Abstract
Fabry disease is a glycosphingolipid storage disorder that is caused by a genetic deficiency of the lysosomal enzyme alpha-galactosidase A (AGA, EC 3.2.1.22). As a result, the glycolipid substrate, globotriaosylceramide (Gb3) accumulates in various cell types throughout the body producing a multisystem disease that affects the vascular, cardiac, renal, and nervous systems. A hallmark of this disorder is neuropathic pain that occurs in up to 80% of Fabry patients and has been characterized as a small fiber neuropathy. The molecular mechanism by which changes in AGA activity produce neuropathic pain is not clear, in part due to a lack of relevant model systems. Using 50B11 cells, an immortalized dorsal root ganglion neuron with nociceptive characteristics derived from rat, we used CRISPR-Cas9 gene editing of the galactosidase alpha (GLA) gene for AGA to create two stable knock-out clones that have the phenotypic characteristics of Fabry cells. The cell lines show severely reduced lysosomal AGA activity in homogenates as well as impaired degradation of Gb3 in cultured cells. This phenotype is stable over long-term culture. Similar to the unedited 50B11 cell line, the clones differentiate in response to forskolin and extend neurites. Flow cytometry experiments demonstrate that the gene-edited cells express TRPV1 pain receptor at increased levels compared to control, suggesting a possible mechanism for increased pain sensitization in Fabry patients. Our 50B11 cell lines show phenotypic characteristics of Fabry disease and grow well under standard cell culture conditions. These cell lines can provide a convenient model system to help elucidate the molecular mechanism of pain in Fabry patients.
Keywords: 50B11 cells, Alpha-galactosidase, CRISPR-Cas9, Dorsal root ganglion, Fabry disease, Neuropathy
Abbreviations: 4-MU, 4-methylumbelliferone; AGA, alpha-galactosidase A; DRG, dorsal root ganglion; ERT, enzyme replacement therapy; Gb3, globotriaosylceramide; GFP, green fluorescent protein; GLA, alpha-galactosidase A gene; HTS, high throughput screening; LR-Gb3, lissamine rhodamine Gb3; NAGA, alpha-N-acetylgalactosaminidase; NGS, normal goat serum; PAM, protospacer adjacent motif; PBS, phosphate buffered saline; rH-AGA, recombinant human-AGA; TLC, thin layer chromatography; TRPV1, transient receptor potential vanilloid family-1
1. Introduction
Glycosphingolipids are important structural components of cellular membranes. During the normal course of membrane turnover in cells, glycolipids are sequentially degraded by a series of hydrolytic enzymes found in lysosomes. Reduced activity of any one of these enzymes prevents further degradation of the glycolipid molecule with a corresponding accumulation of the lipid substrate.
Fabry disease is an X-linked glycosphingolipid storage disorder caused by a deficiency of the enzyme alpha-galactosidase A (AGA, EC 3.2.1.22). This deficiency results in the intracellular accumulation of the glycosphingolipid globotriaosylceramide (Gb3) and, to a lesser extent, other lipid analogs with alpha-galactose moieties, such as globotriaosylsphingosine (lysoGb3), a deacylated derivative of Gb3. The chronic accumulation of these substrates impairs cellular function in tissues and organs, and leads to a multisystem disease that affects the vascular, cardiac, renal, and nervous systems [1].
To date, over 985 mutations in the galactosidase alpha (GLA) gene (NM_000169.2) for AGA expression have been described [2] and mutations may occur anywhere within the 7 coding exons. Severity of the symptoms of Fabry disease is variable depending on the amount of residual activity in the mutant AGA protein [3]. Over 70% of patients have mutations that result in extremely low or absent AGA enzyme activity in their cells [4]. Known as the “classic” Fabry phenotype, these patients present with the most severe disease and have significant morbidity even in childhood [5]. In addition, because Fabry disease is an X-linked disorder, women who carry a GLA mutation can also experience symptoms of the disease depending on the severity of the mutation and the balance of X-inactivation between normal and mutant alleles in each organ [6].
One of the most debilitating symptoms of Fabry disease is severe neuropathic pain, which can be both chronic and acute. Many patients experience a constant burning pain in the palms of their hands and soles of their feet that is difficult to control with pain medication [7]. In addition, they may also have recurrent attacks of excruciating pain (“Fabry pain crises”) that occur spontaneously or in response to extreme temperatures, fever, fatigue, stress, overheating, or exercise [8]. Enzyme replacement therapy (ERT) with recombinant AGA is currently available to treat Fabry disease. However, in a three year study of ERT in Fabry patients with severe pain, enzyme treatment reduced the severity of the pain but did not completely reverse the neurologic dysfunction [9].
The pathophysiology of pain in Fabry disease is still poorly understood. The peripheral nervous system involvement is a small fiber neuropathy affecting mainly small A delta and C fibers [10]. Neuronal storage of Gb3 has been found in dorsal root ganglia (DRG) in Fabry patients at autopsy [11] and it has been proposed that this accumulation contributes to the peripheral neuropathy [9]. Recently, Choi and co-workers [12] demonstrated that exposure to lysoGb3, a deacylated derivative of the Gb3 molecule found in high concentrations in the plasma of Fabry patients, increased intracellular Ca2+ levels in primary cultures of peripheral sensory neurons from Fabry mice and increased pain response when applied to the foot pads of healthy mice. These results suggest another possible mechanism of pain sensitization in Fabry patients.
With the development of the CRISPR-Cas9 technique for gene editing, it is now possible to target the GLA gene to create genetically engineered cell lines that can be used to study the molecular mechanisms of neuronal dysfunction in Fabry disease compared to normal isogenic controls. 50B11 is an immortalized cell line created by transforming E14.5 rat primary DRG neurons with SV40 large T-antigen and human telomerase reverse transcriptase [13]. When differentiated with forskolin, 50B11 cells express the characteristics of small fiber nociceptor sensory neurons including extension of axonal processes and increased gene expression for neurotrophin receptors and voltage-gated ion channels, including transient receptor potential vanilloid family-1 (TRPV1) receptors [13,14]. Because these cells can be cultured easily using standard laboratory techniques, we used CRISPR-Cas9 gene editing to disrupt the GLA gene for AGA in 50B11 cells to create an in vitro model that can be used to help elucidate the molecular mechanisms of peripheral neuropathy in Fabry disease and can potentially be used to screen for new treatments.
2. Methods
2.1. Cell culture
50B11 cells were a gift from Dr. Ahmet Höke, Department of Neurology, School of Medicine, Johns Hopkins University, Baltimore, MD, USA. They were maintained on Neurobasal medium (Invitrogen, Grand Island, New York) supplemented with 10% heat-inactivated GemCell Super Calf Serum (GCS, Gemini Bioproducts, Sacramento, CA), 0.5 mM glutamine, 2% B-27 supplement, and 0.2% glucose. For some experiments DMEM/F12 medium was used instead of Neurobasal medium. Growth medium was replaced every 2–3 days, and the cells were subcultured when approximately 80% confluent.
2.2. CRISPR-Cas9 gene editing
50B11 cells were transfected with an all-in-one CRISPR-Cas9 plasmid (plasmid #RCP285763CG041B, GeneCopoeia, Rockville, MD) expressing sgRNA targeting exon 1 of the rat GLA gene (GenBank accession number NM_001108820.3). The expression elements of this plasmid are Cas9 nuclease under a U6 promoter, sgRNA with a CMV promoter, the fluorescent marker copGFP as a reporter with a T2A promoter, and a neomycin resistance gene with an IRES2 promoter. 50B11 cells in log-phase growth were seeded in Costar® 12-well multiplates (Corning, Glendale, AZ) in growth medium prepared with DMEM/F12. After overnight incubation, when the cells were 80–90% confluent, two wells were transfected with 760 μg purified plasmid complexed either with Jet Prime Transfection Reagent (Polyplus-transfection Inc., New York, NY) at a 1:3 ratio in complete DMEM/F12 growth medium following manufacturer's directions, or with Lipofectamine 2000 (Life Technologies, Grand Island, NY) at a 1:2.5 ratio in DMEM/F12 medium without serum or B27 supplement according to the manufacturer's directions. After 24 h the medium was changed to complete growth medium. After 48 h both wells showed similar numbers of GFP-positive cells. Transfected wells were trypsinized to single cells, both wells were combined, and the cell suspension was sorted by GFP expression with a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). The GFP-positive cells were recovered, washed twice with phosphate buffered saline (Invitrogen), and seeded in 60 mm dishes at low density in growth medium supplemented with 10 μg/ml gentamicin to maintain selection pressure for the established neuronal phenotype [13]. When colonies became visible, well-isolated colonies were transferred to individual wells of a 24-well plate, expanded in growth medium, and screened for AGA activity. Two colonies with AGA activity <3% (clones D and E) were subjected to a second round of cloning by serial dilution.
2.3. Measurement of AGA activity in cell extracts
A standard fluorometric assay for AGA in cell extracts was performed as previously described [15]. Briefly, cell pellets were resuspended in citrate-phosphate buffer (pH 4.6) containing sodium taurocholate (5 mg/ml) and Triton-X 100 (0.1%). The suspensions were frozen (−20 °C) and thawed once, then centrifuged at 5000 X g for 5 min. Aliquots of the supernatant were incubated for one hour with 4-methylumbelliferyl-alpha-D-galactopyranoside (5 mM, Research Products International, Mount Prospect, IL) in citrate-phosphate buffer (pH 4.6) in the presence of N-acetyl-galactosamine (0.1 M, Research Products International), a specific inhibitor of alpha-N-acetylgalactosaminidase (NAGA, also known as alpha-galactosidase B) [16]. At the end of the incubation period, enzyme activity was stopped with glycine buffer (0.1 N, pH 10.6). Fluorescence in samples was determined using a CytoFluor 4000 plate reader (Applied Biosystems, Foster City, CA) with an excitation filter of 360 nm and emission filter of 490 nm. Amount of product formed was determined using 4-methylumbelliferone (4-MU) standards diluted in 0.1 N glycine stop buffer. Protein levels in extracts were determined using the BCA protein assay kit (Pierce, Rockford, IL) according to the manufacturer's instructions. AGA activity was calculated as nmoles 4-MU formed/h/mg protein and results were compared to untransfected controls included in the same assay.
For determination of NAGA activity, homogenates were assayed as for AGA only with and without inhibitor, and NAGA activity was determined by subtracting the AGA activity (with inhibitor) from total activity (without inhibitor).
For comparison, a third lysosomal hydrolase, beta-hexosaminidase, was assayed at the same time under the same conditions except 2 mM 4-methylumbelliferyl-N-acetyl-beta-D-glucosaminide (Sigma Chemical Company, St. Louis, MO) was used as substrate and incubation was stopped after 20 min.
2.4. Western blot analysis
Cultures were harvested in log-phase growth and cell pellets were extracted as described above for the AGA enzyme assay. The protein content of extracts was determined using a BCA Assay Kit (Pierce). Extracts (10 μg per lane) and molecular weight markers (SeeBlue Plus2, Invitrogen) were separated by SDS/PAGE using 4–12% Bis-Tris polyacrylamide gels, transferred to nitrocellulose membranes, and probed with rabbit anti-AGA antibody (a gift from Transkaryotic Therapies, Inc., Cambridge, MA) at a dilution of 1:5000 in TRIS-buffered saline containing 0.1% Tween-20. Protein bands were labeled using HRP-conjugated goat anti-rabbit IgG antibody (Millipore, Temecula, CA) and they were visualized using ECL substrate (Biotool.com, Houston, TX). Chemiluminescence was detected using an Amersham™ 600 digital imager (GE Healthcare Life Sciences, Pittsburgh, PA). Recombinant human AGA (rH-AGA, Transkaryotic Therapies, Inc.) was used as a positive control at 20 ng per lane.
For loading controls, blots were stripped with OneMinute® Western Blot Stripping Buffer (GM Biosciences, Frederick, MD), washed, and reprobed with goat anti-actin (1–19) antibody (Santa Cruz Biotechnology, Inc., Dallas, TX) at 1:5000, which was detected with HRP-labeled donkey anti-goat IgG (Jackson ImmunoResearch, West Grove, PA), followed by development with ECL substrate and digital imaging using an Amersham™ 600 digital imaging system.
2.5. DNA sequencing
Total DNA was extracted from cultured cells using a Genomic Wizard DNA Extraction Kit (Promega, Madison, WI). A 261 bp segment flanking the CRISPR GLA target site was amplified with Platinum® PCR SuperMix (Invitrogen Life Technologies, Waltham, MA) according to the manufacturer's protocol using the primer pair 5′-TGGAGATAACGGCCCAGTTG-3′ (forward) and 5′-GCGGACACCTACTATGGGC-3′ (reverse). PCR products were purified with a Monarch® PCR & DNA Cleanup Kit, (New England Biolabs, Ipswich, MA), checked for quality by agarose electrophoresis, and then submitted for Sanger sequencing using the forward primer to Eurofins Genomics (Louisville, KY).
2.6. Glycolipid analysis by thin layer chromatography (TLC)
Cells from one 75cm2 flask were trypsinized, washed twice with PBS, pelleted by centrifugation, and dry pellets were stored at −20 °C until use. For TLC analysis, each sample was suspended in 1000 ul of water and immediately transferred to a screw-capped Pyrex glass tube and then an aliquot was removed for protein determination. Total lipids from 4 mg of cellular protein were extracted and analyzed by a modification of the method of van Echten-Deckert [17]. Briefly lipids from the cell suspensions were extracted with 9 ml chloroform-methanol-water 60:30:7 (v/v/v) for 24 h at 50 °C. Denatured protein particles were removed by passing the sample through wadding. Phospholipids were degraded by mild alkaline hydrolysis with 2 ml methanolic sodium hydroxide (50 mM) for 2 h at 37 °C. After neutralization with acetic acid, the lipids were desalted by reversed-phase chromatography using silica gel (LiChroprop RP18, Sigma Aldrich, St. Louis, MO). Lipid samples were applied to high performance silica TLC plates (Millipore-Sigma, St. Louis, MO), and then chromatographed with chloroform-methanol-0.22% aqueous calcium chloride 60:35:8 (v/v/v). After the run, plates were dried, sprayed with copper sulfate solution, and baked at 180 °C in order to visualize the bands. Fibroblasts derived from a Fabry patient (GM-00107, NIGMS Human Genetic Cell Repository, Coriell Institute for Medical Research, Camden, NJ) were included as positive controls. Known amounts of Gb3 glycolipid standard (Matreya, LLC., Pleasant Gap, PA) were also run on each plate to produce a standard curve. Plates were imaged with a UVP Epichemi-3 darkroom (UVP, LLC., Upland, CA) and images were analyzed using GelAnalyzer 2010a software (Gelanalyzer.com). For standardization of Gb3 quantity in cells, proteins were determined using the BCA protein assay kit (Pierce) according to manufacturer's instructions and amount of Gb3 detected was normalized to total amount of protein in the original sample.
2.7. Loading of lissamine rhodamine labeled Gb3 into adherent cells
Gb3 labeled in the ceramide moiety with the fluorescent tag lissamine rhodamine (LR-Gb3) was custom synthesized by Matreya, LLC according to the method of.
Monti [18]. 50B11 cells in culture were loaded with LR-Gb3 as previously described for fibroblasts [19]. Briefly, when cultures were 80%–90% confluent, the culture medium was replaced with serum-free DMEM/F12 supplemented 1% insulin-transferrin‑selenium supplement (ITS, Sigma Chemical Co., St. Louis, MO) and 3.0 nmoles/ml LR-Gb3 complexed with bovine serum albumin. After overnight incubation, the cultures were washed twice with PBS, refed with growth medium, and incubated for an additional 48 h with daily feeding. Live cultures were imaged at 4, 24, and 48 h post-loading using a Keyence BZ-9000 fluorescence microscope with a rhodamine filter set and a 10× objective at standard exposure times.
2.8. Neuronal differentiation
For some experiments, 50B11 cells were differentiated by the addition of forskolin in reduced-serum growth medium (5% GCS) and used within 36 h as described by Chen [13] and Bhattacherjee [14]. In our hands, optimal concentration of forskolin for induction of neurites was 100 uM.
2.9. Immunostaining and flow cytometry
For immunostaining, 50B11 were seeded on 2-well glass slides coated with poly-ornithine and laminin. After overnight incubation cells were refed with differentiation medium and incubated for an additional 24 h. At the end of the incubation, they were fixed with 3% paraformaldehyde, blocked with 10% normal goat serum (NGS) in PBS, and stained by indirect immunofluorescence as previously described [20] using saponin as a permeabilizing agent. For TRPV1 expression, cells were stained with a rabbit polyclonal antibody to TRPV1 (ProteinTech, Rosemont, IL) at a dilution of 1:100 and staining was detected with Alexa Fluor 568 goat anti-rabbit IgG (Molecular Probes, Eugene, OR). For CAS9 expression, 50B11 clones and LA-N-2 neuroblastoma cells stably transfected with the Crispr-CAS9 plasmid as positive controls were stained with a mouse monoclonal antibody to Cas9 (Cell Signaling Technology, Danvers, MA) at a dilution of 1:800 and staining was detected with Alexa Fluor 594 goat anti-mouse IgG (Molecular Probes, Eugene, OR). Slides were imaged with a Keyence BZ-9000 fluorescence microscope using a 20× objective. To control for non-specific staining, all immunostaining experiments included a negative control culture that was stained in parallel substituting antibody dilution buffer for the primary antibody.
For flow cytometry analysis of TRPV1, cells were seeded in 60-mm cell culture dishes and differentiated as described above. After 24 h they were detached with TrypLE (Gibco, Gaithersburg, MD). The cells were washed once with PBS, counted, and divided into 2 aliquots of 1 million cells each in 1.5 ml centrifuge tubes. For surface staining, unfixed cells were stained in ice-cold 3% NGS in PBS as follows: one aliquot of each cell line was resuspended in 100 ul TRPV-1 antibody at a dilution of 1:100 in ice-cold 3% NGS in PBS. As a control for non-specific staining, a second aliquot of each cell line was stained in parallel with an equal concentration of rabbit IgG isotype control (Jackson ImmunoResearch Laboratories, West Grove, PA) substituted for primary antibody. After 30 min incubation on ice, cell were washed twice with 3% NGS and incubated with Alexa 488 goat anti-rabbit antibody (Molecular Probes, Eugene, OR) at a dilution of 1:200 for 30 min on ice. Cells were washed twice with cold staining buffer and resuspended in 500 ul 3% NGS in PBS. Fluorescence intensity of stained cells was measured in duplicate using the FL-1 channel of a FACSCalibur flow cytometer (BD Biosciences) and results were analyzed with Flowing Software (Turku Bioscience).
3. Results
3.1. CRISPR-Cas9 editing of GLA in 50B11 cells effectively reduced endogenous AGA activity and protein expression
To create a model of peripheral neurons with Fabry disease we transfected 50B11 cells with an all-in-one CRISPR-Cas9 plasmid expressing sgRNA targeting exon 1 of the rat GLA gene and a GFP reporter as described in Methods. The presence of the GFP reporter allowed an initial enrichment of positively expressing transfected cells by FACS sorting. Because 50B11 cells are well adapted to in vitro cultivation, we were able to efficiently expand and clone these cells to obtain homogenous populations. Of the 24 colonies first selected, 14 showed AGA activities <50% of untransfected controls, with three showing AGA activities of <10% of untransfected controls. Two colonies measured as 1% and 3% of control AGA activity were further purified by recloning using serial dilution in 96-well plates. Subclones were tested for AGA activity and response to forskolin, and the most responsive subclone from each colony, designated 50B11 clone D4W and 50B11 clone E4, were selected for further analysis. These clones no longer expressed GFP and were negative for CAS9 expression by immunostaining (Supplementary Fig. 1). We used two clones to compare results to untransfected cells (50B11 parent) to control for possible off-target effects due to transfection and cloning.
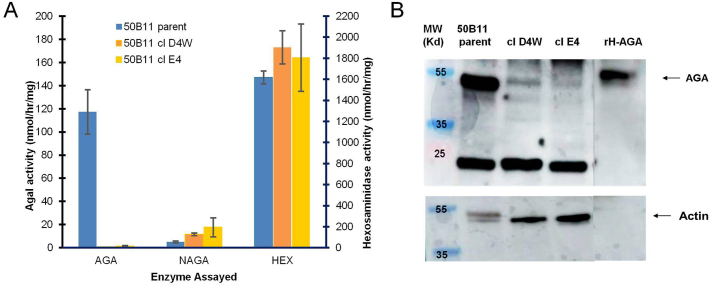
The results of a typical AGA assay for 50B11 clones D4W and E4 are shown in Fig. 1A. AGA activity was reduced to 0.6% and 1.2% of untransfected controls in clone D4W and clone E4, respectively. The specificity of the reduction of AGA activity is shown by normal levels of a second lysosomal enzyme, beta-hexosaminidase. In addition, we measured activity of the lysosomal hydrolase alpha-N-acetylgalactosaminidase (NAGA), also known as alpha-galactosidase B. Although this enzyme primarily hydrolyzes alpha-N-acetylgalactosamine residues, it has a broad substrate specificity that allows it to catalyze alpha-galactoside residues found on Gb3 [21], although with a much lower activity than that of AGA. In our clones, NAGA activity was not reduced and, in contrast, was higher than normal in both 50B11 clones (Fig. 1A), which might represent a compensatory increase in NAGA expression in response to deficient AGA activity.
Fig. 1.
Deficiency of AGA enzyme in CRISPR-Cas9 edited 50B11 cells. (A) AGA enzyme activity in cellular homogenates. Results are expressed as nmol 4mu released/h/mg cell protein. Bars are averages of duplicate cultures +/− SD. Numbers are mean activity. (B) Western blot of AGA protein expression. Amount of immunoreactive AGA was severely reduced in the gene-edited clones. rH-AGA was included as a positive control. Actin levels were used as a loading control.
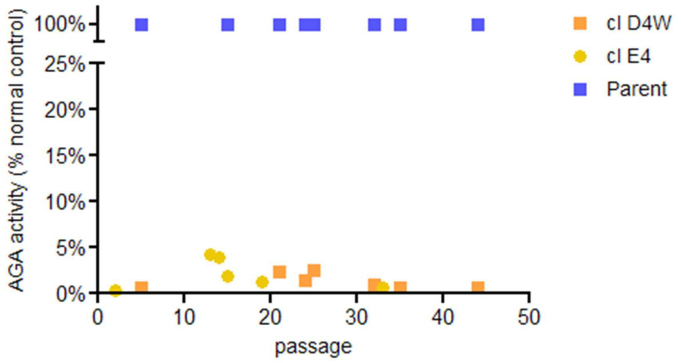
Stability of the knockdown in AGA activity in the two clones during long-term culture was confirmed by repeated AGA measurements over time (Fig. 2). AGA activity relative to untransfected controls was measured as 0.6% in both early passage (p1) and late passage (p44) cultures for clone D4W and as 0.3% in early passage (p2) and 0.7% in late passage (p33) cultures for clone E4. Thus both clones maintained a greater than 99% reduction in AGA activity over many cell divisions.
Fig. 2.
Stability of AGA activity over time in culture. AGA activity was determined for sample cultures at various intervals during long-term culture. Each passage represents a transfer of approximately 1/20 of the cells to a fresh culture flask. AGA activity was determined as described in Methods and results were expressed as a percentage of activity found in normal cells included in the same assay.
To determine whether the lack of AGA activity in our clones was the result of the absence of AGA protein or the expression of a mutant protein, we performed a Western blot of AGA protein levels as described in Methods. The results show that both clones lacked detectable levels of AGA protein compared to untransfected controls (Fig. 1B).
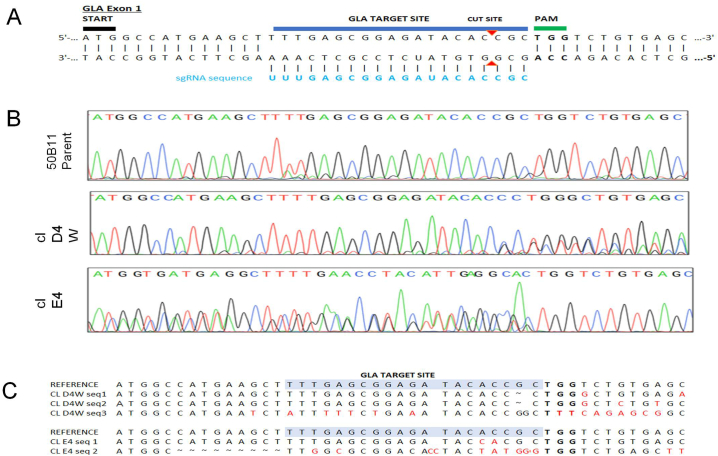
Chromatograms from Sanger sequencing of the PCR products flanking the DNA CRISPR target site showed disruption of the sequence of GLA exon 1 in the chromosomal DNA around the predicted cut site in both clones (Fig. 3B), producing a complex chromatogram trace for each clone. By contrast, Sanger sequencing of the untransfected control, 50B11 parent, produced an even trace with the expected sequence (Fig. 3B). Using the web-based program, CRISP-ID [22], it is possible to deconvolve chromatograms containing mixtures of sequences by comparing the results to the normal control. The probable mutant sequences are shown in Fig. 3C and represent a mixture of insertions, deletions, and base changes, any of which would disrupt the function of the gene. Because 50B11 cells are tetraploid (CellLine Genetics, Madison, WI), the four X-chromosomes present give four potential CRISPR target sites per cell, only one of which will be on the active X chromosome. Therefore the exact mutation resulting in decreasedfunction of AGA cannot be determined.
Fig. 3.
Sequencing of GLA exon 1 target site in 50B11 cells. (A) Sequence for the GLA gene target. The start codon of exon 1 is highlighted by the black bar. The sgRNA sequence (5′-UUUGAGCGGAGAUACACCGC-3′) is labeled in light blue. The DNA sequence under the dark blue bar indicates the DNA target site. The upstream protospacer adjacent motif (PAM) is indicated by a green bar and the double-strand break site is indicated with red triangles. (B) Chromatograms from Sanger sequencing of chromosomal DNA for 50B11 parent and clones D4W and E4. The sequencing chromatograms for each cell line showing the base calls for the highest peak are shown aligned with the target sequence in (A). Both gene-edited clones have complex chromatograms indicating a mixture of mutations while the untransfected controls (50B11 parent) show a normal sequence. (C) Probable sequence changes of clones based on deconvolution of chromatograms using CRISP-ID.
3.2. 50B11 clones D4W and E4 accumulate Gb3 in culture and are slow to degrade labeled Gb3 in intact cells
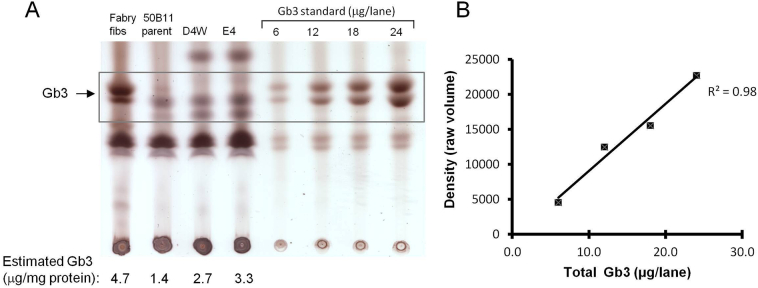
One of the hallmarks of the Fabry phenotype is the accumulation of the glycolipid Gb3 in the cells of Fabry patients. Therefore, we analyzed Gb3 levels in our cell lines by TLC. Under our culture conditions using 10% bovine serum in the culture medium, analysis of the lipid content of the 50B11 clones and the untransfected controls demonstrated detectable levels of Gb3 in all cell lines tested (Fig. 4A).
Fig. 4.
Glycosphingolipid (GSL) storage in 50B11 cells. (A) Extracted GSLs from cultured 50B11 cells were analyzed by TLC using a solvent system of chloroform-methanol-0.22% aqueous calcium chloride (60:35:8, v/v/v) as described in Methods. Fibroblasts (fibs) derived from a Fabry patient were used as positive controls. Known amounts of Gb3 standards were loaded on the same plate to allow estimation of Gb3 quantity in samples by spot densitometry. Gb3 levels for samples shown below the lanes were derived from a standard curve and normalized to the amount of protein in the original sample. (B) Standard curve shows linear correlation of intensity of bands to amount of Gb3 standard loaded.
In standards run simultaneously, density of the spots was proportional to the amount of Gb3 present (Fig. 4B), allowing a semi-quantitative estimate of the amount of Gb3 in the samples. When Gb3 levels in cells were standardized to amount of cellular protein loaded, the 50B11 clones showed approximately a 2-fold increase in the concentration of Gb3 compared to untransfected controls (Fig. 4A).
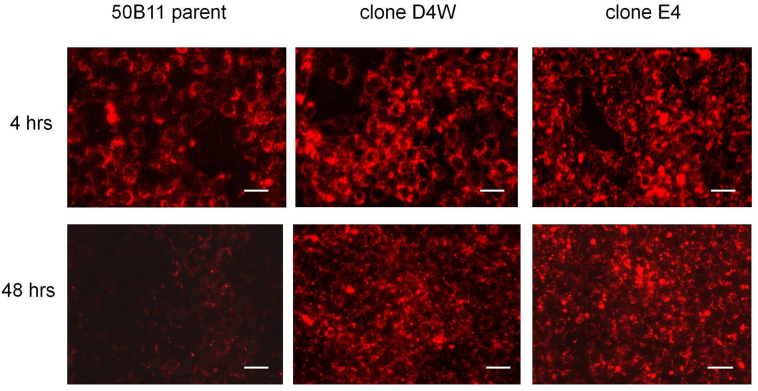
Further evidence for the functional changes in degradation of Gb3 in intact cells is shown in Fig. 5.
Fig. 5.
Uptake and degradation of labeled Gb3 in intact cells. Cells were loaded overnight with fluorescent Gb3 (LR-Gb3) as described in Methods and cultured for 48 h after loading. Live cultures were imaged with identical exposure times at each time point using a Keyence 9000 microscope with a rhodamine filter set. Scale bar = 50 μm.
50B11 cells were loaded overnight with fluorescently-labeled Gb3 as described in Methods. Previously, we demonstrated degradation of this substrate is significantly impaired in fibroblasts and endothelial cells from Fabry patients [19]. Two days after loading, essentially all the labeled Gb3 was cleared in the untransfected controls while the 50B11 clones still showed significant retention of the fluorescent lipid.
3.3. Both the 50B11 parental line and clones D4W and E4 highly express the pain receptor TRPV1 after differentiation
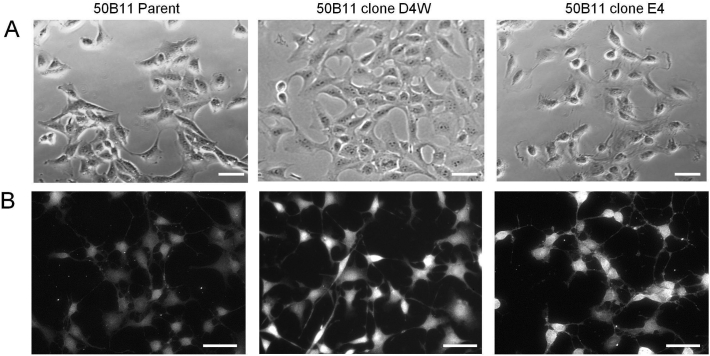
In the absence of forskolin, 50B11 cells appear as relatively undifferentiated cells with occasional processes (Fig. 6A).
Fig. 6.
Differentiation of 50B11 cells and staining with TRPV1. (A) Brightfield images of living undifferentiated 50B11 cell cultures in growth medium without forskolin. Images were taken with a Zeiss Axiovert microscope. Scale bar = 50 μm. (B) 50B11 cells 24 h after the addition of 100 uM forskolin in growth medium containing 5% serum. Cells were fixed and stained with antibody to TRPV1 pain receptor. Note the extension of neurites from the cell bodies and the bright staining for TRPV1 in all cultures. Negative controls in which primary antibody was omitted were blank at this exposure time (image not shown). Scale bar = 50 μm.
Within hours after the addition of forskolin, the cells start to extend neurites and adopt a more bipolar morphology [13]. Both of our 50B11 clones D4W and E4 retained the ability to differentiate in the presence of forskolin forming rounded cell bodies and numerous extensions (Fig. 6B).
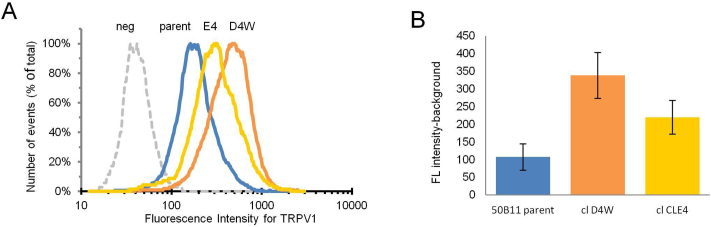
In addition, both the parental 50B11 cell line and the CRISPR-Cas9 edited clones highly express the TRPV1 pain receptor after differentiation as shown by immunostaining with an antibody specific for TRPV1 (Fig. 6B). Semi-quantitative analysis of surface pain receptor expression by flow cytometry showed the staining with TRPV1 antibody and Alexa Fluor 568 secondary antibody was easily detectable above background (Fig. 7A).
Fig. 7.
Flow cytometric quantitation of TRPV1 staining intensity. (A) Histograms showing distribution of fluorescence intensity of surface staining for TRPV1 in 50B11 parent, clone D4W, and clone E4. Dotted line (neg) represents negative control in which primary antibody was omitted. (B) Comparison of the staining intensity for TRPV1 for each cell line. Assay was performed with duplicate samples. Bars are the geometric mean of the fluorescence intensity with background value subtracted. Error bars represent +/− SD. Numbers are data values.
Intensity of staining in both 50B11 clone D4W and clone E4 was increased by 2- to 3-fold relative to the 50B11 parent line (Fig. 7B). However, the differences did not achieve statistical significance using Student's t-test.
4. Discussion
Up to 80% of Fabry disease patients develop a peripheral neuropathy that greatly affects their quality of life [23]. The mechanism of this neuropathy is not well understood, in part because of a lack of cellular models that can be used to study the molecular mechanisms of this neuropathic pain in detail. Several groups have used primary cultures of DRG neurons from Fabry mouse [12,24,25] and rat [26] models to study functional and morphologic changes in isolated neuronal cultures. However, primary DRG cultures are technically challenging, do not proliferate, and require the continuous maintenance of an animal colony for source tissues.
We report here the use of CRISPR-Cas9 gene-editing of 50B11 cells, an immortalized rat DRG cell line with nociceptive properties [13], to create AGA-deficient clones that model the metabolic defect in Fabry disease and can be maintained in long-term culture. Our gene-edited cell lines demonstrate the phenotypic characteristics of cells derived from Fabry patients. As we have shown, our model cells have a specific, severe reduction in lysosomal AGA enzyme activity in cell homogenates, as well as reduced expression of AGA protein by Western blot. The 50B11 clones accumulate the AGA enzyme's glycolipid substrate, Gb3, under standard culture conditions and can take up, but not degrade, exogenous Gb3 when it is added to intact cells.
50B11 cells can be rapidly differentiated into a more neuronal phenotype by incubating the cells in growth medium with forskolin [13]. This treatment results in the development of neurites and increased expression of receptors characteristic of nociceptive, small-diameter sensory neurons, including expression of TRPV1 [13]. The TRPV1 receptor, also known as the capsaicin receptor, is found in the plasma membrane and functions in the perception of pain and heat [27].
Previous groups have demonstrated increased levels of TRPV1 expression in whole DRG from Fabry mice by immunohistochemistry [25] and in primary DRG cultures from mouse [25,28] using immunostaining and Western blot. We demonstrated that our genetically-modified clones differentiate in response to forskolin and express TRPV1. Consistent with findings from mouse models, we also found increased expression of this pain receptor in our cell lines. However, further experiments are needed to determine the significance of this increase in our cell lines.
Unlike many neuronal cell lines and primary DRG cultures, 50B11 cells grow well in standard medium, do not need special substrates, and can be differentiated easily; therefore, they are very well suited for high throughput screening (HTS) of potential modulators and treatments for neuropathic pain [29]. As we have shown, the stability of the genetic editing in our model cells is maintained over long-term culture, allowing the expansion of the cells in large numbers needed for HTS. The development and characterization of two clonal cell lines, as well as the availability of the isogenic 50B11 normal control, allows comparison of morphological and functional differences between normal and affected cells and comparison of findings between affected cell lines. In addition, comparison of transcriptomic profiles among the cell lines could yield valuable insights into pathogenic mechanisms.
Development of a neuronal model of Fabry disease with both defects in glycolipid degradation and expression of nociceptive receptors is important to understanding the pathophysiology of this disease. The establishment of these new cell models will help further the understanding and treatment of the neuropathic pain that affects Fabry patients and will help lessen the burden of this disease.
The following is the supplementary data related to this article.
50B11 clone D4W and clone E4 cells and stably-transfected LA-N-2 control cells were seeded on 2‑well chamber slides. All cultures were stained for Cas9 protein (red) as described in Methods. GFP protein is constitutively expressed in transfected cells (green). Nuclei were stained with DAPI to demonstrate the presence of cells in the field (blue). Cells were imaged with a 20X objective in a Keyence-9000 microscope at equal exposure times for each channel for controls and clones. There were no detectable cells that were positive for Cas9 protein or GFP expression in either of the 50B11 clones. Scale bar = 50 microns.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by the NINDS Intramural Research Program and NIDDK Intramural Research Program at the National Institutes of Health, Bethesda, MD, and by Pfizer Inc., New York, NY (grant number #51755565 to UHS). The authors also gratefully acknowledge the help and advice of Prof. Dr. Konrad Sandhoff, The Life & Medical Sciences Institute, University of Bonn, Bonn, Germany.
References
- 1.Tuttolomondo A., et al. Anderson-Fabry disease: a multiorgan disease. Curr. Pharm. Des. 2013;19:5974–5996. doi: 10.2174/13816128113199990352. [DOI] [PubMed] [Google Scholar]
- 2.Stenson P.D., et al. The human gene mutation database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017;136:665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Germain D.P., et al. Use of a rare disease registry for establishing phenotypic classification of previously unassigned GLA variants: a consensus classification system by a multispecialty Fabry disease genotype–phenotype workgroup. J. Med. Genet. 2020;57:542–551. doi: 10.1136/jmedgenet-2019-106467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duro G., et al. Mutations in the GLA gene and LysoGb3: is it really Anderson-Fabry disease? Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19123726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ries M., et al. Pediatric Fabry disease. Pediatrics. 2005;115:e344–e355. doi: 10.1542/peds.2004-1678. [DOI] [PubMed] [Google Scholar]
- 6.Deegan P.B., Bähner F., Barba M., Hughes D.A., Beck M. In: Fabry Disease: Perspectives from 5 Years of FOS. Mehta A., Beck M., Sunder-Plassmann G., editors. Oxford PharmaGenesis; 2006. Fabry disease in females: clinical characteristics and effects of enzyme replacement therapy. [PubMed] [Google Scholar]
- 7.Schuller Y., Linthorst G.E., Hollak C.E.M., Van Schaik I.N., Biegstraaten M. Pain management strategies for neuropathic pain in Fabry disease - a systematic review. BMC Neurol. 2016;16:25. doi: 10.1186/s12883-016-0549-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacDermot K., Holmes A., Miners A. Anderson-Fabry disease: clinical manifestations and impact of disease in a cohort of 98 hemizygous males. J. Med. Genet. 2001;38:750–760. doi: 10.1136/jmg.38.11.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiffmann R. Neuropathy and Fabry disease: pathogenesis and enzyme replacement therapy. Acta Neurol. Belg. 2006;106:61. [PubMed] [Google Scholar]
- 10.Biegstraaten M., et al. Small fiber neuropathy in Fabry disease. Mol. Genet. Metab. 2012;106:135–141. doi: 10.1016/j.ymgme.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Gadoth N., Sandbank U. Involvement of dorsal root ganglia in Fabry’s disease. J. Med. Genet. 1983;20:309–312. doi: 10.1136/jmg.20.4.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi L., et al. The Fabry disease-associated lipid Lyso-Gb3 enhances voltage-gated calcium currents in sensory neurons and causes pain. Neurosci. Lett. 2015;594:163–168. doi: 10.1016/j.neulet.2015.01.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W., Mi R., Haughey N., Oz M., Höke A. Immortalization and characterization of a nociceptive dorsal root ganglion sensory neuronal line. J. Peripher. Nerv. Syst. 2007;12:121–130. doi: 10.1111/j.1529-8027.2007.00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattacherjee A., Liao Z., Smith P.G. Trophic factor and hormonal regulation of neurite outgrowth in sensory neuron-like 50B11 cells. Neurosci. Lett. 2014;558:120–125. doi: 10.1016/j.neulet.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaneski C.R., Brady R.O., Hanover J.A., Schueler U.H. Development of a model system for neuronal dysfunction in Fabry disease. Mol. Genet. Metab. 2016;119:144–150. doi: 10.1016/j.ymgme.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayes J.S., Scheerer Julia B., Sifers Richard N., Donaldson Mark L. Differential assay for lysosomal alpha-galactosidases in human tissues and its application to Fabry’s disease. Clin. Chem. Acta. 1981;112:247–251. doi: 10.1016/0009-8981(81)90384-3. [DOI] [PubMed] [Google Scholar]
- 17.van Echten-Deckert G. Sphingolipid extraction and analysis by thin-layer chromatography. Methods Enzymol. 2000;312:64–79. doi: 10.1016/s0076-6879(00)12900-3. [DOI] [PubMed] [Google Scholar]
- 18.Monti E., et al. Uptake and metabolism of a fluorescent sulfatide analogue in cultured skin fibroblasts. Biochim. Biophys. Acta (BBA) - LIPIDS Lipid Metabolism. 1992;1124:80–87. doi: 10.1016/0005-2760(92)90129-j. [DOI] [PubMed] [Google Scholar]
- 19.Kaneski C.R., Schiffmann R., Brady R.O., Murray G.J. Use of lissamine rhodamine ceramide trihexoside as a functional assay for alpha-galactosidase a in intact cells. J. Lipid Res. 2010;51:2808–2817. doi: 10.1194/jlr.D007294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schueler U., et al. A short synthetic peptide mimetic of apolipoprotein A1 mediates cholesterol and globotriaosylceramide efflux from fabry fibroblasts. JIMD Rep. 2015 doi: 10.1007/8904_2015_507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asfaw B., et al. Defects in degradation of blood group a and B glycosphingolipids in Schindler and Fabry diseases. J. Lipid Res. 2002;43:1096–1104. doi: 10.1194/jlr.m100423-jlr200. [DOI] [PubMed] [Google Scholar]
- 22.Dehairs J., Talebi A., Cherifi Y., Swinnen J.V. CRISP-ID: decoding CRISPR mediated indels by sanger sequencing. Sci. Rep. 2016;6:28973. doi: 10.1038/srep28973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morand O., et al. Symptoms and quality of life in patients with fabry disease: results from an international patient survey. Adv. Ther. 2019;36:2866–2880. doi: 10.1007/s12325-019-01061-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann L., et al. Characterization of small fiber pathology in a mouse model of Fabry disease. eLife. 2018;7 doi: 10.7554/eLife.39300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakomá J., et al. Increased expression of Trpv1 in peripheral terminals mediates thermal nociception in Fabry disease mouse model. Mol. Pain. 2016;12 doi: 10.1177/1744806916663729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller J.J., et al. Neuropathic pain in a Fabry disease rat model. JCI Insight. 2018;3 doi: 10.1172/jci.insight.99171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi S.-I., Lim J.Y., Yoo S., Kim H., Hwang S.W. Emerging role of spinal cord TRPV1 in pain exacerbation. Neural Plasticity. 2016;2016:1–10. doi: 10.1155/2016/5954890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Üçeyler N., Biko L., Hose D., Hofmann L., Sommer C. Comprehensive and differential long-term characterization of the alpha-galactosidase a deficient mouse model of Fabry disease focusing on the sensory system and pain development. Mol. Pain. 2016;12 doi: 10.1177/1744806916646379. 174480691664637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melli G., Höke A. Dorsal root ganglia sensory neuronal cultures: a tool for drug discovery for peripheral neuropathies. Expert Opin. Drug Discovery. 2009;4:1035–1045. doi: 10.1517/17460440903266829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
50B11 clone D4W and clone E4 cells and stably-transfected LA-N-2 control cells were seeded on 2‑well chamber slides. All cultures were stained for Cas9 protein (red) as described in Methods. GFP protein is constitutively expressed in transfected cells (green). Nuclei were stained with DAPI to demonstrate the presence of cells in the field (blue). Cells were imaged with a 20X objective in a Keyence-9000 microscope at equal exposure times for each channel for controls and clones. There were no detectable cells that were positive for Cas9 protein or GFP expression in either of the 50B11 clones. Scale bar = 50 microns.