Summary
Hearing loss is one of the top contributors to years lived with disability and is a risk factor for dementia. Molecular evidence on the cellular origins of hearing loss in humans is growing. Here, we performed a genome-wide association meta-analysis of clinically diagnosed and self-reported hearing impairment on 723,266 individuals and identified 48 significant loci, 10 of which are novel. A large proportion of associations comprised missense variants, half of which lie within known familial hearing loss loci. We used single-cell RNA-sequencing data from mouse cochlea and brain and mapped common-variant genomic results to spindle, root, and basal cells from the stria vascularis, a structure in the cochlea necessary for normal hearing. Our findings indicate the importance of the stria vascularis in the mechanism of hearing impairment, providing future paths for developing targets for therapeutic intervention in hearing loss.
Keywords: hearing loss, ARHL, cochlea, genetics, GWAS, stria vascularis, spindle cells, root cells, basal cells, hair cells
Introduction
The number of people with mild-to-complete hearing impairment is projected to increase to an estimated 2.45 billion worldwide by 2050, principally driven by age-related hearing impairment (ARHI).1 Hearing impairment is ranked third for causes of global years lived with disability (YLDs) across all ages and the leading cause of YLDs in those older than 70, as compared with all other disease categories.1 The overall global cost of unaddressed hearing loss exceeds $981 billion annually.2 ARHI has been associated with social withdrawal, depression, anxiety, as well as cognitive decline and dementia.3 There is no preventive treatment for hearing decline and therapeutics are currently available only in the form of hearing aids or cochlear implants. Moreover, the impact of untreated hearing loss remains underestimated as governmental and industry incentives are still very low in comparison to other diseases of equal prevalence.4
Hearing thresholds tend to deteriorate gradually with age and ARHI is typically more pronounced in the higher frequencies. Knowledge of the pathophysiological mechanisms of hearing loss derives primarily from animal studies (particularly mouse models5), as well as clinical research on specific families with hearing loss.6 Hearing loss is moderately heritable, with recent studies attributing 36%–70% of the variation in the heritability of hearing impairment to additive genetic effects.7,8 Large genome-wide association studies (GWASs) have been recently conducted: UK Biobank (n = 87,056 individuals with self-reported hearing difficulty) revealed 44 independent loci associated with self-reported hearing difficulty and confirmed that hearing loss is a complex polygenic disorder.9 A combined Icelandic cohort and UK Biobank (n = 121,934 individuals identified through pure-tone audiograms and self-reported hearing difficulty) yielded another 21 novel associations of which 13 were rare variants.10 Kalra et al. performed a multi-trait analysis of GWASs (MTAG)11 using UKB data from up to 337,000 participants with different hearing phenotypes and identified 8 novel hits supported with transcription data.12 However, many loci were not replicated, which may be explained by differences in phenotyping (ICD diagnoses, self-report, hearing thresholds assessed by audiometry), imbalanced sample size with UK Biobank predominating, statistical power, or ancestral differences between samples. While early-onset genetic hearing loss is determined by monogenic factors, ARHI appearing in late adulthood develops from the interaction of environmental and polygenic factors.13
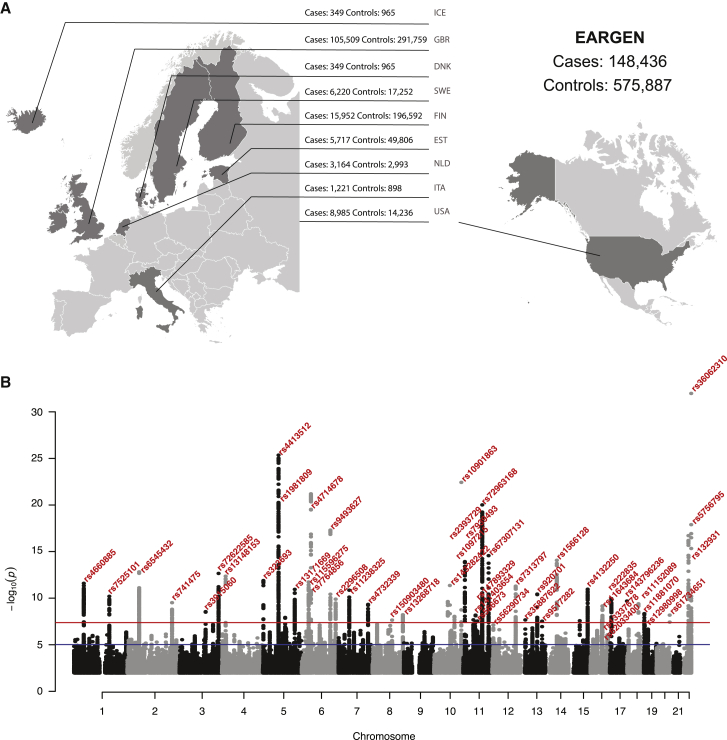
In order to gain fundamental knowledge on the genetic basis of hearing loss, we conducted a meta-analysis of 17 hearing loss GWASs using both ICD diagnoses and self-reported hearing loss. The latter has been demonstrated to be a good proxy for formal hearing assessment.14 The study comprised 147,997 affected individuals and 575,269 control subjects including 60,941 affected individuals that were not in our previously published GWAS meta-analysis.9 We compiled a dataset comprising multiple different European and US population-based cohorts (Figure 1A, Table S1).
Figure 1.
GWAS meta-analysis for ARHI (n = 723,266)
(A) Origin of the datasets used for the meta-analysis of the ARHI GWAS: 8 European countries and the United States.
(B) Manhattan plot displays all associations per variant ordered according to their genomic position on the x axis and showing the strength of the association with the −log10 transformed p values on the y axis. The threshold for genome-wide significance (p < 5 × 10−8) is indicated by the red line, while the blue line represents the suggestive threshold (p < 1 × 10−5).
Material and methods
Study design and phenotyping
Adult male and female participants were included from the following 17 population-based cohort studies: Age, Genes/Environment Susceptibility - Reykjavik (AGES; n = 3,134), the Danish Twin Registry (DTR; n = 1,314), the Estonian Genome Center at the University of Tartu (EGCUT; n = 55,523), FinnGen (n = 212,544), Framingham Heart Study (FHS; n = 2,536), Health Aging and Body Composition (HABC; n = 1,288), Italian Network of Genetic Isolates - Friuli Venezia Giulia (INGI-FVG; n = 339), the Rotterdam Study (RS, cohorts 1–3; n = 6,157), the Salus in Apulia study (SA; n = 1,780; formerly known as Great Age study), Screening Across the Lifespan Twin (SALT; n = 9,565, and SALTY - young; n = 5,133), Screening Twin Adults: Genes and Environment (STAGE; n = 8,345), TwinsUK (n = 5,125), UK Biobank (UKBB; n = 392,143), and the Women’s Genome Health Study (WGHS; n = 18,340). All participants provided written informed consent; ethical approval was obtained locally. The declaration of Helsinki was adhered to. Phenotype was based on ICD9 & 10 diagnoses of hearing loss (EGCUT and FinnGen) or questions on hearing loss (all other cohorts). Mean age overall was 59.6. A detailed description of phenotype definition case control distribution and age range for each study can be found in Table S1. Study details and cohort descriptions are available in supplemental methods. An umbrella ethics license was granted by the local ethics committee, Regionala etikprövningsnämnden in Stockholm (2015/2129-31/1). Informed consent was obtained from all participants. Individual ethical license from all contributing cohorts are included in supplemental methods.
Genome-wide association studies and meta-analysis
GWASs have been carried out for each cohort locally and summary statistics were collected for each study. Standardized quality control was performed using EasyQC software,15 followed by meta-analysis using METAL software.16 Briefly, the quality control steps with EasyQC were excluding monomorphic SNPs, SNP missingness <0.05, filtering out duplicate SNPs and SNPs with imputation score <0.5. After harmonizing allele coding and marker names, uniformed summary statistics were produced. LD score regression17 was applied to estimate the impact of population stratification and other confounders on test-statistic inflation for cohorts with sample size n > 5,000 for QC-ed and harmonized GWAS data. The genome build was hg19.
Meta-analysis of GWAS summary statistics was conducted using an inverse-variance-weighted fixed-effect model in METAL.16 To control for population stratification and other confounders, individual cohorts were adjusted for either genomic control (cohort with sample size >5,000) or intercept. A genome-wide significance threshold was defined as p < 5 × 10−8. Conditional and joint association analysis (COJO) was carried out to reveal independent lead SNPs for genome-wide significant loci using GCTA software.18 We randomly selected 50,000 individuals of European ancestry from UK Biobank as a reference sample for COJO. We examined whether the SNPs have been previously associated with hearing loss in a large-scale GWAS. A locus was designated “new” when LD with previous associated variants was <0.6. In the case of missense SNPs, the Combined Annotation Dependent Depletion (CADD) score (GRCh37-v1.6) was used to estimate the deleterious effect of the SNP.19
Gene prioritization and pathway analysis
For gene prioritization in the genome-wide significant loci, we used MAGMA20 (significance threshold p < 2.66 × 10−6) implemented in FUMA21 and VEGAS2 software22 (Bonferroni corrected p < 0.05). We used the offline version of VEGAS2 and analyzed the most associated 10 SNPs flanking 10 kb upstream and downstream the genes. The list of genes was obtained from the VEGAS2 website (https://vegas2.qimrberghofer.edu.au/glist-hg19) and included 26,056 genes. Given that the number of genes in the output would depend on the analysis parameters, we chose to correct for multiple testing assuming 20,000 independent tests by Bonferroni approach.
Genes within 500 Mb from the top SNP were checked for any known association with hearing loss in either humans or mice. For that purpose, existing literature and the website of the International Mouse Phenotyping Consortium (www.mousephenotype.org) were consulted.23 The Shared Inner Ear Laboratory Database (SHIELD)24 was used to examine whether candidate genes were expressed in inner and outer hair cells of the cochlea in adult mice (P25–P30), designated positive when expression levels (measured by fluorescent intensity readings) exceeded 10.9, as described in the referenced paper.25
We used VEGAS2 for pathway analysis based on the results of gene prioritization as described above and the list of pathways provided as part of VEGAS2 distribution (https://vegas2.qimrberghofer.edu.au/biosystems20160324.vegas2pathSYM).26
Variant analysis
All variants identified in genes of interest in the gnomAD v.2.1.1 were downloaded, and minor allele frequencies (MAFs) were filtered to select for variants that were common (MAF ≥1%) in the non-Finnish European (EUR) population. Transcripts were selected based on consensus with the Deafness Variation Database transcript catalog,27 where possible, or otherwise we used the longest transcript according to Ensembl. Variant effects were analyzed using an array of partially orthogonal computational prediction algorithms, PolyPhen-2,28 CADD,19 DANN,29 PROVEAN,30 REVEL,31 VEST3,32 and Eigen,33 that consider genetic, evolutionary, structural, and biochemical information to infer variant pathogenicity and deleteriousness. The individual algorithmic assessments were aggregated into a consensus ensemble score normalized to the range from zero (variant unanimously predicted to be deleterious) to one (unanimously predicted to be benign). The secondary structure of GJB2 was obtained from the Protein Data Bank (PDB: 2ZW3) and the structural consequences of GJB2 variants were modeled using PyMOL v..1.1.
Fine-mapping was carried out using CAUSALdb-finemapping-pip pipeline (https://github.com/mulinlab/CAUSALdb-finemapping-pip). We analyzed 1 Mb regions surrounding lead SNPs in each genome-wide significant locus. The output includes credible sets and posterior probabilities for SNPs to be causal as per PAINTOR, CAVIARBF, and FINEMAP algorithms.
Genetic correlations
In LD Hub34 we used LD score regression to estimate the genetic correlation between hearing loss and a range of other disorders and traits, to evaluate the extent of shared genetic architecture based on common gene variants and hypothesize about association with potential risk factors. After excluding all UK Biobank-based phenotypes, the list comprised 256 phenotypes. Significance was thus set at p < 2 × 10−04 after Bonferroni correction.
In the internal genome-wide association library (Omnibus data) collated by the Psychiatric Genomics Consortium, we used LDSC to get SNP-based genetic correlation between hearing loss and psychiatric and anthropometric traits.
Expression data sets
We obtained expression specificity data in 37 GTEx v835 human tissues processed by previous research.36 Briefly, the following tissue filters were applied: (1) tissues with fewer than 100 donors, (2) non-natural tissues (e.g., cancer tissue and cell lines), and (3) testis tissues (expression outlier). Since GTEx does not contain cochlear data, we sought expression data from 36,616 cells originating from 2 datasets from adult mouse cochlea (post-natal day 60, CBA/CaJ) published by Milon et al.37 (one from the stria vascularis and the other from spiral ganglion neuron [SGN]) summing to 15 different cell types (Table S2). Since these two datasets were genotyped using exactly the same technique in the same technical infrastructure, we merged them first by aggregating the count per gene per cell type and normalized to 1 TPM per cell type to account for the variation of cell counts per cell type in each dataset, while preserving the relative expression pattern per cell type (Table S2). Monocytes and neutrophils were found in both the stria vascularis and in the SGN data, and the correlations of normalized expression between the two datasets were high (0.90 for monocytes and 0.98 for neutrophils based on 15,798 genes), supporting the appropriateness of the merging step. Additionally, since the data from Milon et al. did not contain data from the organ of Corti (e.g., hair cells and and Deiters’ cells), we relied on single-cell data extracted by Ranum et al.38 (post-natal day 15, C3HeB/FeJ). We note that the murine cochlea is functionally mature at P14. Finally, we also used expression data from Zeisel et al.39 consisting in 160,796 cells of 39 broad cell types sampled from 19 regions in the entire mouse neural system (post-natal day P12–P30, as well as 6 and 8 weeks old, CD-1 and Swiss) that were processed to get cell type expression specificity36 (Table S2). Only genes with 1:1 orthology between human and mouse were preserved for calculating the expression specificity.
Calculation of cell-type expression specificity
We processed and calculated the expression specificity as previously described.36 Briefly, in each tissue expression dataset (i.e., organ of Corti,38 stria vascularis/SGNs,37 and neural tissue39), we first aggregated the count per gene per cell type and excluded genes that are (1) not expressed in any cell type, (2) with duplicated identifier, or (3) not 1:1 orthologous between mouse and human. We then normalized the expression to 1 TPM (transcripts per million) per cell type. Next, gene expression specificity was calculated per gene per cell type as:
Specificity ranges from 0 to 1; a higher value indicates that the gene is more specific to the corresponding cell types compared to its expression profiles across all included cell types. We selected genes with the top 10% specificity values in each cell type as the gene list for the cell type that was used for the heritability enrichment analyses.
SNP-heritability enrichment
We used MAGMA (v.1.08)20 and partitioned LDSC40 to evaluate whether the top 10% specifically expressed genes per tissue/cell type were enriched of the SNP-based h2 of hearing loss.
MAGMA evaluated whether the cell-type-specific genes were enriched in hearing loss gene-level associations in two steps. In the first step (SNP-wise gene analysis), we filtered out SNPs with minor allele frequency <1% and poor imputation quality (INFO < 0.6) from the ARHI summary statistics and calculated the p value for per gene association with hearing loss using SNP p values (35 kb upstream and 10 kb downstream window per gene).36 In the second step (gene-set analysis), the p values were converted to Z-scores, and one-sided tests were performed to compare whether the Z-scores in the gene set (i.e., cell-type-specific genes) were higher than those not in the gene set, which indicated enrichment of SNP-heritability in the gene set.20,41
We then applied partitioned LDSC adjusting for the baseline annotations.42 Partitioned LDSC evaluates whether the per-SNP heritability is higher in the SNPs in an SNP list (in our study the SNPs within ±100 kb of the cell-type-specific genes) compared to the other SNPs.40 We calculated p values from one-sided Z score coefficient for tissue/cell-type-specific genes.
Results
We curated summary statistics from 17 independent cohorts, a total of 723,266 individuals of European descent comprising 147,997 hearing loss cases (20.5%) and 575,269 controls (79.5%). Affected individuals were defined by either clinical diagnosis of hearing loss (ICD9 and 10; FinnGen and EGCUT, 37% of participants) or self-reported hearing impairment (all other cohorts, 63% of participants). We completed a genome-wide meta-analysis of 8,244,938 imputed SNPs that passed quality control (QC) and identified 48 significant loci (p < 5 × 10−8). There was no evidence of residual population stratification in the results of meta-analysis (λGC = 1.2764, λGC scaled to 1,000 cases and 1,000 controls = 1.001173). LD score regression intercept is 1.0039 (0.0095), indicating that the inflation of the GWAS test statistics was due to polygenicity (Figure S1). SNP heritability (h2) on the observed scale was h2 = 0.0252 (SE = 0.0013) and on the liability scale was estimated to be between 0.033 (SE = 0.002) and 0.061 (SE = 0.003) given the case/control ratio in our sample (20%) and populational prevalence in the range of 5%–40%. Next, we employed conditional and joint analysis (COJO) to identify lead independent signals in the loci (Table S3). Of 48 lead significant SNPs, 10 were considered novel associations (Table 1), defined as LD < 0.6. LDSC genetic correlations attributable to genome-wide SNPs (rg) were estimated across all hearing loss cohorts (Table S4). Regional locus zoom plots and forest plots of significant loci are presented in Figures S2 and S3, respectively.
Table 1.
Summary statistics of significantly associated loci identified in the genome-wide association meta-analysis of AHRI
| SNP | Chr | Pos (hg19) | EA | OA | EAF | Beta | SE | p value | Directiona | Locus annotation |
|---|---|---|---|---|---|---|---|---|---|---|
| rs4660885 | 1 | 46243756 | a | G | 0.4344 | −0.007 | 9.00E−04 | 3.74E−12 | --+--?----?-+---- | IPP-[x]-MAST2 |
| rs7525101 | 1 | 165109131 | t | C | 0.4424 | 0.006 | 9.00E−04 | 8.64E−11 | ++++-?++++?--++++ | PBX1---[x]-LMX1A |
| rs6545432 | 2 | 54817683 | a | G | 0.5091 | 0.007 | 9.00E−04 | 2.36E−13 | ++++++?-++?++++++ | [SPTBN1] intronic |
| rs741475 | 2 | 208087139 | t | C | 0.5771 | −0.006 | 9.00E−04 | 4.02E−10 | -----?-+--?------ | KLF7-[x]--CREB1 |
| rs3915060 | 3 | 121712980 | t | C | 0.7272 | −0.006 | 1.00E−03 | 3.96E−09 | +-+--?+-+-?+++--- | [ILDR1] intronic |
| rs72622585b | 3 | 181992315 | t | C | 0.8252 | 0.009 | 1.30E−03 | 3.41E−13 | ++-++--+++???-+++ | SOX2---[x]---ATP11B |
| rs13148153 | 4 | 17517558 | t | c | 0.1342 | 0.010 | 1.40E−03 | 2.64E−12 | +++++?++++???-+++ | [CLRN2] intronic |
| rs323693 | 5 | 2562593 | t | c | 0.882 | −0.010 | 1.40E−03 | 1.91E−12 | --++-?--++?------ | IRX4---[x]--IRX2 |
| rs1981809 | 5 | 72920029 | t | c | 0.4526 | −0.009 | 9.00E−04 | 1.36E−20 | -----+?-+-?+-+--- | UTP15-[x]-ARHGEF28 |
| rs4413512b | 5 | 73077349 | a | g | 0.5289 | −0.010 | 9.00E−04 | 1.28E−25 | -----?--+-?-+---- | [ARHGEF28] intronic |
| rs13171669 | 5 | 148601243 | a | g | 0.5682 | −0.006 | 9.00E−04 | 1.61E−11 | -----?+++-?------ | [ABLIM3] intronic |
| rs115596275 | 6 | 32420218 | c | g | 0.0213 | 0.024 | 3.50E−03 | 2.73E−12 | ?+???++-?+???++++ | HLA-DRA-[x]-HLA-DRB5 |
| rs7764856b | 6 | 32680640 | a | t | 0.3435 | 0.007 | 1.00E−03 | 1.10E−10 | ?+-??+++?-???++++ | HLA-DQB1-[x]-HLA-DQA2 |
| rs4714678 | 6 | 43342591 | a | g | 0.4031 | −0.009 | 9.00E−04 | 7.20E−20 | +-++--+---?+----- | ZNF318-[x]-ABCC10 |
| rs9493627 | 6 | 133789728 | a | g | 0.3191 | 0.009 | 1.00E−03 | 9.56E−18 | ++++-+?+-+?-+++++ | [EYA4] G>S |
| rs2296508 | 6 | 158497717 | t | c | 0.4795 | −0.006 | 9.00E−04 | 4.34E−10 | --+++??-+-?++---- | [SYNJ2] V>V |
| rs11238325 | 7 | 50853151 | t | c | 0.7315 | 0.007 | 1.00E−03 | 1.97E−11 | ++++-?++++?+-++++ | [GRB10] intronic |
| rs4732339 | 7 | 138491839 | a | g | 0.5864 | 0.006 | 9.00E−04 | 6.10E−10 | ++++-?++-+?+-++++ | TMEM213-[x]-KIAA1549 |
| rs150903480 | 8 | 91376248 | a | g | 0.0114 | −0.025 | 4.40E−03 | 2.70E−08 | ?-?+-+--+-?--+--- | [LINC00534] |
| rs13268718 | 8 | 141687200 | t | g | 0.5072 | −0.005 | 9.00E−04 | 7.47E−09 | +++-++-++-?--++-- | [PTK2] intronic |
| rs2393729 | 10 | 63837016 | t | c | 0.4218 | −0.006 | 9.00E−04 | 3.07E−10 | -++--??-+-?+++--- | [ARID5B] intronic |
| rs143282422 | 10 | 73377112 | a | g | 0.0112 | 0.032 | 4.60E−03 | 6.27E−12 | ?+?++?++++???++++ | [CDH23] A>T |
| rs1097215 | 10 | 94787804 | a | g | 0.4752 | −0.005 | 9.00E−04 | 1.11E−08 | -+-+----+-?--+--- | [EXOC6] intronic |
| rs10901863 | 10 | 126812270 | t | c | 0.2683 | 0.011 | 1.10E−03 | 9.30E−23 | ++-++?++-+?++++++ | [CTBP2] 5′ UTR |
| rs7939493 | 11 | 8073610 | a | t | 0.1911 | −0.009 | 1.20E−03 | 2.47E−14 | -++--?-+---??---- | [TUB] intronic |
| rs141403654 | 11 | 47715487 | a | t | 0.9837 | −0.022 | 3.90E−03 | 2.52E−08 | +-?--?-+-------+- | [AGBL2] intronic |
| rs147893329b | 11 | 57735006 | c | g | 0.0107 | 0.028 | 4.80E−03 | 8.17E−09 | +??++?++--+++++++ | CTNND1--[x]-OR9Q1 |
| rs566673 | 11 | 66401373 | t | g | 0.5339 | −0.005 | 9.00E−04 | 3.41E−08 | --+-++?+--------- | RBM14-[x]-RBM4 |
| rs72963168 | 11 | 88943035 | t | c | 0.7254 | −0.009 | 1.00E−03 | 3.73E−19 | -++--?------+---- | [TYR] intronic |
| rs67307131 | 11 | 118480223 | t | c | 0.654 | −0.008 | 1.00E−03 | 4.62E−15 | -+--+?---?-??+-?- | [PHLDB1] intronic |
| rs7313797b | 12 | 109896165 | t | c | 0.5604 | −0.006 | 9.00E−04 | 7.38E−12 | ++++++?++-?------ | [KCTD10] intronic |
| rs35887622b | 13 | 20763620 | a | g | 0.9854 | −0.022 | 3.90E−03 | 2.59E−08 | ?-?-+++++-?------ | [GJB2] M>T |
| rs920701 | 13 | 76417101 | t | c | 0.6357 | −0.006 | 1.00E−03 | 5.06E−11 | -+++--?-+-?------ | [LMO7] intronic |
| rs9517282b | 13 | 99059183 | a | c | 0.548 | −0.005 | 9.00E−04 | 3.54E−08 | -+-+-?----?-+---- | [FARP1] intronic |
| rs1566128 | 14 | 52514981 | a | g | 0.4126 | 0.007 | 9.00E−04 | 1.42E−14 | +--+++?+++?-+++++ | [NID2] intronic |
| rs4132250 | 15 | 89229000 | c | g | 0.778 | 0.007 | 1.10E−03 | 3.18E−11 | +++++?++-+?++++++ | ISG20-[x]--ACAN |
| rs62033400 | 16 | 53811788 | a | g | 0.6044 | 0.005 | 9.00E−04 | 4.52E−08 | ---+++-+-++-+-+++ | [FTO] intronic |
| rs11643684 | 16 | 55490167 | t | g | 0.2031 | −0.007 | 1.10E−03 | 2.26E−09 | --+--?------+---- | IRX6--[x]-MMP2 |
| rs13337678b | 16 | 56379937 | t | c | 0.5711 | −0.005 | 9.00E−04 | 3.72E−08 | ++-++??----+----- | [GNAO1] 3′ UTR |
| rs222835 | 17 | 7134129 | a | g | 0.4247 | 0.006 | 9.00E−04 | 4.81E−10 | -++++??+++?-+++++ | [DVL2] intronic |
| rs143796236 | 17 | 79495969 | t | c | 0.0076 | 0.035 | 5.60E−03 | 2.73E−10 | ?+?++?+??-???++++ | [FSCN2] H>Y |
| rs11152089 | 18 | 52625943 | t | c | 0.2134 | 0.007 | 1.10E−03 | 9.24E−10 | +-++++++++?++-+++ | [CCDC68] 5′ UTR |
| rs11881070 | 19 | 2389140 | t | c | 0.2882 | −0.006 | 1.00E−03 | 5.72E−09 | ----+??---?+-+--- | SPPL2B-[x]-TMRPS9 |
| rs12980998b | 19 | 4217510 | a | t | 0.8135 | −0.007 | 1.20E−03 | 1.02E−07 | +---+?-?+-???---- | [ANKRD24] T>S |
| rs61734651b | 20 | 61451332 | t | c | 0.0721 | 0.011 | 1.90E−03 | 8.16E−09 | ?+-+-+++++?++++++ | [COL9A3] R>W |
| rs5756795 | 22 | 38122122 | t | c | 0.5419 | −0.008 | 9.00E−04 | 3.65E−17 | +-+++-----?++---- | [TRIOBP] F>I |
| rs132931 | 22 | 38487526 | a | g | 0.5869 | −0.007 | 0.0009 | 1.59E−14 | ++---+?---?------ | [BAIAP2L2] intronic |
| rs36062310 | 22 | 50988105 | a | G | 0.0427 | 0.027 | 0.0023 | 4.25E−32 | ++++++---+?+-++++ | [KLHDC7B] V>M |
48 loci significantly (p < 5 × 10−8) associated with hearing loss. Abbreviations: Chr, chromosome; Pos, genomic position (bp); EA, effect allele; OA, other allele; EAF, effect allele frequency; Beta, effect size for EA; SE, standard error of effect size. Missense SNPs are listed in bold, with corresponding amino acid change.
Summary of effect direction for each study: + is risk increasing, - is risk decreasing, ? indicated the SNP was not present in sample cohort sequence: AGES, SA, FVG, RS2, RS3, DTR, HABC, FHS, RS1, EGC, SALT, STAGE, SALTY, TWINSUK, WGHS, FinnGen, UKBB. Locus annotation: single dash (-), <100 kb; double dash (--), 100–500 kb; triple dash (---), >500 kb.
No previous association with hearing loss in a GWAS.
Gene prioritization and pathway analysis
We used MAGMA v.1.08 and VEGAS2 for gene-set analysis and prioritization of genes at associated loci (Figure S4). Genes were examined for their relationship with hearing loss in human or mice. Seventeen loci were in or near genes with known associations to hearing loss (Table S5). Pathway analysis using VEGAS2 revealed strong enrichment in sensory perception of mechanical stimulus, sensory perception of sound, actin binding, and negative regulation of actin filament polymerization (Table S6). Interestingly, sensory perception pathways included KCNQ4, OTOF, POU4F3, PDH15, and GRIN2B, genes known to play a role in many different aspects of hearing function. Additional fine-mapping analysis identified credible sets of SNPs for each locus with 95% probability of being causal (total of 5,605 SNPs, Table S7).
Missense SNPs
Eight SNPs encoded missense mutations (Table 2). The proportion of missense SNPs overall was 17%, which is significantly higher than the average of 5.4% found in other GWAS results (GWAS Catalog accessed 19th October 2021; 1,107 studies with at least 10 genome-wide significant loci were included; Fisher exact test p = 0.005). Four of the identified genes have an established connection to deafness: EYA4 (Deafness, autosomal dominant 10, DFNA10 [MIM: 601316]), CDH23 (DFNB12 [MIM: 601543]), GJB2 (DFNA3A [MIM 601544] and DFNB1A [MIM: 220290]), and TRIOBP (DFNB28 [MIM: 609823]).43 Another three are related to hearing loss in mice: FSCN2,44 ANKRD24, and KLHDC7B (https://www.mousephenotype.org/about-impc/). Mutations in COL9A3 cause autosomal-recessive Stickler syndrome,45 a disorder affecting connective tissue (such as the spiral ligament) and commonly leading to hearing loss. With one exception (ANKRD24 [CADD score = 0.241]), CADD scores of the missense SNPs (18.45 to 31) were among the top ∼1%–0.1% of deleterious variants in the human genome. We conducted an additional in silico functional analysis on these 8 missense variants and the results provide strong evidence that variants in FSCN2 and COL9A3 are highly deleterious and likely impact gene function (Table S8). High-resolution protein structures were only available for GJB2; however, the utilized algorithms were not unanimous regarding the functional consequences of the identified variant GJB2 p.Met34Thr (rs35887622) (Figure S5A). The GJB2 transporter, also named connexin-26 (Cx26), is a hexamer with the altered amino acid being located at the core of the channel (Figure S5A). The amino acid exchange results in a substitution of hydrophilic arginine for hydrophobic methionine, which alters the surface energy of the channel and likely affects substrate translocation (Figure S5B).
Table 2.
Missense SNPs in genes associated with hearing loss
|
SNP |
rs9493627 |
rs143282422 |
rs35887622 |
rs143796236 |
rs12980998 |
rs61734651 |
rs5756795 |
rs36062310 |
|---|---|---|---|---|---|---|---|---|
| Gene | EYA4 | CDH23 | GJB2 | FSCN2 | ANKRD24 | COL9A3 | TRIOBP | KLHDC7B |
| SNP characteristics | ||||||||
| Chr | 6 | 10 | 13 | 17 | 19 | 20 | 22 | 22 |
| Pos (hg19) | 133789728 | 73377112 | 20763620 | 79495969 | 4217510 | 61451332 | 38122122 | 50988105 |
| Locus | DFNA10 | DFNB12 |
DFNA3A DFNB1A |
– | – | – | DFNB28 | – |
| Alleles (major>minor) | G>A | G>A | A>G | C>T | A>T | C>T | T>C | G>A |
| MAF | 0.319 | 0.011 | 0.015 | 0.008 | 0.187 | 0.072 | 0.458 | 0.043 |
| AA change | p.Gly277Ser | p.Ala366Thr | p.Met34Thr | p.His138Tyr | p.Thr785Ser | p.Arg103Trp | p.Phe1187Leu | p.Val1145Met |
| Pathogenicity score | 0.29 | 0.43 | 0.71 | 0 | 1 | 0 | 1 | 0.71 |
| Phenotype | hearing loss, AD | hearing loss, AR/Usher syndrome | hearing loss, AD and AR | hearing loss in mice44 | abnormal ABR in mice23 | Stickler syndrome, AR | hearing loss, AR | abnormal ABR in mice23 |
| Gene characteristics | ||||||||
| Transcript | NM_004100.5 | NM_022124.6 | NM_00400.6 | NM_001077182.3 | NM_133475.1 | NM_001853.4 | NM_001039141.3 | – |
| Gene length (bp) | 5,699 | 10,085 | 2,250 | 1,665 | 4,026 | 2,485 | 10,129 | 2,990 |
| Translation length | 639 | 3,359 | 226 | 492 | 1,146 | 684 | 2,365 | 594 |
| Number of exons | 20 | 70 | 2 | 5 | 22 | 32 | 24 | 1 |
| Total variants gnomAD v2.1.1 | 1,081 | 6,088 | 345 | 836 | 1,793 | 2,251 | 3,281 | 609 |
| All ≥1% (total) | 14 | 84 | 4 | 1 | 48 | 54 | 38 | 7 |
| MAF ≥1% in EUR | 10 | 60 | 1 | 6 | 36 | 53 | 38 | 7 |
| Total unique gnomAD SNPs in same exon as GWAS SNP | 0 | 1 | 0 | 1 | 6 | 1 | 14 | 6 |
| Pathogenicity | ||||||||
| DM | 52 | 353 | 270 | 0 | 0 | 9 | 49 | 0 |
| DM? | 8 | 67 | 77 | 0 | 0 | 1 | 14 | 1 |
| SUM | 60 | 420 | 347 | 0 | 0 | 10 | 63 | 1 |
| Exon containing SNP | 11 | 11 | 2 | 1 | 18 | 5 | 7 | 1 |
Abbreviations: Chr, chromosome; Pos, genomic position; MAF, minor allele frequency in the current study; AA change, amino acid change; AD, autosomal dominant; AR, autosomal recessive; bp, base pair; EUR, European (non-Finnish); MAF, minor allele frequency; SNP, single-nucleotide polymorphism; DM, disease causing mutation. Pathogenicity score is estimated from an aggregated score detailed in Table S6. Aggregated pathogenicity score is normalized from 0 (variant predicted to be deleterious) to 1 (predicted to be benign). Phenotype: in humans, except where noted otherwise. DM?, likely disease causing mutation based on the Human Gene Mutation Database (HGMD) Professional version 2021.3.46
Genetic correlations
A significant positive genetic correlation was found between hearing loss and insomnia, depressive symptoms, neuroticism, and obesity (LD Hub, Figure S6, Table S9). A significant negative genetic correlation was found with subjective well-being. No significant correlation was established between hearing loss and several neurological disorders or medical conditions. Additionally, we implemented the same approach to investigate SNP-based correlation between hearing loss and other traits using recent results from the Psychiatric Genomics Consortium and found significant genetic correlation with major depressive disorder, autism spectrum disorder, alcohol dependence, neuroticism, attention deficit hyperactivity disorder, and smoking initiation/smoking (Figure S7, Table S10).
GTEx tissue enrichment analysis and cell-type specificity of hearing loss genetic associations
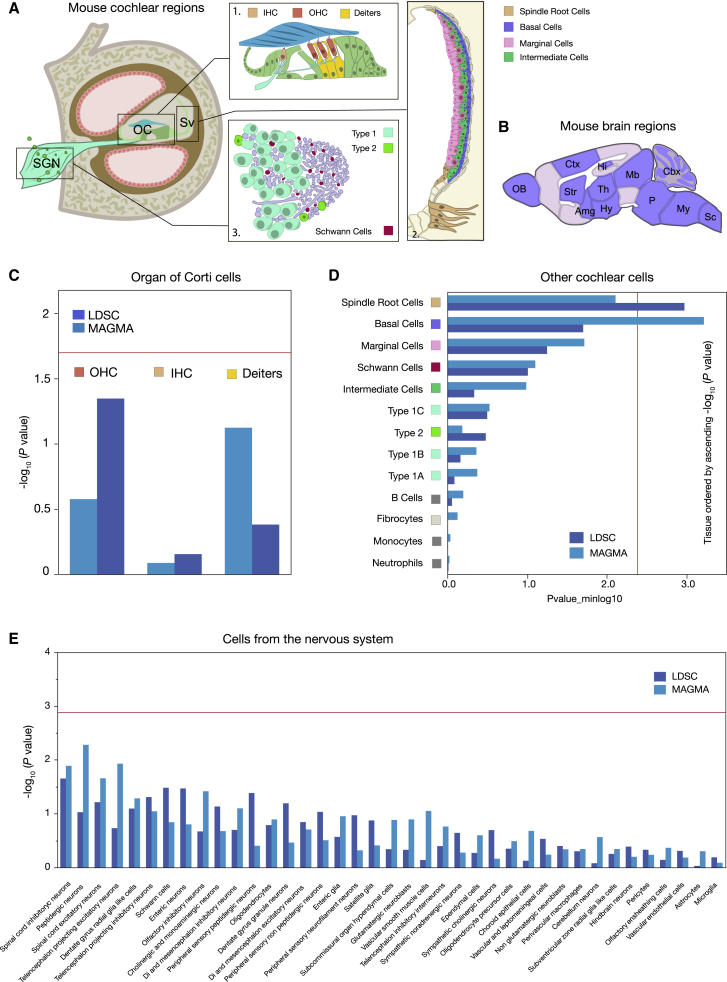
We next attempted to identify the specific somatic tissues implicated by our GWAS and define a spatial topography of hearing loss heritability-associated gene expression. Expression datasets for multiple human tissues GTEx v.8 do not contain inner ear tissue and, unsurprisingly, LDSC and MAGMA revealed no significant enrichment of SNP-h2 in GTEx tissues (Figure S8; Table S11). We relied on mouse cochlear37 and brain39 cell-specific expression profiles to determine cell types matching the common variants identified. This approach enabled us to prioritize cells that are fundamental to the etiology of hearing loss. Mouse cochlea scRNA-seq originated from Milon et al.,37 which included the spiral ganglion region and the stria vascularis (total of 36,616 cells; Figure 2A, Table S2), but since this dataset did not include scRNA-seq data from cells of the organ of Corti (that harbor hair cells and Deiters’ cells), we included an additional dataset from Ranum et al. containing Deiters’ cells and inner and outer hair cells from the mouse cochleae38 (total of 3,189 cells; Figure 2A; Table S2). Since these two cochlea studies used different methods, these were analyzed separately. Nervous system scRNA-seq included 39 broad cell types from 19 regions of the mouse central, peripheral, and enteric nervous system (total of 160,796 cells; Figure 2B).39
Figure 2.
Evaluation of enrichment of common-variant hearing loss GWAS results in scRNA-seq mouse datasets
Schematic of the mouse cochlea (A) and the mouse brain (B) regions used for the enrichment analysis. Abbreviations: Amg, amygdala; Cbx, cerebellum; Ctx, cerebral cortex; DC, Deiters’ cells; Hi, hippocampus; Hy, hypothalamus; IHC, inner hair cells; Mb, midbrain; My, medulla; OB, olfactory bulb; OC, organ of Corti; OHC, outer hair cells; P, pons; Sc, spinal cord; Str, striatum; Th, thalamus. OC is magnified in box 1 and illustrates the IHC, OHC, and DC, whose enrichment is shown in (C). A color box links a specific cell to the schematic. The red line is the Bonferroni significance threshold (−log10 p value 1.77). The enrichment analysis using cells from the stria vascularis (box 2) and the spiral ganglion neuron region (box 3) reveals a significant enrichment for spindle root cells and basal cells (D). All type 1 spiral ganglion neurons (type 1a, b, c) were all labeled the same color for sake of clarity. Given the broad and scarce distribution of immune cells (monocytes, neutrophils, and B cells), these are not shown on the schematic. The red line shows the Bonferroni significance threshold (−log10 p value 2.42) (E). Mouse nervous system cell type enrichment showing no significant enrichment. The red line shows the Bonferroni significance threshold (−log10 p value 2.89). Images from (A) and (B) were reproduced from previous work47, 48, 49 with permission from Nature Springer.
If hearing loss is associated with a particular cell type, we would expect more of the genome-wide association signal to be concentrated in genes with greater specificity for that cell type. To show evidence connecting hearing loss GWASs to cell type, we used two different methods accounting for gene size and linkage disequilibrium: LDSC,40 assessing the enrichment of the common SNP heritability of hearing loss in the most cell-type-specific genes and MAGMA,20 evaluating whether gene-level genetic association with hearing loss linearly increases with cell-type expression specificity. We found no enrichment in cells from the organ of Corti (Deiters’ cells, inner and outer hair cells; Figure 2C, Table S12). Arguably, the lack of results in this enrichment analysis could be due to the fact that specificity is a relative measure between these three cell types. This assumes that the “effective” genes have similar expression pattern in all three cell types and therefore would not be captured by the specificity measure. However, this reasoning is not supported by current findings since Deiters’ cells and hair cells differ considerably in gene expression signature.38 When assessing the enrichment in SGN and cells from the cochlear lateral wall (stria vascularis), LDSC analysis revealed the involvement of spindle cells of the stria vascularis and root cells of the outer sulcus, whereas MAGMA analysis highlighted the involvement of basal cells of the stria vascularis in hearing loss (Figure 2D, Table S13). Here, spindle and root cells could not be distinguished molecularly one from another, which is why they were labeled “spindle root cells.” In contrast, no enrichment was found in any cell type from the mouse nervous system (Figure 2E, Table S14). These findings strongly support a prominent role of the stria vascularis in hearing loss.
To further gain insights into the potential molecular mechanisms involved in strial dysfunction, we investigated the top 10% specifically expressed genes in basal (342 genes, Table S15) and spindle root (380 genes, Table S16) cells. In basal cells, 10 genes were associated with SNPs that were GWAS significant, but none of them were found in the significant pathways associated with hearing loss and listed in Table S6. Among these genes, the evidence for an involvement in hearing loss is sparse. For instance, NID2, which has been associated in humans with the Landau-Kleffner syndrome, a rare language disorder with suspicions of hearing loss.50 PC encodes a pyruvate carboxylase that requires biotin and ATP for catalyzing gluconeogenesis. Pyruvate carboxylase deficiency is a rare severe metabolic disease that in some cases can be manifested with hearing loss.51 CCS encodes a copper ion binding protein and its mutations are associated with disfunctions of copper metabolism resulting in Wilson disease, a rare inherited disorder that causes excess accumulation of copper in several organs.52 Individuals with Wilson disease display abnormal auditory brainstem responses.53 Similarly, AHDC1 is most probably involved in DNA binding, and loss-of-function mutations result in Xia-Gibbs syndrome—a neurodevelopmental disorder with rare presentation of hearing loss.54 In spindle root cells, EYA4 and HOMER2 were identified in pathways that were found significant in VEGAS2—sensory perception of sound and sensory perception of mechanical stimulus, respectively. The contribution of EYA4 variants to hearing loss has been well established.55 HOMER2 is involved in intracellular homeostasis of calcium and cytoskeletal organization and has been previously associated with hearing loss,56 but its function within the stria vascularis remains unknown. TMPRSS9 encodes a membrane-bound serine polyprotease involved in the proliferation of inner ear progenitor cells in the mouse cochlea.57 GAS2 encodes an actin filament binding protein that plays a role in cell shape and regulating microfilament rearrangements. In mice, Gas2 is expressed in supporting cells but also in the stria vascularis from the post-natal cochlea and its disruption causes hearing loss in mice and human.58 Taken together, these findings suggest that dysfunctions in the stria vascularis involve a large range of molecular mechanisms, globally impacting strial function.
Discussion
The present genome-wide meta-analysis is among the largest conducted in hearing genetics to date and provides an association catalog that helps to refine the fundamental basis of hearing loss. We find evidence of association for 48 common genetic loci, of which 10 were novel and highlight the role of genes expressed in cochlear lateral wall, consisting of the spiral ligament and stria vascularis as an important contributor to hearing loss. We employed a pragmatic, clinically informed approach by including cohorts that met empirical criteria for sufficient genetic and phenotypic similarity, based on both self-report and medical registries. We previously verified a high genetic correlation between objective measures of hearing loss and questionnaires.14 Second, our findings point to multiple genes that have been reported to cause Mendelian forms of hearing loss previously, including EYA4, CDH23, TRIOBP, and GJB2, the latter being the most commonly reported gene in autosomal-recessive non-syndromic hearing loss.59 This study is part of an expanding number of hearing loss GWASs with ever increasing sample size and the emerging functional and cellular bioinformatics tools enable us to unravel more of its pathophysiological pathways. Our study confirms as do other recent GWASs that hearing loss is driven by multiple common variants in known hearing genes.
Our results allow us to draw several broad conclusions. Of importance, a large proportion of potentially disruptive missense variants were found in contrast to other disease-related GWASs (Fischer’s exact test; p = 0.005; Figure S9), suggesting that a burden of common and rare yet impactful variants may drive the risk of hearing loss.
Second, our results do not point toward a large involvement of the brain within hearing loss. Although this is not entirely unexpected, proficient hearing acuity requires the functional integration of signals that are captured at the level of the cochlea, which are transduced to provide signal down the VIIIth cranial nerve and further propagate via the brainstem toward the thalamus and the auditory cortex. Abnormal CNS streaming of signal to noise has been implicated in ARHI60 and is supported by the association with cognitive decline, but our GTEx and scRNA-seq analyses revealed no enrichment of GWA signals in the brain, nor in its regions or its major cell types.
Third, we found significant positive genetic correlations with depressive symptoms, obesity, and smoking, but not with Alzheimer disease. The latter is interesting as hearing impairment is an established risk factor for cognitive decline and dementia,61 suggesting that hearing impairment rather than shared underlying genetic factors contribute to the development of dementia. A more complete analysis of the drivers of this relationship is needed, but the inference is that shared environmental factors contributing to hearing loss and dementia will predominate, rather than shared genetic factors.
Fourth, our findings in hearing loss pathway analyses implicate the processes involved in cytoskeleton organization and actin binding, two broad features of the mechano-transduction apparatus of the sensory hair cells.62 Indeed, these findings are consistent with a recent GWAS performed on the UKBB that localized a number of lead SNPs in cells from the post-natal mouse cochleae using scRNA-seq data12 or in the human cochlea (mainly in type I SGN, or hair cells) using immunohistochemistry on samples collected from individuals with life-threatening posterior cranial fossa meningioma compressing the brain stem.63 These included EYA4, LMX1A, PTK2/FAK, UBE3B, MMP2, SYNJ2, GRM5, TRIOBP, LMO-7, and NOX4. Consistent with their findings, we also identified SPTBN1, a mouse ortholog Spectrin expressed in the cuticular plate at the base of the stereocilia, deletion of which causes profound deafness.64 Interestingly, synaptic plasticity genes were also found such CTBP2, which is an important marker of the pre-synaptic machinery—namely the synaptic ribbon—gathering the glutamate-filled vesicles prior to their release. Alterations in ribbon abundance has been associated with cochlear synaptopathy in mouse models of noise-induced hearing loss.65, 66, 67, 68 In these models a decrease in ribbon abundance in the absence of hearing loss (the so-called hidden hearing loss) is thought to be associated with problems in speech in noise recognition and tinnitus.69,70
Fifth, although lead association SNPs are related to sensory hair cell function and their involvement in hearing loss is well established, our cell-specific enrichment analysis revealed hearing loss being also driven, at least in part, by basal cells and spindle cells in the stria vascularis and root cells in the outer sulcus. Indeed, the lead SNPs mainly related to hair cell and auditory neuron function represent the tip of the iceberg (e.g., STBPN1, CLRN2, EYA4, SYNJ2, CDH23, CTBP2, LMO7, FSCN2, ANKRD24, TRIOBP, and BAIAP2L2) influencing hearing loss with the greatest probability, while the whole iceberg is pictured by the contribution of all GWAS signals, pointing to genes expressed in cells from the lateral wall, namely basal cells and spindle cells of the stria vascularis and root cells in the outer sulcus (e.g., EYA4, MMP2, GJB2, and GJB6). These three cell types are primarily involved in endolymph ion homeostasis.47,71, 72, 73 The basal cells are coupled to each other by tight junctions to prevent leakage of ions74 and to keep the stria vascularis separate from the spiral ligament. In addition, the spindle cells have recently been shown through gene regulatory networks to have a role in responses to inflammation.47 However, we were unable to differentiate the spindle cells from the root cells unlike Gu et al.47 who used single-nucleus RNA-seq and could identify a differential expression between these two cell types.
Our findings are in opposition to a recent study suggesting that outer and inner hair cell loss is the main contributor to ARHI and that strial tissue loss does not correlate with audiologic patterns of ARHI.75 The human otopathologic analysis of this study focused on cellular loss of the stria vascularis and may have captured loss of basal cells that cover the full extension of stria. However, cellular loss of the other two cell types is not covered by this analysis. The spindle cells reside at the edges of the stria and the root cells are outside of the stria in the outer sulcus region. Furthermore, genetically caused functional loss cannot be recognized at the histopathologic level and may precede cellular loss. The precise contribution of the sensory cell and stria vascularis mechanisms to the development of hearing loss needs to be further elucidated using molecular techniques and at a time before cell death is apparent.
The lack of cochlear tissue in the GTEx biobank is indeed a major limitation to all genetic studies of hearing and communication disorders that needs to be addressed. Having access to eQTL data from human cochlear tissue would also provide significant advances in understanding the biology of hearing loss. These limitations were partially addressed in our study by gathering a unique combination of datasets of mouse cochlear scRNA-seq. Given the complexity of the organ and its ossification, the number of compartments, and the variety of constituent cell types, such comprehensive knowledge was not available until recently. Using expression data from rodents to infer on human auditory physiology may be seen as a limitation, since mouse expression data are not fully representative of human cells. However, from a total of 48 loci we identified, 18 harbor genes that have been associated with hearing loss. From these 18 loci, 16 are related to genes disruption of which causes hearing loss in mice. Thus, these findings strongly argue in favor of the translational reliability of the present findings.
We also note that part of the scRNA-seq mouse data used here was generated from 10X Genomics, which has a sequencing resolution that may have yielded insufficient number of genes to reveal enrichments (e.g., when compared to new methodologies such as Smart-Seq2). For instance, using 1,100 proprioceptive neurons and Smart-seq2, Wu et al. detected 11,000 genes per cell and identified 8 cell types,76 while previous studies using 10X Genomics favoring a higher number of cells but with lower coverage could not differentiate proprioceptive neurons in any subtypes.77 In the study from Milon et al., only one type of fibrocyte was identified, whereas there are 5 known types of fibrocytes (type I–V) present in both man and experimental animals, but that have quite different function and molecular expression.78,79 Thus, new sequencing technologies may offer increased resolution and statistical power to reveal more accurate predictions of the involvement of more specific cells in hearing loss.
Hearing loss is a heterogeneous disorder with many contributing factors during life. A potential implication for future genetic studies is the elucidation of the bulk of common variants using a cost-effective shortcut involving on-line self-reports combined with auditory tests, such as automated speech and noise test.80 The use of online assessment would allow for a comprehensive recording of phenotypes and environmental exposures in millions of individuals. Conversely, current clinical audiologic phenotyping is mostly limited to pure tone thresholds and by far not exhaustive. Carefully phenotyped individuals with hearing loss in combination with next-generation sequencing may increase the resolution of the genetic coverage and reduce the sample size to thousands, something that has been shown highly effective in the context of schizophrenia.81
Conclusions
This study of hearing loss identified 48 associated loci, including 10 novel associations and 8 missense SNPs. Our work highlights the role of the cochlear lateral wall including the stria vascularis and the outer sulcus as a contributor to hearing loss. The results provide a valuable resource for the selection of promising genes for further functional validation in pre-clinical models and define targets for screening purposes, drug development, gene therapy, or stratification approaches. We believe such experiments will serve as a solid foundation for ultimately improving therapies against hearing loss.
Consortia
The Estonian Biobank Research Team is composed of Andres Metspalu, Mari Nelis, Reedik Mägi, and Tõnu Esko.
Acknowledgments
This study was supported by the GENDER-Net Co-Plus Fund (GNP-182), the European Union's Horizon 2020 Research and Innovation Programme, Grant Agreement No 848261 and No 722046 to C.R.C. C.R.C. received additional funding from Forschung Für Leben, Svenska Läkaresällskapet (SLS-779681), Hörselforskningsfonden (503), and Tysta Skolan. J.H.L. received funding from Swedish Brain Foundation (Hjärnfonden) and Swedish Research Council. F.L. received funding from Knut and Alice Wallenberg Foundation, Tysta Skolan, Swedish Research Council, StratRegen, and Swedish Brain Foundation. R.H. received funding from NIDCD/NIH R01DC013817, NIDCD/NIH R01DC019370, the Hearing Restoration Project, and CDMRP/DOD W81XWH-21-1-0578. S.S. received funding from the National Heart, Lung, and Blood Institute contract for the Framingham Heart Study (contract No. N01-HC-25195, No. HHSN268201500001I, and No. 75N92019D00031), the National Institute on Aging (R01 AG054076, R01 AG049607, U01 AG052409, R01 AG059421, RF1 AG063507, RF1 AG066524, U01 AG058589), and the National Institute of Neurological Disorders and Stroke (R01 NS017950 and UH2 NS100605).
Additional funding was obtained from the German Research Foundation through the Collaborative Research Center 889 (BV), Academy of Finland (grant #312073, J.K.); R01 AG059727, P30 AG066546, and UF1 NS125513 (S.L.S.B.); and Beneficentia Stiftung and D70-RESRICGIROTTO (G.G.). TwinsUK is funded by the Wellcome Trust, Medical Research Council, European Union, Chronic Disease Research Foundation (CDRF), Zoe Global Ltd and the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Center based at Guy's and St Thomas' NHS Foundation Trust in partnership with King’s College London. Hearing research within The Rotterdam Study has been founded by the Dutch Hearing Health Foundation.
Author contributions
Cohort-specific data collection, I.M.L., L.M., P.K.E.M., E.S., T.P., S.P., C.C.M., A.B.S.G., H.L., A.T., C.L.S., D.I.C.; analysis, N.T., M.B.F., L.B., S.Y., E.B.R.T., K.K., Y.Z., M.N., D.I.C., F.M.K.W., A.G., J.K., E.S., R.E., M.P.C., S.C., G.G.N., H.L., N.L.H.C., F.C.; critical revisions of the manuscript, V.G., L.J.L., J.H.L., R.H., T.P., E.S., J.R., A.M., J.K., A.B.V., H.L., A.T., A.G.; writing editing, Y.G., H.J.H., L.B., M.B.F., A.P.N., C.R.C., B.C.O., N.T., C.B., A.K.K., B.C., S.Y., E.S., J.R., A.M., J.K., A.B.V., C.C.M., A.B.S.G., D.I.C., F.M.K.W.; overseeing project, L.B., A.P.N., C.R.C., B.C.O., P.F.S., S.S., L.M., P.K.E.M., J.v.B.H., J.M.F., E.S., T.P., E.S., M.A.N.; study concept and design, Y.G., H.J.H., C.M.L., V.G., A.G., J.M.S., J.v.M., A.C., K.C., V.M.L., C.S., L.J.L., F.L., J.H.L., R.H., L.B., M.B.F., R.S., G.G., N.Q., A.P.N., C.R.C., B.C.O., N.T., P.F.S., S.S., E.S., J.R., A.M., J.K., A.B.V.
Declaration of interests
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. C.R.C. is supported by the UK National Institute for Health Research (NIHR) Biomedical Research Center but the views expressed herein are his own and do not represent those of NIHR nor the UK Department of Health and Social Care. Y.Z. and V.M.L. are co-founders and shareholders of PersoMedix AB. In addition, V.M.L. is CEO and shareholder of HepaPredict AB and discloses support by the Robert Bosch Foundation, Merck KGaA, and Eli Lilly and Company. J.H.L. is co-founder and share-holder of Oscellaria AB. H.L. receives support from a consulting contract between Data Tecnica International and the National Institute on Aging (NIA), National Institutes of Health (NIH). M.A.N. received a competitive contract awarded to Data Tecnica International LLC by the National Institutes of Health to support open science research, and he also currently serves on the scientific advisory board for Clover Therapeutics and is an advisor to Neuron23 Inc as a data science fellow. P.F.S. is consultant and shareholder of Neumora.
Published: May 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.04.010.
Contributor Information
Christopher R. Cederroth, Email: christopher.cederroth@ki.se.
Estonian Biobank Research Team:
Supplemental information
Data and code availability
The GWAS summary statistics are deposited and available in Zenodo (https://zenodo.org/record/5769707#.Ybm6v33MKhx) and codes are available on GitHub (https://github.com/translational-audiology-lab/GWAS_ARHL).
References
- 1.GBD 2019 Hearing Loss Collaborators Hearing loss prevalence and years lived with disability, 1990-2019: findings from the Global Burden of Disease Study 2019. Lancet. 2021;397:996–1009. doi: 10.1016/S0140-6736(21)00516-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDaid D., Park A.L., Chadha S. Estimating the global costs of hearing loss. Int. J. Audiol. 2021;60:162–170. doi: 10.1080/14992027.2021.1883197. [DOI] [PubMed] [Google Scholar]
- 3.Li C.M., Zhang X., Hoffman H.J., Cotch M.F., Themann C.L., Wilson M.R. Hearing impairment associated with depression in US adults, national Health and nutrition examination survey 2005-2010. JAMA Otolaryngol. Head Neck Surg. 2014;140:293–302. doi: 10.1001/jamaoto.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cederroth C.R., Canlon B., Langguth B. Hearing loss and tinnitus--are funders and industry listening? Nat. Biotechnol. 2013;31:972–974. doi: 10.1038/nbt.2736. [DOI] [PubMed] [Google Scholar]
- 5.Bowl M.R., Dawson S.J. The mouse as a model for age-related hearing loss - a mini-review. Gerontology. 2014;61:149–157. doi: 10.1159/000368399. [DOI] [PubMed] [Google Scholar]
- 6.Vona B., Doll J., Hofrichter M.A.H., Haaf T., Varshney G.K. Small fish, big prospects: using zebrafish to unravel the mechanisms of hereditary hearing loss. Hearing Res. 2020;397:107906. doi: 10.1016/j.heares.2020.107906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kvestad E., Czajkowski N., Krog N.H., Engdahl B., Tambs K. Heritability of hearing loss. Epidemiology. 2012;23:328–331. doi: 10.1097/ede.0b013e318245996e. [DOI] [PubMed] [Google Scholar]
- 8.Wolber L.E., Steves C.J., Spector T.D., Williams F.M.K. Hearing ability with age in northern European women: a new web-based approach to genetic studies. PLoS One. 2012;7:e35500. doi: 10.1371/journal.pone.0035500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wells H.R.R., Freidin M.B., Zainul Abidin F.N., Payton A., Dawes P., Munro K.J., Morton C.C., Moore D.R., Dawson S.J., Williams F.M.K. GWAS identifies 44 independent associated genomic loci for self-reported adult hearing difficulty in UK biobank. Am. J. Hum. Genet. 2019;105:788–802. doi: 10.1016/j.ajhg.2019.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivarsdottir E.V., Holm H., Benonisdottir S., Olafsdottir T., Sveinbjornsson G., Thorleifsson G., Eggertsson H.P., Halldorsson G.H., Hjorleifsson K.E., Melsted P., et al. The genetic architecture of age-related hearing impairment revealed by genome-wide association analysis. Commun. Biol. 2021;4:706. doi: 10.1038/s42003-021-02224-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turley P., Walters R.K., Maghzian O., Okbay A., Lee J.J., Fontana M.A., Nguyen-Viet T.A., Wedow R., Zacher M., Furlotte N.A., et al. Social Science Genetic Association Consortium Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 2018;50:229–237. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalra G., Milon B., Casella A.M., Herb B.R., Humphries E., Song Y., Rose K.P., Hertzano R., Ament S.A. Biological insights from multi-omic analysis of 31 genomic risk loci for adult hearing difficulty. PLoS Genet. 2020;16:e1009025. doi: 10.1371/journal.pgen.1009025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vuckovic D., Mezzavilla M., Cocca M., Morgan A., Brumat M., Catamo E., Concas M.P., Biino G., Franze A., Ambrosetti U., et al. Whole-genome sequencing reveals new insights into age-related hearing loss: cumulative effects, pleiotropy and the role of selection. Eur. J. Hum. Genet. 2018;26:1167–1179. doi: 10.1038/s41431-018-0126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cherny S.S., Livshits G., Wells H.R.R., Freidin M.B., Malkin I., Dawson S.J., Williams F.M.K. Self-reported hearing loss questions provide a good measure for genetic studies: a polygenic risk score analysis from UK Biobank. Eur. J. Hum. Genet. 2020;28:1056–1065. doi: 10.1038/s41431-020-0603-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winkler T.W., Day F.R., Croteau-Chonka D.C., Wood A.R., Locke A.E., Magi R., Ferreira T., Fall T., Graff M., Justice A.E., et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 2014;9:1192–1212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Schizophrenia Working Group of the Psychiatric Genomics Consortium, Patterson N., Daly M.J., Price A.L., Neale B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra A., Macgregor S. VEGAS2: software for more flexible gene-based testing. Twin Res. Human Genet. 2015;18:86–91. doi: 10.1017/thg.2014.79. [DOI] [PubMed] [Google Scholar]
- 23.Dickinson M.E., Flenniken A.M., Ji X., Teboul L., Wong M.D., White J.K., Meehan T.F., Weninger W.J., Westerberg H., Adissu H., et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen J., Scheffer D.I., Kwan K.Y., Corey D.P. SHIELD: an integrative gene expression database for inner ear research. Database (Oxford) 2015;2015:bav071. doi: 10.1093/database/bav071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H., Pecka J.L., Zhang Q., Soukup G.A., Beisel K.W., He D.Z.Z. Characterization of transcriptomes of cochlear inner and outer hair cells. J. Neurosci. 2014;34:11085–11095. doi: 10.1523/jneurosci.1690-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra A., MacGregor S. A novel approach for pathway analysis of GWAS data highlights role of BMP signaling and muscle cell differentiation in colorectal cancer susceptibility. Twin Res. Hum. Genet. 2017;20:1–9. doi: 10.1017/thg.2016.100. [DOI] [PubMed] [Google Scholar]
- 27.Azaiez H., Booth K.T., Ephraim S.S., Crone B., Black-Ziegelbein E.A., Marini R.J., Shearer A.E., Sloan-Heggen C.M., Kolbe D., Casavant T., et al. Genomic landscape and mutational signatures of deafness-associated genes. Am. journal Hum. Genet. 2018;103:484–497. doi: 10.1016/j.ajhg.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quang D., Chen Y., Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carter H., Douville C., Stenson P.D., Cooper D.N., Karchin R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics. 2013;14:S3. doi: 10.1186/1471-2164-14-s3-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ionita-Laza I., McCallum K., Xu B., Buxbaum J.D. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 2016;48:214–220. doi: 10.1038/ng.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng J., Erzurumluoglu A.M., Elsworth B.L., Kemp J.P., Howe L., Haycock P.C., Hemani G., Tansey K., Laurin C., Pourcain B.S., et al. Early Genetics and Lifecourse Epidemiology EAGLE Eczema Consortium LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups-Analysis Working Group, Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; et al. (2017). Genetic effects on gene expression across human tissues. Nature 550, 204-213, 10.1038/nature24277. [DOI]
- 36.Bryois J., Skene N.G., Hansen T.F., Kogelman L.J.A., Watson H.J., Liu Z., Eating Disorders Working Group of the Psychiatric Genomics Consortium, International Headache Genetics Consortium, International Headache Genetics Consortium, 23andMe Research Team, et al. Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson's disease. Nat. Genet. 2020;52:482–493. doi: 10.1038/s41588-020-0610-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milon B., Shulman E.D., So K.S., Cederroth C.R., Lipford E.L., Sperber M., Sellon J.B., Sarlus H., Pregernig G., Shuster B., et al. A cell-type-specific atlas of the inner ear transcriptional response to acoustic trauma. Cell Rep. 2021;36:109758. doi: 10.1016/j.celrep.2021.109758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ranum P.T., Goodwin A.T., Yoshimura H., Kolbe D.L., Walls W.D., Koh J.Y., He D.Z.Z., Smith R.J.H. Insights into the biology of hearing and deafness revealed by single-cell RNA sequencing. Cell Rep. 2019;26:3160–3171.e3. doi: 10.1016/j.celrep.2019.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeisel A., Hochgerner H., Lonnerberg P., Johnsson A., Memic F., van der Zwan J., Haring M., Braun E., Borm L.E., La Manno G., et al. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014.e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.R., Anttila V., Xu H., Zang C., Farh K., et al. Schizophrenia Working Group of the Psychiatric Genomics Consortium. The RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown M.B. 400: a method for combining non-independent, one-sided tests of significance. Biometrics. 1975;31:987–992. doi: 10.2307/2529826. [DOI] [Google Scholar]
- 42.Gazal S., Loh P.R., Finucane H.K., Ganna A., Schoech A., Sunyaev S., Price A.L. Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat. Genet. 2018;50:1600–1607. doi: 10.1038/s41588-018-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Camp, G., and Smith, R.J.H. Hereditary Hearing Loss Homepage: hereditaryhearingloss.Org.
- 44.Shin J.B., Longo-Guess C.M., Gagnon L.H., Saylor K.W., Dumont R.A., Spinelli K.J., Pagana J.M., Wilmarth P.A., David L.L., Gillespie P.G., et al. The R109H variant of fascin-2, a developmentally regulated actin crosslinker in hair-cell stereocilia, underlies early-onset hearing loss of DBA/2J mice. J. Neurosci. 2010;30:9683–9694. doi: 10.1523/jneurosci.1541-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faletra F., D'Adamo A.P., Bruno I., Athanasakis E., Biskup S., Esposito L., Gasparini P. Autosomal recessive Stickler syndrome due to a loss of function mutation in the COL9A3 gene. Am. journal Med. Genet. 2014;164:42–47. doi: 10.1002/ajmg.a.36165. [DOI] [PubMed] [Google Scholar]
- 46.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A.D., Cooper D.N. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu S., Olszewski R., Taukulis I., Wei Z., Martin D., Morell R.J., Hoa M. Characterization of rare spindle and root cell transcriptional profiles in the stria vascularis of the adult mouse cochlea. Sci. Rep. 2020;10:18100. doi: 10.1038/s41598-020-75238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skene N.G., Bryois J., Bakken T.E., Breen G., Crowley J.J., Gaspar H.A., Giusti-Rodriguez P., Hodge R.D., Miller J.A., Munoz-Manchado A.B., et al. Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 2018;50:825–833. doi: 10.1038/s41588-018-0129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lang H. In: The Primary Auditory Neurons of the Mammalian Cochlea. Dabdoub A., Fritzsch B., Popper A., Fay R., editors. Springer Handbook of Auditory Research; 2016. Loss, degeneration, and preservation of the spiral ganglion neurons and their processes. [Google Scholar]
- 50.Appleton R.E. The Landau-Kleffner syndrome. Arch. Dis. Child. 1995;72:386–387. doi: 10.1136/adc.72.5.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang D., De Vivo D. In: GeneReviews((R)) Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., Mirzaa G.M., Amemiya A., editors. 1993. Pyruvate carboxylase deficiency. (Seattle (WA) [Google Scholar]
- 52.Czlonkowska A., Litwin T., Dusek P., Ferenci P., Lutsenko S., Medici V., Rybakowski J.K., Weiss K.H., Schilsky M.L. Wilson disease. Nat. Rev. Dis. primers. 2018;4:21. doi: 10.1038/s41572-018-0018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Butinar D., Trontelj J.V., Khuraibet A.J., Khan R.A., Hussein J.M., Shakir R.A. Brainstem auditory evoked potentials in Wilson's disease. J. Neurol. Sci. 1990;95:163–169. doi: 10.1016/0022-510x(90)90239-j. [DOI] [PubMed] [Google Scholar]
- 54.Chander V., Wangler M., Gibbs R., Murdock D. In: GeneReviews((R)) Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., Mirzaa G.M., Amemiya A., editors. 1993. Xia-gibbs syndrome. (Seattle (WA) [Google Scholar]
- 55.Wells H.R.R., Newman T.A., Williams F.M.K. Genetics of age-related hearing loss. J. Neurosci. Res. 2020;98:1698–1704. doi: 10.1002/jnr.24549. [DOI] [PubMed] [Google Scholar]
- 56.Azaiez H., Decker A.R., Booth K.T., Simpson A.C., Shearer A.E., Huygen P.L.M., Bu F., Hildebrand M.S., Ranum P.T., Shibata S.B., et al. HOMER2, a stereociliary scaffolding protein, is essential for normal hearing in humans and mice. PLoS Genet. 2015;11:e1005137. doi: 10.1371/journal.pgen.1005137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Q., Chen J., Gao X., Ding J., Tang Z., Zhang C., Chen J., Li L., Chen P., Wang J. Identification of stage-specific markers during differentiation of hair cells from mouse inner ear stem cells or progenitor cells in vitro. Int. J. Biochem. Cell Biol. 2015;60:99–111. doi: 10.1016/j.biocel.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 58.Chen T., Rohacek A.M., Caporizzo M., Nankali A., Smits J.J., Oostrik J., Lanting C.P., Kucuk E., Gilissen C., van de Kamp J.M., et al. Cochlear supporting cells require GAS2 for cytoskeletal architecture and hearing. Dev. Cell. 2021;56:1526–1540.e7. doi: 10.1016/j.devcel.2021.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan D.K., Chang K.W. GJB2-associated hearing loss: systematic review of worldwide prevalence, genotype, and auditory phenotype. The Laryngoscope. 2014;124:E34–E53. doi: 10.1002/lary.24332. [DOI] [PubMed] [Google Scholar]
- 60.Presacco A., Simon J.Z., Anderson S. Evidence of degraded representation of speech in noise, in the aging midbrain and cortex. J. Neurophysiol. 2016;116:2346–2355. doi: 10.1152/jn.00372.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loughrey D.G., Kelly M.E., Kelley G.A., Brennan S., Lawlor B.A. Association of age-related hearing loss with cognitive function, cognitive impairment, and dementia: a systematic review and meta-analysis. JAMA Otolaryngol. Head Neck Surg. 2018;144:115–126. doi: 10.1001/jamaoto.2017.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drummond M.C., Belyantseva I.A., Friderici K.H., Friedman T.B. Actin in hair cells and hearing loss. Hearing Res. 2012;288:89–99. doi: 10.1016/j.heares.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu W., Johansson A., Rask-Andersen H., Rask-Andersen M. A combined genome-wide association and molecular study of age-related hearing loss in H. sapiens. BMC Med. 2021;19:302. doi: 10.1186/s12916-021-02169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y., Qi J., Chen X., Tang M., Chu C., Zhu W., Li H., Tian C., Yang G., Zhong C., et al. Critical role of spectrin in hearing development and deafness. Sci. Adv. 2019;5:eaav7803. doi: 10.1126/sciadv.aav7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Furman A.C., Kujawa S.G., Liberman M.C. Noise-induced cochlear neuropathy is selective for fibers with low spontaneous rates. J. Neurophysiol. 2013;110:577–586. doi: 10.1152/jn.00164.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kujawa S.G., Liberman M.C. Adding insult to injury: cochlear nerve degeneration after "temporary" noise-induced hearing loss. J. Neurosci. 2009;29:14077–14085. doi: 10.1523/jneurosci.2845-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cederroth C.R., Park J.S., Basinou V., Weger B.D., Tserga E., Sarlus H., Magnusson A.K., Kadri N., Gachon F., Canlon B. Circadian regulation of cochlear sensitivity to noise by circulating glucocorticoids. Curr. Biol. 2019;29:2477–2487.e6. doi: 10.1016/j.cub.2019.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meltser I., Cederroth C.R., Basinou V., Savelyev S., Lundkvist G.S., Canlon B. TrkB-mediated protection against circadian sensitivity to noise trauma in the murine cochlea. Curr. Biol. : CB. 2014;24:658–663. doi: 10.1016/j.cub.2014.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Monaghan J.J.M., Garcia-Lazaro J.A., McAlpine D., Schaette R. Hidden hearing loss impacts the neural representation of speech in background noise. Curr. Biol. 2020;30:4710–4721.e4. doi: 10.1016/j.cub.2020.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schaette R., McAlpine D. Tinnitus with a normal audiogram: physiological evidence for hidden hearing loss and computational model. J. Neurosci. 2011;31:13452–13457. doi: 10.1523/jneurosci.2156-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Christov F., Nelson E.G., Xu L.J., Lopez I.A., Ishiyama A., Gluth M.B. Histology of the cochlear outer sulcus cells in normal human ears, presbycusis, and meniere's disease. Otol. Neurotol. 2020;41:e507–e515. doi: 10.1097/mao.0000000000002535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jagger D.J., Forge A. The enigmatic root cell - emerging roles contributing to fluid homeostasis within the cochlear outer sulcus. Hearing Res. 2013;303:1–11. doi: 10.1016/j.heares.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 73.Eckhard A., Gleiser C., Rask-Andersen H., Arnold H., Liu W., Mack A., Muller M., Lowenheim H., Hirt B. Co-localisation of K(ir)4.1 and AQP4 in rat and human cochleae reveals a gap in water channel expression at the transduction sites of endocochlear K(+) recycling routes. Cell Tissue Res. 2012;350:27–43. doi: 10.1007/s00441-012-1456-y. [DOI] [PubMed] [Google Scholar]
- 74.Kitajiri S.I., Furuse M., Morita K., Saishin-Kiuchi Y., Kido H., Ito J., Tsukita S. Expression patterns of claudins, tight junction adhesion molecules, in the inner ear. Hearing Res. 2004;187:25–34. doi: 10.1016/s0378-5955(03)00338-1. [DOI] [PubMed] [Google Scholar]
- 75.Wu P.Z., O'Malley J.T., de Gruttola V., Liberman M.C. Age-related hearing loss is dominated by damage to inner ear sensory cells, not the cellular battery that powers them. J. Neurosci. 2020;40:6357–6366. doi: 10.1523/jneurosci.0937-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu H., Petitpre C., Fontanet P., Sharma A., Bellardita C., Quadros R.M., Jannig P.R., Wang Y., Heimel J.A., Cheung K.K.Y., et al. Distinct subtypes of proprioceptive dorsal root ganglion neurons regulate adaptive proprioception in mice. Nat. Commun. 2021;12:1026. doi: 10.1038/s41467-021-21173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharma N., Flaherty K., Lezgiyeva K., Wagner D.E., Klein A.M., Ginty D.D. The emergence of transcriptional identity in somatosensory neurons. Nature. 2020;577:392–398. doi: 10.1038/s41586-019-1900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kusunoki T., Cureoglu S., Schachern P.A., Baba K., Kariya S., Paparella M.M. Age-related histopathologic changes in the human cochlea: a temporal bone study. Otolaryngol. Head Neck Surg. 2004;131:897–903. doi: 10.1016/j.otohns.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 79.Furness D.N., Lawton D.M., Mahendrasingam S., Hodierne L., Jagger D.J. Quantitative analysis of the expression of the glutamate-aspartate transporter and identification of functional glutamate uptake reveal a role for cochlear fibrocytes in glutamate homeostasis. Neuroscience. 2009;162:1307–1321. doi: 10.1016/j.neuroscience.2009.05.036. [DOI] [PubMed] [Google Scholar]
- 80.Smits C., Theo Goverts S., Festen J.M. The digits-in-noise test: assessing auditory speech recognition abilities in noise. J. Acoust. Soc. America. 2013;133:1693–1706. doi: 10.1121/1.4789933. [DOI] [PubMed] [Google Scholar]
- 81.Halvorsen M., Huh R., Oskolkov N., Wen J., Netotea S., Giusti-Rodriguez P., Karlsson R., Bryois J., Nystedt B., Ameur A., et al. Increased burden of ultra-rare structural variants localizing to boundaries of topologically associated domains in schizophrenia. Nat. Commun. 2020;11:1842. doi: 10.1038/s41467-020-15707-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GWAS summary statistics are deposited and available in Zenodo (https://zenodo.org/record/5769707#.Ybm6v33MKhx) and codes are available on GitHub (https://github.com/translational-audiology-lab/GWAS_ARHL).