Abstract
We developed a new screening method for potential wood preservatives based on decolorization of the dye Remazol Brilliant Blue R by extracellular oxidative agents produced by wood decay fungi. Oxidative biodegradation of lignin yielded decolorized zones around and under fungal cultures on a dyed agar medium. Inhibitory effects were detected by direct observation and measurement of the decolorized zones.
The cycle time (several weeks) of typical screening methods limits the search for new wood preservatives. Standard soil block and agar block methods require 8 to 24 weeks (1) and relatively large amounts of test material. To circumvent these problems, wood preservative laboratories commonly use an agar plate method (2) that measures fungal growth on malt agar medium with the added biocide. This test yields a MIC, the lowest concentration that totally inhibits fungal growth. Although wood preservative laboratories use the agar plate method as a screening test, the inhibition of wood biodegradation cannot be evaluated using this method. Evaluating the effects of potential wood preservatives on wood biodegradation may offer the opportunity to discriminate between compounds that inhibit specific biochemical mechanisms and those that are simply gross metabolic toxins. Extracellular oxidative agents produced by wood decay fungi are hypothesized to play an important role in wood biodegradation (5, 6). Several investigators reported correlations between the decolorization of dyes and the ligninolytic ability of the microorganisms (7–12). Therefore, model synthetic dyes such as Remazol Brilliant Blue R could be used to detect lignocellulolytic activity of fungi (7–10, 12).
We developed a rapid, easily scored single-dye test that could be used for primary or secondary screening of potential wood preservatives.
White-rot fungi (Trametes versicolor ATCC 42462 and Irpex lacteus ATCC 11245), brown-rot fungi (Gloeophyllum trabeum ATCC 11539 and Postia placenta ATCC 11538), and a soft-rot fungus (Chaetomium globosum ATCC 6205) were selected as model organisms to evaluate the method. The fungal strains were maintained and grown on malt extract agar (Difco, Detroit, Mich.) or potato dextrose agar (Difco) (3). The agar plate method was performed on malt extract agar medium. The dye decolorization method was performed on mineral salts agar medium (4) containing a 10-g/liter mixture (3:7) of lignin (catalog no. 37,095-9; Aldrich, Milwaukee, Wis.) and α-cellulose (catalog no. C-6429; Sigma, St. Louis, Mo.) as a sole source of carbon supplemented with 10 ml of a vitamin solution per liter. The vitamin stock solution contained the following (in mg/liter): d-biotin, 2; d-pantothenic acid hemicalcium salt, 0.2; folic acid, 0.2; niacinamide, 40; thiamine hydrochloride, 40; p-aminobenzoic acid, 20; and riboflavin, 20. Experimental and untreated control plates were stained using a 0.01% (wt/vol) aseptically prepared solution of the dye Remazol Brilliant Blue R (catalog no. R-8001; Sigma), which was added after sterilization. Plates were inoculated using a plug of mycelium from an actively growing fungal culture. Untreated controls were grown without preservatives. Fungi were incubated for 3 to 14 days at 26°C.
Didecyldimethylammonium chloride (DDAC) (Lonzagroup), chromated copper arsenate (CCA; type C) (Osmose Wood Preserving, Inc., Buffalo, N.Y.), and an ammoniacal copper–DDAC mixture in a 2:1 CuO/DDAC weight ratio (ACQ; type B) were added to agar medium at 50°C before inoculation. Three sets of experiments were performed independently. Each biocide concentration for each fungal species was run in triplicate. Controls had no biocides added to the media. Uninoculated stained plates with biocides were used as stability controls for the dye decolorization method. Data were statistically treated using the Microsoft Excel 97 correlation data analysis function.
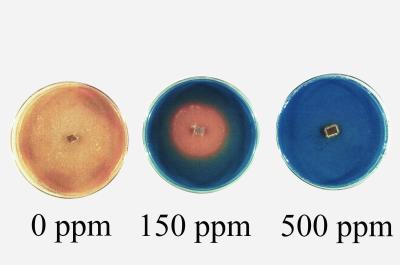
Our screen is based on the ability of wood decay fungi to generate extracellular oxidative agents that change the color of the Remazol Brilliant Blue R as a result of a redox reaction. Normal growth results in a dye decolorization around and under areas of growth on stained media. Potential wood preservatives repress normal growth and oxidative activity, and no dye decolorization occurs. Dye decolorization zones develop in 3 to 7 days on control medium and in 5 to 14 days on media with biocides (Fig. 1). The lowest concentration that totally inhibited dye decolorization formation was called the minimum concentration to inhibit decolorization (MCID) (Fig. 1). The diameter of dye decolorization was measured in millimeters per day, and the radial extension rate was calculated.
FIG. 1.
Dye decolorization development in 7 days with DDAC (T. versicolor).
The MCID data generated with the dye decolorization method correlate with experimental and literature MIC data for the agar plate method (Table 1; Fig. 2) (2). Analysis of growth curves from the agar plate method and radial extension curves from the dye decolorization method showed that the growth rate correlated with decolorization in untreated controls. These data imply that the growth rate should correlate with decolorization after biocide treatment. Although the numerical values of the MCID and MIC data were not always equivalent, the data from the two methods were closely correlated. The dye decolorization method successfully measured the inhibitory effects of various biocides with a sensitivity greater than or equal to that of the standard agar plate method. This method also can be used to screen for microorganisms that degrade lignin extracellularly and that change the color of Remazol Brilliant Blue R as a result of a redox reaction.
TABLE 1.
Correlation between MCID and MIC
| Organism | Preservative | Concn (ppm) | Correlation coefficienta |
|---|---|---|---|
| G. trabeum | None (control) | 0 | 0.9939 |
| CCA | 10 | 0.9802 | |
| 50 | 0.9318 | ||
| I. lacteus | None (control) | 0 | 0.9899 |
| ACQ | 200 | 0.9677 | |
| 625 | 0.9445 | ||
| P. placenta | None (control) | 0 | 0.9467 |
| CCA | 10 | 0.9974 | |
| C. globosum | None (control) | 0 | 0.9997 |
| CCA | 10 | 0.9954 | |
| T. versicolor | None (control) | 0 | 0.8582 |
| DDAC | 10 | 0.9999 | |
| 50 | 0.9999 |
Calculated with Microsoft Excel 97 correlation data analysis function.
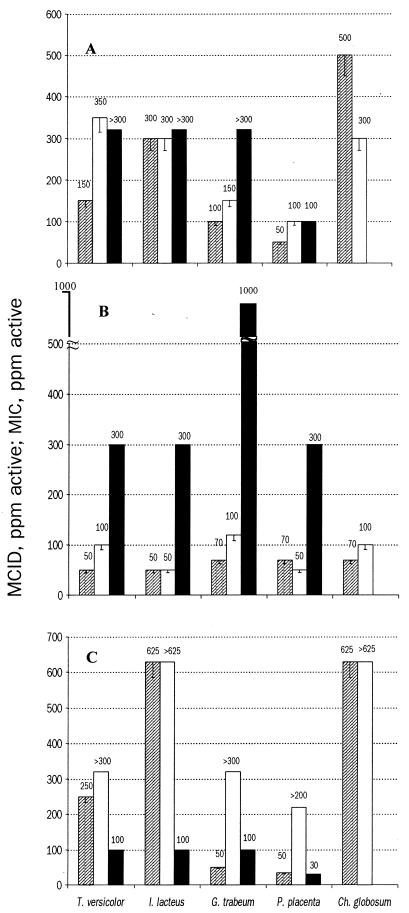
FIG. 2.
MCID and MIC for DDAC (A), CCA (B), and ACQ (C), generated using different methods. Symbols: ▨, dye decolorization method; □, agar plate method; ■, agar plate method (literature data) (2).
The results from the dye decolorization method were reproducible (Fig. 2). The MCID values were similar across all three independent experiments. The error bars (Fig. 2) show the next lowest dilution to the observed MCID and MIC. This concentration is the highest concentration at which growth was observed. Therefore, the error bars show the potential MCID and MIC range.
Reproducible, sensitive, and easily validated methods for screening wood preservatives are rare. The dye decolorization method correlates to known methods, gives reproducible data, and can be used for determining MCIDs for wood rot fungi. Based on our results it is clear that different biocides have different inhibitory mechanisms.
REFERENCES
- 1.American Wood Preservers' Association. AWPA standards 1997. Granbury, Tex: American Wood Preservers' Association; 1997. pp. 316–325. [Google Scholar]
- 2.Archer K, Nicholas D D, Schultz T P. Screening of wood preservatives: comparison of the soil-block, agar-block, and agar-plate tests. Forest Products J. 1995;45:86–89. [Google Scholar]
- 3.Atlas R M. Handbook of microbiological media. Boca Raton, Fla: CRC Press; 1997. p. 802. [Google Scholar]
- 4.Atlas R M. Handbook of microbiological media. Boca Raton, Fla: CRC Press; 1997. p. 1115. [Google Scholar]
- 5.Blanchette R A. Degradation of the lignocellulose complex in wood. Can J Bot. 1995;73:999–1010. [Google Scholar]
- 6.Dutton M V, Evans C S. Oxalate production by fungi: its role in pathogenicity and ecology in the soil environment. Can J Microbiol. 1996;42:881–895. [Google Scholar]
- 7.Glenn J K, Gold M H. Decolorization of several polymeric dyes by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1983;46:1741–1747. doi: 10.1128/aem.45.6.1741-1747.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ollikka P, Alhonmäki K, Leppänen V, Glumoff T, Raijola T, Suominen I. Decolorization of azo, triphenyl methane, heterocyclic, and polymeric dyes by lignin peroxidase isoenzymes from Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:4010–4016. doi: 10.1128/aem.59.12.4010-4016.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasti M B, Crawford D L. Relationships between the abilities of Streptomycetes to decolorize three anthrone-type dyes and to degrade lignocellulose. Can J Microbiol. 1991;37:902–907. [Google Scholar]
- 10.Raghukumar C, D'Souza T M, Thorn R G, Reddy C A. Lignin-modifying enzymes of Flavodon flavus, a basidiomycete isolated from a coastal marine environment. Appl Environ Microbiol. 1999;65:2103–2111. doi: 10.1128/aem.65.5.2103-2111.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez A, Perestelo F, Carnicero A, Regalado V, Perez R, De la Fuente G, Falcon M A. Degradation of natural lignins and lignocellulosic substrates by soil-inhabiting fungi imperfecti. FEMS Microbiol Ecol. 1996;21:213–219. [Google Scholar]
- 12.Vyas B R, Molitoris H P. Involvement of extracellular H2O2-dependent ligninolytic activity of the white rot fungus Pleurotus ostreatus in the decolorization of Remazol Brilliant Blue R. Appl Environ Microbiol. 1995;61:3919–3927. doi: 10.1128/aem.61.11.3919-3927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]