Abstract
Fluorescent Pseudomonas strains were isolated from 38 undisturbed pristine soil samples from 10 sites on four continents. A total of 248 isolates were confirmed as Pseudomonas sensu stricto by fluorescent pigment production and group-specific 16S ribosomal DNA (rDNA) primers. These isolates were analyzed by three molecular typing methods with different levels of resolution: 16S rDNA restriction analysis (ARDRA), 16S-23S rDNA intergenic spacer-restriction fragment length polymorphism (ITS-RFLP) analysis, and repetitive extragenic palindromic PCR genomic fingerprinting with a BOX primer set (BOX-PCR). All isolates showed very similar ARDRA patterns, as expected. Some ITS-RFLP types were also found at every geographic scale, although some ITS-RFLP types were unique to the site of origin, indicating weak endemicity at this level of resolution. Using a similarity value of 0.8 or more after cluster analysis of BOX-PCR fingerprinting patterns to define the same genotypes, we identified 85 unique fluorescent Pseudomonas genotypes in our collection. There were no overlapping genotypes between sites as well as continental regions, indicating strict site endemism. The genetic distance between isolates as determined by degree of dissimilarity in BOX-PCR patterns was meaningfully correlated to the geographic distance between the isolates' sites of origin. Also, a significant positive spatial autocorrelation of the distribution of the genotypes was observed among distances of <197 km, and significant negative autocorrelation was observed between regions. Hence, strong endemicity of fluorescent Pseudomonas genotypes was observed, suggesting that these heterotrophic soil bacteria are not globally mixed.
“Everything is everywhere, the environment selects” (1, 3) has been a fundamental paradigm in microbial ecology for nearly a century. However, little emphasis has been given to the study of bacterial biogeography, and everything and everywhere were never defined. Improved resolution of this principle has important implications for a broad range of topics, ranging from evolution to the search for new pharmaceuticals to quarantine.
There have been a large number of studies on the population genetics of human and animal pathogenic or commensal bacteria, such as Escherichia coli, Haemophilus influenzae, Neisseria meningitidis, Staphylococcus aureus and Streptococcus pneumoniae, to elucidate epidemiological patterns (6, 25, 28, 29, 31, 39, 40, 48). The results from those studies, obtained mainly by multilocus enzyme electrophoresis (MLEE) (38), suggest that bacterial populations are clonal and in linkage disequilibrium; i.e., genetic recombination occurs too infrequently to destroy genetic linkage, so that these bacterial taxa are comprised of a limited number of clones with worldwide distribution. This clonal paradigm has reinforced the century-old principle of global distribution. Although those studies provide important insights into bacterial population genetics, the distribution patterns of those host-associated bacteria were inevitably affected by human activity. Recently, MLEE analysis of the genus Rhizobium showed that these populations are in linkage disequilibrium on a worldwide scale but are approaching linkage equilibrium (panmictic) on a local scale (10, 43). A high diversity of clones even in a single nodule of a plant host was also found (43). Similarly, MLEE analysis of the local populations of two free-living bacteria, Bacillus subtilis and Burkholderia cepacia, showed that they approach linkage equilibrium (18, 50). These findings suggest that limited geographic migration between populations contributes substantially to linkage disequilibrium, rather than the infrequent recombination within populations.
Several workers have begun to explore directly the question of whether free-living bacteria are cosmopolitan or are endemic. Four (2, 5, 22, 45) of the six studies (2, 5, 12, 14, 22, 45) were with organisms that live at temperature extremes (thermophiles or psychrophiles), since the transport of viable propagules through temperate regions was reasoned to be problematic. Hence, endemic species would be more likely. Castenholz (5) found that certain thermophilic cyanobacteria were not found in North American hot springs but were found in those of Alaska and Iceland, inconsistent with the cosmopolitan hypothesis. In contrast, two studies of thermophilic archaeal groups showed >70% DNA-DNA hybridization for isolates from Alaska versus European hyperthermal marine environments (45) and from North Sea versus Italian thermal habitats (2). Staley's group, studying sea ice psychrophiles, has found the same genera in the Arctic and Antarctic but did not yet have sufficient data to conclude whether the isolates show cosmopolitan or endemic features (44). A previous study from this laboratory on mesophiles showed that genotypes of 3-chlorobenzoate-degrading bacteria from pristine and undisturbed soils were not globally dispersed (12).
While the above-described studies have provided preliminary insight into the degree of bacterial endemism, they have focused on specialized populations with a variety of different methods and criteria. To more fully address the cosmopolitan versus endemism hypothesis for free-living soil bacteria, we identified a bacterial group as an initial focal group for study. Our criteria for this group were that its members are easily recovered by at least semiselective cultivation, can be reliably confirmed as a particular taxon, are known heterotrophic free-living colonizers of soil, and do not have long-term survival stages and whose isolation is dependent on chromosomal (not transmissible plasmid) traits. We chose fluorescent Pseudomonas (sensu stricto) strains. This group includes, among others, P. fluorescens, P. chlororaphis, P. aureofaciens, and P. putida. A total of 248 confirmed fluorescent Pseudomonas isolates were analyzed by three molecular typing methods to give different levels of genetic resolution. Our results show strict endemicity at the genotype level but not at coarser levels of resolution.
MATERIALS AND METHODS
Soil and sampling design.
Soil samples were previously collected below the soil surface (5 to 10 cm) from pristine ecosystems in six regions on five continents. All soils were classified as luvisols (Table 1) and were collected in the spring (moist season) for the respective hemisphere. The soil moisture and pH were very similar for the sites more intensively sampled. The details of sampling methods and soil characterization were described by Fulthorpe et al. (13). For this study, we selected for the isolation of fluorescent Pseudomonas 59 soil samples, using a hierarchical geographic strategy scaling from samples along 200-m transects to multiple sites in the same regions to different continents (Table 1). Transect samples were collected 0, 5, 10, 15, 20, 25, 50, 55, 75, 95, 100, 125, 150, and 175 m from transect sample 0. Five main sites (BN, JD, HG, CL, and WV) were chosen according to climate and dominant vegetation. Sites BN and JD in southwestern Australia were the closest sites with the most similar vegetation, Eucalyptus, and served as the origin of the geographic scale for this study. Five additional forested Mediterranean sites were selected in California, Chile, and South Africa, and three sites were selected from the boreal forest ecosystem in northern Canada.
TABLE 1.
Description of sampling sites and soil samples used
| Climate and soil type | Regiona | Sitea | Geographic coordinatesb | No. of transect samples | Dominant vegetation |
|---|---|---|---|---|---|
| Mediterranean, chromic luvisols | Southwestern Australia (A) | Bridgetown (BN) | 34°00′S, 116°15′E | 10 | Eucalyptus |
| Jarrahdale (JD) | 32°23′S, 116°07′E | 10 | Eucalyptus | ||
| California (U) | Hillgate (HG) | 40°10′N, 122°29′W | 10 | Quercus dauglasii, Q. agrifolia | |
| Cloverdale (CL) | 40°29′N, 122°29′W | 10 | Quercus lobata, Q. agrifolia, Arctostaphylos manzanita | ||
| Central Chile (C) | Rio Clarillo (RC) | 33°51′S, 70°29′W | 2 | Acacia, Cryptocaria, Lithraea | |
| La Campana (LC) | 32°57′S, 71°05′W | 1 | Cryptocaria, Acacia, Lithraea, Chusquea | ||
| South Africa (F) | Mooreesburg (MB) | 33°04′S, 18°40′E | 2 | Renosterveld | |
| Boreal forest, albic luvisols | Saskatchewan, Canada (S) | Waitville (WV) | 53°41′N, 105°22′W | 10 | Picea glauca, Pinus banksiana, Betula papyrifera |
| Waskesieu (WK) | 54°00′N, 106°35′W | 2 | Picea glauca, Pinus banksiana, Betula papyrifera | ||
| Porcupine (PC) | 52°39′N, 102°23′W | 2 | Populus tacamahaca |
The letters in parentheses are those used in the isolate codes (see Results).
Determined by a geographic positioning system.
Isolation.
Soil suspensions were homogenized with phosphate-buffered saline solution, serially diluted, and plated on both Cetrimide agar (Difco, Detroit, Mich.) and Pseudomonas isolation agar (Difco). The plates were incubated at 30°C for 48 h. Colonies producing fluorescent pigments (on Pseudomonas isolation agar) or nonfluorescent pigments (on Cetrimide agar) were selected and purified on nutrient agar (Difco). To further select and confirm isolates as fluorescent Pseudomonas, all isolates were individually screened for fluorescent pigment production after growth on Pseudomonas medium F (Difco) in wells of microtiter plates. Fluorescent-pigment-producing strains were then additionally screened by PCR with a primer pair diagnostic for Pseudomonas (sensu stricto) 16S rRNA genes (49). Template DNA solutions were prepared by boiling (23, 46). PCRs were performed by the method of Widmer et al. (49). Representative amplified 16S rRNA genes were sequenced by using the dye terminator sequencing procedure at the Michigan State University (East Lansing) DNA Sequencing Facility and analyzed using the Ribosomal Database Project II online analysis program (http://www.cme.msu.edu/RDP/analyses.html).
rep-PCR genomic fingerprinting.
Repetitive extragenic palindromic PCR (rep-PCR) genomic fingerprinting of the fluorescent Pseudomonas isolates was carried out with a BOX-A1R primer (BOX-PCR) according to the protocol of Rademaker et al. (35). DNA solutions prepared by the boiling methods were used for PCR templates. PCR products were resolved on 20-cm-long 1.5% agarose gels (Gibco BRL) in 0.5× Tris-acetate-EDTA buffer at 2 V/cm for 16 h in a cold room (4°C). A 1-kb DNA size ladder (Gibco BRL) was used at both ends and in the middle of the gels. The gels were stained with ethidium bromide.
rDNA RFLP analysis.
The 16S rRNA gene and the 16S-23S intergenic spacer (ITS) region were amplified by PCR with primers pA and pH (24) and G1 and L1 (19), respectively. The forward primer, G1, was designed to anneal to a highly conserved region immediately adjacent to the 16S-23S spacer, which is located about 30 to 40 nucleotides upstream from the spacer boundary (19). The reverse primer, L1, was designed to anneal to the most conserved 23S sequence immediately following the spacer, which is located about 20 bases downstream from the spacer boundary (19). Almost full-length 16S ribosomal DNA (rDNA) was amplified with primers pA and pH. PCRs were performed by the methods of Jensen et al. (19) for the ITS region (ITS-restriction fragment polymorphism analysis [ITS-RFLP]) and Massol-Deya et al. (24) for the 16S rRNA gene (amplified 16S rDNA restriction analysis [ARDRA]), respectively.
PCR products were digested with restriction endonucleases DdeI, HaeIII, HhaI, and MspI (16S rDNA only), as recommended by the manufacturer (Gibco BRL). The digests were resolved by electrophoresis with 2% MetaPhore agarose gels (FMC) in Tris-borate-EDTA buffer at 5 V/cm for 2 h. A 100-bp DNA ladder (Gibco BRL) was run on both sides and in the central lane of each gel.
Reference strains were purchased from American Type Culture Collection; they were P. fluorescens (ATCC 12983), P. aureofaciens (ATCC 13985), P. chlororaphis (ATCC 43928), and P. aeruginosa (ATCC 19429).
Computer-assisted analysis of BOX-PCR fingerprints and ITS-RFLP and ARDRA patterns.
Gel images were digitized using GelPrint 2000I equipped with a charge-coupled device video camera (BioPhotonics, Ann Arbor, Mich.) and stored as TIFF files. These digitized images were converted, normalized with the above-mentioned DNA size markers, and analyzed with GelCompar software (version 4.0; Applied Maths, Kortrijk, Belgium). The rolling-disk background subtraction method was applied.
For BOX-PCR fingerprint analysis, similarity matrices of whole densitometric curves of the gel tracks were calculated by using the pairwise Pearson's product-moment correlation coefficient (r value) (16, 33) as recommended by Rademaker et al. (35). It is insensitive to the relative concentration of bands between fingerprints, discontinuous noise, and overall intensity of the fingerprint. For ITS-RFLP and ARDRA patterns, a band-matching algorithm (band-matching tolerance of 1.0%) was used to calculate pairwise similarity matrices with the Dice coefficient (9). Cluster analyses of similarity matrices were performed by the unweighted pair group method using arithmetic averages (UPGMA) (42).
Spatial autocorrelation analysis.
The mulitivariate Mantel test (17) was used to test the spatial autocorrelation (7). Euclidean (geographic) distances were computed among soil samples based on the geographic coordinates obtained by a geographic positioning system. The distances were divided into d classes. Similarity matrices (S) of soil samples for genotype, ITS type, and ARDRA type distribution pattern were calculated using the Dice coefficient. The standardized Mantel statistic (rM) (17) was computed between similarity matrix S and geographic distance matrix Xd using a multidimensional analysis software package (R package). The statistic was tested for significance (α = 0.05) using 999 permutations and plotted against distance classes d to form the Mantel correlogram (21, 32, 41). The progressive Bonferroni method (α′ = α/k = 0.05/d, where k is the number of independent tests) was used to test the global significance of spatial correlograms.
RESULTS
Isolation.
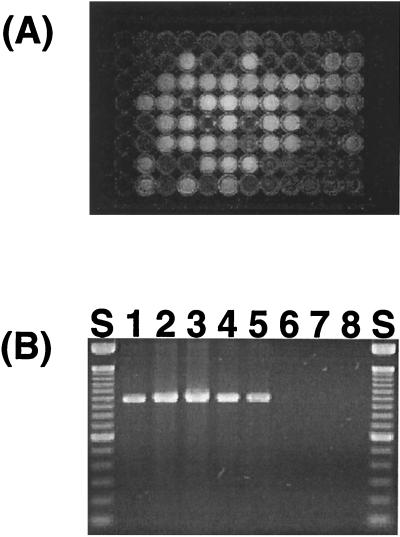
The two commercial selective media, Pseudomonas isolation agar and Cetrimide agar, were not selective enough to differentiate fluorescent Pseudomonas from some nonfluorescent bacteria. Moreover, the fact that many fluorescent Pseudomonas strains produce levans and water-soluble pigments made it difficult to select fluorescent Pseudomonas strains precisely. Hence, approximately 4,000 presumptive colonies were chosen and purified. All isolates were viable on both media. Approximately 300 isolates (7.5%) showed obvious fluorescence under UV light (Fig. 1A). Almost all the fluorescent isolates produced PCR products of the expected size (950 to 1,000 bp) product when amplified with the fluorescent Pseudomonas-specific primers (Fig. 1B). Partial sequencing (ca. 450 bp) of the PCR products from randomly selected strains confirmed that all of them belong to the P. fluorescens intrageneric group (26), which includes most fluorescent-pigment-producing Pseudomonas strains (Table 2).
FIG. 1.
(A) Microtiter plate under UV light showing production of fluorescent pigments by fluorescent Pseudomonas isolates. (B) PCR amplification product from fluorescent Pseudomonas isolates with fluorescent-Pseudomonas-specific PCR primers Ps-for and Ps-rev. Lane S, 100-bp DNA size marker (Gibco-BRL); lane 1, P. aeruginosa; lanes 2, 3, 4, and 5; fluorescent Pseudomonas isolates C-RC-3-403, S-WV-I-406, A-BN-I-405, and A-BN-5-408, respectively; lanes 6 and 7, nonfluorescent isolates; lane 8, E. coli DH5α.
TABLE 2.
Taxa in Ribosomal Database Project II with 16S rRNA gene sequences most similar to those of randomly selected isolates
| Isolate | Best matches | Similarity (%) |
|---|---|---|
| ABN-5-408 | P. corrugata ATCC 29736T | 99.7 |
| AJD-9-402 | P. syringae LMG 1247T | 97.8 |
| AJD-10-402 | P. fulva IAM 1592T | 99.4 |
| AJD-15-125 | P. rhodesiae CIP 104664T | 99.4 |
| UCL-AC-408 | P. rhodesiae CIP 104664T | 99.4 |
| UCL-9-101 | P. veronii CIP 104663T | 99.4 |
| SPC-3-401 | P. veronii CIP 104663T | 98.4 |
| SPC-9-302 | P. corrugata ATCC 29736T | 98.1 |
We obtained 248 confirmed fluorescent Pseudomonas isolates. These isolates were assigned unique codes based on the transect sample, site, and region and the isolation medium. For example, code CRC-3-401 means the first isolate (the last two digits) which was isolated from the transect sample 3 of Rio Clarillo (RC) site in central Chile (C) by using Pseudomonas isolation agar (even number in the first digit, Pseudomonas isolation agar; odd number, Cetrimide agar).
rep-PCR genomic fingerprinting.
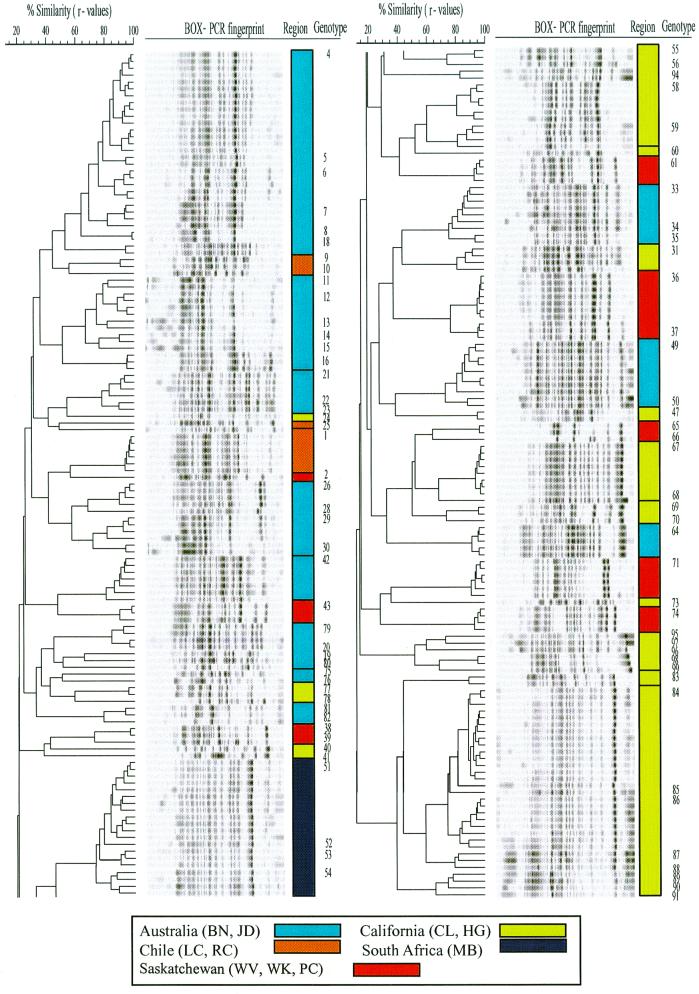
To analyze the reproducibility of BOX-PCR fingerprinting, several isolates were subjected several times to BOX-PCR. A comparison of the resulting fingerprint patterns resolved on the independent gels yielded similarity coefficients (r values) of >0.9. This reproducibility is consistent with those from other studies (35, 47). Hence, we chose a similarity value of 0.8 or more to indicate strains of the same genotype. Cluster analysis resulted in a total of 85 unique fluorescent Pseudomonas genotypes out of 248 isolates (Fig. 2). We obtained 33 genotypes comprising 103 isolates from Australia, 31 genotypes comprising 77 isolates from California, 6 genotypes comprising 14 isolates from Chile, 11 genotypes comprising 34 isolates from Saskatchewan, and 4 genotypes comprising 20 isolates from South Africa (Table 3).
FIG. 2.
Product moment-UPGMA cluster analysis of BOX-PCR fingerprints of fluorescent Pseudomonas isolates. On the scale, r values are expressed as percentages.
TABLE 3.
Geographic distribution pattern of fluorescent Pseudomonas genotypes
| Region | No. of sites | No. of transect samples | No. of isolates | No. of genotypes | Genotype overlap between:
|
||
|---|---|---|---|---|---|---|---|
| Regions | Sites | Transect samples | |||||
| Australia | 2 | 17 | 103 | 33 | No | No | Yes (4) |
| California | 2 | 11 | 78 | 32 | No | No | Yes (3) |
| Chile | 2 | 3 | 13 | 5 | No | No | No |
| Saskatchewan | 3 | 6 | 34 | 11 | No | No | No |
| South Africa | 1 | 1 | 20 | 4 | No | No | NAa |
| Global (total) | 10 | 38 | 248 | 85 | No | No | Yes (7) |
NA, not applicable.
The geographic distribution shows that the same genotype was found only in other samples of the same transect and not at other sites in the region or in other continental regions (Table 3). However, the majority (91.8%) of genotypes were not found in more than one transect sample of one site. Only seven genotypes (genotypes 4, 26, 31, 42, 49, 84, and 86) were repeatedly isolated from different transect samples; genotypes 4 and 26, genotypes 42 and 49, genotypes 84 and 86, and genotypes 31 were found only at sites BN, JD, CL, and HG, respectively.
rDNA ITS-RFLP analysis.
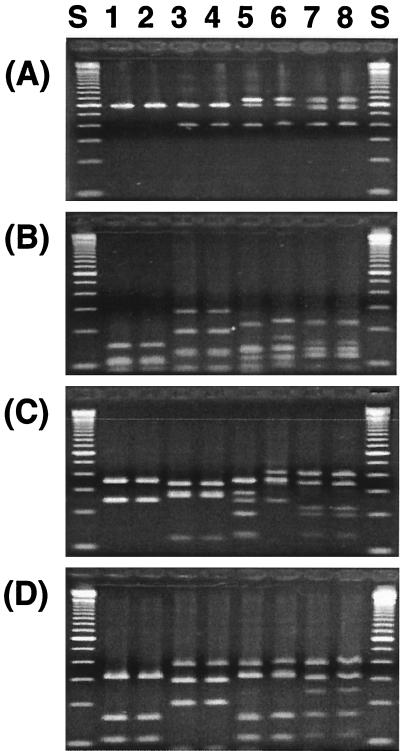
Some genotypes showed a single PCR-amplified ITS product of approximately 600 bp; however, in many cases, multiple ITS amplification products (400 to 800 bp) were observed (Fig. 3A), although all tested genotypes shared the common 600-bp band. The isolates repeatedly produced multiple bands of the same size after reselection of the isolates for purity. When the ITS-PCR products were digested (Fig. 3B to D), the sum of the sizes of restriction fragments did not agree with the sum of the sizes of undigested ITS amplification products. These results indicate the existence of at least two types of ITS regions in the same bacterial genome and that some restriction sites were present in one type of ITS region and absent in the other.
FIG. 3.
Gel electrophoresis showing length polymorphism of PCR-amplified 16S-23S rDNA ITS regions from fluorescent Pseudomonas genotypes (A) and restriction patterns of PCR-amplified 16S-23S rDNA ITS regions digested with DdeI (B), HaeIII (C), and HhaI (D). Lane S, 100-bp DNA size marker (Gibco-BRL); lanes 1, 2, 3, 4, 5, 6, 7, and 8, genotypes 19, 20, 11, 12, 4, 28, 27, and 78, respectively.
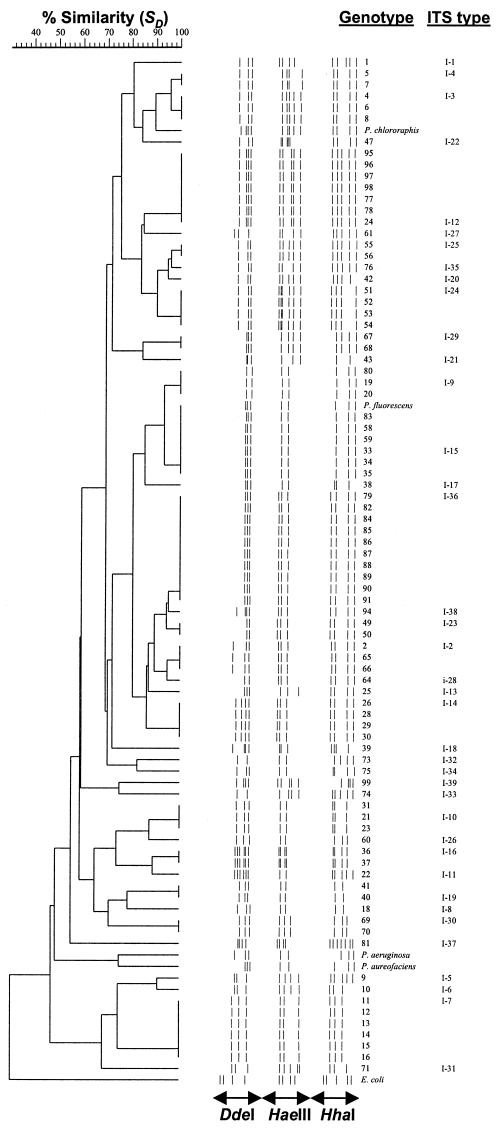
The combined DdeI, HaeIII, and HhaI restriction patterns of the amplified rDNA ITS regions were used for cluster analysis (Fig. 4). Fragments of over 100 bp were included for calculating similarity matrices. The cluster analysis revealed 39 ITS-RFLP types in our collection. All the fluorescent Pseudomonas genotypes clustered with the four Pseudomonas reference strains, making a coherent cluster at a similarity value (SD) of 47%. The E. coli reference strain formed a well-defined separate branch.
FIG. 4.
Dice-UPGMA cluster analysis of combined DdeI, HaeIII, and HhaI restriction patterns of amplified 16S-23S rDNA ITS regions of fluorescent Pseudomonas genotypes. The individual RFLPs shown are derived from the actual restriction fragments. Each ITS type is indicated by a code.
The geographic distribution pattern of ITS-RFLP types showed overlaps in types at every geographic scale (between transect samples, sites, and regions) (Table 4). Many ITS-RFLP types (13 ITS-RFLP types comprising 60% of the genotypes) were repeatedly found in more than one transect sample. ITS-RFLP types I-3, I-9, I-14, I-21, I-23, I-24, I-25, and I-29 were obtained from the genotypes isolated from different transect samples (overlap between transect samples), and ITS-RFLP types I-2 and I-19 were repeatedly obtained from the genotypes isolated from different sites (overlap between sites). Three ITS-RFLP types (I-10, I-15, and I-36) comprising 22.3% of genotypes were found in three, two, and two different regions (overlap between regions), respectively. The majority of ITS-RFLP types, comprising 40% of the genotypes, showed unique ITS-RFLP patterns; they were found in only one transect sample.
TABLE 4.
Overlapping pattern of 16S-23S rDNA ITS-RFLP types and their genotypes at each geographic scale
| Overlap between: | No. of ITS-RFLP types | No. (%) of:
|
|
|---|---|---|---|
| Genotypes | Isolates | ||
| Regions | 3 | 19 (22.3) | 66 (26.6) |
| Sites | 2 | 5 (5.9) | 6 (2.4) |
| Transects | 8 | 27 (31.8) | 89 (35.9) |
| Unique ITS-RFLP type | 26 | 34 (40.0) | 87 (35.1) |
| Total | 39 | 85 (100) | 248 (100) |
ARDRA.
All isolates gave a single amplification product, and the sum of the sizes of restriction fragments agreed well with the size of the undigested amplification product. All genotypes showed similar and simple 16S rDNA RFLP patterns with four or five bands (data not shown). Cluster analysis revealed four ARDRA types. All of the genotypes and all reference Pseudomonas strains except P. aeruginosa and E. coli produced a monomorphic restriction pattern when digested with HhaI. DdeI differentiated ARDRA type B from types A, C, and D; HaeIII differentiated ARDRA type A from types B, C, and D; and MspI differentiated ARDRA type D from types A, B, and C. Except for P. aeruginosa, the other three reference Pseudomonas strains, which belong to the P. fluorescens intrageneric group (26), and all of our collection made a coherent cluster at a similarity value (SD) of 92%, indicating high similarity in 16S rDNA sequences. A majority of the genotypes (94.1%) belonged to ARDRA types A and C, and these ARDRA types were repeatedly found in every region studied.
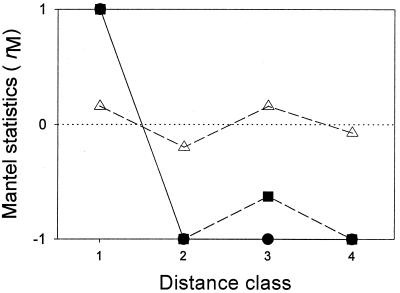
Spatial autocorrelogram.
Since endemism studies can be highly sensitive to sampling size and isolate recovery, we used a multivariate structure function, the Mantel correlogram (21, 32, 41) to describe the spatial structure and to test the presence of spatial autocorrelation (7, 41) in the distribution pattern of fluorescent Pseudomonas strains. The null hypothesis Ho is that geographic distances among sampling points are not correlated to geographic distribution pattern similarities of genotypes in corresponding samples (correlation parameter [ρ] = 0). The alternative hypothesis H1 is either ρ > 0 or ρ < 0.
Geographic distances between the pairs of soil samples were divided into four distance classes with equal frequency of the pairs to take the advantage that the tests of significance have the same power across all distance classes (21). This resulted in four distance classes: class 1 (∼197 km, 175 pairs), class 2 (∼1.09 × 104 km, 176 pairs), class 3 (∼1.59 × 104 km, 176 pairs), and class 4 (∼2.20 × 104 km, 176 pairs).
After examination of the significance of the standardized Mantel statistics (rM) (17) at α = 0.05 using 999 permutations, the null hypothesis Ho was rejected in favor of the alternative hypothesis H1, indicating the existence of a certain geographic pattern in the distribution of fluorescent Pseudomonas genotypes. Significant (P = 0.001) positive spatial autocorrelation was observed for the distribution pattern of genotypes at distance class 1, and significant (P = 0.012 to 0.020) negative spatial autocorrelation was observed at the rest of the distance classes, indicating the prevalence of the genotype in the soil samples within distance class 1 (∼197 km) and the absence in the rest of the distances (Fig. 5).
FIG. 5.
Mantel correlograms for the spatial autocorrelation analysis of fluorescent Pseudomonas (circles, genotype; squares, ITS-RFLP type; triangles, ARDRA type). Standard Mantel statistics (rM) are plotted against the distance classes. Closed symbols represent significant autocorrelation at the α = 0.05 level.
DISCUSSION
We used three methods to explore the level of genetic difference that might be discernible on a geographic scale. From coarse to fine level of resolution these were (i) ARDRA, (ii) 16S-23S rDNA ITS-RFLP, and (iii) rep-PCR genomic fingerprinting with the BOX primers. The ARDRA patterns were very similar for all isolates regardless of origin. No endemicity was seen at this coarse level of resolution, as expected since the rRNA operon is highly conserved. Similar results have also been reported from the analyses of 16S rDNA sequences of other bacterial groups. Members of proteobacterial clusters SAR11, SAR83, and SAR86 and cyanobacterial cluster SAR6-SAR7 were found in both the Atlantic and Pacific Oceans (8, 11, 15, 27, 36, 37), and freshwater bacteria with nearly identical 16S rDNA sequences were found in three distant and isolated lake regions (51).
The ITS-RFLP analysis, however, showed a weak level of endemicity, since 3 of the 39 patterns were found in two, and in one case three, continental regions. The 19 genotypes in these three cosmopolitan ITS groups showed as much genotype diversity as was found among the genotypes with unique ITS-RFLP patterns. The ITS regions, which are less conserved than the 16S rRNA gene, are still more highly conserved parts of the genome. Our results suggest that strong endemicity for this bacterial group does not occur at this level. Other work suggests that ITS-RFLP resolution corresponds roughly to the level of subspecies (4, 20, 30, 47).
We did, however, observe strong endemicity at the level of genome structure as observed by rep-PCR. According to the work of Rademaker et al. (34) with Xanthomonas strains, the similarity coefficients (approximately 0.6 to 1.0) generated by BOX-PCR fingerprinting correlated well with percent DNA-DNA hybridization (approximately 70 to 100). They found that a similarity coefficient of 0.8 corresponded to DNA-DNA hybridization values of 85 to 100%. Similarity values of <0.6 did not show significant correlation to DNA-DNA hybridization.
Our key finding is that the same genotype was found only in other samples of the same transect and not at other sites in the region or in other continental regions. This indicates some mixing and dispersal of the genotypes within a site but not between sites and regions. Isolates found at different sites or in different regions showed significantly different BOX-PCR fingerprint patterns. Even when a cutoff value 0.6 was applied to define clusters, no overlapping clusters were observed between sites or regions. The strict site endemicity observed suggests a high degree of genomic diversity of fluorescent Pseudomonas and, hence, that geographic isolation plays an important role in bacterial diversification.
Additionally, we observed significant spatial autocorrelation in the distribution of genotypes at distance class 1 (<197 km). Although this geographic distance (197 km) is based only on dividing geographic distances between all pairs of soil samples with equal frequency to achieve the same statistical power across all distance classes, distance class 1 includes all pairs of samples within transects. The remaining distance classes showed significant negative autocorrelation; hence, the overall shape of the correlogram was attributed to a structure with single bump (21), and the extent of the zone of the influence of this single bump covers all of the study area. This means that a particular genotype can be repeatedly found within a site and cannot be found out of this range at a significance level of α = 0.05. This inference is supported indirectly by the following estimate of genetic distance versus geographic distance.
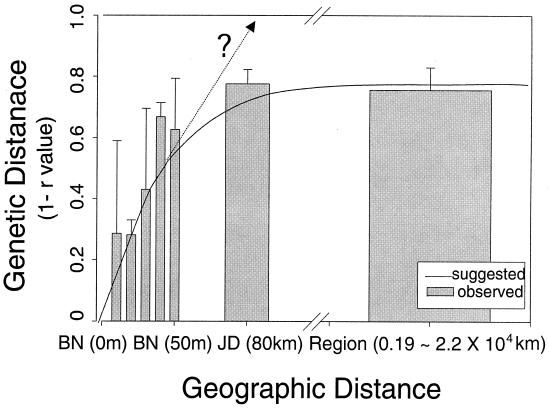
We explored the relationship between the genetic distance based on BOX-PCR fingerprinting and the corresponding geographic distance (Fig. 6). We used genotype 4 as the reference genotype, as it was a dominant genotype in our reference site BN in Australia, and calculated coefficients of similarity to every other genotype in the transect, to all transect samples of Australia site JD, and to all other transect samples in different regions. When those values were plotted against geographic distance, a relationship to genetic distance was revealed. The genetic distance increased with the geographic distance (r = +0.61; P < 0.01) up to, and is saturated roughly at, the geographic scale of site. This saturation is likely due to the method, since the saturation value corresponds to the upper limit of the resolution of BOX-PCR fingerprinting. A similar increase in genetic distance with geographic distance is seen if the JD site is resolved on the scale of the BN site (Fig. 6). The tendency that genetic distance increases with geographic distance also indirectly supports the bacterial endemism hypothesis, indicating that it becomes harder to find the same or similar genotypes with increasing geographic distances. The question mark in Fig. 6 indicates that the true genetic distance may continue to diverge but that this level of resolution is beyond what is detectable by BOX-PCR. The high level of genotype diversity found within the same ITS groups suggests that another measure of genetic diversity is needed to bridge the gap between these two methods.
FIG. 6.
Relationship between geographic distance and genetic distance. Geographic distances are based on the distance from the reference site BN (transect sample 0). Error bars indicate the range of the values of genetic distance. The question mark indicates that genetic distance may continue to diverge if the method's resolution did not saturate.
This study indicates that soil fluorescent Pseudomonas populations are endemic at genotype level and at a scale of <197 km (distance class 1) but only marginally at the subspecies level, if estimated from the ITS-RFLP results. Hence, this study begins to provide some definition to “everything” and “everywhere” in the classic Beijerinck-Bass-Becking statement. That soil heterotrophic bacteria in undisturbed sites are not globally mixed suggests that bacterial diversification is actively ongoing.
ACKNOWLEDGMENTS
We thank R. Lenski for helpful discussion and J. L. Rademaker and F. J. deBruijn for help with rep-PCR pattern analysis.
This research was supported by the Center for Microbial Ecology under NSF grant DEB-9120006 and by DEB-0075564.
REFERENCES
- 1.Baas-Becking L G M. Geobiologie of Inleiding Tot de Milieukunde. The Hague, The Netherlands: W. P. van Stockum & Zoon N. V.; 1934. [Google Scholar]
- 2.Beeder J, Nielsen R K, Rosnes J T, Torsvik T, Lien T. Archaeoglobus fulgidus isolated from hot North Sea oil field waters. Appl Environ Microbiol. 1994;60:1227–1234. doi: 10.1128/aem.60.4.1227-1231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beijerinck M W. De infusies en de ontdekking der backteriën, Jaarboek van de Koninklijke Akademie v. Wetenschappen. Amsterdam, The Netherlands: Müller; 1913. [Google Scholar]
- 4.Bourque S N, Valero J R, Lavoie M C, Levesque R C. Comparative analysis of the 16S to 23S ribosomal intergenic spacer sequences of Bacillus thuringiensis strains and subspecies and of closely related species. Appl Environ Microbiol. 1995;61:1623–1626. doi: 10.1128/aem.61.4.1623-1626.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castenholz R W. Endemism and biodiversity of thermophilic cyanobacteria. Nova Hedwigia Beiheft. 1996;112:33–47. [Google Scholar]
- 6.Caugant D A, Mocca L F, Frasch C E, Froholm L O, Zollinger W D, Selander R K. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein pattern. J Bacteriol. 1987;169:2781–2792. doi: 10.1128/jb.169.6.2781-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cliff A D, Ord J K. Spatial autocorrelation. London, United Kingdom: Pion; 1973. [Google Scholar]
- 8.deLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 9.Dice L R. Measures of the amount of ecological association between species. J Ecol. 1945;26:297–302. [Google Scholar]
- 10.Eardly B D, Materon L A, Smith N H, Johnson D A, Rumbaugh M D, Selander R K. Genetic structure of natural populations of the nitrogen-fixing bacterium Rhizobium meliloti. Appl Environ Microbiol. 1990;56:187–194. doi: 10.1128/aem.56.1.187-194.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulthorpe R R, Rhodes A N, Tiedje J M. High levels of endemicity of 3-chlorobenzoate-degrading soil bacteria. Appl Environ Microbiol. 1998;64:1620–1627. doi: 10.1128/aem.64.5.1620-1627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fulthorpe R R, Rhodes A N, Tiedje J M. Pristine soils mineralize 3-chlorobenzoate and 2,4-dichlorophenoxyacetate via different microbial populations. Appl Environ Microbiol. 1996;62:1159–1166. doi: 10.1128/aem.62.4.1159-1166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Pichel F, Prufert-Bebout L, Muyzer G. Phenotypic and phylogenetic analyses show Microcoleus chthonoplastes to be a cosmopolitan cyanobacterium. Appl Environ Microbiol. 1996;62:3284–3291. doi: 10.1128/aem.62.9.3284-3291.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 16.Häne B G, Jäger K, Drexler H G. The Pearson product-moment correlation coefficient is better suited for identification of DNA fingerprint profiles than band matching algorithms. Electrophoresis. 1993;14:967–972. doi: 10.1002/elps.11501401154. [DOI] [PubMed] [Google Scholar]
- 17.Hubert L J. Combinatorial data analysis: association and partial association. Psychometrika. 1985;50:449–467. [Google Scholar]
- 18.Istock C A, Duncan K E, Ferguson N, Zhou X. Sexuality in a natural population of bacteria, Bacillus subtilis challenges the clonal paradigm. Mol Ecol. 1992;1:95–103. doi: 10.1111/j.1365-294x.1992.tb00161.x. [DOI] [PubMed] [Google Scholar]
- 19.Jensen M A, Webster J A, Straus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laguerre G, Mavingui P, Allard M R, Charnay M P, Louvrier P, Mazurier S I, Rigottier-Gois L, Amarger N. Typing of rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Appl Environ Microbiol. 1996;62:2029–2036. doi: 10.1128/aem.62.6.2029-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legendre P, Legendre L. Numerical ecology. Amsterdam, The Netherlands: Elsevier Science; 1998. pp. 707–785. [Google Scholar]
- 22.L'Haridon S, Reysenbach A L, Prieur D, Jeanthon C. Hot subterranean biosphere in a continental oil reservoir. Nature. 1995;377:223–224. [PubMed] [Google Scholar]
- 23.Manceau C, Horvais A. Assessment of genetic diversity among strains of Pseudomonas syringae by PCR-restriction fragment length polymorphism analysis of rRNA operons with special emphasis on P. syringae pv. Tomato Appl Environ Microbiol. 1997;63:498–505. doi: 10.1128/aem.63.2.498-505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massol-Deya A A, Odelson D A, Hickey R F, Tiedje J M. Bacterial community fingerprinting of amplified 16S and 16S–23S ribosomal DNA gene sequences and restriction endonuclease analysis (ARDRA) In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–8. [Google Scholar]
- 25.Milkman R. Electrophoretic variation in Escherichia coli from natural sources. Science. 1973;182:1024–1026. doi: 10.1126/science.182.4116.1024. [DOI] [PubMed] [Google Scholar]
- 26.Moore E R B, Mau M, Arnscheidt A, Böttger E C, Hutson R A, Collins M D, van de Peer Y, de Wachter R, Timmis K N. The determination and comparison of the 16S rRNA gene sequences of species of the genus Pseudomonas (sensu stricto) and estimation of the natural intrageneric relationship. Syst Appl Microbiol. 1996;19:478–492. [Google Scholar]
- 27.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 28.Musser J M, Kroll J S, Granoff D M, Moxon E R, Brodeur B R, Campos J, Dabernat H, Frederiksen W, Hamel J, Hammond G, et al. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev Infect Dis. 1990;12:75–111. doi: 10.1093/clinids/12.1.75. [DOI] [PubMed] [Google Scholar]
- 29.Musser J M, Schlievert P M, Chow A W, Ewan P, Kreiswirth B N, Rosdahl V T, Naidu A S, Witte W, Selander R K. A single clone of Staphylococcus aureus causes the majority of cases of toxic shock syndrome. Proc Natl Acad Sci USA. 1990;87:225–229. doi: 10.1073/pnas.87.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Normand P, Ponsonnet C, Nesme X, Neyra M, Simonet P. ITS analysis of prokaryotes. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 1–12. [Google Scholar]
- 31.Ochman H, Whittam T S, Caugant D A, Selander R K. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983;129:2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- 32.Oden N L, Sokal R R. Directional autocorrelation: an extension of spatial correlograms to two dimensions. Syst Zool. 1986;35:608–617. [Google Scholar]
- 33.Pearson K. On the coefficient of racial likeness. Biometrika. 1926;18:105–117. [Google Scholar]
- 34.Rademaker J L W, Hoste B, Louws F J, Kersters K, Swings J, Vauterin L, Vauterin P, deBruijn F J. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int J Syst Evol Microbiol. 2000;50:665–677. doi: 10.1099/00207713-50-2-665. [DOI] [PubMed] [Google Scholar]
- 35.Rademaker J L W, Louws F J, de Bruijn F J. Characterization of the diversity of ecologically important microbes by rep-PCR fingerprinting. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual, suppl. 3. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–26. [Google Scholar]
- 36.Rappé M S, Kemp P F, Giovannoni S J. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr. 1997;42:811–826. [Google Scholar]
- 37.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selander R K, Levin B R. Genetic diversity and structure in Escherichia coli populations. Science. 1980;210:545–547. doi: 10.1126/science.6999623. [DOI] [PubMed] [Google Scholar]
- 40.Smith J M, Dowson C G, Spratt B G. Localized sex in bacteria. Nature. 1991;349:29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- 41.Sokal R R. Spatial data analysis and historical processes. In: Diday E, editor. Data analysis and informatics. IV. Amsterdam, The Netherlands: North-Holland; 1986. pp. 29–43. [Google Scholar]
- 42.Sokal R R, Sneath P H A. Principle of numerical taxonomy. San Francisco, Calif: Freeman; 1963. pp. 181–185. [Google Scholar]
- 43.Souza V, Nguyen T T, Hudson R R, Pinero D, Lenski R E. Hierarchical analysis of linkage disequilibrium in Rhizobium populations: evidence for sex? Proc Natl Acad Sci USA. 1992;89:8389–8393. doi: 10.1073/pnas.89.17.8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staley J T, Gosink J J. Poles apart: biodiversity and biogeography. Annu Rev Microbiol. 1999;53:189–215. doi: 10.1146/annurev.micro.53.1.189. [DOI] [PubMed] [Google Scholar]
- 45.Stetter K O, Hubert R, Blöchl E, Kurr M, Eden R D. Hyperthermophilic archaea are thriving in deep North Sea and Alaskan oil reservoirs. Nature. 1993;365:743–745. [Google Scholar]
- 46.Valsecchi E. Tissue boiling: a short-cut in DNA extraction for large-scale population screenings. Mol Ecol. 1998;7:1243–1245. doi: 10.1046/j.1365-294x.1998.00379.x. [DOI] [PubMed] [Google Scholar]
- 47.Vinuesa P, Rademaker J L, de Bruijn F J, Werner D. Genotypic characterization of Bradyrhizobium strains nodulating endemic woody legumes of the Canary Islands by PCR-restriction fragment length polymorphism analysis of genes encoding 16S rRNA (16S rDNA) and 16S–23S rDNA intergenic spacers, repetitive extragenic palindromic PCR genomic fingerprinting, and partial 16S rDNA sequencing. Appl Environ Microbiol. 1998;64:2096–2104. doi: 10.1128/aem.64.6.2096-2104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whittam T S, Ochman H, Selander R K. Multilocus genetic structure in natural populations of Escherichia coli. Proc Natl Acad Sci USA. 1983;80:1751–1755. doi: 10.1073/pnas.80.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Widmer F, Seidler R J, Gillevet P M, Watrud L S, Di Giovanni G D. A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Appl Environ Microbiol. 1998;64:2545–2553. doi: 10.1128/aem.64.7.2545-2553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wise M G, Shimkets L J, McArthur J V. Genetic structure of a lotic population of Burkolderia (Pseudomonas) cepacia. Appl Environ Microbiol. 1995;61:1791–1798. doi: 10.1128/aem.61.5.1791-1798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zwart G, Hiorns W D, Methe B A, van Agterveld M P, Huismans R, Nold S C, Zehr J P, Laanbroek H J. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst Appl Microbiol. 1998;21:546–556. doi: 10.1016/S0723-2020(98)80067-2. [DOI] [PubMed] [Google Scholar]