Abstract
Achieving global elimination of hepatitis C virus requires a substantial scale‐up of testing. Point‐of‐care HCV viral load assays are available as an alternative to laboratory‐based assays to promote access in hard to reach or marginalized populations. The diagnostic performance and lower limit of detection are important attributes of these new assays for both diagnosis and test of cure. Therefore, our objective was to determine an acceptable LLoD for detectable HCV viraemia as a test for cure, 12 weeks post‐treatment (SVR12). We assembled a global data set of patients with detectable viraemia at SVR12 from observational databases from 9 countries (Egypt, the United States, United Kingdom, Georgia, Ukraine, Myanmar, Cambodia, Pakistan, Mozambique) and two pharmaceutical‐sponsored clinical trial registries. We examined the distribution of HCV viral load at SVR12 and presented the 90th, 95th, 97th and 99th percentiles. We used logistic regression to assess characteristics associated with low‐level virological treatment failure (defined as <1000 IU/mL). There were 5973 cases of detectable viraemia at SVR12 from the combined data set. Median detectable HCV RNA at SVR12 was 287,986 IU/mL. The level of detection for the 95th percentile was 227 IU/mL (95% CI 170–276). Females and those with minimal fibrosis were more likely to experience low‐level viraemia at SVR12 compared to men (adjusted odds ratio AOR = 1.60 95% confidence interval [CI] 1.30–1.97 and those with cirrhosis (AOR = 1.49 95% CI 1.15–1.93). In conclusion, an assay with a level of detection of 1000 IU/mL or greater may miss a proportion of those with low‐level treatment failure.
Keywords: diagnostics, HCV, limit of detection, point‐of‐care testing
Abbreviations
- DAA
direct‐acting antiviral
- FIB4
Fibrosis‐4
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- HIV
human immunodeficiency virus
- LLoD
limit of detection
- LLV
low‐level viraemia
- MSF
Médecins Sans Frontières
- NAAT
Nucleic Acid Amplification Testing
- WHO
World Health Organization
1. INTRODUCTION
Chronic hepatitis C virus (HCV) infection is a major cause of progressive liver disease and associated morbidity and mortality globally. 1 In 2021, there were an estimated 58 million persons with a chronic infection, with a disproportionately high burden in low‐ and middle‐income countries. 1 Short‐course curative direct‐acting antiviral (DAA) regimens have transformed opportunities for treatment scale‐up and elimination. 1 , 2 , 3 , 4 In 2016, the World Health Organization (WHO) launched the Global Health Sector Strategy for elimination of viral hepatitis as a public health threat, with ambitious targets for elimination of HCV including a 90% reduction in new infections and a 65% reduction in HCV‐related mortality by 2030. 5 , 6
In order to meet the 2030 global targets for HCV elimination, there is a need to substantially scale‐up access to testing and treatment, with simplified service delivery models and diagnostic innovations to expand access. 7 A key step in the care cascade is the use of HCV viral load assays to confirm presence of viraemic infection, and then a test of cure following treatment. 8 The 2017 WHO viral hepatitis testing guidelines recommended a laboratory‐based PCR Nucleic Acid Amplification Testing (NAAT), or a core HCV antigen assay with comparable clinical sensitivity, as preferred strategies for diagnosis of viraemic HCV infection, and laboratory‐based PCR assays as a test of cure at SVR12. 9 , 10 Point‐of‐care HCV viral load assays are now available as an alternative to laboratory‐based NAAT assays to promote access, especially in hard to reach or marginalized populations. A previous multi‐cohort analysis examined the distribution of HCV viral load at diagnosis in 66,640 individuals from 12 countries and established that 97% had a viral load greater than 1318 IU/mL and 95% had a viral load greater than 3,311 IU/mL. 11 The key laboratory‐based assays (Abbott Real time HCV PCR, Alinity m HCV RT‐PCR, Abbott Real time HCV PCR) have an analytical sensitivity or LloD of between 5–15 IU/mL, and key PoC assays: HCV RNA PoC GeneXpert assays are 10 IU/mL for venous blood, 12 , 13 or 100 IU/mL using fingerstick capillary blood. All these assays are therefore acceptable for diagnosis of HCV viraemic infection.
Currently, the European Association for the Study of the Liver Diseases, IDSA‐AASLD HCV guidance panel, all recommend a minimum LLoD of 1000 IU/mL for HCV diagnosis, but none yet specify minimal test characteristics for test of cure. 14 , 15 While some small studies have examined the distribution of viral load at end of treatment—including a cohort of eight patients from the United States, 16 a cohort of 14 patients in Germany 17 and 330 treatment failures in an analysis of 34 phase 2/3 clinical trials. 18 The latter study in clinical trials identify that 97% had a viral load >10,000 IU/mL 12 weeks post‐treatment, and just 0.9% of patients had a viral load less than 1000 IU/mL (77, 405 and 680 IU/mL). To date, there have been no real world, global analyses of distribution of viral load in those with detectable viraemia at SVR12.
Our primary objective was to determine the LLoD for an HCV RNA assay to detect 90%, 95%, 97% and 99% of treatment failures at 12 weeks post‐treatment in a large multi‐cohort data set, and to assess the characteristics associated with low‐level viraemia (<1000 IU/mL) at SVR12. These findings will help inform global policy as well as guide manufacturers as to whether existing platforms and assays meet requirements for their use both in diagnosis and as a test of cure, and for future development of testing technologies.
2. METHODS
2.1. Data sources: Observational cohorts and clinical trials registries
We assembled a data set of patients with detectable HCV viral load at week 12 after completion of DAA treatment from clinical observational cohorts in nine countries, in addition to international clinical trial registry databases from two pharmaceutical companies. We identified potential cohorts for inclusion from four sources: (1) cohorts included in a previously published analysis of 12 countries for LoD at diagnosis 11 ; (2) cohorts that had previous collaborative projects with WHO or were known by our research team; (3) a PubMed literature search using the search terms ‘HCV SVR’ and ‘cohort study’; and (4) conference abstracts from 2018 to 2020 at the American Association for the Study of Liver Diseases and the European Association for the Study of the Liver. We sent our study protocol (Appendix S1) and an invitation to join a research collaborative to the principal investigators of each identified cohort. We approached 19 cohorts and registries with HCV‐infected patients who had received treatment.
Eleven observational cohorts representing nine countries and two clinical trial registries agreed to collaborate and share data. To be included in the global data set, cohorts and trial databases were required to have the following patient‐level data: Detectable HCV RNA test at 12 weeks post‐treatment and linked demographic data per protocol (see Appendix S1). Observational cohorts were characterized by one of the following: (1) registries from country‐wide national HCV ministries of health (Georgian and Egypt national programs); (2) large healthcare systems (United States Department of Veteran Affairs or the UK); (3) non‐governmental organizations with programmatic data including across multiple countries (Médecins Sans Frontières (MSF) sites in Mozambique, Cambodia and Pakistan); or (4) grant‐funded research projects (Myanmar and Ukraine).
We also included clinical trials databases from two pharmaceutical companies who were responsible for originator DAAs (Gilead and AbbVie). We pooled data from relevant DAA trials into a single repository. Patient eligibility criteria varied by study trial, and the majority required a pre‐treatment viral load over 1000 IU/mL for enrolment and censored individuals who experienced on‐treatment virological failure. Both clinical trial databases were used individually to determine LLoD and the distribution of HCV RNA at treatment failure based on summary data at SVR12 assessment. However, only one database was able to provide patient‐level data to contribute to the multivariable regression analyses of factors associated with low‐level viraemia (LLV).
2.2. Characteristics of study cohorts
Most of the included cohorts have been well described in the literature. 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 Table 1 summarizes key characteristics of these cohorts, including number of HCV‐treated patients, gender, age, and genotype distribution, DAA regimens and the proportion who achieved SVR following treatment. There was a high degree of heterogeneity in patient characteristics across cohorts, reflecting varying HCV epidemic profiles in different countries.
TABLE 1.
Characteristics of treated individuals from cohorts in five countries (United States, Egypt, UK, Georgia and Myanmar) and from clinical trials from two pharmaceutical companies
| US Veterans Affairs20, b | Egypt21, a | United Kingdom22, b | Georgia19, a | Myanmar23, d | Ukraine24, d | Médecins Sans Frontières** (multi‐country) c |
Clinical Trial Registry 125, e (multi‐country) |
Clinical Trial Registry 226, e (multi‐country) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | |
| Total | 174,889 | 337,042 | 24,592 | 52,856 | 763 | 868 | 2,904 | 5,033 | 624 | |||||||||
| Age (years) | ||||||||||||||||||
| Mean ± SD | 60 ± NA | 50 ± 11 | 51 ± 12 | NA | 38 ± 11 | 39 (35–45)* | 50 ± 12 | NA | 54 ± NA | |||||||||
| Sex | ||||||||||||||||||
| Male | 169,642 | 97% | 182,008 | 54% | 17,288 | 70% | 41,175 | 78% | 618 | 81% | 573 | 66% | 1183 | 41% | 2774 | 55% | 374 | 60% |
| Female | 5247 | 3% | 155,034 | 46% | 7304 | 30% | 11,681 | 22% | 145 | 19% | 295 | 34% | 1721 | 59% | 2259 | 45% | 250 | 40% |
| Per cent SVR at 12 weeks | >90% | 68%–99% | 95% | 98% | 78%–100% | 96% | 96% | 90%–100% | 99% | |||||||||
| HCV genotype | ||||||||||||||||||
| 1 | 139,911 | 80% | 12,134 | 4% | 13,353 | 54% | 23,309 | 62% | 69 | 9% | 643 | 74% | 941 | 32% | 2,388 | 47% | 328 | 53% |
| 2 | 20,987 | 12% | 1348 | <1% | 1057 | 4% | 10,518 | 11% | 8 | 1% | 20 | 2% | 116 | 4% | 1,054 | 21% | 104 | 17% |
| 3 | 12,242 | 7% | 674 | <1% | 8878 | 36% | 18,024 | 27% | 366 | 48% | 191 | 22% | 483 | 17% | 1,140 | 23% | 0 | 0% |
| 4–6/missing | 1749 | 1% | 322,886 | 96% | 1303 | 5% | 1004 | 0% | 320 | 42% | 14 | 2% | 1364 | 47% | 451 | 9% | 76 | 12% |
This table represents the general population of those treated in each location, but is not representative of the treated population that yielded the detectable viral loads in this study.
Abbreviations: HCV, hepatitis C virus; NA, not available; SD, standard deviation; SVR, sustained virologic response.
Registries from country‐wide national HCV ministries of health.
Large health care systems.
Non‐governmental organizations with programmatic data including across multiple countries This was the only sample for which we had data on individuals who attained SVR as well as those with detectable viral at 12 weeks, and present the characteristics of that cohort here.
Specific funded research projects.
clinical trials registries from two large pharmaceutical companies (Gilead and AbbVie).
Age data from Ukraine are reported as median and interquartile range.
Data are routinely collected from MSF program sites in Mozambique, Cambodia and Pakistan.
2.3. Data concatenation
We modified a protocol for data concatenation from the previous study of viral load at diagnosis, 27 to create comparable variables across a global data set. 27 First, we requested a core set of demographic and clinical variables for each included individual: age (dichotomized as under 60 or 60 years or older based on the format of data received), sex, HCV genotype, fibrosis stage, type and duration of DAA regimen, and presence of human immunodeficiency virus (HIV) or hepatitis B virus (HBV) co‐infection. Second, we harmonized data for three key variables—fibrosis stage; HCV genotype; and HCV DAA treatment regimens. We standardized fibrosis stage by calculating the Fibrosis‐4 (FIB4) score, which correlates well with staging based on transient elastography and liver biopsy. 28 , 29 , 30 We assigned the corresponding Metavir state to the FIB4 scores: FIB4 score <1.45, (Metavir stage F0‐F1); FIB4 1.45–3.25 (Metavir stage F2─F3); and FIB4 scores above 3.25 (Metavir stage F4). We imputed genotype data for one country, Egypt, which has predominantly genotype 4 infection, and where genotyping is no longer performed routinely. 31 We used the distribution of genotypes from the literature as probabilities of having each given genotype and employed a Markov chain Monte Carlo technique to stochastically assign a genotype to each individual in the Egyptian cohort. All other cohorts had patient‐level data on genotype. Finally, since more than 20 different DAA treatment regimens were in use during our data collection period, we assigned these regimens to three categories based on the time period of introduction: early era DAAs (sofosbuvir/ribavirin and sofosbuvir/simeprevir with or without ribavirin), mid‐era (sofosbuvir/daclatasvir, sofosbuvir/ledipasvir and elbasvir/grazoprevir‐ or ombitasvir/paritaprevir‐containing regimens) and recent‐era (glecaprevir/pibrentasvir and sofosbuvir/velpatasvir regimens).
2.4. Statistical analyses
We evaluated the distribution of HCV viral load at 12 weeks post‐treatment assessment among all patients in the combined data set using both standard IU/mL measures and normalized using a log10 transformation, to allow for better visualization of the viral load distribution. From these measures, we identified the lower 95th, 97th and 99th percentiles of the HCV RNA distribution. To estimate the 95% confidence interval for each viral load threshold, we use the method described by Hahn and Meeker, 1991, which corrects for a non‐normal distribution of values. 32
We next identified the subgroup of individuals who had HCV RNA that is detectable, but <1000 IU/mL at the time of SVR assessment. Such individuals were defined as having ‘low‐level viremia’ (LLV) at the time of treatment failure. We chose the threshold of <1000 IU/mL because that is the LLoD for HCV core antigen assay and a reasonable proxy for newer platforms being developed that have potential to be near point‐of‐care in use and so promote access to diagnosis and treatment. 33 , 34 We described the characteristics of this subgroup and employed logistic regression to assess factors associated with having LLV compared to no LLV among those with detectable viral load at SVR12. Analyses were conducted using SAS (version 9.4; SAS Institute; Cary, NC). The Boston University Institutional Review Board ruled this study not human subjects research.
2.5. Sensitivity analyses
We conducted two additional subgroup and sensitivity analyses. First, we examined whether results differed between clinical trial and observational cohorts to determine whether conclusions potentially differ in investigational and real‐world settings. Second, we examined the HCV RNA viral load at SVR24 after treatment completion to assess whether checking for cure at 24 weeks rather than 12 weeks would provide similar conclusions. This was undertaken in the cohort from Georgia, the US Department of Veterans Affairs, and one of the clinical trial cohorts that reported patient HCV RNA test results at both 12‐ and 24 weeks post‐treatment.
3. RESULTS
3.1. Characteristics of treatment failures
Overall, our cohort consisted of 5973 cases of detectable viraemia following HCV treatment from Egypt (3264), the United States (1125), Georgia (1041), the United Kingdom (131), Myanmar (84), Cambodia (40), Pakistan (27), Ukraine (17), Mozambique (3) and clinical trial registries (241). Table 2 summarizes the characteristics of the individuals included in this analysis. Most individuals were under the age of 60 years (65%), with the exception of the U.S. Veterans Affairs cohort, of which 78% were 60 years or older. The majority of patients in the Mozambique, Myanmar and Ukraine cohorts were HIV co‐infected, but less than 10% were HIV co‐infected in the other cohorts and trial databases. Forty‐one per cent of the cohort had a fibrosis stage of F4 indicating advanced liver disease/cirrhosis. The genotype distribution across all cohorts was GT 1 (23%), 2 (5%), 3 (12%) and 4 (53%). Most of the cohort (70%) received 12 weeks of DAA treatment while 24% received 24 weeks. Most individuals received a mid‐era DAA regimen (66%), followed by early era (27%) and recent‐era (6%) DAAs (Table 2).
TABLE 2.
Characteristics of patients with detectable viraemia 12 weeks post‐treatment overall and from each observational cohort or Clinical Trial Registry
| Total cohort | Clinical Trial Registry 1 e | Clinical Trial Registry 2 e | US Veterans Affairs b | Egypt a | UK b | Georgia a | Myanmar d | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Baseline VL (IU/ml), millions | 2.6 | 6.1 | 9.6 | 8.2 | 5.5 | 5.9 | 4.3 | 5.8 | 2.1 | 6.5 | 1.7 | 3.5 | 1.9 | 4.7 | 4.9 | 7.7 |
| VL at 12‐week SVR (IU/ml), millions f | 1.8 | 5.5 | 6.5 | 7.5 | 4.2 | 6.4 | 2.7 | 6.2 | 1.6 | 5.9 | 1.1 | 2.5 | 0.9 | 2.6 | 1.7 | 2.8 |
| VL at 12‐week SVR log (IU/ml) | 5.1 | 1.4 | 6.4 | 0.7 | 6.0 | 1.3 | 5.7 | 1.1 | 5.0 | 1.4 | 4.9 | 1.5 | 4.6 | 1.5 | 4.8 | 1.7 |
| Treatment duration, days | 107 | 41 | NA | 99 | 44 | 98 | 29 | 106 | 37 | 86 | 17 | 126 | 56 | 84 | 0 | |
| n | % | Only RNA data | n | % | n | % | n | % | n | % | n | % | N | % | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample of post‐treatment viraemic patients | 5,973 | 100% | 68 | 100% | 173 | 100% | 1125 | 100% | 3264 | 100% | 131 | 100% | 1041 | 100% | 84 | 100% |
| Demographic characteristics | ||||||||||||||||
| Sex | ||||||||||||||||
| Male | 4,344 | 73% | 158 | 91% | 1106 | 98% | 1943 | 60% | 113 | 86% | 892 | 86% | 82 | 98% | ||
| Female | 1,561 | 26% | 15 | 9% | 19 | 2% | 1321 | 40% | 18 | 14% | 149 | 14% | 2 | 2% | ||
| Age (years) | ||||||||||||||||
| <60 | 3,880 | 65% | 126 | 73% | 252 | 22% | 2388 | 73% | 101 | 77% | 859 | 83% | 81 | 96% | ||
| >60+ | 2,009 | 34% | 31 | 18% | 873 | 78% | 876 | 27% | 30 | 23% | 182 | 17% | 3 | 4% | ||
| Missing | 16 | 0% | 16 | 9% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | ||
| HIV antibody coinfection | 154 | 3% | 13 | 8% | 38 | 3% | 0 | 0% | 7 | 5% | 0 | 0% | 75 | 89% | ||
| HBsAg‐positive | 119 | 2% | 0 | 0% | 76 | 7% | 9 | 0% | 1 | 1% | 25 | 2% | 5 | 6% | ||
| Disease characteristics | ||||||||||||||||
| Fibrosis FIB4 | ||||||||||||||||
| F0–F1 (FIB4 < 1.45) | 1,539 | 26% | 27 | 16% | 193 | 17% | 991 | 30% | 2 | 2% | 252 | 24% | 54 | 64% | ||
| F2–F3 (3.25 ≥ FIB4 ≥ 1.45) | 1,764 | 30% | 50 | 29% | 403 | 36% | 967 | 30% | 15 | 11% | 294 | 28% | 21 | 25% | ||
| F4 (FIB4 > 3.25) | 2,473 | 41% | 60 | 35% | 529 | 47% | 1294 | 40% | 74 | 56% | 486 | 47% | 9 | 11% | ||
| Missing | 129 | 2% | 36 | 21% | 0 | 0% | 12 | 0% | 40 | 31% | 9 | 1% | 0 | 0% | ||
| Genotype g | ||||||||||||||||
| 1a | 733 | 12% | 49 | 28% | 659 | 59% | 0 | 0% | 25 | 19% | 0 | 0% | 0 | 0% | ||
| 1b | 177 | 3% | 16 | 9% | 156 | 14% | 0 | 0% | 5 | 4% | 0 | 0% | 0 | 0% | ||
| 1 (unknown subtype) | 478 | 8% | 0 | 0% | 44 | 4% | 109 | 3% | 8 | 6% | 294 | 28% | 2 | 2% | ||
| 2 | 315 | 5% | 16 | 9% | 81 | 7% | 0 | 0% | 4 | 3% | 212 | 20% | 0 | 0% | ||
| 3 | 734 | 12% | 89 | 51% | 160 | 14% | 23 | 1% | 83 | 63% | 324 | 31% | 36 | 43% | ||
| 4 | 3,154 | 53% | 2 | 1% | 17 | 2% | 3132 | 96% | 3 | 2% | 0 | 0% | 0 | 0% | ||
| 5 | 2 | 0% | 1 | 1% | 1 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | ||
| 6 | 30 | 1% | 0 | 0% | 1 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | ||
| Mixed genotype | 37 | 1% | 0 | 0% | 6 | 1% | 0 | 0% | 3 | 2% | 0 | 0% | 28 | 33% | ||
| Unknown | 227 | 4% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 211 | 20% | 0 | 0% | ||
| Treatment characteristics | ||||||||||||||||
| Treatment duration | ||||||||||||||||
| 6/8 weeks h | 46 | 1% | 41 | 24% | 0 | 0% | 0 | 0% | 5 | 4% | 0 | 0% | 0 | 0% | ||
| 12 weeks | 4,197 | 70% | 77 | 45% | 886 | 79% | 2406 | 74% | 106 | 81% | 563 | 54% | 84 | 100% | ||
| 16 or 20 weeks | 187 | 3% | 17 | 10% | 97 | 9% | 0 | 0% | 1 | 1% | 72 | 7% | 0 | 0% | ||
| 24 weeks | 1,423 | 24% | 37 | 21% | 142 | 13% | 858 | 26% | 4 | 3% | 370 | 36% | 0 | 0% | ||
| 48 weeks | 37 | 1% | 1 | 1% | 0 | 0% | 0 | 0% | 0 | 0% | 36 | 3% | 0 | 0% | ||
| Missing | 15 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 15 | 11% | 0 | 0% | 0 | 0% | ||
| DAA era i | ||||||||||||||||
| Early | 1,613 | 27% | 83 | 48% | 146 | 13% | 878 | 27% | 3 | 2% | 488 | 47% | 0 | 0% | ||
| Mid | 3,918 | 66% | 22 | 13% | 798 | 71% | 2386 | 73% | 123 | 94% | 517 | 50% | 0 | 0% | ||
| Recent | 374 | 6% | 68 | 39% | 181 | 16% | 0 | 0% | 5 | 4% | 36 | 3% | 84 | 100% | ||
| Ukraine e | Cambodia c | Mozambique c | Pakistan c | |||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Baseline VL (IU/ml), millions | 3.3 | 4.2 | 3.5 | 5.3 | 0.7 | 1.0 | 0.3 | 0.2 |
| VL at 12‐week SVR (IU/ml), millions f | 2.0 | 3.2 | 2.7 | 4.7 | 0.3 | 0.5 | 0.5 | 0.7 |
| VL at 12‐week SVR log (IU/ml) | 4.7 | 1.9 | 5.4 | 1.5 | 3.9 | 2.3 | 4.9 | 1.3 |
| Treatment duration, days | 84 | 0 | 84 | 0 | 84 | 0 | 121 | 43 |
| n | % | n | % | n | % | n | % | |
|---|---|---|---|---|---|---|---|---|
| Sample of post‐treatment viraemic patients | 17 | 100% | 40 | 100% | 3 | 100% | 27 | 100% |
| Demographic characteristics | ||||||||
| Sex | ||||||||
| Male | 11 | 65% | 26 | 65% | 3 | 100% | 10 | 37% |
| Female | 6 | 35% | 14 | 35% | 0 | 0% | 17 | 63% |
| Age (years) | ||||||||
| <60 | 17 | 100% | 28 | 70% | 3 | 100% | 25 | 93% |
| 60+ | 0 | 0% | 12 | 30% | 0 | 0% | 2 | 7% |
| Missing | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
| HIV antibody coinfection | 13 | 76% | 5 | 13% | 3 | 100% | 0 | 0% |
| HBsAg‐positive | 2 | 12% | 1 | 3% | 0 | 0% | 0 | 0% |
| Disease characteristics | ||||||||
| Fibrosis FIB4 | ||||||||
| F0–F1 (FIB4 < 1.45) | 13 | 76% | 4 | 10% | 3 | 100% | 0 | 0% |
| F2–F3 (3.25 ≥ FIB4 ≥ 1.45) | 3 | 18% | 10 | 25% | 0 | 0% | 1 | 4% |
| F4 (FIB4 > 3.25) | 1 | 6% | 19 | 48% | 0 | 0% | 1 | 4% |
| Missing | 0 | 0% | 7 | 18% | 0 | 0% | 25 | 93% |
| Genotype g | ||||||||
| 1a | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
| 1b | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
| 1 (unknown subtype) | 10 | 59% | 10 | 25% | 1 | 33% | 0 | 0% |
| 2 | 0 | 0% | 1 | 3% | 0 | 0% | 1 | 4% |
| 3 | 7 | 41% | 0 | 0% | 0 | 0% | 12 | 44% |
| 4 | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
| 5 | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
| 6 | 0 | 0% | 29 | 73% | 0 | 0% | 0 | 0% |
| Mixed genotype | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
| Unknown | 0 | 0% | 0 | 0% | 2 | 67% | 14 | 52% |
| Treatment characteristics | ||||||||
| Treatment duration | ||||||||
| 6/8 weeks h | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
| 12 weeks | 17 | 100% | 40 | 100% | 3 | 100% | 15 | 56% |
| 16 or 20 weeks | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
| 24 weeks | 0 | 0% | 0 | 0% | 0 | 0% | 12 | 44% |
| 48 weeks | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
| Missing | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
| DAA era i | ||||||||
| Early | 0 | 0% | 0 | 0% | 0 | 0% | 15 | 56% |
| Mid | 17 | 100% | 40 | 100% | 3 | 100% | 12 | 44% |
| Recent | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
Abbreviations: DAA, direct‐acting antiviral; FIB4, fibrosis 4; HBsAg, hepatitis B virus surface antigen; HIV, human immunodeficiency virus; SD, standard deviation; SVR, sustained virologic response; VL, viral load.
Registries from country‐wide national HCV ministries of health.
Large health care systems.
Non‐governmental organizations with programmatic data including across multiple countries.
Specific funded research projects.
Clinical trial registries from two large pharmaceutical companies (Gilead and AbbVie).
Genotype is imputed for Egypt based on previously identified distributions; the majority are genotype 4, and genotyping is no longer undertaken routinely.
4 individuals had 6 weeks of treatment in a clinical trial.
Early era DAAs (sofosbuvir/ribavirin and sofosbuvir/simeprevir with or without ribavirin), mid‐era (sof/daclatasvir, sof/ledipasvir and elbasvir/grazoprevir‐, or ombitasvir/paritaprevir‐containing regimens) and recent era (glecaprevir/pibrentasvir and sof/velpatasvir regimens).
Three individuals had a recorded VL > 1,000,000,000 and were not included in the mean due to likely data entry error.
3.2. HCV viral load distribution and limit of detection analysis
The median detectable HCV RNA at SVR12 was 287,986 IU/mL (approximately 5.5 log10) (IQR = 1,323,500 IU/mL with 25th percentile = 26,500 and 75% percentile = 1,350,000) (Figure 1). 90% of those with detectable viraemia at SV12 had a viral load greater than 1133 (95% CI 940–1390), 95% greater than 227 IU/mL (95% CI 170–276), 97% greater than 70 IU/mL (95% CI 48–86) and 99% greater than 19 IU/mL (95% CI 16–23). Five hundred seventy‐four individuals (10%) were defined as having LLV, meaning that a hypothetical assay with LLoD of 1000 IU/mL would miss approximately 10% of treatment failures in this setting.
FIGURE 1.
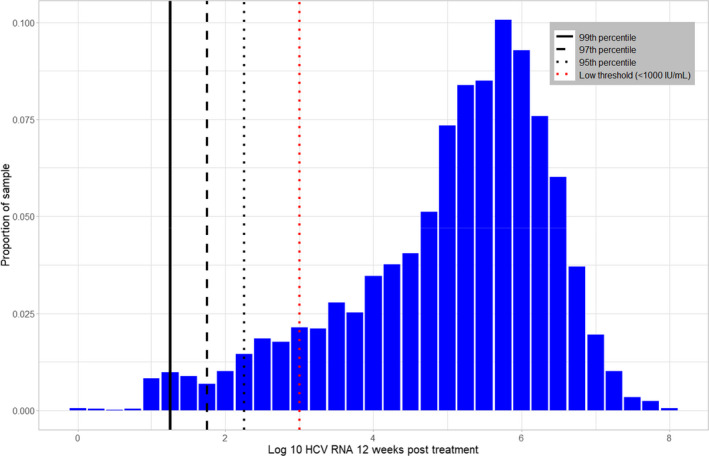
Each bar represents the proportion of the sample with a given log RNA value at time of detection 12 weeks post‐treatment. The labels on the x‐axis are the end point of each bar. For example, the tallest bar, with a label of 5.75–6.00, shows that just over 10% of the cohort has a log RNA value between 5.75 and 6.00
3.3. Factors associated with low‐level viraemia (<1000 IU/mL) at SVR12
We examined baseline demographic, clinical and treatment characteristics associated with LLV (<1000 IU/mL) compared to non‐LLV and present these results in Table 3. In multivariable logistic regression analysis adjusting for demographic (age, sex, data source, HIV/HBV co‐infection) and disease (fibrosis stage, genotype, regimen duration, DAA treatment era) characteristics, females had higher odds of experiencing LLV (odds ratio [OR] = 1.60, 95% CI 1.30–1.97). Compared to cirrhosis (F4), no or minimal fibrosis (F0‐F1) was associated with higher odds of having a low detectable viral load (OR = 1.49, 95% CI 1.15–1.93), as was genotype 3 (OR = 1.69, 95% CI 1.18–2.41). Finally, we found that compared to early era DAA regimens, mid‐era DAA regimens were associated with a lower likelihood of low‐viraemia detection (OR = 0.55, 95% CI 0.40–0.75).
TABLE 3.
Multivariable analysis of factors associated with of low‐level viraemia treatment failure (RNA detectable <1000 IU/mL) at 12 weeks post‐treatment
| Variable |
Mid/high viraemia (≥1000 IU/mL) |
Low‐level viraemia (<1000 IU/mL) | Adjusted odds ratio* | 95% confidence interval | ||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| Total** | 5331 | (100.0%) | 574 | (100.0%) | ||
| Demographic characteristics | ||||||
| Age (years) | ||||||
| <60 | 3451 | (64.7%) | 429 | (74.7%) | Reference | |
| 60+ | 1865 | (35.0%) | 144 | (25.1%) | 0.98 | (0.79–1.23) |
| Missing | 15 | (0.3%) | 1 | (0.2%) | 1.67 | (0.19–15.01) |
| Sex | ||||||
| Male | 3963 | (74.3%) | 381 | (66.4%) | Reference | |
| Female | 1368 | (25.7%) | 193 | (33.6%) | 1.60 | (1.30–1.97) |
| HIV | ||||||
| Co‐infected | 130 | (2.4%) | 124 | (4.2%) | ‐ | ‐ |
| Negative | 4319 | (80.0%) | 352 | (61.2%) | ‐ | ‐ |
| Missing | 950 | (17.6%) | 198 | (34.5%) | ‐ | ‐ |
| HBV (HBsAg positive) | ||||||
| Co‐infected | 110 | (2.0%) | 9 | (1.6%) | 0.99 | (0.48–2.03) |
| Negative | 5221 | (98.0%) | 565 | (98.4%) | Reference | |
| Disease characteristics | ||||||
| Fibrosis stage | ||||||
| F0‐F1 | 1193 | (22.4%) | 179 | (31.2%) | 1.49 | (1.15–1.93) |
| F2‐F3 | 1682 | (31.6%) | 158 | (27.5%) | 1.05 | (0.82–1.33) |
| F4 | 2336 | (43.8%) | 228 | (39.7%) | Reference | |
| Missing | 120 | (2.3%) | 9 | (1.6%) | 0.52 | (0.23–1.15) |
| HCV Genotype | ||||||
| 1 (all subtypes) | 1308 | (24.5%) | 80 | (13.9%) | Reference | |
| 2 | 276 | (5.2%) | 39 | (6.8%) | 1.36 | (0.83–2.24) |
| 3 | 641 | (12.0%) | 93 | (16.2%) | 1.69 | (1.18–2.41) |
| 4 | 2883 | (54.1%) | 274 | (47.7%) | 1.22 | (0.66–2.25) |
| 5/6/mixed | 59 | (1.1%) | 7 | (1.2%) | 1.16 | (0.42–3.20) |
| Missing | 164 | (3.1%) | 81 | (14.1%) | 4.18 | (2.72–6.42) |
| Treatment characteristics | ||||||
| Regimen duration | ||||||
| 8 weeks | 44 | (0.8%) | 2 | (0.4%) | 1.30 | (0.23–7.42) |
| 12 weeks | 3784 | (71.0%) | 413 | (72.0%) | Reference | |
| 16/20 weeks | 175 | (3.3%) | 12 | (2.1%) | 1.64 | (0.82–3.27) |
| 24 weeks | 1278 | (24.0%) | 145 | (25.3%) | 1.42 | (0.70–2.89) |
| 48 weeks | 36 | (0.7%) | 1 | (0.2%) | 0.21 | (0.03–1.77) |
| Missing | 14 | (0.3%) | 1 | (0.2%) | 0.59 | (0.07–5.38) |
| DAA treatment era*** | ||||||
| Early | 1433 | (26.9%) | 180 | (31.3%) | Reference | |
| Mid | 3559 | (66.8%) | 359 | (62.4%) | 0.55 | (0.40–0.76) |
| Recent | 338 | (6.3%) | 36 | (6.3%) | 0.69 | (0.38–1.25) |
We included a fixed effect of each data source to control for source‐specific heterogeneity. This is not meant to represent a causal relationship between a geographic area and LLV, but rather this fixed effect is a proxy for variables we do not have access too such as the difference among HCV epidemics, local and national policies, and other factors that may affect who fails treatment. Bolded values for adjusted odds ratios denote statistically significant findings at p<.05.
Abbreviations: DAA, direct‐acting antiviral; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV, human immunodeficiency virus.
Every variable is controlled for simultaneously in an adjusted model, reference categories are indicated.
The multivariable regression model includes a total of 5905 observations, as it excludes 68 observations from the Clinical Trial Registry 1 which only contained RNA information.
Early era DAAs (sofosbuvir/ribavirin and sofosbuvir/simeprevir with or without ribavirin), mid‐era (sofosbuvir/daclatasvir, sofosbuvir/ledipasvir, and elbasvir/grazoprevir‐, or ombitasvir/paritaprevir‐containing regimens) and recent‐era (glecaprevir/pibrentasvir and sofosbuvir/velpatasvir regimens).
3.4. Sensitivity analyses of distribution of viral load
3.4.1. Clinical trial and observational cohorts
We found a higher distribution of detectable viral loads from the two clinical trial registries. The median viral load was 2,344,229 (IQR = 5,911,542 IU/mL with 25th percentile = 545,000 and 75% percentile = 6,456,542), and 90% of those with detectable viraemia at SVR12 had a viral load greater than 98,420 (95% CI 17,600–199,962), 95% greater than 4030 IU/mL (95% CI 24–4100), 97% greater than 923 IU/mL (95% CI 24–4030) and 99% greater than 24 IU/mL (95% CI 14–24), respectively (Table 4). The distribution of viral load from non‐pharmaceutical trials (observational databases) had median viral load of 264,809 (IQR = 1,196,500 IU/mL with 25th percentile = 23,500 and 75% percentile = 1,220,000), and 90% of those with detectable viraemia at SVR12 had a viral load greater than 1062 (95% CI 816–1300), 95% greater than 214 IU/mL (95% CI 166–266), 97% greater than 69 IU/mL (95% CI 48–85) and 99% greater than 19 (95% CI 16–22) (Table 4).
TABLE 4.
Stratified limit of detection for data from trial registries and non‐trial sources
| HCV RNA on detection Percentile | Overall | Clinical trial registries | Non‐registry data | |||
|---|---|---|---|---|---|---|
| Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | |
| 90th | 1133 | (940–1390) | 98,420 | (17,600–199,962) | 1062 | (816–1300) |
| 95th | 227 | (170–276) | 4030 | (24–4100) | 214 | (166–266) |
| 97th | 70 | (48–86) | 923 | (24–4030) | 69 | (48–85) |
| 99th | 19 | (16–23) | 24 | (14–24) | 19 | (16–22) |
3.4.2. SVR12 and SVR24
We identified 432 individuals with 24 weeks post‐treatment HCV RNA data, including 231 individuals from the U.S. Veterans Affairs cohort, 128 from the Georgia cohort and 73 from the clinical trials cohort. In this sample at SVR12, 95% of individuals had a detectable HCV RNA above 200 IU/mL, 97% above 119 IU/mL and 99% above 24 IU/mL. We graphed the 12‐ and 24‐week viral load by individual in the 65 individuals in the clinical trials cohort with data at both time points (eFigure S2). In this sample, the median change in viral load from 12 to 24 weeks was 128,848 IU/mL. We did not find evidence of different prevalence of LLV at 12 and 24 weeks: at both the 12‐ and 24‐week assessments for cure, approximately 11% of those detectable had LLV of <1000 IU/mL. So, while the mean increase from 12 to 24 weeks is large, this was largely attributable to increases in those with already high viral loads.
4. DISCUSSION
The analysis of a data set of 5973 cases of detectable viraemia following treatment with a wide range of different DAA treatment regimens from nine different country observational cohorts and two international clinical trials databases shows that 95% of HCV treatment failures identified 12 weeks after the end of DAA therapy have a VL greater than 227 IU/mL (2.36 log IU/mL) and 97% have greater than 70 IU/mL (95% CI 48–86). There were important differences in the distribution of viral load at treatment failure in those who were participants in clinical trials compared to observational studies. The median viral load at treatment failure was nearly 10‐fold higher (2,344,229 IU/mL in clinical trial registries vs. 264,809 IU/mL in observational cohorts) and 95% had a viral load greater than 4030 IU/mL (95% CI 24–4100) compared to 214 IU/mL (95% CI 166–266). Just 3% of clinical trial participants had viral load under 1000 IU/mL, compared to 10% in those from observational databases. This is broadly consistent with results from an analysis of 34 phase 2/3 clinical trials which showed less than 1% had had viral load under 1000 IU/mL. 18 The reasons for this higher viral load in treatment failures among trial participants may relate to the more stringent selection criteria for clinical trials and exclusion of those with LLV (only those with VL >1000 IU/ml were enrolled, and on‐treatment failures were excluded from analysis). This highlights importance of reporting analyses separately for clinical trial and observational databases.
We also found that several independent demographic, clinical and treatment characteristics were associated with LLV (<1000 IU/mL) including female sex (relative to male), fibrosis stage F0‐F1 (compared to F4, and medications from early DAA treatment era (vs. mid), although the biological reasons for these associations are not clear. In a sensitivity analysis, using data from SVR week 24 yielded similar results on viral load distribution or predictors of low‐level viraemia.
There are several practical implications for these findings. Currently, there are five assays that have WHO prequalification for HCV confirmatory viral load: three laboratory‐based assays (Abbott Real time HCV PCR, Alinity m HCV RT‐PCR and Abbott Architect HCV Ag) and two point‐of‐care assays (Xpert HCV viral load and GeneDrive HCV). The majority of existing lab‐based assays with a LloD of between 5 and 15 IU/mL as well as approved PoC assays (HCV RNA PoC GeneXpert assays has a LLOD of 10 IU/mL for venous blood, 12 , 13 or 100 IU/mL using fingerstick capillary blood) would detect more than 97% of treatment failures and is also therefore appropriate for testing for HCV cure. The recently WHO prequalified portable PoC Genedrive instrument has a reported LLoD of 2362 IU/mL 35 as well as existing HCV core antigen (ref) with a LLOD of 1000 IU/mL. A clinical trial is currently underway evaluating the Truenat HCV RNA assay from Molbio Diagnostics that uses capillary blood and a battery‐powered mobile platform (LLoD is not yet available). 36 , 37 Currently, the European Association for the Study of the Liver Diseases, IDSA‐AASLD HCV guidance panel, recommend a minimum LLoD of 1000 IU/mL for HCV diagnosis, with no specification of minimal test characteristics for test of cure. 14 End users should be aware that some low‐level virological failures may therefore be missed may therefore be missed with an assay with a LLoD greater than 1000 IU/mL and that there will need to be a trade‐off with the convenience of lower cost and more available viral load assays that may have a higher limit of detection (LLoD) and thus lower analytic sensitivity than standard laboratory‐based assays. The results of our analysis provide an additional valuable evidence base for guidance panels and regulatory authorities to assess use of platforms for monitoring of SVR12 as well as diagnosis. Given the differences in viral load distribution between clinical trials and observational studies, more work is needed to better understand which data sources should be used to inform WHO LOD standards.
The primary strength of this study is that the analysis was based on the largest global data set to date of nearly 6000 of HCV treatment failures. The high cure rate associated with pan‐genotypic DAAs, routinely exceeding 90%, has previously made it difficult to assemble a large enough cohort, representing different geographic regions with different genotypes, range of stage of disease and use of different DAA regimens with adequate rates of follow‐up SVR measurement—to reflect real‐life distribution of viral loads at treatment failure. We had data from both clinical trials with high level of follow‐up, as well as from observational cohorts reflecting real‐world treatment experience.
There are several limitations to the data and analysis. First, we are not able to measure all potential factors contributing to HCV RNA level at treatment failure, such as risk characteristics (injection drug behaviours, sex work, etc.), but the initial analysis did not show that drug regimen, genotype or stage of disease were important determinants of low‐level treatment failure. It is important to study and understand those unmeasured confounders, because if the underlying causal relationship is between a measurable or identifiable trait and the likelihood of having LLV at the time of treatment failure, then it may be possible to tailor guidance to identify venues or subgroups of people in whom it is still appropriate to employ available, close to patient assays to test for HCV cure. Second, our data set only included those who initiated treatment and returned for follow‐up HCV RNA testing 12 weeks post‐treatment. It is likely that those who fail to return for SVR may be at higher risk of treatment failure, and it is unclear whether they will be at lower or higher risk for LLV. However, this primarily affects the observational cohort and not clinical trial registry data. Third, more than 70% of our global data set cohort of treatment failures came from either Egypt (predominantly genotype 4) or the United States (predominantly genotype 1). We were not able to assemble a cohort of individuals with HCV treatment failure that represented all HCV genotypes, and all stages of disease. Finally, our global data set included data from both national and health system‐wide observational databases reflecting real‐world treatment experience, as well as from clinical trials with strict inclusion and exclusion criteria.
This study assessed the distribution of detectable viral loads 12 weeks following the end of treatment for HCV infection in an international cohort to inform the lower limit of detection of viral load assays for test of cure to identify treatment failure as well as for diagnosis of chronic hepatitis C infection. Based on a combined data set of clinical trials and observational data, a LLoD of 227 IU/mL (4030 IU/mL in the clinical trials subsample) would identify 95% of patients with a detectable viral load 12 weeks after treatment. While more than 10 times higher than the analytical sensitivity of laboratory‐based NAATs, it is more than 10 times lower than the LLoD for HCV diagnosis. These findings demonstrate it might be prudent and necessary to consider different LLoD standards for HCV diagnosis and for test of cure. Development of a point‐of‐care HCV test for cure with a low enough limit of detection to identify 95% of patients and is affordable, is an important aspect of expanding access to HCV treatment and a vital component of the WHO’s HCV elimination targets.
CONFLICTS OF INTEREST
The authors report no other funding or conflicts of interest.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Georgia National Hepatitis C Elimination Program and Hepatology and Gastroenterology Department of the Medical Center Mrcheveli, Tbilisi Georgia for their contribution. The authors thank Liyun Ni, Anand Chokkalingam and Betty Chiang of Gilead Sciences for the provision of data and thoughtful comments on the draft manuscript. Further, the authors wish to acknowledge the role of the HCV Research UK (Funded by the Medical Research Foundation [award number C0365]) in collecting and making available the data used in the generation of this publication and the United States Department of Veteran Affairs and the Government of Egypt for the provision of data for this project. Part of the data used for this study was provided by Medecins sans Frontieres and Epicentre. This project was supported through a grant from UNITAID, and the FIND contribution was supported by UNITAID as part of HEAD‐Start (Hepatitis Elimination through Access to Diagnostics). The National Institute on Drug Abuse (grant P30DA040500, P30AI042853) funded this project. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the United States Centers for Disease Control and Prevention.
Morgan JR, Marsh E, Savinkina A, et al. Determining the lower limit of detection required for HCV viral load assay for test of cure following direct‐acting antiviral‐based treatment regimens: Evidence from a global data set. J Viral Hepat. 2022;29:474–486. doi: 10.1111/jvh.13672
REFERENCES
- 1. World Health Organization . Global progress report on HIV, viral hepatitis and sexually transmitted infections, 2021. 2021. https://www.who.int/publications/i/item/9789240027077
- 2. Lancaster K, Rhodes T, Rance J. "Towards eliminating viral hepatitis": Examining the productive capacity and constitutive effects of global policy on hepatitis C elimination. Int J Drug Policy. 2020;80:102419. doi: 10.1016/j.drugpo.2019.02.008 [DOI] [PubMed] [Google Scholar]
- 3. Morgan JR, Kim AY, Naggie S, Linas BP. The effect of shorter treatment regimens for hepatitis C on population health and under fixed budgets. Open Forum Infect Dis. 2018;5(1):ofx267. doi: 10.1093/ofid/ofx267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Linas BP, Morgan JR, Pho MT, et al. Cost effectiveness and cost containment in the era of interferon‐free therapies to treat hepatitis C virus genotype 1. Open Forum Infect Dis. 2017;4(1):ofw266. doi: 10.1093/ofid/ofw266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization . Global health sector streategy on viral hepatitis 2016‐2021: towards ending viral hepatitis. http://apps.who.int/iris/bitstream/10665/246177/1/WHO‐HIV‐2016.06‐eng.pdf?ua=1
- 6. World Health Organization . Global hepatitis report, 2017. http://apps.who.int/iris/bitstream/10665/255016/1/9789241565455‐eng.pdf?ua=1
- 7. World Health Organization . Progress report on HIV, viral hepatitis and sexually transmitted infections. WHO; 2019. [Google Scholar]
- 8. Morgan JR, Servidone M, Easterbrook P, Linas BP. Economic evaluation of HCV testing approaches in low and middle income countries. BMC Infect Dis. 2017;17(Suppl 1):697. doi: 10.1186/s12879-017-2779-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Easterbrook PJ, Group WHOGD . Who to test and how to test for chronic hepatitis C infection – 2016 WHO testing guidance for low‐ and middle‐income countries. J Hepatol. 2016;65(1 Suppl):S46‐66. doi: 10.1016/j.jhep.2016.08.002 [DOI] [PubMed] [Google Scholar]
- 10. Isenhour CJ, Hariri SH, Hales CM, Vellozzi CJ. Hepatitis C antibody testing in a commercially insured population, 2005–2014. Am J Prev Med. 2017;52(5):625‐631. doi: 10.1016/j.amepre.2016.12.016 [DOI] [PubMed] [Google Scholar]
- 11. Freiman JM, Wang J, Easterbrook PJ, et al. Deriving the optimal limit of detection for an HCV point‐of‐care test for viraemic infection: Analysis of a global dataset. J Hepatol. 2019;71(1):62‐70. doi: 10.1016/j.jhep.2019.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cepheid . Xpert® HCV Viral Load. https://www.cepheid.com/en/tests/Virology/Xpert‐HCV‐Viral‐Load
- 13. McHugh MP, Wu AHB, Chevaliez S, Pawlotsky JM, Hallin M, Templeton KE. Multicenter evaluation of the Cepheid Xpert hepatitis C virus viral loaD Assay. J Clin Microbiol. 2017;55(5):1550‐1556. doi: 10.1128/JCM.02460-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. European Association for the Study of the Liver . Electronic address eee, European Association for the Study of the L . EASL recommendations on treatment of hepatitis C 2018. J Hepatol. 2018;69(2):461‐511. doi: 10.1016/j.jhep.2018.03.026 [DOI] [PubMed] [Google Scholar]
- 15. AASLD/IDSA/IAS–USA . Recommendations for testing, managing, and treating hepatitis C. www.hcvguidelines.org
- 16. Pal V, Ancha N, Mann J, Modi AA. Effect of low positive end of treatment viral load with direct‐acting antiviral therapy on sustained virologic response. Can J Gastroenterol Hepatol. 2020;2020:8815829. doi: 10.1155/2020/8815829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maasoumy B, Buggisch P, Mauss S, et al. Clinical significance of detectable and quantifiable HCV RNA at the end of treatment with ledipasvir/sofosbuvir in GT1 patients. Liver Int. 2018;38(11):1906‐1910. doi: 10.1111/liv.13932 [DOI] [PubMed] [Google Scholar]
- 18. Harrington PR, Komatsu TE, Sun H, Naeger LK. Hepatitis C virus RNA levels following virologic failure with direct‐acting antivirals: implications for lower sensitivity diagnostic assays. Clin Infect Dis. 2020;70(2):327‐330. doi: 10.1093/cid/ciz385 [DOI] [PubMed] [Google Scholar]
- 19. Tsertsvadze T, Gamkrelidze A, Chkhartishvili N, et al. Three years of progress toward achieving hepatitis C elimination in the country of Georgia, April 2015‐March 2018. Clin Infect Dis. 2020;71(5):1263‐1268. doi: 10.1093/cid/ciz956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Belperio PS, Chartier M, Ross DB, Alaigh P, Shulkin D. Curing hepatitis C virus infection: best practices from the U.S. department of veterans affairs. Ann Intern Med. 2017;167(7):499‐504. doi: 10.7326/M17-1073 [DOI] [PubMed] [Google Scholar]
- 21. Omran D, Alboraie M, Zayed RA, et al. Towards hepatitis C virus elimination: Egyptian experience, achievements and limitations. World J Gastroenterol. 2018;24(38):4330‐4340. doi: 10.3748/wjg.v24.i38.4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harris H, Costella A, Mandal S, contributors. Hepatitis C treatment monitoring in England: Content, completeness and preliminary findings from the Hepatitis C patient registry and treatment outcome system. Public Health England; 2018. [Google Scholar]
- 23. Hlaing NKT, Mitrani RA, Aung ST, et al. Safety and efficacy of sofosbuvir‐based direct‐acting antiviral regimens for hepatitis C virus genotypes 1–4 and 6 in Myanmar: Real‐world experience. J Viral Hepatitis. 2017;24(11):927‐935. doi: 10.1111/jvh.12721 [DOI] [PubMed] [Google Scholar]
- 24. Antoniak S, Charles Chasela C, Freiman MJ, et al. Treatment and Cost ‐outcomes of a simplified antiretroviral treatment strategy for hepatitis C among HCV and HIV co‐infected patients in Ukraine. medRxiv 2021. doi: 10.1101/2021.03.19.21253780 [DOI]
- 25. Morgan JR, Savinkina A, Pires Dos Santos AG, Xue Z, Shilton S, Linas B. HCV viral load greater than 1000 IU/ml at time of virologic failure in direct‐acting antiviral‐treated patients. Adv Ther. 2021;38(3):1690‐1700. doi: 10.1007/s12325-021-01647-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feld JJ, Jacobson IM, Hezode C, et al. Sofosbuvir and velpatasvir for HCV genotype 1, 2, 4, 5, and 6 infection. N Engl J Med. 2015;373(27):2599‐2607. doi: 10.1056/NEJMoa1512610 [DOI] [PubMed] [Google Scholar]
- 27. Freiman JM, Tran TM, Schumacher SG, et al. Hepatitis C core antigen testing for diagnosis of hepatitis C virus infection: a systematic review and meta‐analysis. Ann Intern Med. 2016;165(5):345‐355. doi: 10.7326/M16-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bachofner JA, Valli PV, Kroger A, et al. Direct antiviral agent treatment of chronic hepatitis C results in rapid regression of transient elastography and fibrosis markers fibrosis‐4 score and aspartate aminotransferase‐platelet ratio index. Liver Int. 2017;37(3):369‐376. doi: 10.1111/liv.13256 [DOI] [PubMed] [Google Scholar]
- 29. Sanchez‐Conde M, Montes‐Ramirez ML, Miralles P, et al. Comparison of transient elastography and liver biopsy for the assessment of liver fibrosis in HIV/hepatitis C virus‐coinfected patients and correlation with noninvasive serum markers. J Viral Hepatitis. 2010;17(4):280‐286. doi: 10.1111/j.1365-2893.2009.01180.x [DOI] [PubMed] [Google Scholar]
- 30. Xiao G, Yang J, Yan L. Comparison of diagnostic accuracy of aspartate aminotransferase to platelet ratio index and fibrosis‐4 index for detecting liver fibrosis in adult patients with chronic hepatitis B virus infection: a systemic review and meta‐analysis. Hepatology. 2015;61(1):292‐302. doi: 10.1002/hep.27382 [DOI] [PubMed] [Google Scholar]
- 31. Sievert W, Altraif I, Razavi HA, et al. A systematic review of hepatitis C virus epidemiology in Asia, Australia and Egypt. Liver Int. 2011;31:61‐80. doi: 10.1111/j.1478-3231.2011.02540.x [DOI] [PubMed] [Google Scholar]
- 32. Hahn GJ, Meeker WQ. Statistical intervals: a guide for practitioners. Wiley; 1991. [Google Scholar]
- 33. Rockstroh JK, Feld JJ, Chevaliez S, et al. HCV core antigen as an alternate test to HCV RNA for assessment of virologic responses to all‐oral, interferon‐free treatment in HCV genotype 1 infected patients. J Virol Methods. 2017;245:14‐18. doi: 10.1016/j.jviromet.2017.03.002 [DOI] [PubMed] [Google Scholar]
- 34. Medici MC, Furlini G, Rodella A, et al. Hepatitis C virus core antigen: analytical performances, correlation with viremia and potential applications of a quantitative, automated immunoassay. J Clin Virol. 2011;51(4):264‐269. doi: 10.1016/j.jcv.2011.05.003 [DOI] [PubMed] [Google Scholar]
- 35. Llibre A, Shimakawa Y, Mottez E, et al. Development and clinical validation of the Genedrive point‐of‐care test for qualitative detection of hepatitis C virus. Gut. 2018;67(11):2017‐2024. doi: 10.1136/gutjnl-2017-315783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. ClinicalTrials.gov . Truenat™ HCV RNA assay evaluation. https://clinicaltrials.gov/ct2/show/NCT04236973
- 37. Lee DJ, Kumarasamy N, Resch SC, et al. Rapid, point‐of‐care diagnosis of tuberculosis with novel Truenat assay: Cost‐effectiveness analysis for India's public sector. PLoS One. 2019;14(7):e0218890. doi: 10.1371/journal.pone.0218890 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


