between cases and controls (2 upregulated and 8 downregulated). Interestingly, the promoters of both upregulated genes (LSP1 and ZFP57) were significantly hypomethylated, and those of four downregulated genes (FTCD, KRTAP5-11, MGAT3 and RMRP) were significantly hypermethylated in this study, indicating that DNA methylation is altered by cannabis exposure and this is translated into transcriptional changes. Interestingly, in the brain, chronic exposure to cannabis has been shown to decrease PKA activity (Kesner and Lovinger, 2021). PKA signaling is involved in numerous functions including glucose and lipid metabolism, transcription factor regulation, cell adhesion and calcium homeostasis, and alterations to its transcription or activity can have significant downstream effects (Sassone-Corsi, 2012). In this study, we observed decreased expression of a catalytic subunit of PKA (PRKACG) in patients who had used cannabis when compared to those who had not, however this downregulation was not caused by alterations in DNA methylation. The decrease in expression is likely in response to cannabis through signaling via the CBRs and downstream GPCRs, much like that observed in the brain (Kesner and Lovinger, 2021). Further exploration of the functional impact of these genes, other potential mechanisms of action, as well as a larger impact on transcription in the context of oocyte development and folliculogenesis is ongoing. Finally, two genes critical to granulosa cell function, AMH and CYP19A1, were significantly downregulated in patients who used cannabis compared to controls; however, there were no observed changes to DNA methylation in either of these genes, indicating that cannabis alters the expression of these genes through a DNA methylation-independent mechanism (Fig. 5).
Our study is limited by the probe-based enrichment method we employed because it tends to enrich CpG associated with a higher CpG density, and may miss CpG associated with less dense CpG regions. Furthermore, since the present study was cross-sectional in design, we do not know how exposure over time, frequency of exposure, route of exposure (inhalation or ingestion) or type of cannabis product consumed (flower, oil or refined product) might influence the methylome. However, by establishing correlations between methylome alterations and FF concentrations of PCs, we were able to strengthen our findings and eliminate the bias that may have been introduced by all the above. In addition, FF was not measured for the presence of other psychoactive drugs, and even though none of the patients reported concomitant tobacco use or the use of other psychoactive drugs or prescription medications, self-reporting alone cannot rule out the exposure of the follicle to these substances, and this is a limitation of our study. This study is also limited by its small sample size; however, by matching case and control patients by several confounding variables, we were able to minimize the inter-patient variability. This study was not designed, and was hence underpowered, to determine the effect cannabis exposure has on IVF outcomes. Finally, this study was not designed to examine the reversibility of the epigenetic modifications or their functional consequences. To further our understanding of the functional effects of cannabis on the developing oocyte, as well as the impact on IVF outcomes, we are currently conducting a larger scale study.
Conclusions
To our knowledge, this is the first study profiling the methylome of human granulosa cells exposed to cannabis. We have described key genes involved in GPCR signaling, cell proliferation, apoptosis, immune response, metabolism and histone modifications that were affected. Moreover, we have characterized a concentration-dependent relationship between cannabis concentration and DNA methylation as well as alterations to gene expression directly related to changes in DNA methylation. As cannabis legalization increases worldwide and with its use increasing in women of reproductive age, it is critical that we expand our understanding of the effects cannabis has on the developing ovarian follicle and the oocyte itself. If future studies point to the heritability and functional consequences of these epigenetic modifications, these findings must be considered for drafting cannabis policy decisions, for public education, and for consulting patients.
Supplementary data
Supplementary data are available at Molecular Human Reproduction online.
Data availability
The data underlying this article are available in the article and in its online supplementary material. Raw sequencing data can be made available by contacting the corresponding author.
Supplementary Material
Acknowledgements
The authors would like to thank CReATe Fertility Centre patients for the donation of their material, the CReATe BioBank and its personnel in the provision of samples and associated de-identified data for this study, Ashley St. Pierre at the Analytical Facility for Bioactive Molecules for conducting the LC-MS/MS and analysis, Nicholas Khuu at the Princess Margaret Genomics Centre for preparing and sequencing the DNA methylation libraries, and Zhibin Lu at the University Hospital Network Bioinformatics and HPC Core for conducting the differential methylation analysis. Finally, we would like to thank all embryologists, nurses and staff for helping with data collection.
Authors’ roles
N.F.W., B.A.W. and C.L.L. designed this study. N.F.W. and B.A.W. analyzed the data, interpreted the results and drafted the manuscript. B.A.W. performed the experiments. J.M. performed the statistical analysis. S.J. collected, processed and released samples and de-identified clinical data. All authors read and approved the final version of the paper.
Funding
All funding was provided by the CReATe Fertility Centre through the reinvestment of clinical earnings.
Conflict of interest
The authors declare no conflicts of interest.
Contributor Information
Noga Fuchs Weizman, CReATe Fertility Centre, Toronto, ON, Canada; Racine IVF Unit, Lis Maternity Hospital, Tel Aviv Sourasky Medical Center, Affiliated to the Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel.
Brandon A Wyse, CReATe Fertility Centre, Toronto, ON, Canada.
Sahar Jahangiri, CReATe Fertility Centre, Toronto, ON, Canada; CReATe BioBank, Toronto, Canada.
Clifford L Librach, CReATe Fertility Centre, Toronto, ON, Canada; CReATe BioBank, Toronto, Canada; Department of Obstetrics and Gynecology, University of Toronto, Toronto, ON, Canada; Department of Physiology, University of Toronto, Toronto, ON, Canada.
References
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE.. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 2012;13:R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (November 2020, date last accessed).
- Bai H, Li Y, Gao H, Dong Y, Han P, Yu H.. Histone methyltransferase SMYD3 regulates the expression of transcriptional factors during bovine oocyte maturation and early embryonic development. Cytotechnology 2016;68:849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balikova M, Hlozek T, Palenicek T, Tyls F, Viktorinova M, Melicher T, Androvicova R, Tomicek P, Roman M, Horacek J.. [Time profile of serum THC levels in occasional and chronic marihuana smokers after acute drug use—implication for driving motor vehicles]. Soud Lek 2014;59:2–6. [PubMed] [Google Scholar]
- Battista N, Bari M, Maccarrone M.. Endocannabinoids and reproductive events in health and disease. Handb Exp Pharmacol 2015;231:341–365. [DOI] [PubMed] [Google Scholar]
- Battista N, Pasquariello N, Di Tommaso M, Maccarrone M.. Interplay between endocannabinoids, steroids and cytokines in the control of human reproduction. J Neuroendocrinol 2008;20(Suppl 1):82–89. [DOI] [PubMed] [Google Scholar]
- Bottino C, Peserico A, Simone C, Caretti G.. SMYD3: an oncogenic driver targeting epigenetic regulation and signaling pathways. Cancers (Basel) 2020;12:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brents LK. Marijuana, the endocannabinoid system and the female reproductive system. Yale J Biol Med 2016;89:175–191. [PMC free article] [PubMed] [Google Scholar]
- Cavalcante RG, Sartor MA.. annotatr: genomic regions in context. Bioinformatics 2017;33:2381–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi S, Rossi G, Oddi S, Di Nisio V, Maccarrone M.. Role of major endocannabinoid-binding receptors during mouse oocyte maturation. Int J Mol Sci 2019;20:2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cylwa R, Kiełczewski K, Machnik M, Oleksiewicz U, Biecek P.. KRAB ZNF explorer-the online tool for the exploration of the transcriptomic profiles of KRAB-ZNF factors in The Cancer Genome Atlas. Bioinformatics 2020;36:980–981. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElSohly MA, Radwan MM, Gul W, Chandra S, Galal A.. Phytochemistry of Cannabis sativa L. Prog Chem Org Nat Prod 2017;103:1–36. [DOI] [PubMed] [Google Scholar]
- Fonseca BM, Correia-Da-Silva G, Almada M, Costa MA, Teixeira NA.. The endocannabinoid system in the postimplantation period: a role during decidualization and placentation. Int J Endocrinol 2013;2013:510540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Weizman N, Wyse BA, Szaraz P, Defer M, Jahangiri S, Librach CL.. Cannabis alters epigenetic integrity and endocannabinoid signaling in the human follicular niche. Hum Reprod 2021;36:1922–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera L, Ottolenghi C, Garcia-Ortiz JE, Pellegrini M, Manini F, Ko MS, Nagaraja R, Forabosco A, Schlessinger D.. Mouse ovary developmental RNA and protein markers from gene expression profiling. Dev Biol 2005;279:271–290. [DOI] [PubMed] [Google Scholar]
- Howlett AC. The cannabinoid receptors. Prostaglandins Other Lipid Mediat 2002;68-69:619–631. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AM, Bartolomei MS.. Evolving imprinting control regions: KRAB zinc fingers hold the key. Genes Dev 2019;33:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukic AM, Weinberg CR, Baird DD, Wilcox AJ.. Lifestyle and reproductive factors associated with follicular phase length. J Womens Health (Larchmt) 2007;16:1340–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juknat A, Rimmerman N, Levy R, Vogel Z, Kozela E.. Cannabidiol affects the expression of genes involved in zinc homeostasis in BV-2 microglial cells. Neurochem Int 2012;61:923–930. [DOI] [PubMed] [Google Scholar]
- Kesner AJ, Lovinger DM.. Cannabis use, abuse, and withdrawal: cannabinergic mechanisms, clinical, and preclinical findings. J Neurochem 2021;157:1674–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klonoff-Cohen HS, Natarajan L, Chen RV.. A prospective study of the effects of female and male marijuana use on in vitro fertilization (IVF) and gamete intrafallopian transfer (GIFT) outcomes. Am J Obstet Gynecol 2006;194:369–376. [DOI] [PubMed] [Google Scholar]
- Krueger F. Trim Galore! 2012. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (November 2020, date last accessed).
- Krueger F, Andrews SR.. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011;27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo C, Hsueh A. Ovarian Kaleidoscope Database. 2000. [DOI] [PubMed]
- Levin ED, Hawkey AB, Hall BJ, Cauley M, Slade S, Yazdani E, Kenou B, White H, Wells C, Rezvani AH. et al. Paternal THC exposure in rats causes long-lasting neurobehavioral effects in the offspring. Neurotoxicol Teratol 2019;74:106806. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang Q, Ge P, Zeng C, Lin F, Wang W, Zhao J.. Is a prognostic lncRNA for patients with recurrent glioblastoma. Dis Markers 2020;2020:8888085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Cardona AP, Sanchez-Calabuig MJ, Beltran-Brena P, Agirregoitia N, Rizos D, Agirregoitia E, Gutierrez-Adan A.. Exocannabinoids effect on in vitro bovine oocyte maturation via activation of AKT and ERK1/2. Reproduction 2016;152:603–612. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccarrone M. Endocannabinoid signaling in female reproductive events: a potential therapeutic target? Expert Opin Ther Targets 2015;19:1423–1427. [DOI] [PubMed] [Google Scholar]
- Machnik M, Cylwa R, Kiełczewski K, Biecek P, Liloglou T, Mackiewicz A, Oleksiewicz U.. The expression signature of cancer-associated KRAB-ZNF factors identified in TCGA pan-cancer transcriptomic data. Mol Oncol 2019;13:701–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misner MJ, Taborek A, Dufour J, Sharifi L, Khokhar JY, Favetta LA.. Effects of delta-9 tetrahydrocannabinol (THC) on oocyte competence and early embryonic development. Front Toxicol 2021;3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller BA, Daling JR, Weiss NS, Moore DE.. Recreational drug use and the risk of primary infertility. Epidemiology 1990;1:195–200. [DOI] [PubMed] [Google Scholar]
- Murphy SK, Itchon-Ramos N, Visco Z, Huang Z, Grenier C, Schrott R, Acharya K, Boudreau M-H, Price TM, Raburn DJ. et al. Cannabinoid exposure and altered DNA methylation in rat and human sperm. Epigenetics 2018;13:1208–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne AJ, Pearson JF, Noble AJ, Gemmell NJ, Horwood LJ, Boden JM, Benton MC, Macartney-Coxson DP, Kennedy MA.. Genome-wide DNA methylation analysis of heavy cannabis exposure in a New Zealand longitudinal cohort. Transl Psychiatry 2020;10:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontis J, Planet E, Offner S, Turelli P, Duc J, Coudray A, Theunissen TW, Jaenisch R, Trono D.. Hominoid-specific transposable elements and KZFPs facilitate human embryonic genome activation and control transcription in naive human ESCs. Cell Stem Cell 2019;24:724–735.e725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J.. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 2019;47:W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A.. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotermann M. Analysis of Trends in the Prevalence of Cannabis Use and Related Metrics in Canada. Ottawa, Ontario, Canada: Statistics Canada, 2019, 3–13. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J. et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 2010;463:237–240. [DOI] [PubMed] [Google Scholar]
- Sagvekar P, Kumar P, Mangoli V, Desai S, Mukherjee S.. DNA methylome profiling of granulosa cells reveals altered methylation in genes regulating vital ovarian functions in polycystic ovary syndrome. Clin Epigenetics 2019;11:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M, Mirabella M, De Fino C, Bianco A, Lucchini M, Losavio F, Sabino A, Nociti V.. Sativex® effects on promoter methylation and on CNR1/CNR2 expression in peripheral blood mononuclear cells of progressive multiple sclerosis patients. J Neurol Sci 2017;379:298–303. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P. The cyclic AMP pathway. Cold Spring Harb Perspect Biol 2012;4:a011148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrott R, Rajavel M, Acharya K, Huang Z, Acharya C, Hawkey A, Pippen E, Lyerly HK, Levin ED, Murphy SK.. Sperm DNA methylation altered by THC and nicotine: vulnerability of neurodevelopmental genes with bivalent chromatin. Sci Rep 2020;10:16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopp G, Richter B, Potsch L.. [Serum cannabinoid levels 24 to 48 hours after cannabis smoking]. Arch Kriminol 2003;212:83–95. [PubMed] [Google Scholar]
- Smart R, Caulkins JP, Kilmer B, Davenport S, Midgette G.. Variation in cannabis potency and prices in a newly legal market: evidence from 30 million cannabis sales in Washington state. Addiction 2017;112:2167–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobocinska J, Molenda S, Machnik M, Oleksiewicz U.. KRAB-ZFP transcriptional regulators acting as oncogenes and tumor suppressors: an overview. Int J Mol Sci 2021;22:2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Substance Abuse and Mental Health Services Administration (SAMHSA). Results from the 2013 National Survey on Drug Use and Health: Summary of National Findings. Rockville, MD: Oxford University Press, 2014. [Google Scholar]
- Sun X, Dey SK.. Endocannabinoid signaling in female reproduction. ACS Chem Neurosci 2012;3:349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szutorisz H, Hurd YL.. Epigenetic effects of cannabis exposure. Biol Psychiatry 2016;79:586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Coluccio A, Thorball CW, Planet E, Shi H, Offner S, Turelli P, Imbeault M, Ferguson-Smith AC, Trono D.. ZNF445 is a primary regulator of genomic imprinting. Genes Dev 2019;33:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AH, Ang C, Bell SC, Konje JC.. The role of the endocannabinoid system in gametogenesis, implantation and early pregnancy. Hum Reprod Update 2007;13:501–513. [DOI] [PubMed] [Google Scholar]
- Treinen KA, Sneeden JL, Heindel JJ.. Specific inhibition of FSH-stimulated cAMP accumulation by delta 9-tetrahydrocannabinol in cultured rat granulosa cells. Toxicol Appl Pharmacol 1993;118:53–57. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Han B, Compton WM, McCance-Katz EF.. Self-reported medical and nonmedical cannabis use among pregnant women in the United States. JAMA 2019;322:167–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xie H, Dey SK.. Endocannabinoid signaling directs periimplantation events. AAPS J 2006;8:E425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CT, Szutorisz H, Garg P, Martin Q, Landry JA, Sharp AJ, Hurd YL.. Genome-wide DNA methylation profiling reveals epigenetic changes in the rat nucleus accumbens associated with cross-generational effects of adolescent THC exposure. Neuropsychopharmacology 2015;40:2993–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bao X, Peng Z, Wang L, Du L, Niu W, Sun Y.. Comprehensive analysis of genome-wide DNA methylation across human polycystic ovary syndrome ovary granulosa cell. Oncotarget 2016;7:27899–27909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Bam M, Nagarkatti PS, Nagarkatti M.. RNA-seq analysis of δ9-tetrahydrocannabinol-treated T cells reveals altered gene expression profiles that regulate immune response and cell proliferation. J Biol Chem 2016;291:15460–15472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Hegde VL, Rao R, Zhang J, Nagarkatti PS, Nagarkatti M.. Histone modifications are associated with Δ9-tetrahydrocannabinol-mediated alterations in antigen-specific T cell responses. J Biol Chem 2014;289:18707–18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn NL, Bartolomei MS, Blendy JA.. Multigenerational and transgenerational inheritance of drug exposure: the effects of alcohol, opiates, cocaine, marijuana, and nicotine. Prog Biophys Mol Biol 2015;118:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material. Raw sequencing data can be made available by contacting the corresponding author.