Abstract
Background
Perturbagen analysis of Crohn’s disease (CD) ileal gene expression data identified small molecules including eicosatetraynoic acid (ETYA), which may exert an antifibrotic effect. We developed a patient-specific human intestinal organoid (HIO) model system to test small molecule regulation of mitochondrial and wound-healing functions implicated in stricturing behavior.
Methods
HIOs were made from CD induced pluripotent stem cells with and without a loss-of-function haplotype in the DUOX2 gene implicated in ileal homeostasis and characterized under basal conditions and following exposure to butyrate and ETYA using RNA sequencing, flow cytometry, and immunofluorescent and polarized light microscopy. Mitochondrial activity was measured using high-resolution respirometry and tissue stiffness using atomic force microscopy.
Results
HIOs expressed core mitochondrial and extracellular matrix (ECM) genes and enriched biologic functions implicated in CD ileal strictures; ECM gene expression was suppressed by both butyrate and ETYA, with butyrate also suppressing genes regulating epithelial proliferation. Consistent with this, butyrate, but not ETYA, exerted a profound effect on HIO epithelial mitochondrial function, reactive oxygen species production, and cellular abundance. Butyrate and ETYA suppressed HIO expression of alpha smooth muscle actin expressed by myofibroblasts, type I collagen, and collagen protein abundance. HIOs exhibited tissue stiffness comparable to normal human ileum; this was reduced by chronic ETYA exposure in HIOs carrying the DUOX2 loss-of-function haplotype.
Conclusions
ETYA regulates ECM genes implicated in strictures and suppresses collagen content and tissue stiffness in an HIO model. HIOs provide a platform to test personalized therapeutics, including small molecules prioritized by perturbagen analysis.
Keywords: small molecule, NADPH oxidase, reactive oxygen species, fibrosis
INTRODUCTION
Biologic therapy targeting tumor necrosis factor α (TNFα) is associated with improved growth, higher rates of steroid-free remission, and lower rates of penetrating complications in children with Crohn’s disease (CD).1,2 However, a subset of patients progress to stricturing complications within 5 years of diagnosis, implicating additional pathogenic drivers. We have previously reported that ileal expression of genes regulating mitochondrial cellular respiration (RESP) is reduced while expression of genes regulating extracellular matrix (ECM) production and wound healing is increased at diagnosis in those who ultimately develop an ileal stricture.1 Consistent with this, increased expression of an inflammatory macrophage and fibroblast gene signature at diagnosis was associated with lower rates of response to anti-TNF therapy.3 We conducted a perturbagen bioinformatics analysis using the CD ileal gene signature associated with future stricturing to identify novel small molecules predicted to have an anti-inflammatory and antifibrotic effect in this context.4 Results of this analysis prioritized testing the long-chain fatty acid eicosatetraynoic acid (ETYA), which may exert an antifibrotic effect via cyclooxygenase and lipoxygenase inhibition.4 Prior experimental studies have also suggested a potential anti-inflammatory and antifibrotic role for the short-chain fatty acid butyrate in the gut.5-7 However, whether ETYA or butyrate would induce these predicted molecular or functional effects in a relevant patient-derived model system was not known.
The dual oxidase 2 (DUOX2) epithelial nicotinamide adenine dinucleotide phosphate oxidase is induced in the ileum by pathobionts which are expanded at diagnosis in pediatric CD.8 DUOX2 produces hydrogen peroxide at the enterocyte apical membrane to directly control microbes, and DUOX2-dependent reactive oxygen species (ROS) also serve as intracellular signaling molecules to regulate additional host antimicrobial pathways.9 We reported a profound induction of ileal DUOX2 expression in treatment-naïve pediatric CD patients, in association with increased abundance of Enterobacteriaceae, and associated host innate inflammatory responses.8 Studies in animal models and enteroids have shown that in this context DUOX2 likely functions to constrain pathogenic microbes, thereby maintaining mucosal homeostasis.10 Consistent with this, rare loss-of-function DUOX2 genetic mutations are now recognized as a risk factor for inflammatory bowel disease, with data implicating dysregulation of host-microbe interactions.11 Whether more common DUOX2 gene variants would influence the response to small molecules predicted to regulate metabolic and wound healing functions implicated in stricture formation was not known.12
Human intestinal organoids (HIOs) derived from induced pluripotent stem cells (iPSCs) have emerged as a powerful tool for testing genetic mechanisms of disease, as well as for testing effects of microbial metabolites and therapeutic small molecules relevant to tissue fibrosis.13,14 This is because HIOs contain both epithelial and stromal cells highly relevant to these processes, and demonstrate profibrotic changes in gene expression in response to cytokines including transforming growth factor β (TGFβ).15 In the current study, we developed an HIO model to test effects of the microbial metabolite butyrate, and the long-chain fatty acid ETYA, on mitochondrial cellular respiration, ECM gene expression, and tissue stiffness in the context of CD patient DUOX2 genetic variation. HIOs expressed core mitochondrial and ECM genes implicated in CD strictures. Consistent with our perturbagen analysis, ETYA suppressed ECM gene expression and reduced HIO collagen content, with a reduction in tissue stiffness observed in organoids carrying a common DUOX2 loss-of-function haplotype.12
METHODS
Whole Exome Sequencing and Ileal Biopsy Bulk RNA Sequencing
Pediatric CD patients for whole exome sequencing (WES) and pretreatment ileal biopsy bulk RNA sequencing (RNASeq) at diagnosis were selected from the multicenter Crohn’s and Colitis Foundation–sponsored Risk Stratification in Pediatric Crohn’s Disease (The RISK Study) inception cohort study, as previously reported.1,4,16 For the current report we utilized our publicly available WES data, with a focus on DUOX2 gene missense mutations validated by Sanger sequencing, and our previously reported ileal bulk RNASeq data, with a focus on gene signatures linked to future stricturing.4,16
HIO Generation and Quality Control
For generation of HIOs, iPSCs were differentiated as previously described with minor modifications.17 The DUOX2 reference genotype (DUOX2ref) and DUOX2 H678R;R701Q;P982A loss-of-function haplotype (DUOX2hapl)12 iPSC lines tested negative for mycoplasma. The karyotype of each iPSC line matched each patient’s peripheral blood mononuclear cell karyotype. The short tandem repeat or electropherogram for each iPSC line matched each patient’s peripheral blood mononuclear cells. Stemness analysis showed over 95% + cells for SSEA4 and Tra-160 for both iPSC lines. Functional pluripotency was assessed in undifferentiated and d14 embryoid bodies as previously described.18 Briefly, definitive endoderm was generated by exposure to recombinant human activin A for 3 days (100 ng/mL). Hindgut endoderm was generated by exposure to recombinant 500 ng/mL FGF4 and 3 µM CHIR99021 for 4 days. Spontaneously derived spheroids were then embedded in Matrigel (BD Biosciences, San Jose, CA, USA; catalog # 354234) and incubated in media containing recombinant 50 ng/mL human epidermal growth factor, 100 ng/mL hNoggin, and 5% Respondin-2 conditioned media, with complete media changes every 3 days. HIOs were passaged and re-embedded in Matrigel after 1 week in culture. Approximately 28 days after embedding, HIOs were harvested and used for assays. We examined HIOs under basal conditions, after exposure to 10 mM butyrate or 50 μM ETYA between days 25 and 28 of culture, and after exposure to 50 μM ETYA between days 16 and 28 of culture. To generate single cells, HIOs were transferred into a 5-mL tube, intermittently vortexed for 30 to 60 seconds, washed with ice-cold phosphate-buffered saline, and incubated in 1 mL of Tryple Express (Gibco, Grand Island, NY, USA) with vigorous shaking at 37°C for 10 minutes. The cell suspension was filtered over a 40-µm filter, washed, and used for assays.
Total ROS Detection
ROS staining was performed on the previous single-cell isolations with flow cytometry using the ROS and superoxide kit (Enzo Life Sciences, Farmingdale, NY, USA; cat #ENZ-51010) per manufacturer’s protocol. Briefly, the cells were stained for CD90 (BD Biosciences, Franklin Lakes, NJ, USA) and EpCAM (BioLegend, San Diego, CA, USA) for 30 minutes at room temperature. Cells were washed and oxidative stress with superoxide detection reagents were added, and cells were stained for 30 minutes at 37°C with slow shaking. The ROS inducer pyocyanin was used in all groups for positive induction of ROS. Directly after the cells were placed on ice and acquired on a Canto flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA), and data were analyzed using De Novo software, Flow Cytometry System Express Version 7 (De Novo Software, Pasadena, CA, USA).
JC-1 Mitochondrial Membrane Potential Measurement
JC-1 staining was performed on the previous single-cell isolations with flow cytometry using JC-1 reagent (Molecular Probes, Eugene, OR, USA) reagent according to the manufacturer’s instructions. In brief, JC-1 dye was added at 1 mcM to washed cells, and incubated for 20 minutes at 37°C, 5% CO2. Cells were washed and CD90 APC-Cy7 (BD Biosciences, Franklin Lakes, NJ, USA) and EpCAM APC (BioLegend) antibodies were added for an additional 30 minutes at room temperature. Cells were washed, cells placed on ice and acquired on a Canto flow cytometer, and data were analyzed using De Novo software. The mitochondrial membrane potential (MMP) was calculated as the ratio of PE-MFI/FITC-MFI in EpCAM+ and CD90+ cells. As a positive control for the specificity of the assay, we used 50 mcM of CCCP to depolarize the MMP measured using JC-1 dye.
High-Resolution Respirometry
The Oxygraph-2k (O2k; Oroboros Instruments, Innsbruck, Austria) was used for measurements of respiration. Each chamber was air-calibrated in Mir05 respiration medium (0.5 mM EDTA, 3 mM MgCl2, 60 mM k-lactobionic acid, 20 mM taurine, 10 mM KH2PO4, 20 mM HEPES, 110 mM D-sucrose, 0.1% bovine serum albumin essentially fatty acid free) before each experiment. All experiments were performed at 37°C. Oxygen concentrations in each chamber never dropped below 80 μM during any experiment. HIOs were homogenized in Mir05 respiration medium, and 100 μL of the tissue homogenate was added to each chamber along with digitonin (10 µg per chamber). Once baseline oxygen levels in each chamber became stable, cytochrome c (10 μM), malate (2 mM), pyruvate (5 mM), adenosine diphosphate (5 mM), and glutamate (10 mM) were added to stimulate respiration through Complex I. Once the oxygen consumption rate plateaued, succinate (10 mM) was added to assess the combined activity of complexes I + II. Next, rotenone (1 mM) was added to inhibit Complex I activity, and additional succinate was added to analyze maximal Complex II activity. Carbonyl cyanide p-trifluoromethoxyphenylhydrazone (0.5 μM) was then added to uncouple the mitochondrial membrane and induce maximal respiration. Respiration rates were normalized to the amount of protein added for each sample. Complex I respiration was defined as the rate of respiration of malate, adenosine diphosphate, pyruvate, and glutamate (first succinate—rotenone). Complex II respiration was defined as respiration after adding the second dose of succinate minus Complex I respiration. Average rates of oxygen consumption [(pmol/(s × mL)/μg protein] + SEM were graphed.
RNA Sequencing
RNA was isolated using the Qiagen AllPrep RNA/DNA Micro Kit (Qiagen, Germantown, MD, USA). The HIO global pattern of gene expression was determined using RNASeq on the Illumina platform (Illumina, San Diego, CA, USA) as previously described.4 Reads were quantified by kallisto using Gencode v24 as the reference genome and transcripts per million, including protein-coding messenger RNA genes with transcripts per million above 1 in 20% of the samples (n = 14 264 genes for the butyrate experiments and n = 13 649 genes for the ETYA experiments) in our downstream analysis. RNASeq samples were stratified into specific subgroups including DUOX2ref or DUOX2hapl HIOs untreated (n = 8 each), pretreated with 10 mM butyrate for 72 hours (n = 4 each), or pretreated with 50 μM ETYA for 12 days (n = 5 each). Differentially expressed genes had fold change ≥1.5 and false discovery rate (FDR) <0.05 using R package DESeq2 version 1.24.0 (R Foundation for Statistical Computing, Vienna, Austria), and importing and summarizing the kallisto output files to gene level with R package tximport version 1.12.3. ToppGene and ToppCluster software were used to perform gene set enrichment analyses, and visualization of the networks was obtained using Cytoscape.v3.0.217.19 For functional enrichments, we included only those genes with -3 < Log2(x) < 3, to avoid potential subject-specific differences.
TaqMan Low Density Array
HIO RNA was extracted using the AllPrep DNA/RNA Micro kit (Qiagen). Complementary DNA was synthesized using SuperScript IV VILO Master Mix (Thermo Fisher Scientific, Waltham, MA, USA). When necessary, preamplification of complementary DNA was performed using TaqMan PreAmp Master Mix and a custom preamp pool (Thermo Fisher Scientific). Real-time polymerase chain reaction (PCR) was performed using a 7900HT Fast Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) with custom TaqMan Low Density Array cards and TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific). Polymerase chain reaction primers are listed in the Supplement. Gene expression data were normalized to expression of GAPDH. Data were analyzed using GraphPad PRISM 9.0 (GraphPad Software, San Diego, CA, USA).20
Histology
Cultured HIOs were fixed with 4% paraformaldehyde, embedded in paraffin, and then sliced on a microtome and mounted on charged slides. Slides were stained with hematoxylin and eosin stain. Immunohistochemistry detection of Sirius red was performed, using Picro Sirius Red Stain Kit (Abcam, Cambridge, MA, USA; cat. #ab150681). Sirius red–stained slides were analyzed used both bright field and polarized microscopy. Immunofluorescence detection of alpha smooth muscle actin, DAPI, pimonidazole (PMDZ), and vimentin (VIM) was performed using anti-vimentin antibody (Sigma-Aldrich, St. Louis, MO, USA; cat. # SAB-4503083), anti-actin antibody (Sigma-Aldrich; cat. #A5228), and anti-PMDZ antibody (Hypoxyprobe, Burlington, MA, USA; cat. #HP1-100), and co-stained with DAPI (Thermo Fisher Scientific; cat. #D1306). Images were acquired using an Olympus BX51 microscope and a DP80 camera or an Olympus BX43 and DP74 camera (Olympus America, Center Valley, PA, USA). CellSens Dimension (Olympus America), NIS-Elements (Nikon Instruments, Melville, NY, USA), and ImageJ version 1.53o (National Institutes of Health, Bethesda, MD, USA) software were employed to measure the frequency or surface area of positive stains in HIOs.
Atomic Force Microscopy
HIOs were measured for stiffness with an atomic force microscope (NanoWizard IV; JPK Instruments, Berlin, Germany). The atomic force microscope head with a silicon nitride cantilever (CSC37, k = 0.3 N/m; MikroMasch, Talinn, Estonia) was mounted on a fluorescence stereo microscope (M205 FA; Leica, Wetzlar, Germany) coupled with a Z axis piezo stage (JPK CellHesion module; JPK Instruments), which allows the indentation measurement up to the depth of ~100 μm. As a substrate for organoids, a Matrigel-coated dish (φ = 34 mm; TPP Techno Plastic Products, Trasadingen, Switzerland) was used. Thereafter, HIOs were deposited to the Matrigel-coated dish and incubated for 1 hour at 37°C. The sample dish was then placed onto the atomic force microscope stage, and force-distance curves in a 4 × 4 matrix in a 25 × 25 μm square were measured from each HIO. Finally, Young’s moduli (E, Pa) of organoids were determined by fitting the obtained force-distance curves with the modified Hertz model.21
Statistical Analysis
Data are shown as the median (interquartile range) or frequency. Frequencies were compared using the Fisher exact test or chi squared test. The Shapiro-Wilk test was used to assess the normality of the data. Pairwise comparisons within the DUOX2ref or DUOX2hapl genotypes were tested using the unpaired t test for data that passed the normality test and the Mann-Whitney test for data that did not pass the normality test. Differences between butyrate- or ETYA-treated samples and untreated samples within the DUOX2ref or DUOX2hapl genotypes were tested using a 1-way analysis of variance with Dunnett’s multiple comparisons test for data that passed the normality test, and using a Kruskal-Wallis test with Dunn’s multiple comparisons test for data that did not pass the normality test in GraphPad PRISM 9.0. No statistical tests for differences were performed between HIOs carrying the DUOX2ref and DUOX2hapl genotypes, as these could be due to other patient-specific genetic differences not controlled for in the design.
Ethical Considerations
The patient-based studies were approved by the Institutional Review Boards at each of the RISK study sites including Emory and Cincinnati Children’s, and consent was obtained from parents and adult subjects and assent from pediatric subjects 11 years of age and older.
RESULTS
DUOX2 Genetic Architecture and Clinical Phenotype in Pediatric CD
We utilized WES to define the frequency of potentially damaging DUOX2 gene variants in 220 children with CD. Supplemental Table 1 shows the frequency of these DUOX2 gene variants in the overall pediatric CD cohort, as well as in children with CD stratified by African American race, and in unaffected African Americans and those with European ancestry reported in dbSNP. Consistent with the dbSNP data, the DUOX2 H678R variant was the most common variant detected in the CD cohort, and was in turn more common in African American patients. Overall, the most common variants included H678R and R701Q, which cause a modest reduction in DUOX2 generation of ROS, and P982A, which does not affect ROS production.12,22 Of these 220 patients, 70 (32%) exhibited at least 1 potentially damaging DUOX2 gene variant. As shown in Supplemental Table 2, a significant number of these patients exhibited carriage of multiple heterozygous DUOX2 variants, with the H678R;R701Q;P982A haplotype associated with a modest reduction in hydrogen peroxide production being the most common.12 Targeted gene sequencing of the DUOX2 gene identified 27 additional patients with a potentially damaging DUOX2 gene variant. Clinical and demographic characteristics of the 220 participants with WES, and 27 additional participants with targeted gene sequencing, are shown in Table 1. Age at diagnosis, sex, ileal location or endoscopic severity, and antimicrobial serologies did not vary with DUOX2 variant carriage (Table 1). Patients with carriage of multiple DUOX2 variants were more likely to be African American (23% vs 4%; P < .001) and to develop stricturing disease complications during follow-up (18% vs 7%; P < .05). A trend toward increased stricturing with carriage of multiple DUOX2 variants was observed within both Caucasians (16% vs 7%; P = .07) and African Americans (30% vs 11%; P = .3). The DUOX2 variants in patients with stricturing behavior included 1 case each with L171P, P234S, H678R, or E1546G carriage; 1 case each with L171P;H678R, H678R;E1017G, H678R;H678R;R701Q;P982A, L171P;P234S;H678R, or H678R;R701Q;P982A;A1277G carriage; and 3 cases with H678R;R701Q;P982A carriage. The rate of internal penetrating complications was low and did not vary with DUOX2 genotype.
Table 1.
Clinical and demographic characteristics stratified by DUOX2 genotype
| No DUOX2 Variant (n = 150) | 1 DUOX2 Variant (n = 52) | 2+ DUOX2 Variants (n = 45) | |
|---|---|---|---|
| Age, y | 8 (7-9) | 10 (8-14)a | 9 (8-10) |
| Male | 93 (62) | 31 (62) | 21 (47) |
| African American | 6 (4) | 5 (10) | 10 (23)b |
| ASCA IgA positive | 48 (32) | 16 (32) | 14 (32) |
| ASCA IgG positive | 19 (14) | 7 (16) | 9 (24) |
| CBir1 positive | 48 (36) | 15 (35) | 20 (53) |
| Ileal involvement | 101 (68) | 34 (68) | 30 (68) |
| Ileal deep ulcer | 38 (31) | 16 (34) | 11 (28) |
| Stricturing behavior | 10 (7) | 4 (8) | 8 (18)a |
| Penetrating behavior | 3 (2) | 2 (4) | 2 (5) |
Values are median (interquartile range) or n (%). Crohn’s disease patients were grouped based on carriage of potentially damaging DUOX2 gene variants determined by whole exome sequencing (n = 220) or targeted gene sequencing (n = 27). Disease behavior was classified through 60-month follow-up from diagnosis.
Abbreviations: ASCA, anti-Saccharomyces cerevisiae antibody; Ig, immunoglobulin.
P < .05 vs group with no DUOX2 variant carriage.
P < .001 vs group with no DUOX2 variant carriage.
HIOs Express a Gene Program Enriched for Cellular Respiration, ECM, and Wound-Healing Pathways Implicated in Strictures
Based on these results, we developed a HIO model system using iPSCs derived from a pediatric CD patient with carriage of the common DUOX2 H678R;R701Q;P982S haplotype (DUOX2hapl) associated with a modest reduction in hydrogen peroxide production12 and a pediatric CD patient with carriage of the reference DUOX2 genotype (DUOX2ref). Both patients were Caucasian, were diagnosed under 10 years of age with ileocolonic involvement, and had experienced persistent inflammatory disease behavior with more than 10 years of follow-up, with complete mucosal healing with medical therapy including immune modulators and anti-TNF at most recent ileocolonoscopy. We used this system to test whether mechanisms previously linked to strictures in patients would vary with butyrate or ETYA exposure. We had previously defined core mitochondrial RESP and ECM wound-healing genes linked to stricturing complications in patients.1 Gene set enrichment analysis of 7938 genes expressed at a moderate to high level (transcripts per million >10 by bulk RNASeq) in DUOX2ref HIOs under basal conditions confirmed enrichment of genes encoding oxidative phosphorylation (FDR = 7.1E-17) and TGFβ signaling (FDR = 8.2E-16) implicated in mitochondrial and ECM function, respectively (Supplemental Table 3). Consistent with this, the majority of both RESP (n = 29, 85%) and ECM (n = 30, 68%) genes differentially expressed in the ileum of patients who subsequently developed strictures were also expressed in DUOX2ref HIOs under basal conditions. These data confirmed that HIOs express a gene program highly relevant to core RESP and ECM genes implicated in ileal strictures.
The Global Pattern of Gene Expression in HIOs Following Butyrate or ETYA Exposure Is Enriched for Suppression of Pathways Regulating ECM Formation
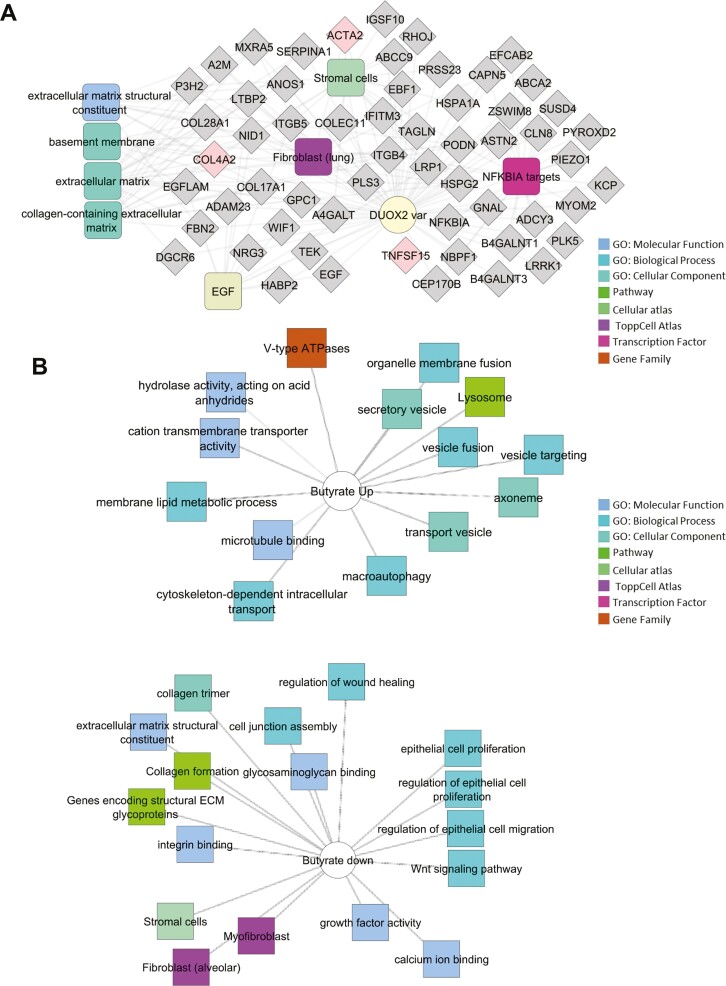
We first asked whether the global pattern of gene expression would vary with DUOX2 genotype in HIOs, either under basal conditions or following exposure to the microbial metabolite butyrate or the long-chain fatty acid ETYA. Butyrate was tested due to prior studies showing that it regulates both cellular mitochondrial function and intestinal fibrosis in murine models,5-7 while ETYA was prioritized by our recent perturbagen analysis of CD patient ileal gene expression data.4 A modest number of genes (n = 344) were differentially expressed between the DUOX2hapl and DUOX2ref HIOs under basal conditions (Supplemental Table 4). The DUOX2hapl HIO transcriptome under basal conditions was notable for upregulation of genes implicated in wound healing and ECM formation (FDR = 3.52E-03) regulated by epidermal growth factor (FDR = 2.33E-05), TNFSF15/TL1A, and the inhibitor of nuclear factor kappa B signaling NFKBIA (FDR = 9.38E-04) (Figure 1A and Supplemental Table 4).23
Figure 1.
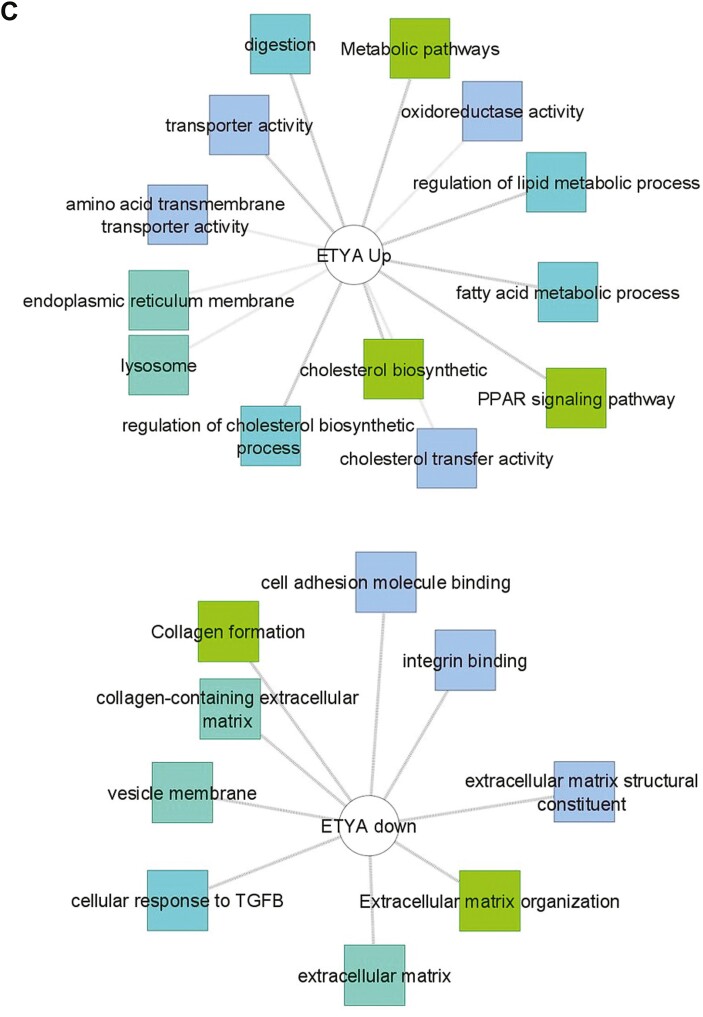
Differentially expressed genes associated with DUOX2 genotype and butyrate or eicosatetraynoic acid (ETYA) exposure. A, Differentially expressed genes between DUOX2ref and DUOX2haplo human intestinal organoids (HIOs) were defined using bulk RNA sequencing (RNASeq) under basal conditions (n = 8 per group, with false discovery rate <0.05 and fold change ≥1.5). Functional annotation enrichment analysis of the 198 upregulated genes in the DUOX2hapl HIOs was completed using ToppGene, ToppCluster, and Cytoscape. Individual genes are shown in gray, with some highlighted in pink, with enriched signaling pathways, cell types, and biologic functions as shown. B, Differentially expressed genes between DUOX2ref HIOs under basal conditions and following 10 mM butyrate exposure for 72 hours were defined using bulk RNASeq (n = 4-8 per group, n = 8052 genes with false discovery rate <0.05 and fold change ≥1.5). Functional annotation enrichment analyses of the up- and downregulated genes were as shown. C, Differentially expressed genes between DUOX2ref HIOs under basal conditions and following 50 μM ETYA exposure for 12 days were defined using bulk RNASeq (n = 5-8 per group, n = 1383 genes with false discovery rate <0.05 and fold change ≥1.5). Functional annotation enrichment analyses of the up- and downregulated genes were as shown. ECM, extracellular matrix; GO, Gene Ontology; PPAR, peroxisome proliferator-activated receptor; TGFB, transforming growth factor β.
Butyrate exposure at a concentration of 10 mM, comparable to that measured in the intestine, for 72 hours had a profound effect on the global pattern of gene expression in both DUOX2ref HIOs (n = 8052 genes) (Figure 1B and Supplemental Table 5) and DUOX2hapl HIOs (n = 7813 genes) (Supplemental Table 6). This was most notable for inductions of genes regulating cellular transporter activity (FDR = 7.18E-06), vesicle localization (FDR = 1.02E-18), and microtubule-based transport (FDR = 2.45E-17) (Figure 1B), and suppression of genes regulating cell proliferation (FDR = 4.44E-19), migration (FDR = 1.92E-19), and ECM formation (FDR = 1.28E-27) (Figure 1B), irrespective of DUOX2 genotype. These data implicated butyrate exposure in regulation of a broad number of genes controlling HIO cellular transport function, proliferation, migration, and ECM production.
ETYA exposure at a concentration of 50 μM, predicted to reverse the ileal gene signature for stricturing, for 12 days had a more specific effect upon the global pattern of gene expression in both DUOX2ref HIOs (n = 1383 genes) (Figure 1C and Supplemental Table 7) and DUOX2hapl HIOs (n = 1155 genes) (Supplemental Table 8). This was most notable for induction of genes regulating peroxisome proliferator-activated receptor (PPAR) (FDR = 7.957E-09) signaling and cholesterol metabolic functions (FDR = 7.90E-23) (Figure 1C and Supplemental Table 7) and suppression of genes regulating ECM formation (FDR = 7.01E-31) (Figure 1C and Supplemental Table 7), irrespective of DUOX2 genotype. This included downregulation of HIO expression of 2 circulating ECM biomarkers for future stricturing identified in the RISK pediatric CD inception cohort study, extracellular matrix protein 1 (ECM1) and collagen type III alpha 1 chain (COL3A1), and C-C motif chemokine ligand 2 (CCL2), a chemokine implicated in inflammatory macrophage-fibroblast interactions and lack of anti-TNF response.3,24,25 Importantly, we did not observe enrichment for suppression of genes involved in cell proliferation or migration, as was noted with butyrate exposure. In this regard, the effects of chronic ETYA exposure appeared to be more specific, implicating upregulation of PPAR signaling and cholesterol metabolism, and suppression of ECM formation.
Butyrate Inhibits Toxin-Induced Epithelial ROS Production
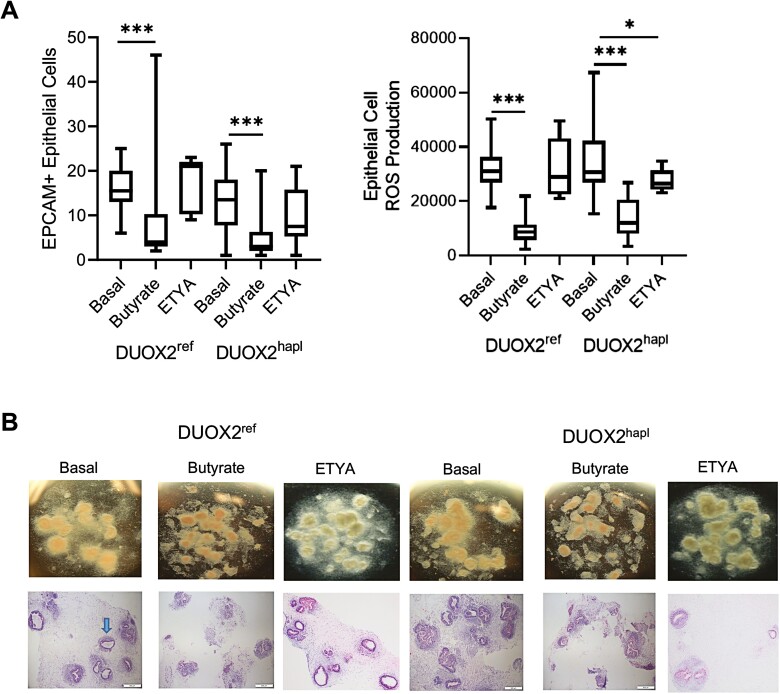
We next tested the effects of butyrate and ETYA exposure upon HIO epithelial cell abundance, morphology, and ROS production. The frequency of epithelial cell adhesion molecule–positive epithelial cells detected by flow cytometry was decreased in both DUOX2ref and DUOX2hapl HIOs following butyrate exposure (Figure 2A). Epithelial cell abundance did not vary with ETYA exposure. Butyrate exposure profoundly inhibited epithelial ROS production in response to the pseudomonas toxin pyocyanin; this was not observed with ETYA exposure (Figure 2A). We did not observe a difference in HIO growth or histology with DUOX2 genetic variation under basal conditions or following ETYA exposure (Figure 2B). However, HIOs appeared more condensed, with a less prominent lumen (arrow), following butyrate exposure. These data confirmed significant effects of butyrate, but not ETYA, upon relative epithelial cell abundance, toxin-induced ROS production, and HIO morphology irrespective of DUOX2 genotype.
Figure 2.
Variation in epithelial cell abundance and reactive oxygen species production with butyrate or eicosatetraynoic acid (ETYA) exposure. A, The frequency of epithelial cell adhesion molecule–positive (EPCAM+) epithelial cells was determined by flow cytometry in DUOX2ref and DUOX2hapl human intestinal organoids (HIOs) under basal conditions and following butyrate (10 mM for 72 hours) or ETYA (50 μM for 72 hours) exposure. Total reactive oxygen species production was measured by flow cytometry in EPCAM+ epithelial cells isolated from DUOX2ref and DUOX2hapl HIOs following pyocyanin stimulation without and with butyrate (10 mM for 72 hours) or ETYA (50 μM for 72 hours) pretreatment. B, HIO morphology was assessed using bright field microscopy and hematoxylin and eosin staining in DUOX2ref and DUOX2hapl HIOs under basal conditions and following butyrate (10 mM for 72 hours) or ETYA (50 μM for 72 hours) exposure. A representative DUOX2ref HIO with associated lumen is indicated by the arrow. n = 12-42 per group, with 6 per group from 2-7 independent experiments. Data are shown as the median (interquartile range) for each group. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 vs HIOs under basal conditions with the same DUOX2 genotype. Images representative of at least 3 replicates in 3 independent experiments are shown. The Shapiro-Wilk test was used to assess the normality of the data. Differences between butyrate- or ETYA-treated samples and untreated samples within the DUOX2ref or DUOX2hapl genotypes were tested using a 1-way analysis of variance with Dunnett’s multiple comparisons test for data that passed the normality test, and using a Kruskal-Wallis test with Dunn’s multiple comparisons test for data that did not pass the normality test.
Butyrate Regulates Mitochondrial Gene Expression and Function
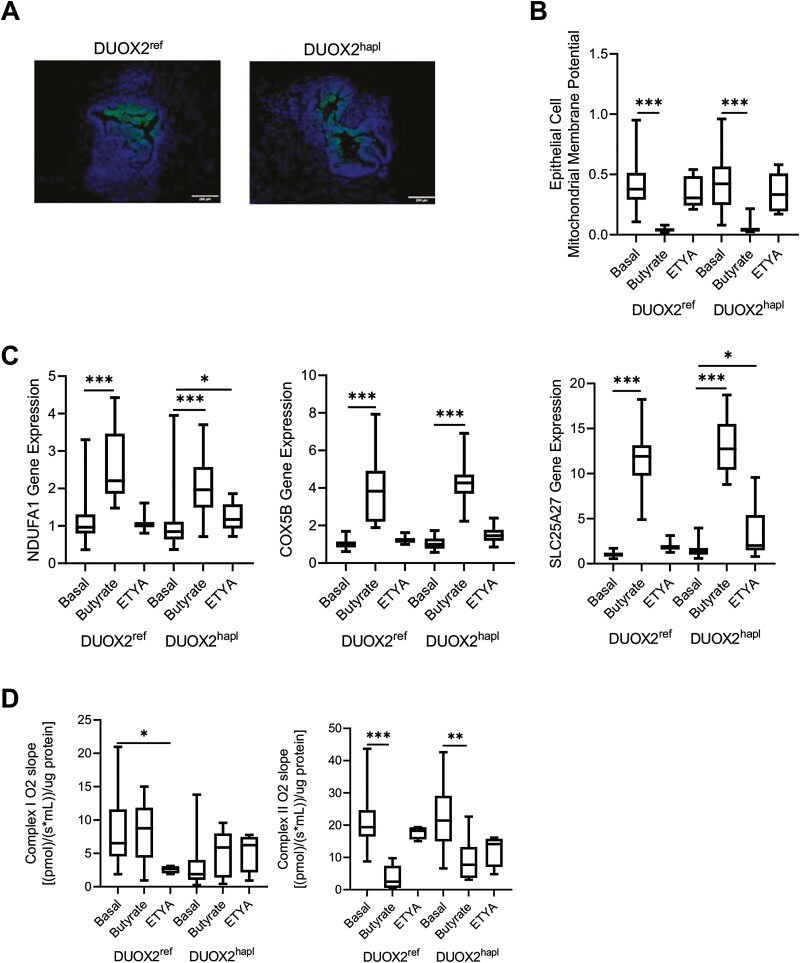
Reduced mitochondrial gene expression was implicated in the development of CD strictures,1 while intact mitochondrial function limits cellular oxidative stress and inflammatory responses.26,27 We therefore next tested for variation in mitochondrial function across the groups. We employed the oxygen-sensitive dye PMDZ, which is retained in tissues at pO2 levels ≤10 mm Hg, to test for relative levels of hypoxia in the HIO cells. Data suggested a relative hypoxia in luminal epithelial cells of both DUOX2ref and DUOX2hapl organoids, similar to the pattern observed in the intestine in vivo (Figure 3A).28 The epithelial MMP was profoundly suppressed by butyrate, with no effect observed following ETYA exposure (Figure 3B). Ileal expression of the mitochondrial uncoupling gene SLC25A27, which reduces the MMP and associated cellular ROS production, was positively associated with time to stricturing in pediatric CD, suggesting a protective effect (r = 0.4926, P = .0465).29 In HIOs, SLC25A27 gene expression was induced by butyrate (Figure 3C), together with expression of mitochondrial complex genes including NDUFA1 and COX5B. This was not observed with ETYA exposure. With the exception of a reduction following ETYA exposure in DUOX2ref HIOs, mitochondrial complex I activity was largely preserved across the butyrate or ETYA treatment groups (Figure 3D). By comparison, mitochondrial complex II activity was profoundly suppressed by butyrate exposure in both HIO lines, in conjunction with downregulation of 3 of the 4 genes that encode mitochondrial complex II (SDHA, SDHB, and SDHD) (Supplemental Table 5). Collectively, these data demonstrated a specific effect of butyrate upon mitochondrial gene expression and function not observed with ETYA, irrespective of DUOX2 genotype.
Figure 3.
Variation in human intestinal organoid (HIO) mitochondrial gene expression and function with butyrate or eicosatetraynoic acid (ETYA) exposure. A, HIO cellular oxygen levels were estimated using the oxygen-sensitive dye pimonidazole, which is retained in tissues (green signal) at pO2 levels ≤10 mm Hg, in DUOX2ref and DUOX2hapl HIOs under basal conditions. Representative images are shown with nuclei stained with DAPI (blue signal). B, The mitochondrial membrane potential was measured by JC-1 staining and flow cytometry in epithelial cell adhesion molecule–positive epithelial cells isolated from DUOX2ref and DUOX2hapl HIOs under basal conditions, or following butyrate (10 mM for 72 hours) or ETYA (50 μM for 72 hours) exposure. C, NDUFA1, COX5B, and SLC25A27 gene expression was measured using real time polymerase chain reaction and RNA isolated from DUOX2ref and DUOX2hapl HIOs under basal conditions, or following butyrate (10 mM for 72 hours) or ETYA (50 μM for 72 hours) exposure. D, HIO mitochondrial complex I and II activity was measured using an Oroboros respirometer in DUOX2ref and DUOX2var HIOs under basal conditions, or following butyrate (10 mM for 72 hours) or ETYA (50 μM for 72 hours) exposure. n = 12-48 per group, with 6 per group from 2-8 independent experiments, for mitochondrial membrane potential and gene expression data. n = 4-23 per group, with 2-4 per group from 2-6 independent experiments, for mitochondrial respiration data. Data are shown as the median (interquartile range) for each group. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 vs HIOs under basal conditions with the same DUOX2 genotype. Images representative of at least 3 replicates in 3 independent experiments are shown. Differences between butyrate- or ETYA-treated samples and untreated samples within the DUOX2ref or DUOX2hapl genotypes were tested using a 1-way analysis of variance with Dunnett’s multiple comparisons test for data that passed the normality test, and using a Kruskal-Wallis test with Dunn’s multiple comparisons test for data that did not pass the normality test.
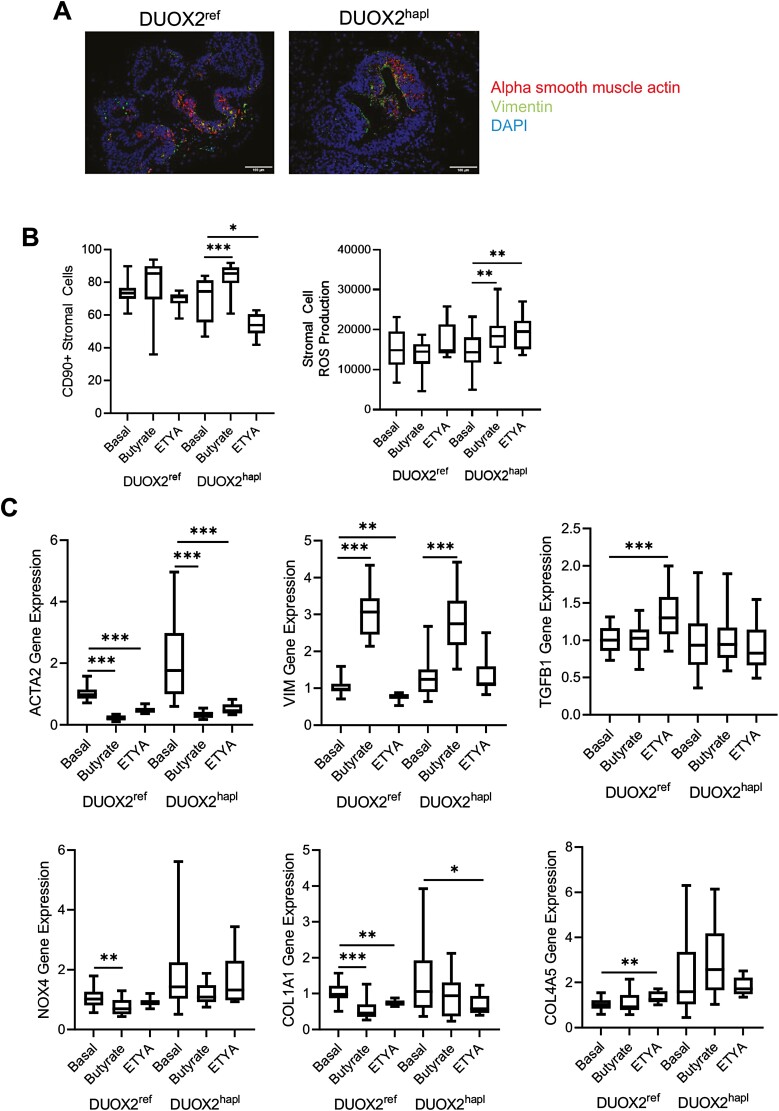
Butyrate and ETYA Regulate Expression of Genes Involved in Wound Healing and ECM Formation
Finally, we tested whether differences in epithelial cell adhesion and stromal cell function suggested by the bulk RNASeq data would be confirmed by real-time PCR, tissue staining, and a measure of HIO stiffness.21 Tissue staining detected stromal cells expressing ACTA2 (myofibroblast) and VIM (fibroblast) but did not reveal a consistent variation in these populations between the DUOX2ref and DUOX2hapl HIOs under basal conditions (Figure 4A). Consistent with the reduction in epithelial cell abundance, we observed an increase in the relative abundance of CD90+ stromal cells following butyrate exposure for 72 hours, which reached significance in the DUOX2hapl HIOs (Figure 4B). By comparison, the relative abundance of CD90+ stromal cells was reduced following ETYA exposure for 72 hours in the DUOX2hapl HIOs (Figure 4B). In contrast to the profound suppression of epithelial ROS production by butyrate (Figure 2A), stromal cell ROS production did not vary across with groups, with the exception of a modest increase in the DUOX2hapl HIOs (Figure 4B). Real-time PCR confirmed a profound suppression of alpha smooth muscle actin (ACTA2) expressed by activated myofibroblasts by both butyrate and ETYA exposure for 72 hours, irrespective of DUOX2 genotype (Figure 4C). By comparison, and consistent with the effect observed for relative CD90+ stromal cell abundance, expression of VIM expressed by fibroblasts increased with butyrate, but not ETYA, exposure. Expression of the type I collagen gene COL1A1 was reduced by both butyrate and ETYA in DUOX2ref HIOs, and by ETYA alone in DUOX2hapl HIOs. Expression of TGFB1, NOX4, and COL4A5 also implicated in wound healing and ECM formation was largely preserved across the groups. These data confirmed effects of butyrate and ETYA exposure on ECM gene expression suggested by the global pattern of gene expression.
Figure 4.
Variation in human intestinal organoid (HIO) stromal cell abundance and extracellular matrix gene expression with butyrate or eicosatetraynoic acid (ETYA) exposure. A, The frequency of HIO alpha smooth muscle actin (ACTA2)–positive (red) and/or vimentin (VIM)–positive (green) stromal cells was determined in DUOX2ref and DUOX2hapl HIOs under basal conditions. Nuclei were stained with DAPI (blue). B, The frequency of CD90+ stromal cells was determined by flow cytometry in DUOX2ref and DUOX2hapl HIOs under basal conditions, or following butyrate (10 mM for 72 hours) or ETYA (50 μM for 72 hours) exposure. Total reactive oxygen species production were measured by flow cytometry in CD90+ stromal cells isolated from DUOX2ref and DUOX2hapl HIOs following pyocyanin stimulation without and with butyrate (10 mM for 72 hours) or ETYA (50 μM for 72 hours) pretreatment. C, Gene expression was measured using real-time polymerase chain reaction and RNA isolated from DUOX2ref and DUOX2hapl HIOs under basal conditions, or following butyrate (10 mM for 72 hours) or ETYA (50 μM for 72 hours) exposure. n = 12-48 per group, with 6 per group from 2-8 independent experiments. Data are shown as the median (interquartile range) for each group. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 vs HIOs under basal conditions with the same DUOX2 genotype. Images representative of at least 3 replicates in 3 independent experiments are shown. Differences between butyrate- or ETYA-treated samples and untreated samples within the DUOX2ref or DUOX2hapl genotypes were tested using a 1-way analysis of variance with Dunnett’s multiple comparisons test for data that passed the normality test, and using a Kruskal-Wallis test with Dunn’s multiple comparisons test for data that did not pass the normality test.
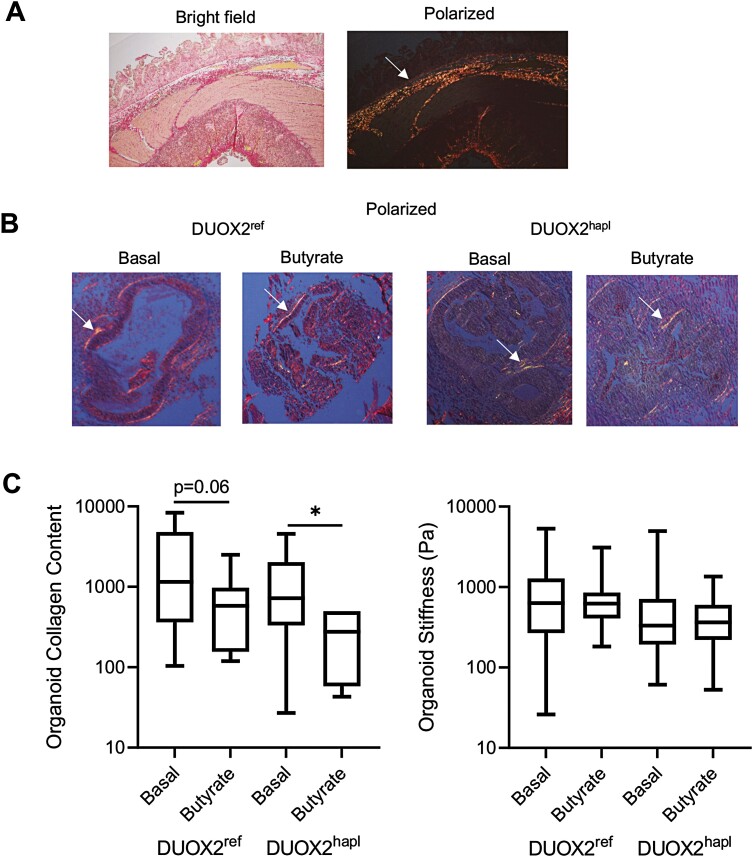
Short-Term Butyrate Exposure Regulates HIO Collagen Content Without Affecting Tissue Stiffness
We utilized Sirius red staining coupled with polarized light microscopy to assess collagen content in the HIO groups. Representative staining for a CD ileal resection specimen is shown in Figure 5A, highlighting the collagen bands detected in the submucosa (arrow). Consistent with this, we observed well-organized collagen bands within the HIOs corresponding to the subepithelial space (arrow, Figure 5B). A reduction in HIO collagen content following 72-hour butyrate exposure was observed which reached significance in DUOX2hapl HIOs (Figure 5C). A prior study confirmed an increase in tissue stiffness within CD ileal strictures relative to adjacent normal bowel, and implicated both tissue fibrosis and smooth muscle hyperplasia.30 We therefore utilized atomic force microscopy to directly measure HIO stiffness, expected to be regulated by combined effects of the cellular actin cytoskeleton, cell-matrix focal adhesion complexes, and the ECM. For these studies, individual HIOs were removed from the surrounding Matrigel, replated, and then tested. As shown in Figure 5C, HIO stiffness did not vary within the DUOX2ref or DUOX2hapl HIOs with the relatively short term 72-hour butyrate exposure. Owing to the profound effects of butyrate upon epithelial cell mitochondrial function and relative abundance, we were not able to test a longer exposure to butyrate. These data confirmed that short-term exposure to butyrate led to a reduction in HIO collagen content, in the absence of a change in tissue stiffness.
Figure 5.
Variation in human intestinal organoid (HIO) collagen content and tissue stiffness with butyrate exposure. A, Collagen content in a representative Crohn’s disease ileal resection specimen was assessed using Sirius red staining and polarized light microscopy. The arrow indicates the submucosal collagen band. B, Collagen content was assessed in DUOX2ref and DUOX2hapl HIOs under basal conditions and following butyrate exposure (10 mM for 72 hours) using Sirius red staining and polarized light microscopy. The arrow indicates the subepithelial collagen band. C, Collagen protein abundance was quantified using Sirius red staining and polarized light microscopy in ImageJ software. Tissue stiffness was determined using atomic force microscopy in in DUOX2ref and DUOX2hapl HIOs under basal conditions and following butyrate exposure (10 mM for 72 hours). n = 6-14 per group, with 2-6 per group from 3 independent experiments for collagen content data. n = 71-105 measurements per group, with 6 per group from 2 independent experiments, for tissue stiffness data. Data are shown as the median (interquartile range) for each group. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 vs HIOs under basal conditions with the same DUOX2 genotype. Images representative of at least 3 replicates in 3 independent experiments are shown. Differences between butyrate-treated samples and untreated samples within the DUOX2ref or DUOX2hapl genotypes were tested using the unpaired t test for data that passed the normality test, and the Mann-Whitney test for data that did not pass the normality test.
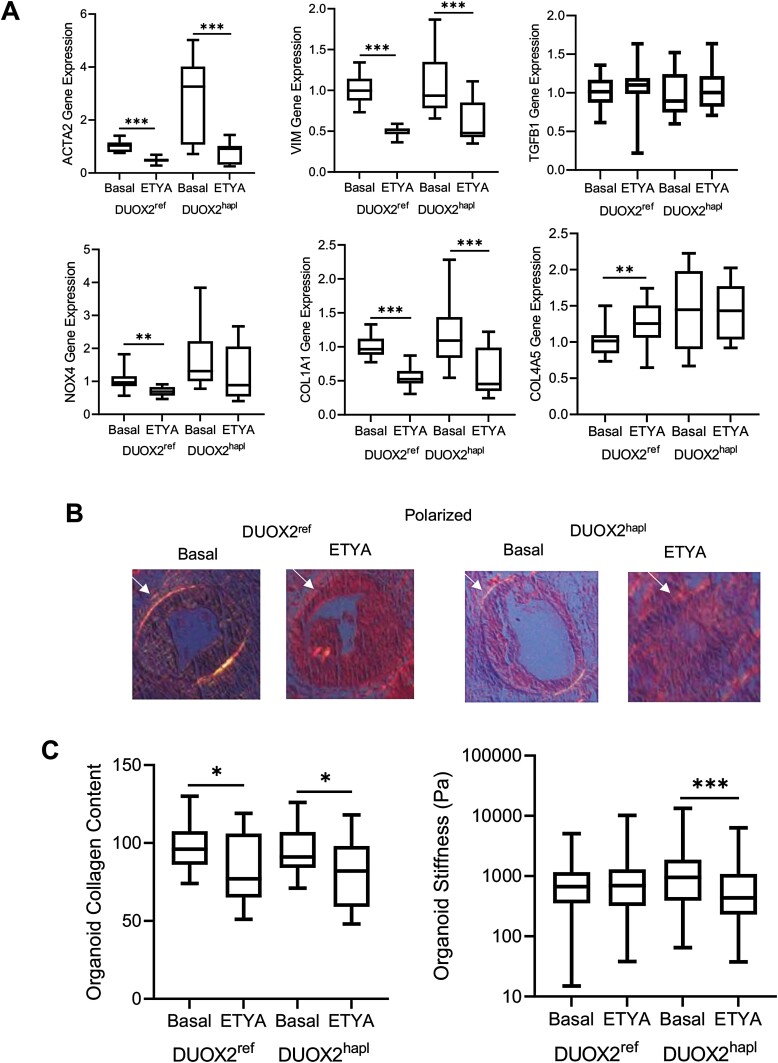
Chronic ETYA Exposure Reduces HIO Collagen Content and Tissue Stiffness
Because ETYA did not affect epithelial mitochondrial function or relative cellular abundance, we were able to test the effects of a chronic 12-day exposure upon ECM gene expression and associated HIO collagen content and stiffness. Real-time PCR confirmed a profound suppression of both alpha smooth muscle actin (ACTA2) expressed by activated myofibroblasts, and VIM expressed by fibroblasts, with chronic ETYA exposure, irrespective of DUOX2 genotype (Figure 6A). Expression of the type I collagen gene COL1A1 was reduced by ETYA in both the DUOX2ref and DUOX2hapl HIOs. Expression of TGFB1, NOX4, and COL4A5 also implicated in wound healing and ECM formation was largely preserved across the groups. We again utilized Sirius red staining coupled with polarized light microscopy to assess collagen content in the HIO groups (Figure 6B). A reduction in HIO collagen content following 12-day ETYA exposure was observed irrespective of DUOX2 genotype (Figure 6C). Under these conditions, HIO stiffness was reduced in the DUOX2hapl HIOs, but not the DUOX2ref HIOs. These data confirmed that chronic exposure to ETYA led to a reduction in core ECM gene expression and HIO collagen content irrespective of DUOX2 genotype, with a significant reduction in tissue stiffness observed in the DUOX2hapl line.
Figure 6.
Variation in human intestinal organoid (HIO) collagen content and tissue stiffness with chronic eicosatetraynoic acid (ETYA) exposure. A, Gene expression was measured using real-time polymerase chain reaction and RNA isolated from DUOX2ref and DUOX2hapl HIOs under basal conditions, or following ETYA (50 μM for 12 days) exposure. B, Collagen content was assessed in DUOX2ref and DUOX2hapl HIOs under basal conditions and following ETYA exposure (50 μM for 12 days) using Sirius red staining and polarized light microscopy. The arrow indicates the subepithelial collagen band. C, Collagen protein abundance was quantified using Sirius red staining and polarized light microscopy in ImageJ software. Tissue stiffness was determined using atomic force microscopy in in DUOX2ref and DUOX2hapl HIOs under basal conditions and following ETYA exposure (50 μM for 12 days). n = 18 per group, with 6 per group from 3 independent experiments, for gene expression data. n = 15-21 per group, with 2-6 per group from 3 independent experiments, for collagen content data. n = 186-206 measurements per group, with 6 per group from 2 independent experiments, for tissue stiffness data. Data are shown as the median (interquartile range) for each group. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 vs HIOs under basal conditions with the same DUOX2 genotype. Images representative of at least 3 replicates in 3 independent experiments are shown. Differences between ETYA-treated samples and untreated samples within the DUOX2ref or DUOX2hapl genotypes were tested using the unpaired t test for data that passed the normality test, and the Mann-Whitney test for data that did not pass the normality test.
DISCUSSION
The DUOX2 nicotinamide adenine dinucleotide phosphate oxidase is expressed at the apical surface of ileal enterocytes where it generates hydrogen peroxide to control microbiota and maintain homeostasis.10 While rare DUOX2 loss-of-function mutations increase risk for inflammatory bowel disease, whether more common variants would also be linked to patient outcomes was not known.11,12 We reported that children with CD who subsequently developed strictures exhibited a reduction in mitochondrial gene expression, and an increase in expression of ECM genes involved in wound healing, in the ileum at diagnosis.1 Our perturbagen analysis of these gene expression data suggested a potential specific antifibrotic effect of the long-chain fatty acid ETYA.4 In the current study, we found that carriage of common DUOX2 gene variants is associated with the development of strictures in pediatric CD. We developed an HIO model system derived from CD patient iPSCs to test potential small molecule regulation of epithelial and stromal cell mechanisms of stricture formation, in the context of a common DUOX2 loss-of-function haplotype (DUOX2hapl). We found that while the microbial metabolite butyrate exerted broad effects upon both epithelial and stromal cell function, ETYA exhibited more specific effects upon ECM gene expression, and associated HIO collagen content and tissue stiffness. These data illustrate a pipeline for utilization of patient-specific gene expression data and intestinal organoids to test small molecule regulation of pathways linked to stricture formation in CD, supporting another recent report that utilized control and CD patient–derived HIOs to test epithelial and mesenchymal responses to TGFβ.15
We recently reported a refined ileal gene signature (n = 518 genes) for future stricturing behavior in children with CD utilizing deeper RNA sequencing than in the initial report.1,4 We conducted a perturbagen bioinformatics analysis using data in the National Institutes of Health’s LINCS Web-based tool to identify small molecules likely to suppress expression of differentially expressed genes regulating inflammatory macrophage and fibroblast functions contained within the CD patient gene signature for stricturing.4 Although the signatures are based on epithelial cell lines, we and others have shown that this approach can be leveraged for discovery of novel therapies for fibrotic disorders, including idiopathic pulmonary fibrosis.31,32 This analysis prioritized 140 small molecules predicted to suppress inflammatory macrophage and fibroblast gene signatures associated with future stricturing.4 For the current study, we focused on testing ETYA as proof of concept for this pipeline given extensive experimental data for its role as a PPAR agonist and inhibitor of fibroblast proliferation.33,34 Future studies employing a more high-throughput HIO system are now indicated to screen these candidate small molecules more broadly.
Enterocyte mitochondrial function plays a critical role in maintaining barrier function and tolerant responses to commensal microbes.26 Previous studies have shown that increased oxidative stress and reduced mitochondrial cellular respiration promote inflammatory responses to commensals.27 Effects of the microbial metabolite butyrate upon cellular respiration in the gut are well established, and we confirmed profound effects upon the epithelial mitochondrial membrane potential, mitochondrial complex II activity, and toxin-induced ROS production in the HIO model system. By comparison, the long-chain fatty acid ETYA, predicted to exert a more specific antifibrotic effect, did not regulate HIO mitochondrial function or ROS production. Ileal SLC25A27 expression was associated with a longer time to stricturing in patients, suggesting a protective effect. It was upregulated by butyrate in the HIO model system, in conjunction with suppression of the mitochondrial membrane potential and microbial toxin–induced ROS production. SLC25A27 encodes mitochondrial uncoupling protein 4 (UCP4), and studies have shown that UCP4 regulates cellular energy metabolism by increasing glucose uptake and shifting the mode of adenosine triphosphate production from mitochondrial respiration to glycolysis.29 Consequently, this shift in energy metabolism reduces ROS production and increases the resistance of cells to oxidative stress. These data support the utility of the HIO model system to test effects of small molecules including butyrate upon mitochondrial function and oxidative stress implicated in barrier function and inflammatory responses to commensals in CD.27
We found that children with CD and heterozygous DUOX2 mutations were more likely to develop strictures. We therefore tested a candidate antifibrotic agent, ETYA, in the context of iPSCs derived from CD patients carrying the reference DUOX2 genotype (DUOX2ref), and a common DUOX2 loss-of-function haplotype (DUOX2hapl)12 in the HIO model system. At the functional level, HIOs exhibited tissue stiffness comparable to normal human ileum; this did not vary with 72-hour butyrate exposure.30 A longer exposure to candidate small molecules may be required to promote HIO remodeling and changes in tissue stiffness, as we did observe a reduction in collagen content following the 72-hour butyrate exposure, which reached significance in the DUOX2hapl HIOs. Consistent with this concept, a chronic 12-day exposure to ETYA did result in a significant reduction in HIO ECM gene expression and collagen content irrespective of DUOX2 genotype, coupled with a reduction in tissue stiffness in the DUOX2hapl HIOs. Our previous analysis suggested that ETYA would exert antifibrotic effects via activation of the peroxisome proliferator-activated receptors PPARA and PPARG, and we confirmed activation of PPAR target genes in the HIO model system.4 Consistent with a prior study in cultured fibroblasts, we confirmed that ETYA induced lipid metabolic genes, while suppressing ECM genes and expression of the myofibroblast marker alpha-smooth muscle actin.34 This included downregulation of the ECM biomarkers ECM1 and COL3A1 previously linked to future stricturing in pediatric CD, supporting the clinical relevance of the HIO model system.24,25 It will be important in future studies to test for variation in ETYA dependent PPAR activation, lipid metabolism, and collagen production by HIO myofibroblasts. While effects of ETYA upon gene expression and HIO collagen content did not vary between the HIO lines, we only observed a reduction in tissue stiffness in the DUOX2hapl line. This may suggest the ability to use the HIO model system to detect personalized differences in important physiologic responses to small molecules. For example, the RNASeq dataset suggested specific effects of ETYA in suppressing genes regulating basement membrane formation (FDR = 1.284E-14), anchoring junctions (FDR = 9.386E-08), and focal adhesion complexes (FDR = 1.823E-06) (Supplemental Table 8) in the DUOX2hapl HIO line. A more high-throughput system involving a much greater number of patient-derived lines will be required to test these potential patient-specific responses.
Our study has several strengths, but also some limitations. Given the 3-fold higher frequency of the common DUOX2 H678R variant in African Americans relative to individuals with European ancestry (Supplemental Table 1), it will be important in future studies to validate the frequency of DUOX2 mutations, and associations with African American race and stricturing behavior, in independent CD cohorts. The clinical outcome of CD needing surgery for complicated disease in African Americans is reported to be worse. Although access to care and racial disparities are postulated as an explanation, our study provides a biological plausibility as well for the differential outcome seen in African Americans with CD and paves the way for future genome enabled clinical trials for preventing strictures based on race.35,36 It is possible that additional genetic differences beyond the defined DUOX2 haplotype could account for differences in HIO function observed. Future studies using CRISPR/Cas9 gene editing will provide isogenic controls for selected DUOX2 mutations. A recent study examined effects of TGFβ upon profibrotic gene expression programs in purified epithelial vs mesenchymal cell populations isolated from control and CD patient–derived HIOs.15 Our analysis of bulk gene expression data will need to be extended with a similar examination of purified cell populations in the future, to test for genes regulating epithelial-to-mesenchymal transition, and mesenchymal ECM expression, more precisely.15 It will be important to develop HIOs that include immune cells such as macrophages also implicated in stricturing behavior; this work is ongoing.3 It is likely that ETYA may exert antifibrotic effects in CD in part by suppressing signals regulating inflammatory macrophage-fibroblast cross-talk, including downregulation of CCL2 identified in the current study.3,4 Finally, in vivo HIO transplantation may ultimately be required to assess the functional consequences of candidate small molecules, in the context of the intestinal luminal contents.37
CONCLUSIONS
We found that DUOX2 mutation carriage is associated with stricturing behavior in pediatric CD, and we developed an HIO model system to test small molecules predicted to have an antifibrotic effect in the context of a common DUOX2 haplotype. HIOs exhibited mitochondrial function and tissue stiffness under basal conditions very similar to normal human ileum. The microbial metabolite butyrate exerted a profound effect upon mitochondrial function, and constrained toxin-induced epithelial ROS production and basal ECM gene expression. By comparison, the long-chain fatty acid ETYA exhibited a more specific suppression of ECM gene expression, and associated HIO collagen content and tissue stiffness, with data suggesting patient-specific effects. Collectively, these data have implications for our understanding of pathways regulating ileal fibrosis and support the use of HIOs as a relevant model system to test candidate small molecules prioritized via the perturbagen bioinformatics pipeline in a personalized manner.
Supplementary Material
Acknowledgments
The following RISK site investigators enrolled patients and collected clinical data and biospecimens: Thomas D. Walters, Anne Griffiths, Robert N. Baldassano, Joshua D. Noe, Jeffrey S. Hyams, Wallace V. Crandall, Barbara S. Kirschner, Melvin B. Heyman, Scott Snapper, Stephen L. Guthery, Marla C. Dubinsky, Neal S. Leleiko, and Anthony R. Otley.
Contributor Information
Ingrid Jurickova, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, the University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Erin Bonkowski, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, the University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Elizabeth Angerman, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, the University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Elizabeth Novak, Division of General and Thoracic Surgery, UPMC Children’s Hospital of Pittsburgh, University of Pittsburgh School of Medicine, Pittsburgh, PA, USA.
Alex Huron, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, the University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Grayce Akers,, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, the University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Kentaro Iwasawa, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, the University of Cincinnati College of Medicine, Cincinnati, OH, USA; Division of Biomedical Informatics, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Tzipi Braun, Department of Pediatrics, Sheba Medical Center, Tel-Aviv University, Tel-HaShomer, Israel.
Rotem Hadar, Department of Pediatrics, Sheba Medical Center, Tel-Aviv University, Tel-HaShomer, Israel.
Maria Hooker, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, the University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Sarah Han, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, the University of Cincinnati College of Medicine, Cincinnati, OH, USA.
David J Cutler, Department of Human Genetics, Emory University, Atlanta, GA, USA.
David T Okou, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Emory University, Atlanta, GA, USA.
Subra Kugathasan, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Emory University, Atlanta, GA, USA.
Anil Jegga, Division of Biomedical Informatics, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, University of Cincinnati College of Medicine, Cincinnati, OH, USA.
James Wells, Division of Developmental Biology, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Takanori Takebe, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, the University of Cincinnati College of Medicine, Cincinnati, OH, USA; Division of Developmental Biology, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, University of Cincinnati College of Medicine, Cincinnati, OH, USA; Institute of Research, Tokyo Medical and Dental University, Japan.
Kevin P Mollen, Division of General and Thoracic Surgery, UPMC Children’s Hospital of Pittsburgh, University of Pittsburgh School of Medicine, Pittsburgh, PA, USA.
Yael Haberman, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, the University of Cincinnati College of Medicine, Cincinnati, OH, USA; Department of Pediatrics, Sheba Medical Center, Tel-Aviv University, Tel-HaShomer, Israel.
Lee A Denson, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Cincinnati Children’s Hospital Medical Center, the University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Author Contributions
Study concept and design: S.K., J.W., T.T., Y.H., L.A.D. Acquisition of data: I.J., E.B., E.A., E.N., A.H., G.A., K.I., M.H., S.H. Analysis and interpretation of data: I.J., E.B., E.A., A.H., G.A., K.I., T.B., R.H., M.H., S.H., D.C., D.O., A.J., Y.H., L.A.D. Drafting of the manuscript: I.J., E.B., E.A., A.H., M.H., S.H., Y.H., L.A.D. Critical revision of the manuscript for important intellectual content: S.K., J.W., T.T., K.M., Y.H., L.A.D. Obtained funding: S.K., T.T., K.M., L.A.D. Administrative, technical, or material support: I.J., E.B., E.A., E.N., A.H., G.A., K.I., M.H., S.H. Study supervision: I.J., L.A.D.
Supported By
This work was supported by the Crohn’s and Colitis Foundation (L.A.D., S.K.); the CureForIBD Foundation (L.A.D.); the Cincinnati Children’s Center for Stem Cell and Organoid Medicine (L.A.D., J.W., T.T.); the Gene Analysis, Pluripotent Stem Cell, and Integrative Morphology cores of the National Institutes of Health–supported Cincinnati Children’s Hospital Research Foundation Digestive Health Center (1P30DK078392); and National Institutes of Health grants R01 DK098231 (L.A.D., S.K.), R56 DK098231 (L.A.D., SK), R21 DK128635 (L.A.D. & TT), and R01 DK120986 (K.M.). Y.H. is also supported in part by the Helmsley Charitable Trust and the European Research Council (Grant # 758313). The study sponsors did not play any role in the study design or in the collection, analysis, and interpretation of data.
Conflicts of Interest
The authors have no financial arrangement(s) with a company whose product figures prominently in the submitted manuscript or with a company making a competing product.
Data Availability
The human whole exome sequencing data are publicly available on dbGaP (study accession number phs001076.v1.p1; https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001076.v1.p1).The human ileal RNA sequencing data are deposited in the SRA database (accession number PRJNA594730; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA594730).
References
- 1. Kugathasan S, Denson LA, Walters TD, et al. . Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet. 2017;389(10080):1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walters TD, Kim MO, Denson LA, et al. . Increased effectiveness of early therapy with anti-tumor necrosis factor-alpha vs an immunomodulator in children with Crohn’s disease. Gastroenterology. 2014;146(2):383–391. [DOI] [PubMed] [Google Scholar]
- 3. Martin JC, Chang C, Boschetti G, et al. . Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 2019;178(6):1493–1508.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haberman Y, Minar P, Karns R, et al. . Mucosal inflammatory and wound healing gene programs reveal targets for stricturing behavior in pediatric Crohn’s disease. J Crohns Colitis. 2020;15(2):273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heerdt BG, Houston MA, Wilson AJ, et al. . The intrinsic mitochondrial membrane potential (Deltapsim) is associated with steady-state mitochondrial activity and the extent to which colonic epithelial cells undergo butyrate-mediated growth arrest and apoptosis. Cancer Res. 2003;63(19):6311–6319. [PubMed] [Google Scholar]
- 6. Pacheco RG, Esposito CC, Muller LC, et al. . Use of butyrate or glutamine in enema solution reduces inflammation and fibrosis in experimental diversion colitis. World J Gastroenterol. 2012;18(32):4278–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Parada Venegas D, De la Fuente MK, Landskron G, et al. . Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. 2019;10:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haberman Y, Tickle TL, Dexheimer PJ, et al. . Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124(8):3617–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lipinski S, Till A, Sina C, et al. . DUOX2-derived reactive oxygen species are effectors of NOD2-mediated antibacterial responses. J Cell Sci. 2009;122(Pt 19):3522–3530. [DOI] [PubMed] [Google Scholar]
- 10. Grasberger H, Gao J, Nagao-Kitamoto H, et al. . Increased expression of DUOX2 Is an epithelial response to mucosal dysbiosis required for immune homeostasis in mouse intestine. Gastroenterology. 2015;149(7):1849–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grasberger H, Magis AT, Sheng E, et al. . DUOX2 variants associate with preclinical disturbances in microbiota-immune homeostasis and increased inflammatory bowel disease risk. J Clin Invest. 2021;131(9):e141676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Muzza M, Rabbiosi S, Vigone MC, et al. . The clinical and molecular characterization of patients with dyshormonogenic congenital hypothyroidism reveals specific diagnostic clues for DUOX2 defects. J Clin Endocrinol Metab. 2014;99(3):E544–E553. [DOI] [PubMed] [Google Scholar]
- 13. Rodansky ES, Johnson LA, Huang S, et al. . Intestinal organoids: a model of intestinal fibrosis for evaluating anti-fibrotic drugs. Exp Mol Pathol. 2015;98(3):346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spence JR, Mayhew CN, Rankin SA, et al. . Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470(7332):105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Estrada HQ, Patel S, Rabizadeh S, et al. . Development of a personalized intestinal fibrosis model using human intestinal organoids derived from induced pluripotent stem cells. Inflamm Bowel Dis. Published online December 17, 2021. doi: 10.1093/ibd/izab292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shaw KA, Cutler DJ, Okou D, et al. . Genetic variants and pathways implicated in a pediatric inflammatory bowel disease cohort. Genes Immun. 2019;20(2):131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCracken KW, Howell JC, Wells JM, et al. . Generating human intestinal tissue from pluripotent stem cells in vitro. Nat Protoc. 2011;6(12):1920–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McCracken KW, Cata EM, Crawford CM, et al. . Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516(7531):400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaimal V, Bardes EE, Tabar SC, et al. . ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res. 2010;38(Web Server issue):W96–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taylor SC, Nadeau K, Abbasi M, et al. . The ultimate qPCR experiment: producing publication quality, reproducible data the first time. Trends Biotechnol. 2019;37(7):761–774. [DOI] [PubMed] [Google Scholar]
- 21. Ouchi R, Togo S, Kimura M, et al. . Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 2019;30(2):374–384.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O’Neill S, Brault J, Stasia MJ, et al. . Genetic disorders coupled to ROS deficiency. Redox Biol. 2015;6:135–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harn HI, Wang SP, Lai YC, et al. . Symmetry breaking of tissue mechanics in wound induced hair follicle regeneration of laboratory and spiny mice. Nat Commun. 2021;12(1):2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ballengee CR, Stidham RW, Liu C, et al. . Association between plasma level of collagen type III alpha 1 chain and development of strictures in pediatric patients with Crohn’s disease. Clin Gastroenterol Hepatol. 2019;17(9):1799–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu J, Lubman DM, Kugathasan S, et al. . Serum protein biomarkers of fibrosis aid in risk stratification of future stricturing complications in pediatric Crohn’s disease. Am J Gastroenterol. 2019;114(5):777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mancini NL, Rajeev S, Jayme TS, et al. . Crohn’s disease pathobiont adherent-invasive E coli disrupts epithelial mitochondrial networks with implications for gut permeability. Cell Mol Gastroenterol Hepatol. 2021;11(2):551–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saxena A, Lopes F, McKay DM.. Reduced intestinal epithelial mitochondrial function enhances in vitro interleukin-8 production in response to commensal Escherichia coli. Inflamm Res. 2018;67(10):829–837. [DOI] [PubMed] [Google Scholar]
- 28. Zheng L, Kelly CJ, Colgan SP.. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: cellular responses to hypoxia. Am J Physiol Cell Physiol. 2015;309(6):C350–C360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu D, Chan SL, de Souza-Pinto NC, et al. . Mitochondrial UCP4 mediates an adaptive shift in energy metabolism and increases the resistance of neurons to metabolic and oxidative stress. Neuromolecular Med. 2006;8(3):389–414. [DOI] [PubMed] [Google Scholar]
- 30. Stidham RW, Xu J, Johnson LA, et al. . Ultrasound elasticity imaging for detecting intestinal fibrosis and inflammation in rats and humans with Crohn’s disease. Gastroenterology. 2011;141(3):819–826.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karatzas E, Bourdakou MM, Kolios G, et al. . Drug repurposing in idiopathic pulmonary fibrosis filtered by a bioinformatics-derived composite score. Sci Rep. 2017;7(1):12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sontake V, Wang Y, Kasam RK, et al. . Hsp90 regulation of fibroblast activation in pulmonary fibrosis. JCI Insight. 2017;2(4):e91454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berg C, Hammarstrom S, Herbertsson H, et al. . Platelet-induced growth of human fibroblasts is associated with an increased expression of 5-lipoxygenase. Thromb Haemost. 2006;96(5):652–659. [PubMed] [Google Scholar]
- 34. McGowan SE, Jackson SK, Doro MM, et al. . Peroxisome proliferators alter lipid acquisition and elastin gene expression in neonatal rat lung fibroblasts. Am J Physiol. 1997;273(6):L1249–L1257. [DOI] [PubMed] [Google Scholar]
- 35. Brant SR, Okou DT, Simpson CL, et al. . Genome-wide association study identifies African specific susceptibility loci in African Americans with inflammatory bowel disease. Gastroenterology. 2017;152(1):206–217.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Somineni HK, Nagpal S, Venkateswaran S, et al. . Whole-genome sequencing of African Americans implicates differential genetic architecture in inflammatory bowel disease. Am J Hum Genet. 2021;108(3):431–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Singh A, Poling HM, Sundaram N, et al. . Evaluation of transplantation sites for human intestinal organoids. PLoS One. 2020;15(8):e0237885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The human whole exome sequencing data are publicly available on dbGaP (study accession number phs001076.v1.p1; https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001076.v1.p1).The human ileal RNA sequencing data are deposited in the SRA database (accession number PRJNA594730; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA594730).