Abstract
The SARS-CoV-2 is an RNA-based virus and the most vital step of its survival is the attachment to hACE2 through its spike protein. Although SARS-CoV-2 has the ability to maintain high accurate replication and it can be accepted as a low mutation risked virus, it already showed more than nine thousand mutations in spike protein, of which 44 mutations are located within a 3.2 Å interacting distance from the hACE2 receptor. Mutations on spike protein, N501Y and N501T raised serious concerns for higher transmissibility and resistance towards current vaccines. In the current study, the mutational outcomes of N501Y and N501T on the hACE2-SARS CoV-2 spike protein complexation were analyzed by employing all-atom classic molecular dynamics (MD) simulations. These simulations revealed that both N501Y and N501T mutations increased the binding strength of spike protein to the host hACE2, predicted by binding free energy analysis via MM/GBSA rescoring scheme. This study highlights the importance of energy-based analysis for identifying mutational outcomes and will shed light on handling long-term and effective treatment strategies including repurposing anti-viral drugs, anti-SARS-CoV-2 antibodies, vaccines, and antisense based-therapies.
Keywords: SARS-CoV-2 mutations, N501Y and N501T mutations, hACE2-SARS-CoV-2 spike protein binding, Effect of mutations on binding energy, Molecular Dynamics simulations
Graphical abstract
1. Introduction
The SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) is an RNA-based virus, which uses RNA as a genetic material instead of DNA [1,2]. The life cycle of these viruses begins with the attachment of viruses to the human host through receptor-ligand interactions. The viral surface receptor, receptor binding domain (RBD) of spike protein, comes in contact with the angiotensin-converting enzyme 2 (hACE2) on the surface of type 2 alveolar epithelium cells of the human upper respiratory tract (Fig. 1 ). After the binding of spike protein to the hACE2, SARS-CoV-2 virions begin to enter host cells either, via endosomes, and or membrane fusion. Following the viral entry, viral RNA is allowed to spread inside the host. Host machinery, ribosomes on the rough ER, translate viral genetic material directly as it is a positive-stranded RNA. Virion precursors are then assembled in the host, which is followed by the transport to the cell surface via small vesicles. SARS-CoV-2 virions released from the host are following the same life cycle, leading to high viral loads in the upper respiratory tract [[3], [4], [5], [6], [7], [8]].
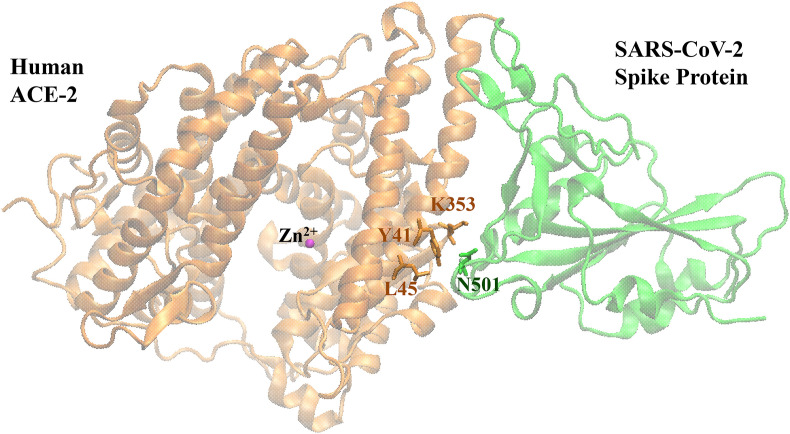
Fig. 1.
X-ray structure of hACE2-SARS CoV-2 spike protein RBD complex (PDB ID: 6M0J). The active site Zn2+ ion of hACE2 was shown with magenta sphere. Residues comprising the hot spot focused on the study were labeled.
Generally, RNA viruses are more open to new mutations as they lack exonuclease-proofreading activity of the virus-encoded RNA polymerases. The only exceptions are Nidoviruses, which.
Comprise the coronaviruses as well [9,10]. SARS-CoV-2, a member of the coronavirus family, has the ability to maintain high accurate replication. Thus, SARS-CoV-2 can be accepted as a low mutation risked virus [1]. However, COVID-19 caused by SARS-CoV-2 infection has affected millions of people around the world. Thereby, the COVID-19 pandemic provided a good environment for the creation of novel mutations. Recent studies have shown the presence of more than nine thousand mutations have raised in spike protein, of which 44 mutations located within a 3.2 Å interacting distance from the hACE2 receptor [9,11,12]. However, there is undoubtedly an increase in the number of single mutations that occurred during pandemic.
Previous studies on MERS-CoV and SARS-CoV and another member of the coronaviruses have demonstrated that single amino acid mutations make them resistant to neutralizing antibodies [[13], [14], [15]]. Furthermore, several mutations on the SARS-CoV-2 spike protein changed the SARS-CoV-2-hACE2 binding affinities and also transmissivity as well [14,16,17]. The bioinformatic analysis also showed that the spike protein-encoding region, RBD in the spike protein in particular, which generates proteins to be utilized in the host attachment, are more prone to new mutations compared to other genomic regions [9,18].
In this study, mutational outcomes of N501Y and N501T, from structural and energetics point of view, on the human hACE2-SARS-CoV-2 spike protein complex will be analyzed using molecular dynamics (MD) simulations. MD simulations will provide valuable data on the dynamics of each complex and binding free energy calculations will shed light on the change on the binding strength of spike protein upon mutations.
2. Materials and methods
2.1. Systems construction
X-ray structure of SARS CoV-2 spike protein RBD complexed with human hACE2 receptor (PDB ID: 6M0J, resolution: 2.45 Å) [19] was used as the initial model. Crystal water molecules were deleted and the most likely protonation state of each titratable residue at physiological pH (∼7.0) was predicted by the PROPKA server [20]. Asn501Tyr and Asn501Thr mutations were performed in YASARA Structure software [21], where the energy minimized conformations of Tyr and Thr residue side chains were obtained.
2.2. Molecular dynamics (MD) simulations and analysis
GROMACS 5.1.4 software [22,23] was utilized for MD simulations and GROMOS 53A6 force field was used [24]. The force field also includes the parameters for Zn2+. Each system, hACE2-WT spike protein, hACE2-N501Y mutant spike protein, and hACE2-N501T mutant spike protein, was placed into the center of the box with dimensions 10.0 x 10.0 × 15.0 nm. These dimensions were chosen to ensure that proteins stay in the box (unit cell) throughout the simulations and does not interact with its copy image dictated by periodic boundary conditions (PBC). The protein amino acid distances at the edges, calculated by the last atom to the solvent atoms at the edge of the unit cell were 1.7 nm, 1.3 nm, 1.3 nm for x, y, and z dimensions, respectively. These distances were much larger than chosen cut-off of 1.2 nm. The boxes were filled with extended simple point charge (SPC/E) model water molecules [25] and 58 Na+, 34 Cl- ions were added to neutralize systems and mimic the physiological environment. WT system comprised of ∼144,577 atoms, N501Y system of 144,590 atoms and N501T of 144,566 atoms. Each starting system was subsequently energy-minimized using the steepest descent method for 50,000 steps with energy minimization tolerance of 2.39 kcal mol−1 nm−1 and step size of 0.01 nm. Then, energy-minimized structures were taken for the production phase. All-atom MD production simulations were run for 100 nano-seconds (ns) for each system with the constant number of particles (N), pressure (P), and temperature (T), i.e. NPT ensemble, for each system. The SETTLE algorithm [26] was employed to constrain the bond length and bond angle of the solvent molecules. LINCS algorithm [27] was utilized to constraint bond lengths of the amino acid residues were constrained using. Long-range electrostatic interactions were treated with Particle-mesh Ewald (PME) method [28]. A constant pressure of 1 bar was applied with a coupling constant of 1.0 pico-seconds (ps) and water molecules/ions were coupled separately to a bath at 300 K with a coupling constant of 0.1 ps. Leap-frog algorithm [29] was used for integrating the equation of motion, which was integrated at 2 femto-seconds (fs) time steps. Tools available in GROMACS and VMD 1.9.1 [30] programs were utilized to analyze trajectories. The most representative structure was obtained by cluster command implemented in GROMACS 5.1.4. It adds a structure to a cluster when its distance to any element of the cluster is less than a cut-off value of 0.3 nm. The center of a cluster is the structure with the smallest average RMSD from all other structures of the cluster.
All results and discussion were based on the data from the entire 100 ns trajectory for each system. For some cases, the most representative structure was mentioned throughout the manuscript, which was obtained from GROMACS software based on equilibrated part (60–100 ns) of RMSD value graphs.
2.3. Binding free energy calculations and solvation energy estimation
Many free energy calculation methods have been developed to calculate the protein-protein binding free energies and determine the hot spots at interface surfaces thus far. Chen et al. studied the rescoring capability of Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) [31]. It was revealed that the use of the MM/GBSA scheme is a more efficient way to predict binding affinities for protein-protein interactions compared to alternative rescoring functions such as MM/PBSA. Therefore, we have decided to implement MM/GBSA scheme as a free energy calculation method. Binding free energy calculations were performed on the HAWKDOCK server (using default parameters), which utilizes MM/GBSA method [32]. As HAWKDOCK server calculates binding free energies both for the total complex and per residue, which helped us analyze contributions of each residue on the binding interface in detail. We provided snapshots (a total of 21 snapshots), obtained from every one ns of the equilibrated time frame of the simulations (80–100 ns) for hACE2-WT and mutant spike proteins to the HAWKDOCK server (MM/GBSA). For WT system, calculations were also repeated with snapshots for each 500 ps of the simulations, for the total of 41 snapshots and the results were similar (Supporting information 2).
We also used PISA server to evaluate the solvation-free energy gain (ΔiG) upon binding and interface surface [33]. These calculations were performed on cluster structures for each system, obtained with clustering method explained above.
3. Results and discussion
Even though the SARS-CoV-2, which is a member of the coronavirus family, has the ability to maintain replication with high accuracy, the current pandemic affecting all the continents, and regions on earth provided a huge chance for the natural selection of favorable mutations. In this study, we analyzed two mutations on the critical virus-binding hotspot (N501Y, and N501T) following computational methodologies and revealed structural details on how the mutations affect the binding energy of spike protein.
3.1. Validation of MD simulation protocol
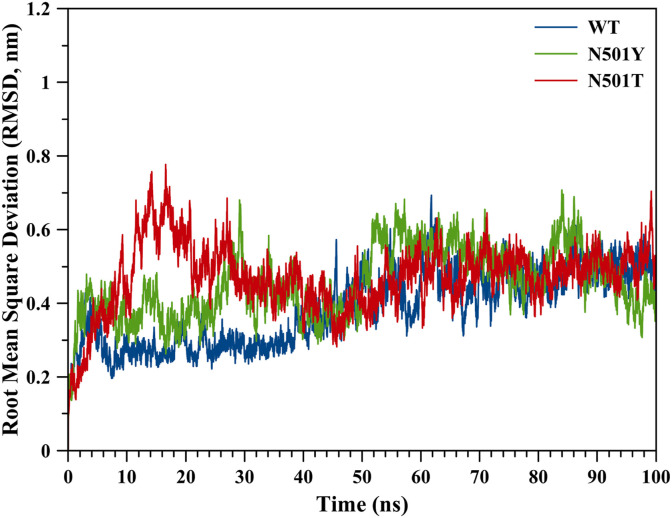
We validated our MD simulation protocols by considering root mean square deviation (RMSD) values for the hACE2-spike protein complex during the simulation. RMSD values can also validate the MD simulation protocols: the smaller the RMSD, the higher accuracy for the reproduction of the crystal structure [34,35]. For this purpose, we obtained RMSD values throughout MD simulations (0–100 ns), which revealed that the protein complex reached equilibrium after ∼55 ns of the simulation (Fig. 2 ).
Fig. 2.
Root mean square deviation (RMSD) values for WT, N501Y and N501T mutant ACE2-spike protein complexes throughout the MD simulation.
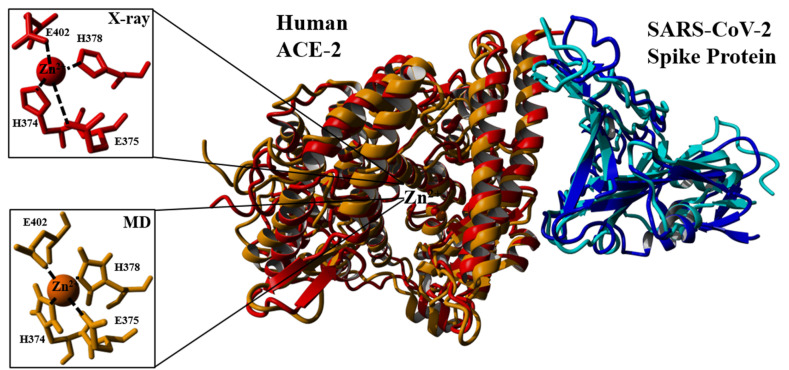
Moreover, to validate the methodology of the MD simulations, the most representative structure of the hACE2-WT spike protein complex from MD simulations and the X-ray structure (PDB ID: 6M0J) was superimposed. The RMSD value was 0.19 nm, which revealed good alignment on the overall complex and the hACE2 active site Zn2+ binding moiety (Fig. 3 ).
Fig. 3.
Superposition of X-ray ACE2 (red) - SARS CoV-2 spike protein (blue) complex with ACE2 (orange) - SARS CoV-2 spike protein (cyan) the most representative structures from MD simulations. Active site Zn2+ ion coordination shells for both structures were provided in the inlet figures. The overall RMSD of the alignment was calculated as 0.19 nm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
As a consequence, we concluded here that our MD simulations produced data in line with the experimental structures. After the validation of the MD simulation protocol, we approached the results of this study from two different perspectives, binding free energy and structural features leading to these energy values.
3.2. Analysis of N501Y, and N501T mutations on the receptor-binding domain (RBD)
First, we analyzed the binding free energy for hACE2 and WT, N501T, and N501Y mutant spike proteins by the MM/GBSA method to predict the experimental strength of hACE2-spike protein interactions [32]. From 80 to 100 ns time frame, snapshots were taken for every 1 ns, providing 21 structures for MM/GBSA analysis. The mean calculated binding energies from 21 snapshots were −61.94 ± 8.69 kcal/mol for WT, −94.16 ± 9.45 kcal/mol for N501Y mutant, and −82.91 ± 10.18 kcal/mol for N501T mutant, where more negative free energies indicated stronger binding. We applied t-test statistics to reveal if the means of binding free energies for WT, N501Y, and N501T were significantly different from each other. The time evolution of binding free energies and t-test statistics were provided in the Supporting Information Fig. 1. N501Y mutant provided much lower free energy than the WT, whereas N501T mutant also yielded much lower binding energies than the WT, but slightly higher free energy than N501Y mutant throughout last 20 ns. This result clearly showed that mutations produced remarkably lower binding free energies.
We have analyzed the individual amino acid contributions to the total binding free energy from both binding interface of hACE2 and RBD of spike protein. 12 hACE2 and RBD amino acids that yielded higher energies were listed in Table 1 and Table 2 , respectively. Upon mutations, amino acids’ contribution profile was significantly altered on the RBD residues, while the same profile stayed similar on the hACE2 residues. However, the sum of energy contributions from both 12 RBD and hACE2 residues increased significantly for both N501Y and N501T mutants compared to those of WT.
Table 1.
Binding energy contributions of top 12 residues of hACE2 domain of spike proteins. (Energies provided in kcal/mol with their mean values ± standard deviation).
| hACE2 | |||||
|---|---|---|---|---|---|
| WT |
N501Y |
N501T |
|||
| Residue | Energy | Residue | Energy | Residue | Energy |
| Y41 | −5.54 ± 0.76 | Y41 | −4.38 ± 0.59 | Y41 | −5.25 ± 0.93 |
| Q24 | −4.92 ± 0.92 | Q24 | −3.78 ± 0.70 | Y83 | −4.29 ± 1.01 |
| Y83 | −2.10 ± 0.56 | Q42 | −3.51 ± 0.95 | D38 | −3.63 ± 2.01 |
| T27 | −2.00 ± 0.80 | L45 | −3.35 ± 0.52 | K31 | −3.2 ± 1.34 |
| Q42 | −1.68 ± 0.479 | Y83 | −2.46 ± 0.18 | Q24 | −3.15 ± 0.67 |
| H34 | −1.49 ± 0.66 | L79 | −2.45 ± 0.30 | L79 | −2.77 ± 0.20 |
| L45 | −1.39 ± 0.21 | K31 | −2.39 ± 1.55 | L45 | −2.27 ± 0.42 |
| F28 | −1.32 ± 0.34 | F28 | −2.21 ± 0.23 | Q42 | −2.18 ± 0.33 |
| D38 | −1.15 ± 1.02 | T27 | −2.20 ± 0.27 | M82 | −2.11 ± 0.35 |
| L79 | −1.06 ± 0.65 | M82 | −1.95 ± 0.28 | F28 | −1.98 ± 0.15 |
| M82 | −0.84 ± 0.26 | D38 | −1.14 ± 1.99 | T27 | −1.91 ± 0.28 |
| K31 | −0.75 ± 0.62 | N61 | −1.09 ± 0.96 | H34 | −1.01 ± 0.40 |
| Sum | −24.2 | −30.9 | −33.8 | ||
Table 2.
Binding energy contributions of top 12 residues of RBD domain of spike proteins. (Energies provided in kcal/mol with their mean values ± standard deviation).
| Spike protein (RBD) | |||||
|---|---|---|---|---|---|
| WT |
N501Y |
N501T |
|||
| Residue | Energy | Residue | Energy | Residue | Energy |
| Y505 | −5.97 ± 0.32 | Q493 | −9.07 ± 1.71 | F486 | −5.46 ± 0.75 |
| F486 | −3.30 ± 0.66 | Y501 | −5.82 ± 1.55 | Y489 | −4.8 ± 0.32 |
| Y449 | −3.00 ± 1.70 | Y449 | −5.23 ± 0.75 | Q498 | −4.71 ± 0.54 |
| G476 | −2.98 ± 1.72 | F486 | −5.21 ± 0.50 | Y449 | −3.8 ± 0.83 |
| N501 | −2.79 ± 0.64 | Y489 | −4.99 ± 0.44 | Q493 | −3.51 ± 0.38 |
| Y489 | −2.36 ± 0.47 | S494 | −4.09 ± 1.95 | V445 | −3.18 ± 0.59 |
| N487 | −2.09 ± 1.07 | V445 | −3.59 ± 1.50 | Y505 | −2.82 ± 0.97 |
| F456 | −1.99 ± 0.30 | L455 | −2.99 ± 0.37 | F456 | −2.76 ± 0.25 |
| A475 | −1.73 ± 0.94 | F456 | −2.46 ± 0.24 | L455 | −2.59 ± 0.59 |
| S477 | −1.72 ± 1.13 | Y505 | −2.12 ± 0.52 | S494 | −1.97 ± 0.29 |
| V445 | −1.58 ± 1.05 | Y495 | −1.99 ± 0.40 | N487 | −1.95 ± 0.91 |
| Q498 | −1.48 ± 0.39 | G496 | −1.66 ± 0.64 | T501 | −1.73 ± 1.52 |
| Sum | −31.0 | −49.2 | −39.3 | ||
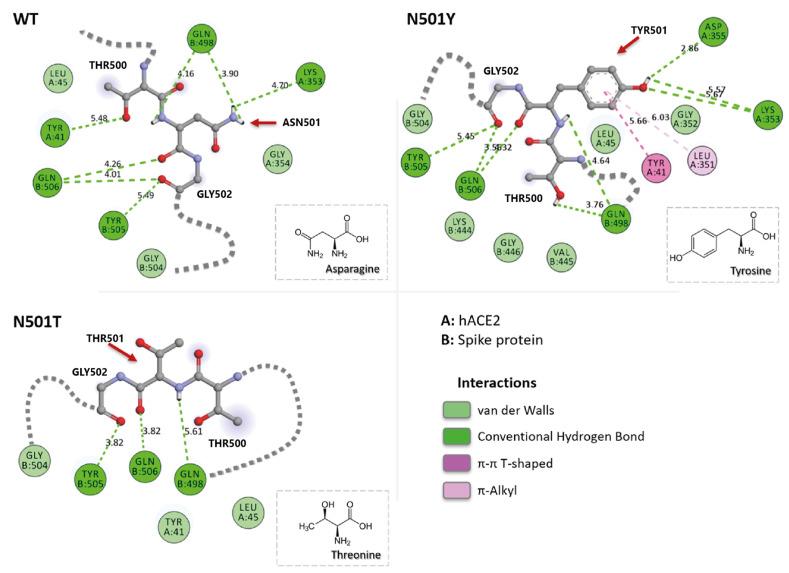
When Asn 501 mutated to Tyr, the energy contribution of this residue increased by 3.03 kcal/mol (lower binding free energy), indicating a stronger interaction with hACE2, whereas Thr mutation significantly decreased the contribution by 1.06 kcal/mol (higher binding free energy). The reason for these alterations was the new environments around these residues. Tyr being an aromatic amino acid, brings the aromatic amino acid residues Tyr 41 (hACE2), Tyr 449 and Tyr 505 (RBM) together bound into highly hydrophobic binding pocket (Fig. 4 ). hACE2 residues Leu 45 and Leu 351 lies in this hydrophobic pocket as well.
Fig. 4.
2-D amino acid interaction maps around N/Y/T 501 positions of WT, N501Y, and N501T mutants. A chain represents hACE2 and B chain represents spike protein (ex. GLN B:506 represents spike protein residue Gln 506).
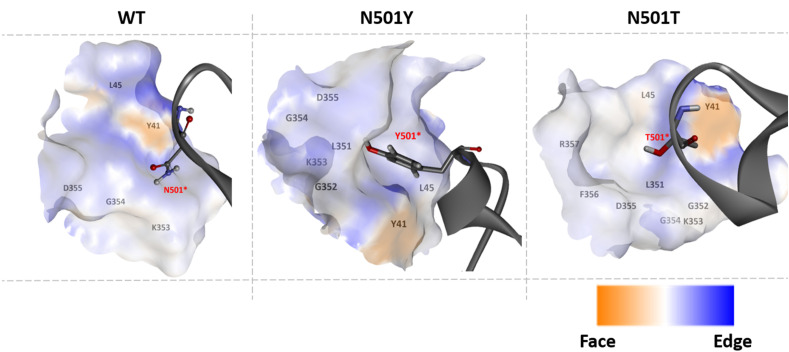
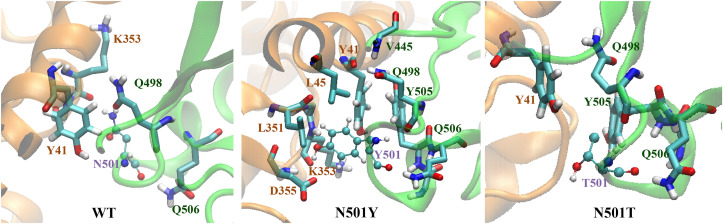
It was clear that Tyr 501 was very well accommodated in highly hydrophobic pocket, creating face to edge (π-π T-shaped) interaction with Tyr 41 (Fig. 5 ). This aromatic interaction was not observed in WT and in N501T mutant. Moreover, there were π-π stacking (face to face) interaction between Tyr 41 (hACE2) and Tyr 505 (Spike), and π-alkyl interaction between Leu 351 (hACE2) and Tyr 501 (Spike) for N501Y mutant (Fig. 6 ).
Fig. 5.
Aromatic environments of residue 501 interaction site for WT, N501Y, and 501T mutants.
Fig. 6.
Zoomed in amino acid interactions around N/Y/T 501 positions of WT, N501Y, and N501T mutants. Residue 501 was labeled in purple. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Tyr 501 created a strong H-bond with Asp 355 (hACE2). On contrary, Thr 501 in N501T mutant did not interact directly with any of hACE2 residues.
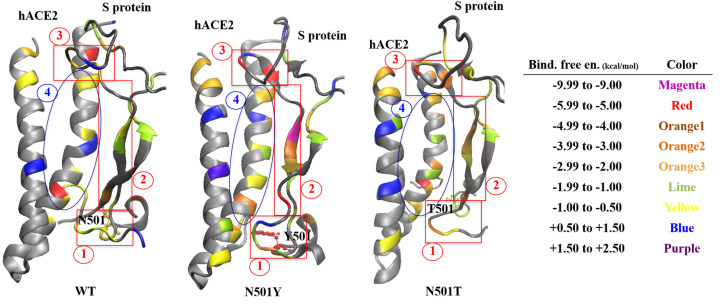
Furthermore, we created binding free energy contribution map through the hACE2-RBD interface, which also reflects the altered contributions shown in Table 1. Both mutations changed contributions mainly on four regions.
Fig. 7 highlights that N501Y mutation resulted in stronger hACE2 interaction in regions 1 (local area of mutation), 2, and 3. On the other hand, N501T mutation did not alter the interaction significantly in region 1, but it led to stronger interaction in regions 3 and 4.
Fig. 7.
Binding free contribution maps for hACE2-Spike protein complexes (WT, N501Y, and N501T).
Although, N501T did not change the energy contribution in the local mutation area (region 1), it significantly increased energy contributions in other regions, which explained the overall increased binding free energy for this mutant.
We also calculated the solvation-free energy gain (ΔiG) upon binding by utilizing the PISA server [36]. The most representative (cluster) structures were used for these calculations. hACE2-N501Y spike protein complex provided 1.4 kcal/mol decrease in ΔiG, while N501T mutant provided 0.5 kcal/mol lower energy compared to hACE2-WT complex (Table 3 ).
Table 3.
Solvation free energy gains and interface areas in Å2 calculated from cluster structures by PISA server. More negative ΔiG indicates stronger binding of proteins.
| Complex | ΔiG (kcal/mol) | Interface surface (Å2) |
|---|---|---|
| WT | −9.1 | 941.2 |
| N501Y | −10.5 | 1114.8 |
| N501T | −9.6 | 956.4 |
This was expected because tyrosine comprised of highly hydrophobic side chain gained more energy upon complexation compared to threonine and asparagine, comprised of the polar side chain and Tyr 501 was accommodated better than Thr 501 and Asn 501 in region 1. Increased total binding interface surface area for the mutants also supports the observed higher binding free energies.
4. Conclusion
In this study, the binding energy-based consequences of two spike protein RBD mutations, N501Y and N501T, were analyzed by utilizing all-atom classical MD simulations. Binding free energy calculations revealed that N501Y mutant spike protein bound to hACE2 with higher affinity, 32.22 kcal/mol lower binding free energy, than the WT protein. Golubchick et al. [37] studied on the N501Y reported increased viral load for the lineage including N501Y mutation in-vitro. Additionally, in-silico studies via FEP simulations by Luan et al. provided increased binding affinity towards hACE2 upon N501Y mutation [38], which is parallel to our results. The calculated binding free energy for N501T mutant spike protein was 20.97 kcal/mol lower than the WT spike protein, but 11.25 kcal/mol higher than the N501Y mutant. Shang et al. also shared an in-vitro data [17] proposing reduced spike binding affinity towards hACE2 upon N501T mutations but, other studies conducted by Wang et al. [39] showed opposite results, leading increase in the RBD and ACE2 binding affinity. Our results for increased binding strength of N501T mutant spike protein was in line with data presented in the study by Wang et al. This study combines the structural and energy-based approaches together for the detailed analysis of N501Y and N501T mutations on SARS-CoV-2 spike protein.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Authors acknowledge the supercomputing facility TRUBA, hosted by TÜBİTAK ULAKBİM, for computational resources used during MD simulations.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmgm.2022.108260.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Burrell C.J., Howard C.R., Murphy F.A. Fenner and White's Medical Virology. Elsevier; 2017. Coronaviruses; pp. 437–446. [DOI] [Google Scholar]
- 2.Wan Y., Shang J., Graham R., Baric R.S., Li F., Wan C.Y. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:127–147. doi: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abedi F., Rezaee R., Hayes A.W., Nasiripour S., Karimi G. MicroRNAs and SARS-CoV-2 life cycle, pathogenesis, and mutations: biomarkers or therapeutic agents? Cell Cycle. 2021;20:143–153. doi: 10.1080/15384101.2020.1867792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020;19 doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valle C., Martin B., Touret F., Shannon A., Canard B., Guillemot J.C., Coutard B., Decroly E. Drugs against SARS-CoV-2: what do we know about theirmode of action? Rev. Med. Virol. 2020;30:1–10. doi: 10.1002/rmv.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiu S., Dick A., Ju H., Mirzaie S., Abdi F., Cocklin S., Zhan P., Liu X. Inhibitors of SARS-CoV-2 entry: current and future opportunities. J. Med. Chem. 2020;63:12256–12274. doi: 10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taka E., Yilmaz S.Z., Golcuk M., Kilinc C., Aktas U., Yildiz A., Gur M. Critical interactions between the SARS-CoV-2 spike glycoprotein and the human ACE2 receptor. bioRxiv. 2020 doi: 10.1101/2020.09.21.305490. 2020.09.21.305490. [DOI] [PubMed] [Google Scholar]
- 8.Kumar V. Understanding the complexities of SARS-CoV2 infection and its immunology: a road to immune-based therapeutics. Int. Immunopharm. 2020;88 doi: 10.1016/j.intimp.2020.106980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J., Wang R., Wang M., Wei G.W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432:5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferron F., Subissi L., De Morais A.T.S., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U.S.A. 2017;115:E162. doi: 10.1073/pnas.1718806115. –E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang R., Hozumi Y., Yin C., Wei G.-W. 2020. Decoding SARS-CoV-2 Transmission and Evolution and Ramifications for COVID-19 Diagnosis, Vaccine, and Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guruprasad L. Human SARS CoV-2 spike protein mutations. Proteins: Struct., Funct., Bioinf. 2021;89:569–576. doi: 10.1002/prot.26042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sui J., Aird D.R., Tamin A., Murakami A., Yan M., Yammanuru A., Jing H., Kan B., Liu X., Zhu Q., Yuan Q.A., Adams G.P., Bellini W.J., Xu J., Anderson L.J., Marasco W.A. Broadening of neutralization activity to directly block a dominant antibody-driven SARS-coronavirus evolution pathway. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Cell Host & Microbe; 2021. Comprehensive Mapping of Mutations in the SARS-CoV-2 Receptor-Binding Domain that Affect Recognition by Polyclonal Human Plasma Antibodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C.C.C.C., Muecksch F., Rutkowska M., Hoffmann H.-H.H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Luchsinger L., Hillyer C.D., Caskey M., Robbiani D.F., Rice C.M., Nussenzweig M.C., Hatziioannou T., Bieniasz P.D. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9:1. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Angyal A., Brown R.L., Carrilero L., Green L.R., Groves D.C., Johnson K.J., Keeley A.J., Lindsey B.B., Parsons P.J., Raza M., Rowland-Jones S., Smith N., Tucker R.M., Wang D., Wyles M.D., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 20.Dolinsky T.J., Nielsen J.E., McCammon J.A., Baker N.A. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krieger E., Vriend G. Models@Home: distributed computing in bioinformatics using a screensaver based approach. Bioinformatics. 2002;18:315–318. doi: 10.1093/bioinformatics/18.2.315. [DOI] [PubMed] [Google Scholar]
- 22.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. doi: 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- 23.Lindahl E., Hess B., van der Spoel D. Gromacs 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 2001;7:306–317. doi: 10.1007/S008940100045. [DOI] [Google Scholar]
- 24.Schmid N., Eichenberger A.P., Choutko A., Riniker S., Winger M., Mark A.E., Van Gunsteren W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 2011;40:843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- 25.Berendsen H.J.C., Grigera J.R., Straatsma T.P. The missing term in effective pair potentials. J. Phys. Chem. 1987;91:6269–6271. doi: 10.1021/j100308a038. [DOI] [Google Scholar]
- 26.Miyamoto S., Kollman P.A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. doi: 10.1002/jcc.540130805. [DOI] [Google Scholar]
- 27.Hess B., Bekker H., Berendsen H.J.C.C., Fraaije J.G. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 28.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:5648. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 29.Hockney R.W., Goel S.P., Eastwood J.W. Quiet high-resolution computer models of a plasma. J. Comput. Phys. 1974;14:148–158. doi: 10.1016/0021-9991(74)90010-2. [DOI] [Google Scholar]
- 30.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 31.Chen F., Liu H., Sun H., Pan P., Li Y., Li D., Hou T. Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein–protein binding free energies and re-rank binding poses generated by protein–protein docking. Phys. Chem. Chem. Phys. 2016;18:22129–22139. doi: 10.1039/C6CP03670H. [DOI] [PubMed] [Google Scholar]
- 32.Weng G., Wang E., Wang Z., Liu H., Zhu F., Li D., Hou T. HawkDock: a web server to predict and analyze the protein-protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gkz397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/J.JMB.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Reva B.A., Finkelstein A.V., Skolnick J. What is the probability of a chance prediction of a protein structure with an rmsd of 6 Å? Folding Des. 1998;3:141–147. doi: 10.1016/S1359-0278(98)00019-4. [DOI] [PubMed] [Google Scholar]
- 35.Sargsyan K., Grauffel C., Lim C. How molecular size impacts RMSD applications in molecular dynamics simulations. J. Chem. Theor. Comput. 2017;13:1518–1524. doi: 10.1021/acs.jctc.7b00028. [DOI] [PubMed] [Google Scholar]
- 36.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Golubchik T., Lythgoe K.A. Early analysis of a potential link between viral load and the N501Y mutation in the SARS-COV-2 spike protein. medRxiv. 2021;1 doi: 10.1101/2021.01.12.20249080. 2021.01.12.20249080. [DOI] [Google Scholar]
- 38.Luan B., Wang H., Huynh T. Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2021;595:1454–1461. doi: 10.1002/1873-3468.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang R., Chen J., Gao K., Wei G.-W. Vaccine-escape and fast-growing mutations in the United Kingdom, the United States, Singapore, Spain, India, and other COVID-19-devastated countries. Genomics. 2021;113:2158–2170. doi: 10.1016/j.ygeno.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.