Abstract
BACKGROUND
Increasing utilization of long-term outpatient ambulatory electrocardiographic (ECG) monitoring continues to drive the need for improved ECG interpretation algorithms.
OBJECTIVE
The purpose of this study was to describe the BeatLogic® platform for ECG interpretation and to validate the platform using electrophysiologist-adjudicated real-world data and publicly available validation data.
METHODS
Deep learning models were trained to perform beat and rhythm detection/classification using ECGs collected with the Preventice BodyGuardian® Heart monitor. Training annotations were created by certified ECG technicians, and validation annotations were adjudicated by a team of board-certified electrophysiologists. Deep learning model classification results were used to generate contiguous annotation results, and performance was assessed in accordance with the EC57 standard.
RESULTS
On the real-world validation dataset, BeatLogic beat detection sensitivity and positive predictive value were 99.84% and 99.78%, respectively. Ventricular ectopic beat classification sensitivity and positive predictive value were 89.4% and 97.8%, respectively. Episode and duration F1 scores (range 0–100) exceeded 70 for all 14 rhythms (including noise) that were evaluated. F1 scores for 11 rhythms exceeded 80, 7 exceeded 90, and 5 including atrial fibrillation/flutter, ventricular tachycardia, ventricular bigeminy, ventricular trigeminy, and third-degree heart block exceeded 95.
CONCLUSION
The BeatLogic platform represents the next stage of advancement for algorithmic ECG interpretation. This comprehensive platform performs beat detection, beat classification, and rhythm detection/classification with greatly improved performance over the current state of the art, with comparable or improved performance over previously published algorithms that can accomplish only 1 of these 3 tasks.
Keywords: Artificial intelligence, BeatLogic, Deep learning, Electrocardiographic interpretation, Preventice Solutions
Introduction
Outpatient ambulatory electrocardiographic (ECG) monitoring has grown in popularity due to technological advancements, which have decreased monitor size, increased battery life, and enabled mobile telemetry. Modern ambulatory ECG monitors allow for up to 30 days of continuous monitoring, producing far too much data for physicians to comprehensively analyze. For this reason, service providers are commonly used to annotate ECG recordings and create reports that summarize and highlight ectopic activity. These reports provide clinical decision support for prescribing physicians. Service providers rely on certified technicians and supporting algorithms to process and annotate the data from ECG monitoring studies. Historically, supporting algorithms have achieved levels of performance well below that of humans1 but high enough to be used for prioritization and conservative filtering of ECG as it is queued for human interpretation.2 Better supporting algorithms have the potential to improve this process by more accurately detecting the presence and absence of cardiac arrhythmias.
Currently, most ECG interpretation algorithms rely on signal processing and classic machine learning; however, recent studies applying deep learning (DL) to aspects of ECG interpretation have generated exciting results.3–8 DL models rely on simple computational units that are stacked in layers and operate on raw data to extract complex features relevant to the classification problem at hand.9 This differs from classic machine learning, in which manual feature discovery and extraction are performed using signal processing. Automated feature discovery with DL generally delivers superior performance in domains where data contain subtle details and complex interactions. These factors make DL well suited for ECG interpretation algorithms. Previous studies applying DL to ECG interpretation performed only beat detection, beat classification, or rhythm classification, and only those created for beat classification tend to follow the American National Standards Institute (ANSI)/Association for the Advancement of Medical Instrumentation (AAMI) EC57 standard10 when evaluating performance. This work details and validates the Preventice BeatLogic® platform, a comprehensive ECG annotation platform that leverages DL for beat and rhythm detection/classification. Performance was measure dusing the EC57 standard and compared to a commercial state-of-the-art ECG interpretation algorithm using real-world gold standard data and also compared to previously published work using publicly available validation datasets.
Methods
Training data
Deidentified ECG recordings from the single-channel Preventice BodyGuardian® Heart (Preventice Solutions, Rochester, MN) ambulatory patch-style monitor were mined from the Preventice ECG monitoring platform using a combination of random selection and targeted mining. Targeted mining ensured sufficient representation of artifact and arrhythmias by selecting ECGs in which normal processing through the Preventice ECG monitoring platform identified the targeted arrhythmia. Targeted arrhythmias included junctional rhythms, heart blocks, and intraventricular conduction delay with occasional ventricular ectopic beats (VEBs). Training data were captured as 20,932 individual records with duration between 15 seconds and 4 minutes. Annotations were made in accordance with standard practice by a dedicated team of Certified Cardiographic Technician (CCT)-certified ECG technicians having experience ranging from 9–30 years. These technicians received specialized training to ensure that annotations were sufficiently detailed and consistent. The final training dataset consisted of 782.44 hours of ECG from 11,008 unique patients (Table 1). Beat and rhythm contents for the training dataset are detailed in the Supplemental Material (Supplemental Tables 1, 2, and 3).
Table 1.
ECG dataset general information
| Records | Patients | Duration (h) | |
|---|---|---|---|
|
| |||
| Training | 20,932 | 11,008 | 782.44 |
| Gold validation | 515 | 505 | 12.79 |
| MIT-BIH | 48 | 47 | 24.07 |
| MIT-BIH 11 | 11 | 11 | 5.52 |
| MIT-AFDB | 23 | 23 | 234.28 |
ECG = electrocardiography.
Validation data
ECG for the gold standard validation dataset (aka gold validation) was selected from a candidate pool of 3000 pseudo-randomly selected deidentified BodyGuardian Heart recordings. The candidate pool contained 120 examples of 25 rhythms that were partially annotated by CCT-certified ECG technicians during normal processing through the Preventice ECG monitoring platform. From the candidate pool, approximately 20 examples of each rhythm were randomly selected from records in which the partial annotations were confirmed by a senior ECG technician. Comprehensive annotation was performed by a team of CCT-certified ECG technicians having experience ranging from9–30 years. Annotations were individually adjudicated by3 board-certified electrophysiologists (EPs), and records with, 100% agreement were adjudicated in a group forum at which time the annotations were adjusted to align with the group consensus. Records were excluded from the validation library if a consensus could not be reached. The gold validation dataset included 515, 1- to 4-minute records from 505 patients (Table 1). No patient overlap was allowed between the training and gold standard validation datasets.
Validation was also performed using the MIT-BIH Arrhythmia Database11 (MIT-BIH) and the MIT Atrial Fibrillation Database12 (AFDB). MIT-BIH consists of 24.07 hours of 2-channel ambulatory ECG from 47 patients (Table 1). In accordance with previously published work, the full database was used to measure beat detection performance, and an 11-record subset was used to measure VEB classification performance. AFDB consists of 234.28 hours of 2-channel ambulatory ECG from 23 patients (Table 1) and was used to measure atrial fibrillation/flutter performance. Beat and rhythm contents for each validation dataset are detailed in the Supplemental Material (Supplemental Tables 1, 2, and 3).
BeatLogic platform
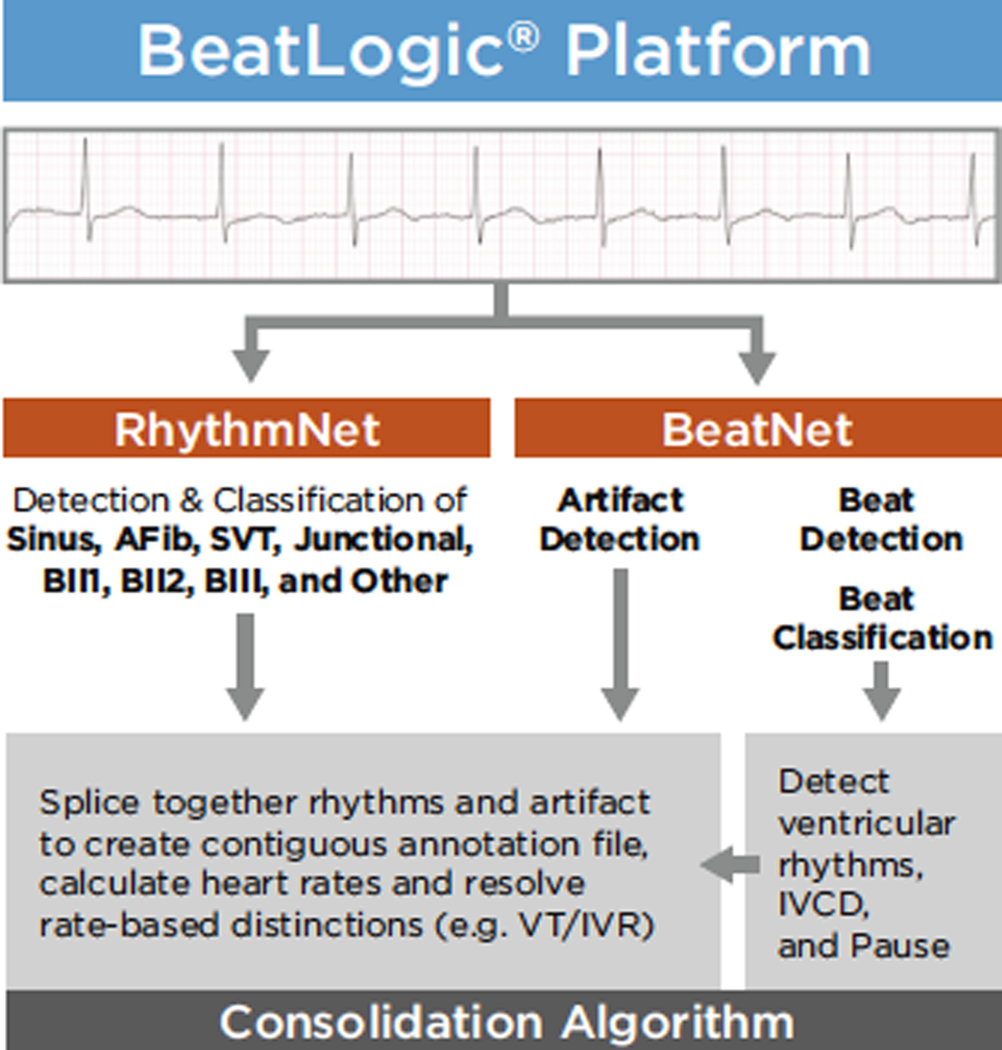
The BeatLogic platform consists of 2 DL models—BeatNet and RhythmNet—the results of which are consolidated using rules-based logic to produce a single contiguous annotation file (Figure 1). BeatNet performs artifact detection, beat detection, and beat classification. RhythmNet performs detection and classification of Sinus rhythm (Sinus), Atrial fibrillation/flutter (AFib), Supraventricular tachycardia (SVT), Junctional rhythm (Junc), Second-degree heart block type 1 (BII1), Second-degree heart block type 2 (BII2), Third-degree heart block (BIII), and Other. The consolidation algorithm generates Ventricular tachycardia (VT), Idioventricular rhythm (IVR), Intraventricular conduction delay (IVCD), Ventricular bigeminy (VBigem), Ventricular trigeminy (VTrigem), and Pause annotations using the BeatNet output and then splices together RhythmNet rhythms, ventricular rhythms, and artifact to create contiguous annotation files.
Figure 1.
BeatLogic platform flowchart. AFib = atrial fibrillation/flutter; BII1 = second-degree heart block type 1; BII2 = second-degree heart block type 2; IVCD = intraventricular conduction delay; IVR = idioventricular rhythm; Junctional = junctional rhythm; Sinus = sinus rhythm; SVT = supraventricular tachycardia; VT = ventricular tachycardia.
DL architecture
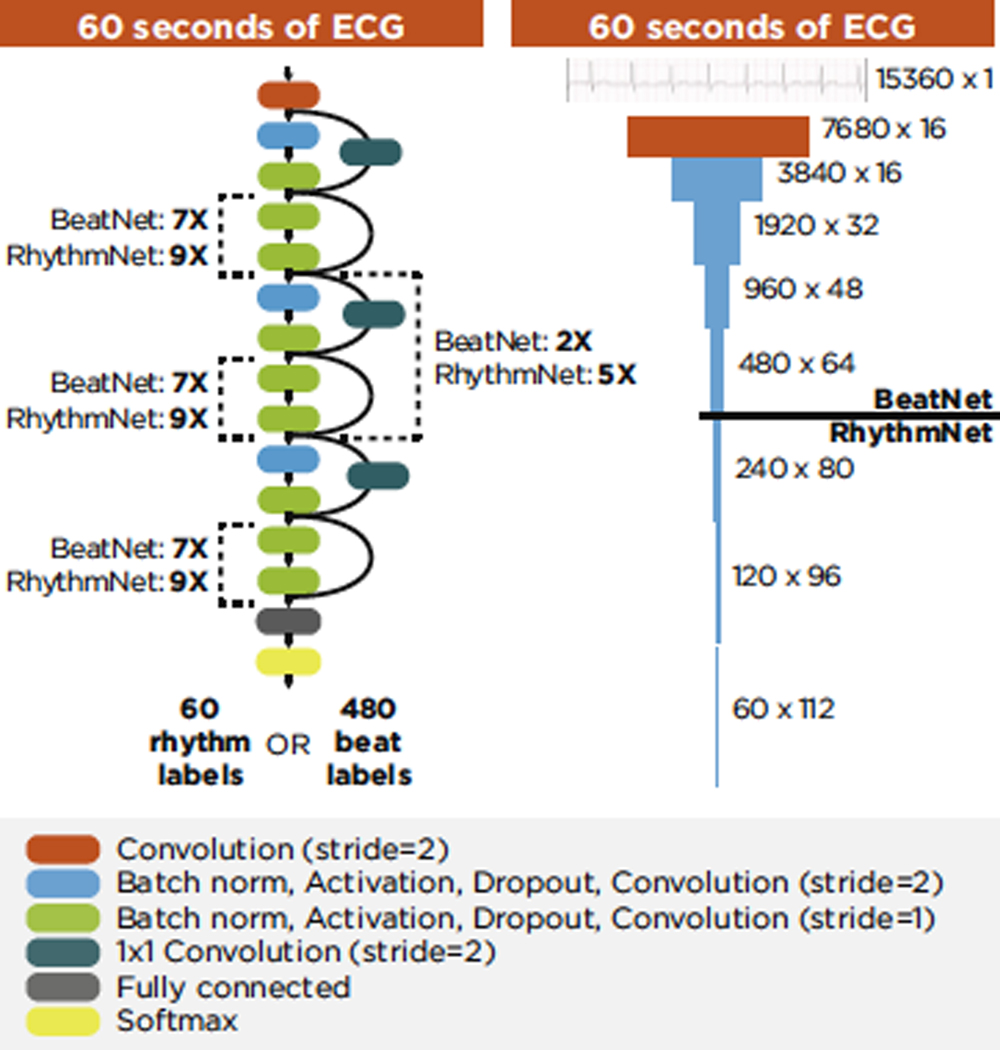
Both DL models rely on a similar architecture, which produces a sequence of classification results from a time series of single-channel ECG voltage values (Figure 2). The architecture is derived from preactivation ResNet,13,14 a popular image classification architecture. Modifications to the architecture included replacing 2-dimensional (2D) convolutions with 1 dimension (1D) and removing the final pooling layer in order to repurpose the 2D image classification design to a 1D sequence-to-sequence classification design. As raw ECG flows through the network, it is compressed in the time dimension and extended depthwise. Compression occurs in the first convolution and at regular intervals throughout the remainder of the network. The input size and number of compression layers determine the model output resolution. The input to both DL models was 15,360 samples (60 seconds), which was compressed 5 times, resulting in 480 sequential outputs (every 0.125 second) for BeatNet, and compressed 8 times, resulting in 60 sequential outputs (every 1 second) for RhythmNet. Both architectures ended with a fully connected layer and softmax activation function, which produced classwise probabilities for each sequential output. The highest probability was selected as the label for each sequential output.
Figure 2.
Deep learning model architecture. ECG = electrocardiogram.
ECG signal processing
ECG recordings were preprocessed using a wavelet high-pass (fc = 0.5 Hz) filter15 to remove baseline wander and 2 second-order Butterworth band-stop (fc = 50 and 60 Hz) filters to remove powerline interference. After filtering, MIT-BIH and AFDB data were resampled to 256 Hz using linear interpolation.
Training record annotations
Training record annotations were generated for each model at the designed output resolution. BeatNet annotations were divided into 480 sequential classification labels consisting of Artifact, Not-a-beat, Ventricular ectopic, Bundle branch block, Normal, and Other. The Normal class included supraventricular ectopic beats, and the Other class included paced and unclassifiable beats. Sections with artifact onset/offset were labeled Artifact; sections with no beat and no artifact were labeled Not-a-beat; and sections in which a beat peak occurred anywhere within the 0.125-second window were labeled with the appropriate beat class label. Training records shorter than 60 seconds were padded using Other. RhythmNet annotations were divided into 60 sequential classification labels consisting of Sinus, AFib, BII1, BII2, BIII, SVT, Junctional, and Other. Rhythm transitions were labele dusing the rhythm that spanned the majority of the 1-second region. Training records shorter than 60 seconds were padded using Other.
Model initialization and training
DL model weights were initialized in accordance with He et al16 and trained using Adam17 to optimize softmax cross-entropy. Padded and Other regions were masked in the training loss calculation. Mini-batch size and initial learning rate were optimized using the hyperparameter tuning process. A development dataset was partitioned from the training data, which was evaluated during training to implement early stopping and at the end of training to compare the performance of models with different hyperparameters. The development dataset contained at least 10 examples of each annotation, and no patient overlap was allowed between the development dataset and there maining training dataset. After each training epoch(1 cycle through the full training dataset), micro-averaged training and development dataset F1 scores were calculated, and the training dataset was randomly shuffled. During training, learning rate was reduced when the training dataset F1 score did not improve for 5 consecutive epochs. Early stopping was invoked when the calculated PQ value18 exceeded a threshold that was set using the hyperparameter tuning process.
Hyperparameter tuning
To fully define the model architecture and training procedure, model hyperparameters were optimized. Because preactivation ResNet was designed for image classification, this base architecture was reparameterized for sequence-to-sequence ECG classification in the context of BeatNet and RhythmNet. Hyperparameter optimization was performed using a combination of grid-search and tree-structured parzen estimator optimization19 (for details see the Supplemental Material and Supplemental Table 4). The optimized BeatNet and RhythmNet models contained 81 and 113 convolutional layers.
State-of-the-art algorithm
The state-of-the-art algorithm was selected from several commercially available Food and Drug Administration (FDA)–cleared options capable of comprehensive beat and rhythm detection/classification. Candidate algorithms were evaluated using the EC57 standard, and the most accurate system was selected. Details of the selected algorithm are proprietary and were not disclosed to the authors for publication; however, the selected algorithm is known to leverage signal processing and classic machine learning techniques that are derived from the current ECG literature.
Validation procedure
Algorithm validation was performed in accordance with the EC57 guidelines.10 EC57 is the FDA-recognized consensus standard and provides detailed instructions for measuring beat and rhythm detection/classification sensitivity (Se), and positive predictive value (PPV). Additionally, F1 scores (0–100) were calculated for each validation metric per Equation 1.
| (Eq. 1) |
Results
Beat detection
On the MIT-BIH dataset, the BeatLogic platform performed equal to or better than 5 of the 8 previously published algorithms, whereas the state-of-the art algorithm outperformed only 1 published algorithm (Table 2). On the gold validation dataset, BeatLogic sensitivity was 99.84%, which exceeded the state-of-the-art algorithm by >4 percentage points. BeatLogic PPV was 99.78%, which exceeded the state-of-the-art algorithm by >3 percentage points (Table 2).
Table 2.
Beat detection performance
| Algorithm | Dataset | Se (%) | PPV (%) | F1 |
|---|---|---|---|---|
|
| ||||
| Pan and Tompkins24 | MIT-BIH | 99.76 | 99.56 | 99.66 |
| Christov25 | MIT-BIH | 99.74 | 99.65 | 99.69 |
| Chiarugi et al26 | MIT-BIH | 99.76 | 99.81 | 99.78 |
| Chouakri et al27 | MIT-BIH | 98.68 | 97.24 | 97.95 |
| Elgendi28 | MIT-BIH | 99.78 | 99.87 | 99.82 |
| State of the art | MIT-BIH | 97.58 | 99.44 | 98.50 |
| BeatLogic | MIT-BIH | 99.60 | 99.78 | 99.69 |
| Martinez et al29 | MIT-BIH VFib excluded | 99.80 | 99.86 | 99.83 |
| Arzeno et al30 | MIT-BIH VFib excluded | 99.68 | 99.63 | 99.65 |
| Zidelmal et al31 | MIT-BIH VFib excluded | 99.64 | 99.82 | 99.73 |
| State of the art | MIT-BIH VFib excluded | 97.58 | 99.57 | 98.56 |
| BeatLogic | MIT-BIH VFib excluded | 99.60 | 99.90 | 99.75 |
| State of the art | Gold validation | 95.79 | 96.32 | 96.05 |
| BeatLogic | Gold validation | 99.84 | 99.78 | 99.81 |
PPV = positive predictive value; Se = sensitivity; VFib = ventricular fibrillation.
VEB classification performance
On the 11-record MIT-BIH data subset for measuring VEB performance, BeatLogic outperformed all other algorithms, achieving an F1 score of 98.4, which is 0.8 points higher than the next highest performing algorithm (Table 3). On the gold validation dataset, BeatLogic outperformed the state-of-the-art algorithm, achieving sensitivity of 89.4% and PPV of 97.8% (Table 3).
Table 3.
Ventricular ectopic beat classification performance
| Algorithm | Dataset | Se (%) | PPV (%) | F1 |
|---|---|---|---|---|
|
| ||||
| de Chazal et al22 | MIT-BIH 11 | 77.5 | 90.6 | 83.5 |
| Jiang and Kong3 | MIT-BIH 11 | 94.3 | 95.8 | 95.0 |
| Ince et al32 | MIT-BIH 11 | 90.3 | 92.2 | 91.2 |
| Kiranyaz et al20 | MIT-BIH 11 | 95.9 | 96.2 | 96.0 |
| Zhang et al8 | MIT-BIH 11 | 97.6 | 97.6 | 97.6 |
| State of the art | MIT-BIH 11 | 73.2 | 96.3 | 83.2 |
| BeatLogic | MIT-BIH 11 | 97.9 | 98.9 | 98.4 |
| State of the art | Gold validation | 36.0 | 51.2 | 42.2 |
| BeatLogic | Gold validation | 89.4 | 97.8 | 93.4 |
Abbreviations as in Table 2.
Rhythm detection and classification
On the AFDB dataset, BeatLogic outperformed the previously published algorithms. The BeatLogic platform achieved episode Se/PPV of 97.7%/99.3% and duration sensitivity/PPV of 97.7%/99.7% (Table 4). On the gold validation dataset, BeatLogic outperformed the state-of-the-art algorithm for all 14 rhythms in measures of episode and duration sensitivity and PPV (Table 4). Three rhythm classes (junctional rhythm, second-degree heart block type 1, third-degree heart block) were not called at all by the state-of-the-art algorithm. State-of-the-art episode and duration F1 scores exceeded 70 for 7 rhythms and exceeded 80 for episode detection of 3 rhythms. State-of-the-art episode and duration F1 scores did not exceed 85 for any rhythm.
Table 4.
Rhythm episode and duration performance
| Episode | Duration | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Rhythm | Dataset | Algorithm | Se (%) | PPV (%) | Fi | Se (%) | PPV (%) | Fi |
|
| ||||||||
| AFib | AFDB | Petrucciet al33 DRR | 92.0 | 78.0 | 84.4 | 89.0 | 90.0 | 89.5 |
| Petrucciet al33 RRP | 91.0 | 92.0 | 91.5 | 93.0 | 97.0 | 95.0 | ||
| State of the art | 63.3 | 100.0 | 77.5 | 65.3 | 99.3 | 78.8 | ||
| BeatLogic | 97.7 | 99.3 | 98.5 | 97.7 | 99.7 | 98.7 | ||
| AFib | Gold validation | State of the art | 67.4 | 78.4 | 72.5 | 71.4 | 80.4 | 75.6 |
| BeatLogic | 96.4 | 98.6 | 97.5 | 97.2 | 99.7 | 98.4 | ||
| Sinus | Gold validation | State of the art | 84.9 | 79.0 | 81.8 | 83.5 | 84.5 | 84.0 |
| BeatLogic | 97.8 | 87.3 | 92.3 | 99.5 | 95.5 | 97.5 | ||
| IVCD | Gold validation | State of the art | 11.5 | 19.2 | 14.4 | 10.8 | 19.0 | 13.8 |
| BeatLogic | 90.1 | 75.4 | 82.1 | 90.8 | 83.1 | 86.8 | ||
| Artifact | Gold validation | State of the art | 51.5 | 56.6 | 53.9 | 69.8 | 51.5 | 59.3 |
| BeatLogic | 79.9 | 79.8 | 79.8 | 90.4 | 65.7 | 76.1 | ||
| Pause | Gold validation | State of the art | 69.8 | 100.0 | 82.2 | 67.6 | 99.9 | 80.6 |
| BeatLogic | 97.7 | 93.2 | 95.4 | 92.0 | 93.7 | 92.8 | ||
| SVT | Gold validation | State of the art | 66.7 | 33.3 | 44.4 | 81.3 | 51.6 | 63.2 |
| BeatLogic | 90.0 | 83.1 | 86.4 | 97.7 | 95.0 | 96.3 | ||
| VT | Gold validation | State of the art | 51.6 | 20.9 | 29.7 | 16.7 | 27.3 | 20.7 |
| BeatLogic | 100.0 | 94.0 | 96.9 | 97.4 | 95.2 | 96.3 | ||
| IVR | Gold validation | State of the art | 61.7 | 33.8 | 43.7 | 60.5 | 28.5 | 38.7 |
| BeatLogic | 83.0 | 98.0 | 89.8 | 63.8 | 96.4 | 76.8 | ||
| Junctional | Gold validation | State of the art | — | — | — | — | — | — |
| BeatLogic | 91.3 | 73.9 | 81.7 | 84.9 | 77.5 | 81.0 | ||
| VBigem | Gold validation | State of the art | 62.3 | 75.3 | 68.2 | 29.1 | 77.5 | 42.3 |
| BeatLogic | 100.0 | 98.6 | 99.3 | 99.2 | 98.7 | 99.0 | ||
| VTrigem | Gold validation | State of the art | 80.6 | 88.9 | 84.5 | 73.0 | 92.5 | 81.6 |
| BeatLogic | 97.2 | 97.3 | 97.3 | 98.4 | 98.4 | 98.4 | ||
| BII1 | Gold validation | State of the art | — | — | — | — | — | — |
| BeatLogic | 56.9 | 93.2 | 70.7 | 72.6 | 97.7 | 83.3 | ||
| BII2 | Gold validation | State of the art | 30.0 | 73.2 | 42.6 | 9.9 | 68.9 | 17.3 |
| BeatLogic | 80.0 | 82.9 | 81.4 | 85.3 | 86.1 | 85.7 | ||
| BIII | Gold validation | State of the art | — | — | — | — | — | — |
| BeatLogic | 98.7 | 95.8 | 97.2 | 93.2 | 97.2 | 95.1 | ||
AFib = atrial fibrillation/flutter; BII1 = second-degree heart block type 1; BII2 = second-degree heart block type 2; BIII = third-degree heart block; DRR = delta-RR; IVCD = intraventricular conduction delay; IVR = idioventricular rhythm; Junctional = junctional rhythm; RRP = RR prematurity; Sinus = sinus rhythm; SVT = supraventricular tachycardia; VBigem = ventricular bigeminy; VT = ventricular tachycardia; VTrigem = ventricular trigeminy; other abbreviations as in Table 2.
BeatLogic episode and duration F1 scores exceeded 70 for all 14 rhythms, exceeded 80 for 11 rhythms, exceeded 90 for 7 rhythms, and exceeded 95 for the following 5 rhythms: atrial fibrillation/flutter, ventricular tachycardia, ventricular bigeminy, ventricular trigeminy, and third-degree heart block. Figures 3 and 4 illustrate results produced by the BeatLogic platform.
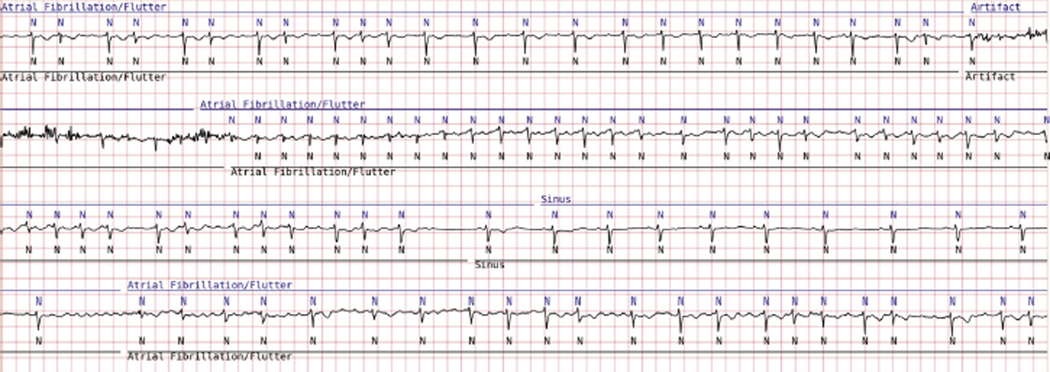
Figure 3.
BeatLogic results (blue) compared with gold validation truth (black) demonstrating beat detection/classification, noise detection, and atrial fibrillation/flutter onset/offset.
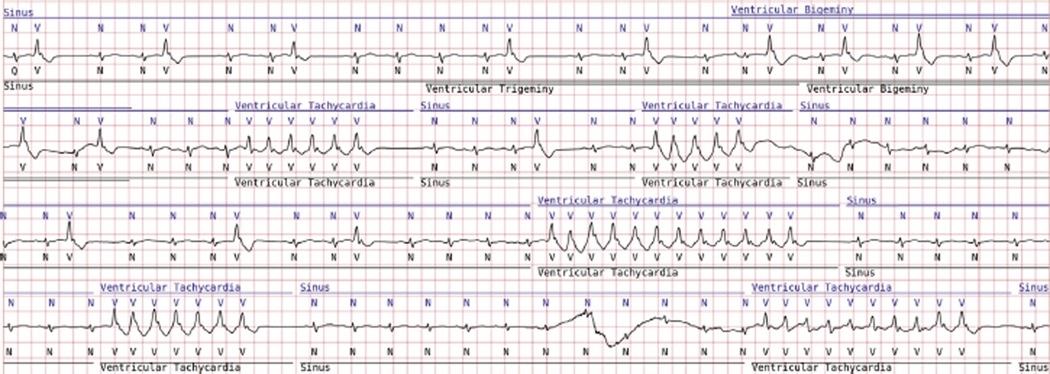
Figure 4.
BeatLogic results (blue) compared with gold validation truth (black) demonstrating beat detection/classification, ventricular bigeminy, and ventricular tachycardia detection. Where ventricular trigeminy transitions to bigeminy, BeatLogic elects to extend the duration of the higher-acuity rhythm.
Discussion
This study is the first to demonstrate a comprehensive DL-based platform capable of performing beat and rhythm detection/classification. With the exception of 3 studies, the BeatLogic platform performed equal to or better than all other algorithms for beat detection, VEB classification, and detection/classification of the 14 evaluated rhythms. This work builds on previous studies using DL for single ECG interpretation tasks5,6,20 but is differentiated by several key factors: (1) the large diverse real-world training dataset; (2) our method for leveraging beat classification results to annotate ventricular rhythms and beat patterns; (3) hyperparameter optimization, which produced very deep networks; (4) the large diverse real-world EP-adjudicated validation dataset; and (5) comparisons to previously published work and to a commercially available state-of-the-art ECG interpretation system.
Data quality and patient diversity
In developing this platform, training data diversity and quality were fundamental to achieving high performance. Initial experiments using publicly available data, which had limited patient and arrhythmia diversity, produced models that performed well on a public data holdout dataset but would not generalize to new patients. BeatLogic training annotations were created and adjudicated by a dedicated team of experienced ECG technicians using a rigorous process designed to ensure quality and consistency. The training dataset was meticulously and continuously grown over several years using a data-driven approach, which identifies algorithm failure modes and addresses them with additional training data.
DL architecture
In designing this system, several DL architecture designs were evaluated. The sequence-to-sequence convolutional network was selected because it achieved better performance at reduced computational cost compared to other architectures we tested. This finding was consistent with that of Hannun et al,6 who used a similar architecture to create a 12-rhythm (including noise) classifier. One major difference in the 2 architectures is the number of convolutional layers (113 for RhythmNet vs 34 from Hannun et al6). Consistent with findings in the image classification domain,13 our optimization results demonstrated a preference for deeper networks with narrow filters. Combining narrow filters with more convolutional layers enables the network to create more complex features without reducing the network receptive field, that is, the region of the input that can affect the value of the output.21 Because deeper networks have larger receptive fields, the model can leverage more contextual information from the 60-second input than can be achieved using a shallow version of the same network. Contextual information is extremely important for human ECG interpretation, so we expect it should be equally important for algorithmic ECG interpretation. Whether Hannun et al6 experimented with deeper networks is unclear; however, benefits from increasing depth receptive field may have been limited by their input data duration, which was 30 seconds.
Ventricular rhythm detection
In contrast to previous studies that used DL rhythm classifiers to detect ventricular rhythms,6 we leverage beat classification results for identifying ventricular rhythms. We selected this approach because it enables detection of standalone VEBs and couplets, but we found it also facilitated superior ventricular rhythm detection performance. Currently used only for ventricular rhythms, this approach could also be utilized for atrial, junctional, and supraventricular rhythms.
Comparisons with the state-of-the-art commercial algorithm
Improvements over the state-of-the-art commercial algorithm demonstrate the unique capacity of DL models to outperform classic machine learning for ECG annotation. Nearly all commercially available algorithms we evaluated performed better on the MIT-BIH and AFDB datasets than on the gold validation dataset. This suggests that these algorithms were tuned to the public datasets, which were not captured using a patch-style monitor. Patch-style recordings present a unique challenge for automated systems due to the short dipole and placement near large muscle groups. This results in lower-amplitude p waves and reduced signal-to-noise ratio. In contrast, BeatLogic DL models were trained using patch-style recordings and in some cases performed better on the MIT-BIH and AFDB datasets than on the gold validation dataset. The different makeup of these datasets prevents strict comparisons; however, these results suggest that the DL models have discovered features that generalize to ECGs recorded using different methods. Because basic techniques used by humans for beat and rhythm detection/classification are generally device agnostic, this finding bodes well for the DL approach.
Comparisons with previously published work
BeatLogic outperformed previously published algorithms capable of performing only a single task. Performance was compared with 14 previously published algorithms, which represents a small proportion of the studies uncovered in our literature search. Studies were excluded for using nonstandard subsets of the MIT-BIH or AFDB database, for using nonstandard analysis techniques, and for allowing training/validation patient overlap. An exception was made for VEB classification performance, for which nearly all studies leveraged training/validation patient overlap to create patient-specific classifiers. In this group, only de Chazal et al22 and BeatLogic generalize without time-consuming patient-specific training. Of the many atrial fibrillation/flutter algorithm studies, we found only one that followed the EC57 standard for measuring performance. Other studies used beat-by-beat analysis, just episode analysis, or arbitrary 1- to 10-second-long segments to calculate sensitivity and PPV. In a recent study, Gusev et al23 demonstrated how these nonstandard validation methods can fail to accurately reflect algorithm performance. Although the widespread use of nonstandard rhythm performance measurement techniques does not invalidate the findings of these studies, it does make their results difficult to interpret in the context of other work.
Study limitations
Patient deidentification prevented characterization of the patient population in this study. We sought to mediate the impact of patient subtypes by leveraging random selection and a large patient population; however, future work incorporating diagnosis status, medication status, body mass index, activity level, and other factors would allow for measuring algorithm performance within specific patient subsets and ensure equal representation in the training dataset. Unfortunately, the proprietary state-of-the-art algorithm used for comparison prevents us from fully describing its underlying algorithms. However, as a commercially available FDA-cleared system, its performance represents a meaningful baseline for contextualizing BeatLogic performance. Notable conditions not represented within the study include pacing and Ventricular Fibrillation (VF). Because remote ambulatory monitor antialiasing filters distort pacer artifacts, detection is commonly performed on-device rather than with downstream annotation algorithms. VF is a critical but rare arrhythmia, and because DL requires many examples for each rhythm our DL models were not trained to detect VF. Instead, downstream systems leverage classic signal processing for VF detection. As with all learning-based algorithms, performance of this system is, in general, limited by the training data volume, diversity, and label consistency. We sought to mediate these limitations through intelligent mining of training records and standardization of the annotation process. Although the impact of these efforts is difficult to quantify, we anticipate that continuous iteration on these approaches will be fundamental to improving the performance of beat and rhythm detection/classification and to expanding the types of rhythms and beats that the platform can accurately identify.
Conclusion
As the popularity of long-term ambulatory ECG monitoring continues to grow, reliance on ECG interpretation algorithms will increase. Initial applications of DL to ECG interpretation focused on only beat detection, beat classification, or rhythm classification have shown promising results. By leveraging high-quality comprehensive training data and multiple DL models to create a system that can perform all 3 tasks, Beat-Logic represents the next stage of advancement for algorithmic ECG interpretation. Real-world gold standard validation demonstrates the superiority of this approach over the current state of the art.
Supplementary Material
Acknowledgments
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. Dr Teplitzky and Mr McRoberts are Preventice Solutions employees and stockholders. Dr Ghanbari is a consultant for Preventice Solutions.
Footnotes
Appendix
Supplementary data
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.hrthm.2020.02.015.
References
- 1.Poon K, Okin PM, Kligfield P. Diagnostic performance of a computer-based ECG rhythm algorithm. J Electrocardiol 2005;38:235–238. [DOI] [PubMed] [Google Scholar]
- 2.Schläpfer J, Wellens HJ. Computer-interpreted electrocardiograms: benefits and limitations. J Am Coll Cardiol 2017;70:1183–1192. [DOI] [PubMed] [Google Scholar]
- 3.Jiang W, Kong SG. Block-based neural networks for personalized ECG signal classification. IEEE Trans Neural Netw 2007;18:1750–1761. [DOI] [PubMed] [Google Scholar]
- 4.Limam M, Precioso F. Atrial fibrillation detection and ECG classification based on convolutional recurrent neural network. Comput Cardiol 2017;44 10.22489/CinC.2017.171-325. [DOI] [Google Scholar]
- 5.Ghiasi S, Abdollahpur M, Madani N, Kiani K, Ghaffari A. Atrial fibrillation detection using feature based algorithm and deep convolutional neural network. Comput Cardiol 2017;44. 10.22489/CinC.2017.159-327. [DOI] [Google Scholar]
- 6.Hannun AY, Rajpurkar P, Haghpanahi M, et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med 2019;25:65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teplitzky BA, McRoberts M. Fully-automated ventricular ectopic beat classification for use with mobile cardiac telemetry. 2018 IEEE 15th International Conference on Wearable and Implantable Body Sensor Networks 2018;58–61. [Google Scholar]
- 8.Zhang C, Wang G, Zhao J, Gao P, Lin J, Yang H. Patient-specific ECG classification based on recurrent neural networks and clustering technique. 2017 13th IASTED International Conference on Biomedical Engineering (BioMed) 2017;63–67. [Google Scholar]
- 9.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–444. [DOI] [PubMed] [Google Scholar]
- 10.ANSI/AAMI (American National Standards Institute/Association for the Advancement of Medical Instrumentation) EC57. Testing and reporting performance results of cardiac rhythm and ST segment measurement algorithms AAMI, 2012. [Google Scholar]
- 11.Moody GB, Mark RG. The impact of the MIT-BIH arrhythmia database. IEEE Eng Med Biol Mag 2001;20:45–50. [DOI] [PubMed] [Google Scholar]
- 12.Moody GB, Mark RG. A new method for detecting atrial fibrillation using R-R intervals. Comput Cardiol 1983;10:227–230. [Google Scholar]
- 13.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Las Vegas, NV: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016. p. 770–778. [Google Scholar]
- 14.He K, Zhang X, Ren S, Sun J. Identity mappings in deep residual networks. In: Leibe B, Matas J, Sebe N, Welling M, eds. Computer Vision—ECCV 2016. Cham: Springer International Publishing; 2016. p. 630–645. [Google Scholar]
- 15.Lenis G, Pilia N, Loewe A, Schulze WHW, Döossel O. Comparison of baseline wander removal techniques considering the preservation of ST changes in the ischemic ECG: a simulation study. Comput Math Methods Med 2017;2017:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He K, Zhang X, Ren S, Sun J. Delving deep into rectifiers: surpassing human-level performance on ImageNet Classification. 2015 IEEE Int Conf Comput Vision (ICCV) 2015;1026–1034. [Google Scholar]
- 17.Kingma DP, Ba J. Adam: a method for stochastic optimization. arXiv:14126980 [cs.LG]. November 20, 2019]. Available at: http://arxiv.org/abs/1412.6980. [Google Scholar]
- 18.Prechelt L. Early stopping—but when? In: Orr GB M €uller K-R, eds. Neural Networks: Tricks of the Trade. Berlin, Heidelberg: Springer Berlin Heidelberg; 1998. p. 55–69. [Google Scholar]
- 19.Bergstra JS, Bardenet R, Bengio Y, Kégl B. Algorithms for hyper-parameter optimization. In: Shawe-Taylor J, Zemel RS, Bartlett PL, Pereira F, Weinberger KQ, eds. Advances in Neural Information Processing Systems 24. Red Hook, NY: Curran Associates; 2011. p. 2546–2554. [Google Scholar]
- 20.Kiranyaz S, Ince T, Gabbouj M. Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Trans Biomed Eng 2016;63:664–675. [DOI] [PubMed] [Google Scholar]
- 21.Luo W, Li Y, Urtasun R, Zemel R. Understanding the effective receptive field in deep convolutional neural networks. In: Lee DD, Sugiyama M, Luxburg UV, Guyon I, Garnett R, eds. Advances in Neural Information Processing Systems 29. Red Hook, NY: Curran Associates; 2016. p. 4898–4906. [Google Scholar]
- 22.de Chazal P, O’Dwyer M, Reilly RB. Automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Trans Biomed Eng 2004; 51:1196–1206. [DOI] [PubMed] [Google Scholar]
- 23.Gusev M, Boshkovska M. Performance evaluation of atrial fibrillation detection. 2019 42nd International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO) 2019;342–347. [Google Scholar]
- 24.Pan J, Tompkins WJ. A real-time QRS detection algorithm. IEEE Trans Biomed Eng 1985;BME-32:230–236. [DOI] [PubMed] [Google Scholar]
- 25.Christov II. Real time electrocardiogram QRS detection using combined adaptive threshold. Biomed Eng Online 2004;3:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiarugi F, Sakkalis V, Emmanouilidou D, Krontiris T, Varanini M, Tollis I. Adaptive threshold QRS detector with best channel selection based on a noise rating system. Comput Cardiol 2007;34:157–160. [Google Scholar]
- 27.Chouakri SA, Bereksi-Reguig F, Taleb-Ahmed A. QRS complex detection based on multi wavelet packet decomposition. Appl Math Comput 2011;217:9508–9525. [Google Scholar]
- 28.Elgendi M. Fast QRS detection with an optimized knowledge-based method: evaluation on 11 standard ECG databases. PloS One 2013;8:e73557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez JP, Almeida R, Olmos S, Rocha AP, Laguna P. A wavelet-based ECG delineator: evaluation on standard databases. IEEE Trans Biomed Eng 2004; 51:570–581. [DOI] [PubMed] [Google Scholar]
- 30.Arzeno NM, Deng Z-D, Poon C-S. Analysis of first-derivative based QRS detection algorithms. IEEE Trans Biomed Eng 2008;55:478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zidelmal Z, Amirou A, Adnane M, Belouchrani A. QRS detection based on wavelet coefficients. Comput Methods Programs Biomed 2012;107:490–496. [DOI] [PubMed] [Google Scholar]
- 32.Ince T, Kiranyaz S, Gabbouj M. A generic and robust system for automated patient-specific classification of ECG signals. IEEE Trans Biomed Eng 2009; 56:1415–1426. [DOI] [PubMed] [Google Scholar]
- 33.Petrucci E, Balian V, Filippini G, Mainardi LT. Atrial fibrillation detection algorithms for very long term ECG monitoring. Comput Cardiol 2005; 32:623–626. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.