Abstract
Bacillus anthracis, Bacillus cereus, Bacillus mycoides, Bacillus pseudomycoides, Bacillus thuringiensis, and Bacillus weihenstephanensis are closely related in phenotype and genotype, and their genetic relationship is still open to debate. The present work uses amplified 16S-23S internal transcribed spacers (ITS) to discriminate between the strains and species and to describe the genetic relationships within the “B. cereus group,” advantage being taken of homoduplex-heteroduplex polymorphisms (HHP) resolved by polyacrylamide gel electrophoresis and silver staining. One hundred forty-one strains belonging to the six species were investigated, and 73 ITS-HHP pattern types were distinguished by MDE, a polyacrylamide matrix specifically designed to resolve heteroduplex and single-strand conformation polymorphisms. The discriminating bands were confirmed as ITS by Southern hybridization, and the homoduplex or heteroduplex nature was identified by single-stranded DNA mung bean nuclease digestion. Several of the ITS-HHP types corresponded to specific phenotypes such as B. anthracis or serotypes of B. thuringiensis. Unweighted pair group method arithmetic average cluster analysis revealed two main groups. One included B. mycoides, B. weihenstephanensis, and B. pseudomycoides. The second included B. cereus and B. thuringiensis, B. anthracis appeared as a lineage of B. cereus.
The “Bacillus cereus group,” one of the most homogeneous groups of the genus Bacillus, encompasses six validly described species, B. anthracis, B. cereus, B. mycoides, B. pseudomycoides, B. thuringiensis, and B. weihenstephanensis, all of which have an important impact on human activity (18, 39, 44).
These species are known to be strictly related phylogenetically, as has been shown by DNA-DNA hybridization studies (35, 39, 42–44, 57) and the sequencing of the ribosomal RNA genes (2–4, 39). On the other hand, a marked variability is always observed when large collections of strains are examined by DNA fingerprinting methods that target the whole genome (8–13, 17, 25–29, 36, 58) and/or discrete genes (15, 16, 38, 54, 55, 61). Hence, the phylogenetic and taxonomic relationship among these species is still open to debate. It has been proposed previously that B. anthracis, B. cereus, and B. thuringiensis represent a single species, this conclusion having been reached through genome sizing and mapping (9–13) and, very recently, by multilocus enzyme electrophoresis and the sequencing of nine different DNA loci (27).
The ribosomal operon is a classic molecular marker used to trace genetic relationships and to identify strains rapidly (1). Of all the different regions of the ribosomal operon, the internal transcribed spacers (ITS) between 16S and 23S ribosomal DNA are frequently used as molecular markers to identify microbial species and analyze the phylogenetic relationship between strains (see, for example, references 14, 22, and 24). ITS are generally found in multiple copies in most bacterial genomes (24, 33). Since ITS are hypervariable with respect to adjacent genes, due to a higher mutation rate, they can differentiate between multiple operons in the cell (22–24, 31, 41). Agarose gel ITS-PCR fingerprinting patterns are identical for the six species of the B. cereus group (14, 17) and are thus not suitable for the rapid identification of the species. Since the B. cereus group strains have from 8 to 12 ribosomal operons (34, 39, 49) that can differ in ITS sequence, it might be possible to detect interspecies and intraspecies differences through ITS homoduplex-heteroduplex polymorphisms (ITS-HHP) (6, 31–33). The principle of strain discrimination by ITS-HHP analysis is based on the detection of sequence polymorphisms in the form of heteroduplex DNA bands that are generated during PCR when fragments of different length, which contain homologous regions at the 3′ and 5′ ends, cross-hybridize to form heteroduplex DNA structures. The electrophoretic mobility of these heteroduplex structures is significantly reduced with respect to homoduplex DNA fragments, depending on the amount of single-stranded DNA present in the heteroduplex product and the degree of secondary structures formed within the single-stranded regions (6, 31–33).
The aim of this study was to evaluate whether ITS-HHP can be used advantageously to highlight molecular diversity within the B. cereus group and to address the identification of the species themselves or particular phenotypic groups. Thus, the ITS-PCR products from 141 strains of the six species being investigated (Table 1) were subjected to high-resolution polyacrylamide gel electrophoresis, and it was found that ITS-HHP is a further tool toward a greater understanding of the genetic relationship among the species of the B. cereus group and for addressing the strain identification.
TABLE 1.
Strains analyzed in this study, ITS-HHP pattern type number, and relevant characteristics
| Speciesa (no. of strains) | Hb | Strain | Relevant characteristic(s) of strain (reference[s]) |
|---|---|---|---|
| BA (27) | 30 | Davis TE702c | pXO1−/2+ (51) |
| 31 | 256c, 282c, 582c, 846c, 6445/RA3c, 6445/RA3Rc, 6687/4896/Lc, 6769c, 7611c, 7611Rc, 9240c | All the strains were pXO1+/2+ except strains 6445/RA3R and 7611R (pXO1+/2−) (48, 51). Strains 256, 282, 582, and 846 were isolated in the French Pyrenees in 1994 (Guy Patra, personal communication). All the other strains were isolated in France, in the Pyrenees and Alps, in 1997 (48). | |
| 32 | 170c, 227c, 300c, 376c, 663c, 779c, 832c, 957c, 4229c, 6602c, 7700c, 7702c, Cepanzoc, 2522d, antiMid | All the strains were pXO1+/2+ except 7700 (pXO1−/2−); 957, 7702, and Cepanzo (pXO1+/2−); and 4229 and 6602 (pXO1−/2+) (51). Strains 170, 376, 663, 779, and 832 were isolated in France (Guy Patra, personal communication). | |
| BC (23) | 13 | cer3e | Isolated from candies (17) |
| 14 | po1e | Isolated from ultrahigh-temperature milk (17) | |
| 25 | 360f | ||
| 26 | cer1e | ||
| 27 | cer6e | Isolated from tomato sauce (17) | |
| 28 | co1e | Isolated from pasteurized milk (17) | |
| 29 | 2896f | ||
| 33 | bc2e | ||
| 34 | 345f | ||
| 35 | cer5e | Isolated from rice (17) | |
| 37 | 46321f | ||
| 38 | 487f | ||
| 39 | be1e | Isolated from marble of Ca' d'Oro façade, Venice, Italy (17) | |
| 40 | 31Te | B. cereus type strain | |
| 41 | cer4e | Isolated from rice (17) | |
| 42 | myde | ||
| 43 | 351f | ||
| 44 | my1e | Isolated from ultrahigh-temperature milk (17) | |
| 45 | co2e | ||
| 47 | 6127f | ||
| 49 | 626f | ||
| 50 | 5148g | Same as strain Bc7, produces the bacteriocin cerein 7 (45) | |
| 58 | IO200h | ||
| BM (21) | 3 | G2e | Isolated from garden soil, Italy |
| 15 | 309f, 384f, bmFe | 384 strain growth at 7°C (15) | |
| 16 | NRS273Ti | B. mycoides type strain | |
| 17 | 2048Tf, 229f | 2048T, B. mycoides type strain; growth at 7°C (15) | |
| 18 | B615i | Isolated from soil (43) | |
| 19 | B14828i | ||
| 20 | BmMede | Isolated from alkaline soil, Italy | |
| 21 | BmSe | ||
| 22 | Ndre, Bife | Isolated from soil, Italy | |
| 23 | NRS306i, NRS319i | Isolated from soil (43) | |
| 24 | G1e, Nov1e | G1, isolated from garden soil, Italy; Nov1, isolated from maize rhizosphere, Italy | |
| 36 | A81e, TP2e | Isolated from soil, Italy | |
| 46 | MycHe | ||
| 48 | Nov2e | Isolated from maize rhizosphere, Italy | |
| BP (8) | 7 | BD10i, BD14i | Isolated from soil (43) |
| 8 | A82e | Isolated from soil, Italy | |
| 9 | B346i | Isolated from soil (43) | |
| 10 | CAe | Isolated from soil, Italy | |
| 11 | B617Ti, B618i | B617T, B. pseudomycoides type strain (44); isolated from soil (43) | |
| 12 | TP1e | Isolated from soil, Italy | |
| BT (58) | 51 | BMG1.6h | Isolated from soil, Tunisia |
| 52 | 2046Tf | B. thuringiensis type strain, subsp. thuringiensis | |
| 53 | Bt10h | B. thuringiensis subsp. darmstadiensis | |
| 54 | BUMP30j | Same as strain BST30, B. thuringiensis subsp. sotto (30) | |
| 55 | 5424f | B. thuringiensis subsp. israelensis | |
| 56 | HD868k | B. thuringiensis subsp. tochigiensis; produces the bacteriocin tochicin (46) | |
| 57 | BUMP33j | Isolated from soil, Tunisia | |
| 59 | Bt5e | ||
| 60 | Bt14h, Bt44h | Bt14, B. thuringiensis subsp. israelensis; Bt44, B. thuringiensis subsp. dendrolimus | |
| 61 | Ht39l | Displays entomocidal activity against Culex sp. larvae | |
| 62 | Hc15l, Hc24l, Hc32l, Hc35l | Display entomocidal activity against Culex sp. larvae | |
| 63 | Hc17l | Displays entomocidal activity against Culex sp. larvae | |
| 64 | Hc16l | Displays entomocidal activity against Culex sp. larvae | |
| 65 | Hc13l | Displays entomocidal activity against Culex sp. larvae | |
| 66 | BMG1.3h, BMG1.4h, BMG1.5h | Isolated from surface water, Tunisia | |
| 67 | Ht51l | Displays entomocidal activity against Culex sp. larvae | |
| 68 | Hc45l | Displays entomocidal activity against Culex sp. larvae | |
| 69 | Hc36l | Displays entomocidal activity against Culex sp. larvae | |
| 70 | BX1h, BX3h, BX8h, BX9h, BX11h, BX12h, BX16h, BX19h, BX20h, BX21h, BX22h, BG7h | BX strains were isolated from hypersaline soils in Tunisia and display antifungal activity against Fusarium sp. | |
| 71 | BMG1.2h, BMG1.7h, BMG1.8h, BMG1.9h, BUMP24j, BUMP25j, BUMP26j, BUMP28j, BUMP29j | Isolated from soil, Tunisia; strain BMG1.7 produces the bacteriocin thuricin 7 (Cherif et al., unpublished) | |
| 72 | BTXe, PTBe, BTKe, HD2k | Strain HD2, B. thuringiensis subsp. thuringiensis, produces the bacteriocin thuricin (19) | |
| 73 | Bt1h, Bt7h, Bt9h, Bt33h, Bt55h, HD1m, Ale | Bt1, B. thuringiensis subsp. thuringiensis; Bt7, B. thuringiensis subsp. aizawai; Bt9, B. thuringiensis subsp. tolworthi; Bt33 and HD1, B. thuringiensis subsp. kurstaki; Bt55, B. thuringiensis subsp. galleriae | |
| 74 | BMG1.1h | Isolated from soil, Tunisia | |
| 75 | Bt13h | Bt13, B. thuringiensis subsp. pakistani | |
| BW (4) | 4 | 10201n | Growth at 7°C (39, 50); isolated from milk (39) |
| 5 | 10208n | Growth at 7°C (39, 50); isolated from milk (39) | |
| 6 | 10204Tn, 10202n | 10204T, B. weihenstephanensis type strain; growth at 7°C (39, 50); isolated from milk (39) | |
| BL (1) | 1 | BI32 | |
| PL (1) | 2 | BX7 |
Abbreviations used to indicate the B. cereus group species; BA, B. anthracis; BC, B. cereus; BM, B. mycoides; BP, B. pseudomycoides; BT, B. thuringiensis; BW, B. weihenstephanensis. The outgroup species were B. licheniformis (BL) and Paenibacillus lentimorbus (PL).
H, ITS-HHP type number (Fig. 4).
Institute Pasteur, Paris, France. Total DNA of the B. anthracis strain was kindly provided by M. Mock.
Istituto Zooprofilattico, Milan, Italy. Total DNA of the B. anthracis strains was kindly provided by M. Luini.
Dipartimento di Scienze e Tecnologie Alimentari e Microbiologiche, Milan, Italy.
Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany.
Spanish Type Culture Collection. B. cereus strain 5148 was kindly provided by Federico Uruburu.
Laboratoire de Microbiologie de Tunis. BX strains were isolated and kindly provided by N. Sadfi. The reference strains of B. thuringiensis subspecies Bt1, Bt7, Bt9, Bt10, Bt13, Bt14, Bt33, and Bt55 were kindly provided by A. Boudabous.
Agricultural Research Service Culture Collection, Peoria, Ill. Strains were kindly provided by L. K. Nakamura.
Centre de Biotechnologie de Sfax. BUMP strains were kindly provided by S. Jaoua.
Bacillus Genetic Stock Center. B. thuringiensis strains HD2 and HD868 were kindly provided by D. R. Zeigler.
Strains kindly provided by Hala Khyami-Horani, Department of Biological Sciences, Amman, Jordan.
Strain derived from commercial B. thuringiensis based bioinsecticides.
Weihenstephan Bacillus Collection, Weihenstephan, Germany. B. weihenstephanensis strains were kindly provided by S. Scherer.
Total DNA was extracted from the washed cells by sodium dodecyl sulfate-proteinase K treatment (1a, 15). Amplification of the ITS was performed using the already-described primers S-D-Bact-1494-a-S-20 and L-D-Bact-0035-a-A-15 (17). For each strain, the products of at least two separate amplifications were analyzed individually by electrophoresis.
The ITS-PCR amplification patterns were electrophoresed on standard 2% agarose gels in 0.5× Tris-borate-EDTA buffer and stained for 30 min in an 0.5-mg/liter solution of ethidium bromide (53). Separation in 160- by 200- by 0.75-mm polyacrylamide gels was performed in a Protean II apparatus (Bio-Rad, Milan, Italy) in 1× Tris-borate-EDTA buffer for 10 to 14 h at 100 V. To enhance the heteroduplex separation, the samples were run in an 0.6× MDE gel under the same conditions used for standard polyacrylamide gel electrophoresis. All the polyacrylamide gels were cast vertically over a Gel Bond PAG film (FMC Bioproducts, Milan, Italy) to facilitate manipulation for the subsequent staining procedure. After the run, the gels were stained either with ethidium bromide (53) or by silver staining according to a known procedure (5). After staining, the gels were rinsed for 10 min in a 25% (vol/vol) ethanol–10% (vol/vol) glycerol solution, then covered with an extra cellophane sheet (Sigma, Milan, Italy), and left to dry at room temperature for 48 h.
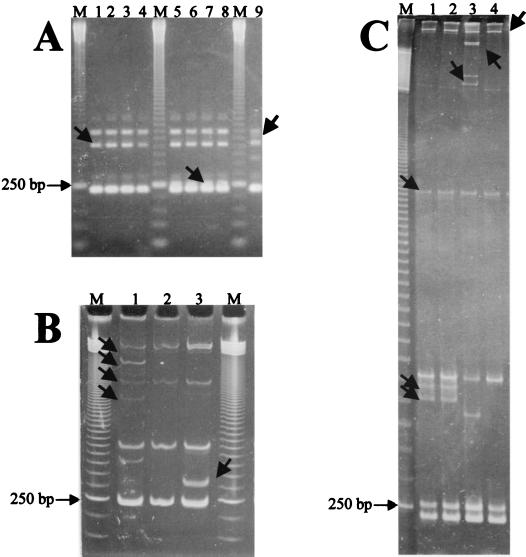
An agarose gel electrophoresis examination of the ITS-PCR products from 50 strains selected randomly from the collection revealed typical B. cereus group ITS-PCR patterns with three bands of about 240, 460, and 530 bp, respectively (14) (Fig. 1A). Only strains of B. anthracis showed different patterns that could be differentiated into three groups (Fig. 1A).
FIG. 1.
Example of ITS-PCR fingerprinting patterns of B. anthracis strains resolved by agarose (A), polyacrylamide (B), and MDE (C) gel electrophoresis, showing the migration shifts of some amplified products. Lanes M, 50-bp ladder. (A) Lanes 1 to 9, B. anthracis strains 256, 282, 582, 846, 7700, 7702, Cepanzo, 4229, and Davis TE702, respectively. (B) Lanes 1 to 3, B. anthracis strains Davis TE702, 282, and Cepanzo, respectively. (C) Lanes 1 to 4, B. anthracis strains 7700, 7702, Davis TE702, and 256, respectively. The 250-bp band of the ladder is indicated. Arrows indicate bands that showed different mobilities among agarose, polyacrylamide, and MDE gel electrophoresis assays.
The band pattern resolution using two other matrices, polyacrylamide and MDE (a particular polyacrylamide specifically designed to enhance the separation of heteroduplex and single-stranded fragments), confirmed the pattern differences in B. anthracis and showed a marked migration shift of several bands (Fig. 1B and C). By comparison with the bands of the 50-bp ladder, only the 240- and 530-bp fragments of the ITS-PCR patterns observed in the agarose gel showed a similar migration. The polyacrylamide gel revealed additional bands, several of which migrated further when the analysis was carried out on the MDE matrix. This behavior was also observed when the ITS-PCR products obtained from independent amplifications were analyzed (data not shown). The differential migration of several bands could be explained by assuming that the separation of these fragments was based not only on molecular size but also on the formation of secondary structures that slow down the fragment migration and that are typical of heteroduplex and single-stranded fragments (21, 31–33).
To verify that all the bands in the pattern, including the putative heteroduplex–single-stranded products, were ITS fragments and not aspecific PCR products, the bands from a polyacrylamide gel were blotted over a nylon membrane (53) and hybridized with a digoxigenin-labeled (14, 16, 17; Boehringer Mannheim user's guide, Boehringer Mannheim GmbH Biochemica, Mannheim, Germany) short ITS fragment from the B. anthracis strain Cepanzo, identified as an ITS by cloning and sequencing (data not shown). The Southern hybridization confirmed that the bands showing migration shifts with respect to the bands observed in the agarose gel were all homologous to the ITS (data not shown).
The nature of the polymorphic bands was investigated by treating the ITS-PCR products with mung bean nuclease. This treatment eliminates the single strands in the heteroduplex products, permitting only homoduplex products to be detected in the gel (31–33). Fifty microliters of the ITS-HHP product was purified through a QIAquick column (Qiagen) and eluted with 80 μl of resuspension buffer (6 mM Tris-HCl [pH 7.5], 6 mM NaCl, and 0.2 mM EDTA). Thirty-eight microliters of the eluted DNA solution, with an added 10 to 15 U of mung bean endonuclease (Pharmacia Biotech, Milan, Italy) diluted 1/10 (vol/vol) just before use in the dilution buffer (10 mM sodium acetate [pH 5], 0.1 mM zinc acetate, 1 mM cysteine, 0.1% [vol/vol] Triton X-100, and 50% [vol/vol] glycerol), was incubated for 30 min at 30°C. The reaction was performed in a final volume of 50 μl containing 10 μl of 5× reaction buffer made of 150 mM sodium acetate (pH 5.0), 250 mM NaCl, 5 mM ZnCl2, and 25% glycerol. To stop the reaction, 100 μl of 0.2% (wt/vol) sodium dodecyl sulfate–74 mM Tris-HCl (pH 9.5)–1.2 M LiCl solution was added; the enzyme was removed by phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol]) treatment. The aqueous phase was extracted with 1 volume of diethyl ether, and the DNA was precipitated with ethanol and resuspended in 10 μl of Tris-EDTA, pH 7.5. The sample was electrophoresed in an MDE gel, and the DNA bands were revealed by silver staining.
Figure 2 shows the results of these experiments on some strains of B. anthracis, B. cereus, B. mycoides, and B. thuringiensis. In all the cases, treatment resulted in the elimination of several bands, especially slowly migrating bands (over 500 to 600 bp), but in the case of the B. anthracis strain Cepanzo, also two bands with an apparent molecular size of 450 and 470 bp were eliminated, suggesting that these bands are heteroduplex fragments whose endonuclease digestion products (of less than 200 bp in molecular size) could be detected in the gel (Fig. 2). All the other bands were homoduplex products, since they could not be digested by the enzymatic treatment.
FIG. 2.
Identification of the heteroduplex bands in the ITS-HHP patterns separated in an MDE gel after mung bean nuclease digestion of the PCR products. The MDE runs for B. anthracis 7700, Davis TE702, and 282; B. cereus 31T; B. mycoides 2048T; and B. thuringiensis Bt7 are shown. Lanes M, 50-bp ladder. Lanes 1, normal ITS-HHP profiles. Lanes 2, the same products of lanes 1 after treatment with mung bean nuclease. The 250-bp band of the ladder is indicated. Arrowheads indicate the heteroduplex products removed by the mung bean nuclease. Open arrows show degradation products of the heteroduplex bands.
The mung bean nuclease experiments showed that the fragments in the pattern around 250 and 500 bp are indeed the main homoduplex fragments and display several length polymorphisms. Some strains, like Davis TE702, showed additional homoduplex bands of intermediate size (ca. 420 bp in the case of Davis TE702) between short (ca. 250 bp) and long (ca. 510 bp) ITS. The size difference, i.e., less than 100 bp, roughly corresponds to the length of a tRNA gene and could be the result of tDNA gene deletion due to ITS rearrangement.
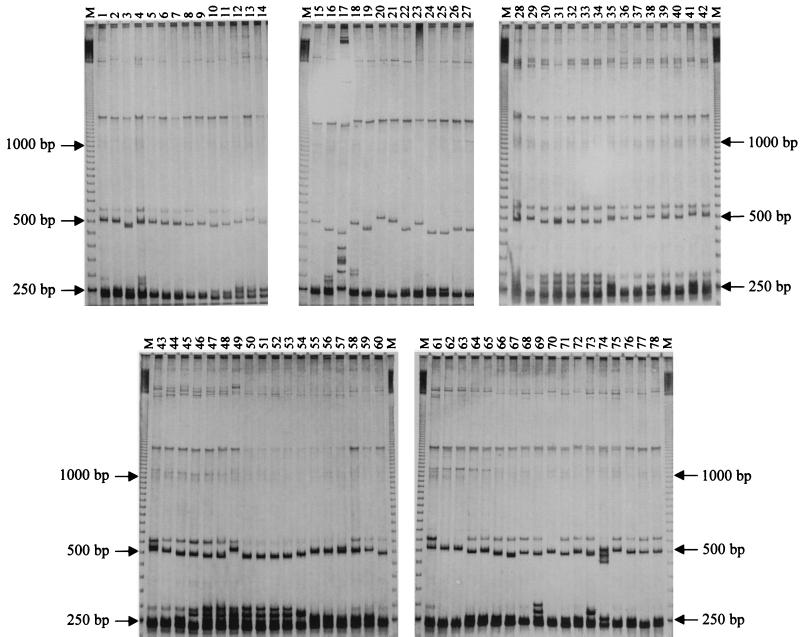
The entire collection of 141 strains of the six species of the B. cereus group plus two outgroup strains (Table 1) was analyzed for ITS-HHP on MDE gels to evaluate the intraspecific and interspecific diversity. To minimize migration shift due to the gel preparation procedure, the strains were loaded on gels prepared from the same MDE master mix and run simultaneously in a four-gel electrophoresis apparatus. The runs were repeated at least three times for each strain even from different PCRs, and the electrophoretic patterns were found to be constant and reproducible. The ITS-HHP patterns were shown to be very discriminative and allowed us to identify 75 pattern types, an unexpected variability in the extremely homogeneous B. cereus group (36) (Table 1). Representative ITS-HHP pattern types are shown in Fig. 3. A total of 72 polymorphic bands was detected. The number of bands per profile varied from 3 to 16, and the apparent molecular size ranged from 200 to more than 2,000 bp.
FIG. 3.
Examples of the ITS-HHP pattern variability observed in the collection examined. For each strain, the ITS-HHP type (H), (Table 1 and Fig. 4) is marked in parentheses below. Lanes M, 50-bp ladder. Lanes 1 to 14, B. cereus 351 (ITS-HHP type H43), myd (H42), co2 (H45), 6127 (H47), 46321 (H37), 2896 (H29), 31T (H40), co1 (H28), cer4 (H41), cer3 (H13), 360 (H25), 487 (H38), cer6 (H27), and bc1 (H39), respectively. Lanes 15 to 18, B. mycoides mych (H46), nov2 (H48), G2 (H3), and NRS273 (H16), respectively. Lane 19, B. pseudomycoides TP1 (H12). Lanes 20 and 21, B. mycoides 309 (H15) and 2048T (H17), respectively. Lane 22, B. pseudomycoides CA (H10). Lane 23, B. mycoides B615 (H18). Lanes 24 to 27, B. pseudomycoides BD14 (H7), BD10 (H7), B617T (H11), and B346 (H9), respectively. Lane 28, B. cereus 5148 (H50). Lanes 29 to 60, B. thuringiensis HD868 (H56), HD2 (H72), HD1 (H73), BTK (H72), PTB (H72), BTX (H72), Ht51 (H67), Ht39 (H61), Hc45 (H68), Hc36 (H69), Hc17 (H63), Hc16 (H64), Hc15 (H62), Hc13 (H65), BMG1.6 (H51), BUMP30 (H54), BUMP33 (H57), BMG1.1 (H74), BX16 (H70), BMG1.7 (H71), BMG1.3 (H66), Bt33 (H73), Bt9 (H73), Bt7 (H73), Bt1 (H73), Bt13 (H75), Bt44 (H60), Bt14 (H60), Bt10 (H53), Bt5 (H59), 5424 (H55), and 2046T (H52), respectively. Lanes 61 to 64, B. weihenstephanensis 10201 (H4), 10202 (H6), 10204T (H6), and 10208 (H5), respectively. Lanes 65 to 73, B. cereus myd (H42), my1 (H44), co2 (H45), co1 (H28), cer5 (H35), cer4 (H41), cer3 (H13), cer1 (H26), and 345 (H34), respectively. Lane 74, B. anthracis 7700 (H32). Lanes 75 to 78, B. cereus 351 (H43), bc2 (H33), 2896 (H29), and 360 (H25), respectively. The 250-, 500-, and 1,000-bp bands of the ladder are indicated.
Computer-assisted analysis of the ITS-HHP was performed using the Diversity Database fingerprinting software (Bio-Rad). The banding patterns were acquired from the dried silver-stained gels with the Gel Doc2000 image system (Bio-Rad), using the white-light transilluminator of the device, and stored on disk as TIFF files. The rolling-disk background subtraction method was applied to each gel, and a database containing all the gel images was created. The bands from all the gels were automatically detected and normalized using the 50-bp DNA ladder (Amersham Pharmacia Biotech) as the molecular size marker. A band set including all the polymorphic fragments was created, and each band in each lane was compared with the band set. The similarity between strains was determined by the band-sharing coefficient calculated by the formula of Jaccard (56), and strain clustering was performed by the unweighted pair group method with arithmetic averages (UPGMA) (56).
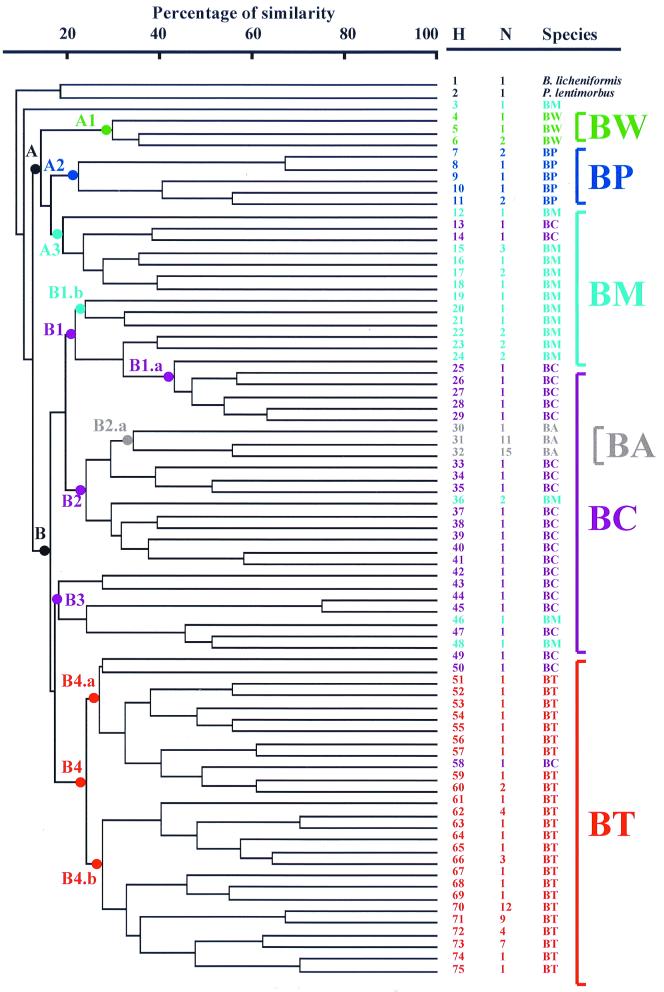
The dendrogram generated by UPGMA cluster analysis is shown in Fig. 4. Seven major subclusters (A1, A2, A3, B1, B2, B3, and B4) could be identified, with an early separation between clusters A and B. Cluster A grouped the strains of B. weihenstephanensis (subcluster A1), B. pseudomycoides (subcluster A2), and B. mycoides (subcluster A3).
FIG. 4.
Genetic relationship among B. cereus group strains as described by the UPGMA cluster analysis of the ITS-HHP patterns. The percentages of similarity among the ITS-HPP patterns were calculated using the Jaccard coefficient. H, ITS-HHP type number (Table 1); N, number of isolates for each ITS-HHP type (Table 1). Abbreviations (and colors) used to indicate the B. cereus group species are as follows: BA (grey), B. anthracis; BC (purple), B. cereus; BM (pale blue), B. mycoides; BP (blue), B. pseudomycoides; BT (red), B. thuringiensis; and BW (green), B. weihenstephanensis. The dots indicated with letter-number designations (see text) and marked with the colors identifying the six B. cereus group species were drawn on the dendrogram nodes where separated clusters and subclusters are evident.
Cluster B was subdivided into four major subclusters. Subcluster B1 included B. mycoides and several strains of B. cereus. Subcluster B2 included mainly B. cereus strains and two soil isolates of B. mycoides that yielded the same ITS-HHP pattern type. In branch B2.a of subcluster B2 (Fig. 4), there was the grouping of the 27 strains of B. anthracis, confirming it to be a relatively uniform and clonal species. The subcluster B3 included several B. cereus strains as well as two strains of B. mycoides. Subcluster B4 included all 58 strains of B. thuringiensis plus three strains of B. cereus. The subcluster B4 was divided into two main groups, B4.a and B4.b.
The 27 B. anthracis strains were grouped in the same three groups observed by agarose gel electrophoresis (Fig. 3 and 4). The simplest profile was one from strains isolated from the recent anthrax outbreaks that occurred in the French Pyrenees and Alps in 1994 and 1997. These strains could be differentiated from the other species of the B. cereus group by the differential migration of the 250-bp bands and/or the 500-bp band. The second group of B. anthracis included the classic strains used for the development of vaccines and several strains isolated from France that showed two additional typical signature bands around 450 bp. Strain Davis TE702 had the most diverse band pattern, with a signature band of about 420 bp and six additional bands in the upper part of the gel.
Our ITS-HHP analysis data indicate that it is easy to discriminate B. anthracis from related species on chromosomally located sequences. The identification of three lineages in the species suggests that ITS-HHP could be of value in tracing the spread of this pathogen and its genomic differentiation. For example, the strains isolated from the anthrax outbreaks in the French Pyrenees and Alps in 1994 and 1997 (48) are different from all the other strains isolated in France, and it can be hypothesized that (i) they are traces of the introduction of a B. anthracis strain quite different from the typical strains found in France, or (ii) they derive from other French strains, differing from them due to the loss of ribosomal operons or to a rearrangement of the ITS sequence. This latter possibility could, perhaps, be clarified through a survey of ITS-HHP on a wider collection of B. anthracis strains. In any case, ITS-HHP analysis provides a tool in addition to amplified fragment length polymorphism and the multiple-locus variable-number tandem repeat analysis applied by Keim et al. (36, 37) to analyze the polymorphism of this species.
The 23 B. cereus strains all had different band patterns characterized by one or two bands around 250 bp, one band around 500 bp (480 to 520 bp), and several other bands of different sizes (Fig. 3). The main differences in the band patterns were due to the presence or absence of the second band at 250 bp and differences in the migration of the 500-bp band.
Most of the 58 B. thuringiensis strains were, in general, characterized by the presence of several bands in the lower part of the profile (below 300 bp), allowing us to distinguish 24 ITS-HHP types (Table 1 and Fig. 3 and 4). Some important B. thuringiensis subspecies known to be genetically related were grouped together by ITS-HHP. For example, strains belonging to the B. thuringiensis subspecies aizawai, galleriae, kurstaki, and tolworthi showed the same ITS-HHP patterns, in agreement with the ribotyping data of Priest et al. (49), the M13 DNA fingerprinting data of Miteva et al. (40), and the DNA-DNA hybridization data of Nakamura (42). The fact that the ITS-HPP patterns of these subspecies were identical reflected their close DNA relatedness (90%) (42), while the B. thuringiensis subspecies darmstadiensis, israelensis, and sotto, which have relatively low DNA-DNA hybridization levels (below 70%) (42), showed different ITS-HPP types and were assigned to a cluster different from that of B. thuringiensis subsp. kurstaki and its related subspecies. The different strains of the B. thuringiensis subspecies thuringiensis were grouped in different branches of the dendrogram, confirming the genetic variability revealed by Priest et al. by ribotyping analysis (49). Interestingly, strains with relevant phenotypic characteristics such as antifungal or antibacterial activity were found to be clustered differently: strains displaying antibacterial activity due to the production of bacteriocins such as B. thuringiensis strain HD868 (46), strain HD2 (19), and strain BMG1.7 (A. Cherif, H. Ouzari, D. Daffonchio, M. Gtari, A. Hassen, S. Jaoua, and A. Boudabous, unpublished data) and B. cereus strain 5148 (45) appeared far apart, while BX strains with anti-Fusarium activity (Sadfi Najla, personal communication) were clustered together. This could be a reflection of the restricted area (Tunisia) from which the anti-Fusarium strains were isolated. In contrast, there was marked variation in the 11 B. thuringiensis strains isolated in Jordan; all displayed anti-Culex activity (Hala Khyami-Horani, personal communication) but were clustered in eight different ITS-HHP types (types 61 to 65 and 67 to 69, Table 1). Probably the bacteriocinogenic and anti-Culex activity of these strains in different genetic backgrounds is due to horizontal gene transfer of the genetic determinants for activity, such as cry genes, that are frequently carried on plasmids and are hence potentially transferable to other strains (19, 20).
The strains of B. mycoides and B. pseudomycoides showed a wide migration shift in the 530-bp band that ranged from 450 to 530 bp (Fig. 3). In the rest of the ITS-HHP pattern, the strains were relatively similar. Strain G2 showed a very different ITS-HHP pattern type with several additional bands in the upper and lower parts of the gel. For the 21 strains of B. mycoides tested, 14 different ITS-HHP types were detected. Six ITS-HHP pattern types were observed among the eight B. pseudomycoides strains examined. Different ITS-HHP types were found for the B. mycoides type strains deriving from different culture collections (strain NRS273T from the Agricultural Research Service Culture Collection and strain 2048T from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Table 1). This could be due to the different manipulations that the two strains had undergone for maintenance in the different collections. B. pseudomycoides was related to B. mycoides, having phenotypic characteristics of rhizoid growth in common. The few strains of B. pseudomycoides analyzed in this study were separated from B. mycoides and grouped in a separate branch of the dendrogram (Fig. 4). In agreement with the DNA-DNA hybridization results of Nakamura and Jackson (43, 44), who found two main groups of strains sharing 75% DNA relatedness, the B. pseudomycoides strains analyzed here were clustered by ITS-HHP analysis in two well-separated groups, the first including strains BD10 and BD14 and the second including the type strain B617T (Table 1 and Fig. 4).
The four strains of B. weihenstephanensis were revealed to have three different ITS-HHP patterns (Table 1 and Fig. 3 and 4).
The study has shown that for strain typing the ITS-HHP approach is advantageous with respect to direct sequence comparison in that it takes into consideration, and highlights, sequence polymorphism among all the amplified operons of the genome. With the ITS-HHP analysis, the ribosomal operon diversity in the B. cereus group appeared greater than previously estimated. ITS-HHP analysis can also be used to address the identification of uncharacterized isolates. The method permits the rapid identification of B. anthracis isolates on the basis of chromosomal traits and independent of the virulence determinants located on the pXO plasmids that may be lost by the bacteria (7, 20, 47, 51, 52, 59, 60). In agreement with recent data (27), the genetic relationships within the B. cereus group profiled by the ITS-HHP analysis further support the idea that B. cereus, B. anthracis, and B. thuringiensis are members of a single species (B. cereus sensu lato) that has achieved specialization by acquiring particular plasmid-encoded phenotypes such as pathogenicity. B. weihenstephanensis, B. mycoides, and B. pseudomycoides are the most divergent species (36).
Acknowledgments
Partial support came from European Community funds in the ambit of the ELECTRO project “Electrochemical treatment of fresh animal manure for reducing environment and health risk” (EC contract FAIR5-PL97-3506) and from the Italian Ministry of the Environment and Italian National Research Council in the ambit of the project “Biodiversità e Organismi Geneticamente Modificati” (contributo di ricerca no. 00.00077.PG04). A.C. was supported by a grant from the Direction Generale de Recherche Scientifique et Technologique of the Ministere de l'Education Superieure of Tunisia.
We thank Claudia Sorlini for making her laboratory available to us to carry out this work.
We thank Michéle Mock and Guy Patra for kindly giving us the total DNA of B. anthracis strains; Abdellatif Boudabous for the reference strains of B. thuringiensis; Samir Jaoua, for the B. thuringiensis BUMP strains; Hala Khyami-Horani for the anti-Culex strains isolated in Jordan; Sadfi Najla for the antifungal strains isolated in Tunisia; Lawrence K. Nakamura for the B. pseudomycoides strains; Siegfried Scherer and Ralf Mayr for providing us with the strains of B. weihenstephanensis; and Federico Uruburu (Colección Española de Cultivos Tipo) and Daniel R. Zeigler (Bacillus Genetic Stock Center), for the generous gift of bacteriocinogenic strains of B. thuringiensis and B. cereus. We are also indebted to Mario Luini and Silvia Grassi, who allowed us to cultivate two B. anthracis strains for DNA extraction at the Istituto Zooprofilattico of Milan. We thank Diego Mora for his help in the Southern hybridization experiments and for helpful discussions. The manuscript was edited by Barbara Carey.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Andreoni V, Moro Luischi M, Cavalca L, Erba D, Ciapellano S. Selenite tolerance and accumulation in the Lactobacillus species. Ann Microbiol. 2000;50:77–88. [Google Scholar]
- 2.Ash C, Collins M D. Comparative analysis of 23S ribosomal RNA gene sequences of Bacillus anthracis and emetic Bacillus cereus determined by PCR-direct sequencing. FEMS Microbiol Lett. 1992;94:75–80. doi: 10.1016/0378-1097(92)90586-d. [DOI] [PubMed] [Google Scholar]
- 3.Ash C, Farrow J A E, Dorsch M, Stackebrandt E, Collins M D. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int J Syst Bacteriol. 1991;41:343–346. doi: 10.1099/00207713-41-3-343. [DOI] [PubMed] [Google Scholar]
- 4.Ash C, Farrow J A E, Wallbanks S, Collins M D. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett Appl Microbiol. 1991;13:202–206. [Google Scholar]
- 5.Bassam B J, Caetano-Anolles G, Gresshoff P M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;80:81–84. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- 6.Baudart J, Lemarchand K, Brisabois A, Lebaron P. Diversity of Salmonella strains isolated from the acquatic environment as determined by serotyping and amplification of the ribosomal DNA spacer regions. Appl Environ Microbiol. 2000;66:1544–1552. doi: 10.1128/aem.66.4.1544-1552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beyer W, Pocivalsek S, Böhm R. Polymerase chain reaction-ELISA to detect Bacillus anthracis from soil samples—limitations of present published primers. J Appl Microbiol. 1999;87:229–236. doi: 10.1046/j.1365-2672.1999.00875.x. [DOI] [PubMed] [Google Scholar]
- 8.Brousseau R, Saint-Onge A, Prefontaine G, Masson L, Cabana J. Arbitrary primer polymerase chain reaction, a powerful method to identify Bacillus thuringiensis serovars and strains. Appl Environ Microbiol. 1993;59:114–119. doi: 10.1128/aem.59.1.114-119.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson C R, Caugant D A, Kolstø A-B. Genotypic diversity among Bacillus cereus and Bacillus thuringiensis strains. Appl Environ Microbiol. 1994;60:1719–1725. doi: 10.1128/aem.60.6.1719-1725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson C R, Grønstad A, Kolstø A-B. Physical maps of the genomes of three Bacillus cereus strains. J Bacteriol. 1992;174:3750–3756. doi: 10.1128/jb.174.11.3750-3756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson C R, Johansen T, Kolstø A-B. The chromosome map of Bacillus thuringiensis subsp. canadensis HD224 is highly similar to that of the Bacillus cereus type strain ATCC 14579. FEMS Microbiol Lett. 1996;141:163–167. doi: 10.1111/j.1574-6968.1996.tb08379.x. [DOI] [PubMed] [Google Scholar]
- 12.Carlson C R, Kolstø A-B. A complete physical map of a Bacillus thuringiensis chromosome. J Bacteriol. 1993;175:1053–1060. doi: 10.1128/jb.175.4.1053-1060.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlson C R, Kolstø A-B. A small (2.4 Mb) Bacillus cereus chromosome corresponds to one conserved region of a larger (5.3 Mb) Bacillus cereus chromosome. Mol Microbiol. 1994;13:161–169. doi: 10.1111/j.1365-2958.1994.tb00411.x. [DOI] [PubMed] [Google Scholar]
- 14.Daffonchio D, Borin S, Consolandi A, Mora D, Manachini P L, Sorlini C. 16S–23S rRNA internal transcribed spacers as molecular markers for the species of the 16S rRNA group I of the genus Bacillus. FEMS Microbiol Lett. 1998;163:229–236. doi: 10.1111/j.1574-6968.1998.tb13050.x. [DOI] [PubMed] [Google Scholar]
- 15.Daffonchio D, Borin S, Consolandi A, Sorlini C. Restriction site insertion-PCR (RSI-PCR) for rapid discrimination and typing of closely related microbial strains. FEMS Microbiol Lett. 1999;180:77–83. doi: 10.1111/j.1574-6968.1999.tb08780.x. [DOI] [PubMed] [Google Scholar]
- 16.Daffonchio D, Borin S, Frova G, Gallo R, Mori E, Fani R, Sorlini C. A randomly amplified polymorphic DNA marker specific for the Bacillus cereus group is diagnostic for Bacillus anthracis. Appl Environ Microbiol. 1999;65:1298–1303. doi: 10.1128/aem.65.3.1298-1303.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daffonchio D, Borin S, Frova G, Manachini P L, Sorlini C. PCR fingerprinting of whole genomes, the spacers between the 16S and 23S rRNA genes and of intergenic tRNA gene regions reveals a different intraspecific genomic variability of Bacillus cereus and Bacillus licheniformis. Int J Syst Bacteriol. 1998;48:107–116. doi: 10.1099/00207713-48-1-107. [DOI] [PubMed] [Google Scholar]
- 18.Drobniewski F A. Bacillus cereus and related species. Clin Microbiol Rev. 1993;6:324–338. doi: 10.1128/cmr.6.4.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Favret M E, Yousten A A. Thuricin: the bacteriocin produced by Bacillus thuringiensis. J Invertebr Pathol. 1989;53:206–216. doi: 10.1016/0022-2011(89)90009-8. [DOI] [PubMed] [Google Scholar]
- 20.Gonzales J M J, Brown B S, Carlton B C. Transfer of Bacillus thuringiensis plasmids coding for δ-endotoxin among strains of Bacillus thuringiensis and Bacillus cereus. Proc Natl Acad Sci USA. 1982;79:6951–6955. doi: 10.1073/pnas.79.22.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grompe M. The rapid detection of unknown mutation in nucleic acids. Nat Genet. 1993;5:111–117. doi: 10.1038/ng1093-111. [DOI] [PubMed] [Google Scholar]
- 22.Gürtler V. The role of recombination and mutation in 16S–23S rDNA spacer rearrangement. Gene. 1999;238:241–252. doi: 10.1016/s0378-1119(99)00224-3. [DOI] [PubMed] [Google Scholar]
- 23.Gürtler V, Rao Y, Pearson S R, Bates S M, Mayall B C. DNA sequence heterogeneity in three copies of the long 16S–23S rDNA spacer of Enterococcus faecalis isolates. Microbiology. 1999;145:1785–1796. doi: 10.1099/13500872-145-7-1785. [DOI] [PubMed] [Google Scholar]
- 24.Gürtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S–23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 25.Helgason E, Caugant D A, Lecadet M M, Chen Y, Mahillon J, Lövgren A, Hegna I, Kvaloy K, Kolstø A-B. Genetic diversity of Bacillus cereus/Bacillus thuringiensis isolates from natural sources. Curr Microbiol. 1998;37:80–87. doi: 10.1007/s002849900343. [DOI] [PubMed] [Google Scholar]
- 26.Helgason E, Caugant D A, Olsen I, Kolstø A-B. Genetic structure of population of Bacillus cereus and B. thuringiensis isolates associated with periodontitis and other human infections. J Clin Microbiol. 2000;38:1615–1622. doi: 10.1128/jcm.38.4.1615-1622.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helgason E, Økstad O A, Caugant D A, Johansen H A, Fouet A, Mock M, Hegna I, Kolstø A-B. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl Environ Microbiol. 2000;66:2627–2630. doi: 10.1128/aem.66.6.2627-2630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson I, Duggleby C J, Turnbull P C B. Differentiation of Bacillus anthracis from other Bacillus cereus group bacteria with the PCR. Int J Syst Bacteriol. 1994;44:99–105. doi: 10.1099/00207713-44-1-99. [DOI] [PubMed] [Google Scholar]
- 29.Jackson P J, Hill K K, Laker M T, Ticknor L O, Keim P. Genetic comparison of Bacillus anthracis and its close relatives using amplified fragment length polymorphism and polymerase chain reaction analysis. J Appl Microbiol. 1999;87:263–269. doi: 10.1046/j.1365-2672.1999.00884.x. [DOI] [PubMed] [Google Scholar]
- 30.Jaoua S, Zouari N, Tounsi S, Ellouz R. Study of the delta-endotoxins produced by three recently isolated strains of Bacillus thuringiensis. FEMS Microbiol Lett. 1996;145:349–354. [Google Scholar]
- 31.Jensen M A, Hubner R J. Use of homoduplex ribosomal DNA spacer amplification products and heteroduplex cross-hybridization products in the identification of Salmonella serovars. Appl Environ Microbiol. 1996;62:2741–2746. doi: 10.1128/aem.62.8.2741-2746.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen M A, Straus N. Effect of PCR conditions on the formations of heteroduplex and single-stranded DNA products in the amplification of bacterial ribosomal DNA spacer regions. PCR Methods Appl. 1993;3:186–194. doi: 10.1101/gr.3.3.186. [DOI] [PubMed] [Google Scholar]
- 33.Jensen M A, Webster J A, Straus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA polymorphism. Appl Environ Microbiol. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansen T, Carlson C R, Kolstø A-B. Variable numbers of rRNA operons in Bacillus cereus strains. FEMS Microbiol Lett. 1996;136:325–328. doi: 10.1111/j.1574-6968.1996.tb08068.x. [DOI] [PubMed] [Google Scholar]
- 35.Kaneko T, Nozaki R, Aizawa K. Deoxyribonucleic acid relatedness between Bacillus anthracis, Bacillus cereus and Bacillus thuringiensis. Microbiol Immunol. 1978;22:639–641. doi: 10.1111/j.1348-0421.1978.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 36.Keim P, Kalif A, Schupp J, Hill K, Travis S E, Richmond K, Adair D M, Hugh-Jones M, Kuske C R, Jackson P. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J Bacteriol. 1997;179:818–824. doi: 10.1128/jb.179.3.818-824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keim P, Price L B, Klevytska A M, Smith K L, Schupp J M, Okinaka R, Jackson P J, Hugh-Jones M E. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J Bacteriol. 2000;182:2928–2936. doi: 10.1128/jb.182.10.2928-2936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y-R, Czajka J, Batt C A. Development of a fluorogenic probe-based PCR assay for detection of Bacillus cereus in nonfat dry milk. Appl Environ Microbiol. 2000;66:1453–1459. doi: 10.1128/aem.66.4.1453-1459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lechner S, Mayr R, Francis K P, Pruss B M, Kaplan T, Wiessner-Gunkel E, Stewart G S, Scherer S. Bacillus weihenstephanensis sp. nov. is a new psychrotolerant species of the Bacillus cereus group. Int J Syst Bacteriol. 1998;48:1373–1382. doi: 10.1099/00207713-48-4-1373. [DOI] [PubMed] [Google Scholar]
- 40.Miteva V, Abadjieva A, Grigorova R. Differentiation among strains and serotypes of Bacillus thuringiensis by M13 DNA fingerprinting. J Gen Microbiol. 1991;137:593–600. [Google Scholar]
- 41.Nagpal M L, Fox K F, Fox A. Utility of 16S–23S rRNA spacer region methodology: how similar are interspace regions within a genome and between strains for closely related organisms? J Microbiol Methods. 1998;33:211–219. [Google Scholar]
- 42.Nakamura L K. DNA relatedness among Bacillus thuringiensis serovars. Int J Syst Bacteriol. 1994;44:125–129. doi: 10.1099/00207713-44-1-125. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura L K, Jackson M A. Clarification of the taxonomy of Bacillus mycoides. Int J Syst Bacteriol. 1995;45:46–49. [Google Scholar]
- 44.Nakamura L K. Bacillus pseudomycoides sp. nov. Int J Syst Bacteriol. 1998;48:1031–1034. doi: 10.1099/00207713-48-3-1031. [DOI] [PubMed] [Google Scholar]
- 45.Oscariz J C, Lasa I, Pisabarro A G. Detection and characterization of cerein 7, a new bacteriocin produced by Bacillus cereus with a broad spectrum of activity. FEMS Microbiol Lett. 1999;178:337–341. doi: 10.1111/j.1574-6968.1999.tb08696.x. [DOI] [PubMed] [Google Scholar]
- 46.Paik H D, Bae S S, Park S H, Pan J G. Identification and partial characterization of tochicin, a bacteriocin produced by Bacillus thuringiensis subsp. tochigiensis. J Ind Microbiol Biotechnol. 1997;19:294–298. doi: 10.1038/sj.jim.2900462. [DOI] [PubMed] [Google Scholar]
- 47.Patra G, Sylvestre P, Ramisse V, Thérasse J, Guesdon J-L. Isolation of a specific chromosomic DNA sequence of Bacillus anthracis and its possible use in diagnosis. FEMS Immunol Med Microbiol. 1996;15:223–231. doi: 10.1111/j.1574-695X.1996.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 48.Patra G, Vaissaire J, Weber-Levy M, Le Doujet C, Mock M. Molecular characterization of Bacillus strains involved in outbreaks of anthrax in France in 1997. J Clin Microbiol. 1998;36:3412–3414. doi: 10.1128/jcm.36.11.3412-3414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Priest F G, Kaji D A, Rosato Y B, Canhos V P. Characterization of Bacillus thuringiensis and related bacteria by ribosomal RNA gene restriction fragment polymorphisms. Microbiology. 1994;140:1015–1022. doi: 10.1099/13500872-140-5-1015. [DOI] [PubMed] [Google Scholar]
- 50.Pruss B M, Francis K P, von Stetten F, Scherer S. Correlation of 16S ribosomal DNA signature sequences with temperature-dependent growth rates of mesophilic and psychrotolerant strains of the Bacillus cereus group. J Bacteriol. 1999;181:2624–2630. doi: 10.1128/jb.181.8.2624-2630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramisse V, Patra G, Garrigue H, Guesdon J-L, Mock M. Identification and characterization of Bacillus anthracis by multiplex PCR analysis of sequences on plasmids pXO1 and pXO2 and chromosomal DNA. FEMS Microbiol Lett. 1996;145:9–16. doi: 10.1111/j.1574-6968.1996.tb08548.x. [DOI] [PubMed] [Google Scholar]
- 52.Ramisse V, Patra G, Vaissaire J, Mock M. The Ba813 chromosomal DNA sequence effectively traces the whole Bacillus anthracis community. J Appl Microbiol. 1999;87:224–228. doi: 10.1046/j.1365-2672.1999.00874.x. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 54.Schraft H, Griffith M W. Specific oligonucleotide primers for detection of lecitinase-positive Bacillus spp. by PCR. Appl Environ Microbiol. 1995;61:98–102. doi: 10.1128/aem.61.1.98-102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schraft H, Steele M, McNab B, Odumeru J, Griffiths M W. Epidemiological typing of Bacillus spp. isolated from food. Appl Environ Microbiol. 1996;62:4229–4232. doi: 10.1128/aem.62.11.4229-4232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sneath P H A, Sokal R R. Numerical taxonomy: the principles and practice of numerical classification. W. H. San Francisco, Calif: Freeman and Co.; 1973. [Google Scholar]
- 57.Sommerville H J, Jones M L. DNA competition experiments within the Bacillus cereus group of bacilli. J Gen Microbiol. 1972;73:257–261. doi: 10.1099/00221287-73-2-257. [DOI] [PubMed] [Google Scholar]
- 58.Stephan R. Randomly amplified polymorphic DNA (RAPD) assay for genomic fingerprinting of Bacillus cereus isolates. Int J Food Microbiol. 1996;31:311–316. doi: 10.1016/0168-1605(96)00966-x. [DOI] [PubMed] [Google Scholar]
- 59.Turnbull P C, Hutson R A, Ward M J, Jones M N, Quinn C P, Finnie N J, Duggleby C J, Kramer J M, Melling J. Bacillus anthracis but not always anthrax. J Appl Bacteriol. 1992;72:21–28. doi: 10.1111/j.1365-2672.1992.tb04876.x. [DOI] [PubMed] [Google Scholar]
- 60.Turnbull P C B. Definitive identification of Bacillus anthracis— a review. J Appl Microbiol. 1999;87:237–240. doi: 10.1046/j.1365-2672.1999.00876.x. [DOI] [PubMed] [Google Scholar]
- 61.Yamada S, Ohashi E, Agata N, Venkateswaran K. Cloning and nucleotide sequence analysis of gyrB of Bacillus cereus, B. thuringiensis, B. mycoides, and B. anthracis and their application to the detection of B. cereus in rice. Appl Environ Microbiol. 1999;65:1483–1490. doi: 10.1128/aem.65.4.1483-1490.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]