Summary
Objective
Endometrial carcinoma (EC) is the most common gynecological malignant disease in high income countries. The 2020 edition of the World Health Organization (WHO) Classification of Tumors of the Female Genital Tract underlines the important clinical implications of the new integrated histo-molecular classification system, in order to correctly define the specific prognostic risk group. This survey analysis will focus on the most commonly adopted immunohistochemical and molecular biomarkers used in daily clinical characterization of a diagnosed endometrial carcinoma in Italian labs.
Methods
An evaluation questionnaire was distributed to 41 Italian pathology laboratories. Normal habits in EC evaluation, especially regarding mismatch repair status (MMR) and microsatellite instability (MSI), were collected. A summary and a descriptive statistical analysis were used to show the current practice of each laboratory.
Results
The analysis of MMR status by immunohistochemistry (IHC) is carried out on the majority of all EC samples. The most frequent strategy for the analysis of MMR status in EC is IHC of four proteins (PMS2, MSH6, MSH2, MLH1). MSI analysis by molecular method in endometrial cancer is rarer and more restricted to some circumstances. Hypermethylation of the MLH1 promoter by methylation-specific PCR and pyrosequencing was analyzed in case of negative expression of MLH1/PMS2. Also, the analysis of p53 in EC is performed in the majority of cases. POLE mutational profiling is adopted only in a limited number of laboratories. Fifty-five percent of Italian laboratories refer to national/international guidelines when analyzing biomarkers in EC (among those, 45% use the ESGO Guidelines, 18% ASCO-CAP, 18% AIOM, 14% WHO, 5% British Association of Gynaecological Pathologist, 5% ESMO, 5% NCCN).
Conclusions
Adoption of guidelines and standardization of pre-analytical and analytical procedures are effective tools for adequate EC prognostic risk stratification and high quality standard of care.
Key words: endometrial cancer (EC), Italian labs, immunohistochemistry (IHC), molecular analysis, guidelines
Introduction
In 2013, The Cancer Genome Atlas (TCGA) Research Network discovered four molecular subtypes of EC - genomic architecture based, with each group having distinct clinical outcomes: POLE/ultra-mutated, MSI/hypermutated, copy-number low/endometrioid and copy-number high/serous-like 1. Following this discovery, the Leiden/PORTEC group and the Vancouver/ProMisE group validated a more pragmatic, cost-effective and clinically applicable molecular classification, using surrogate markers to recapitulate TCGA subtypes. MMR and p53 immunohistochemical assessment has been used as surrogate of molecular testing of MSI and somatic copy-number alteration (SCNA), yielding four molecular subtypes: POLE-mutated, MMR-deficient (MMRd), p53-abnormal (abn) and no specific molecular profile (NSMP) 2-4.
This surrogate marker approach has been demonstrated to be prognostically informative in low-, intermediate-, and high-risk EC. Among high-grade ECs, the POLE-mutated group shows an excellent prognosis, while the p53-abn group shows the poorest prognosis 5-8.
According to the 2020 5th edition of the WHO Female Genital Tumours, the EC molecular classification adds new information to the conventional morphologic features and should be integrated in the standard pathologic report. In view of the important prognostic and therapeutic implications, the 2020 ESGO/ESTRO/ESP guidelines for the EC management defined new prognostic risk groups, integrating molecular markers 5.
Today, morphology represents the framework in which molecular analysis should be interpreted 9-12. Since the great importance given to this new new morpho-molecular pathologic approach, we decided to perform a survey among 41 Italian laboratories in order to investigate differences and similarities in the current pathologic practice regarding biomarker analysis in EC.
Methods
The survey was focused on:
The immunohistochemical and molecular analysis in EC.
The different testing approaches depending on the laboratory setting.
The information about the type of technology used in molecular evaluation.
From 2020 to 2021, we selected 41 Italian pathology laboratories (both public or Academic), equally located in all the country, to which a 12-item questionnaire was distributed. Most of the questions were answered by selecting one choice. A summary and descriptive statistical analysis were used to describe the current practice of each laboratory. In accordance with the journal’s guidelines, we will provide our data for the reproducibility of this study in other centers if requested.
Results
MMR ANALYSIS
How do you perform MMR analysis? Do you routinely use IHC markers or molecular assessment?
Most responders performed MMR analysis by IHC (Fig. 1A). In all, 47.5% of laboratories performed MMR analysis by IHC in all EC samples, and 32.5% in selected cases: morphological ambiguity, suspicion of Lynch Syndrome and early-stage cancers. 20% of centers did not use either IHC or molecular analysis.
Figure 1.
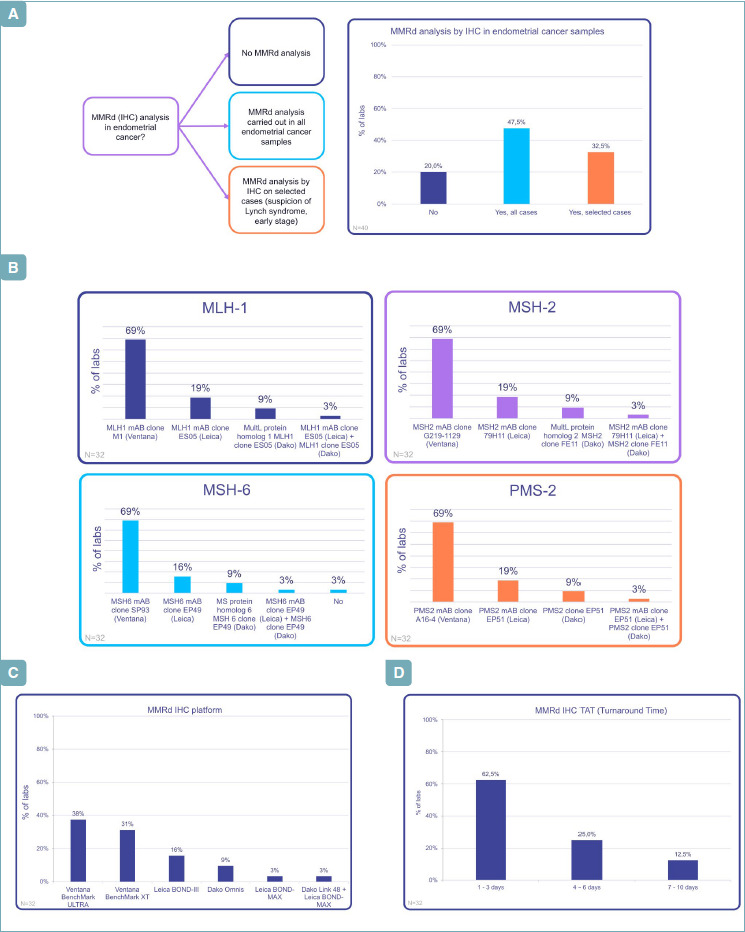
(A) MMRd by IHC in endometrial carcinoma: when? (B) MMRd by IHC in endometrial carcinoma: which antibodies? (C) MMRd by IHC in endometrial carcinoma: which platforms? (D) MMRd by IHC in endometrial carcinoma: TAT
IHC ANALYSIS: ANTIBODIES
In EC do you use an IHC panel? If yes, which clones do you prefer?
97% of Italian laboratories testing MMR used an IHC antibody panel and the most common is Ventana with the following clones (Fig. 1B): MLH1-M1 (69%), MSH2-G219-1129 (69%), MSH6-SP93 (69%), PMS2-A16-4 (69%). Other antibodies were described in Figure 1B. 16-19% of labs perform their analysis using Leica antibodies, as second choice, with the clones: MLH1-ES05, MSH6-EP49, PMS2-EP51, MSH2-79H11.
IHC ANALYSIS: PLATFORMS
What kind of immunohistochemical platform do you routinely use?
Regarding platforms (Fig. 1C), the Ventana platform with Ventana clones was the most commonly adopted by the largest part of Italian laboratories testing MMR. Twelve labs used Ventana BenchMark ULTRA (38%), while in 10 centers (31%) Ventana BenchMark XT was preferred. Leica (Leica BOND-MAX and Leica BOND-III) was chosen in 6 cases (19%), and DAKO platform (Dako Omnis and Dako Link 48+ Leica BOND-MAX) in 5 cases (12%). All (100%) of laboratories used commercial kits of MMR testing.
IHC ANALYSIS: WAITING TIME
How many days does your laboratory take to give you IHC results?
Most responders reported IHC results in a rapid turnaround time (Fig. 1D), within 1-3 days (62.5%). Some laboratories had longer times for analysis from 4-6 days (25%) to 7-10 days (12.5%).
MOLECULAR ANALYSIS: YES, OR NO? WHEN IS IT USED AND HOW?
Do you perform a molecular MSI analysis? If yes, in which cases?
MSI analysis by molecular method was performed in 27 institutions (67.5%). However, IHC for MMR protein expression is confirmed as first choice test. In some cases, the molecular method was restricted to specific circumstances, especially when MMR IHC status was unclear/indeterminate/equivocal, to confirm the IHC result in case of instable phenotype, or in presence of family history of Lynch Syndrome (LS) (Fig. 2A). Among these, in 5% of laboratories, MSI molecular analysis was requested by physician to confirm MMR results. In 13 laboratories (32.5%), there was no availability of molecular evaluation.
Figure 2.
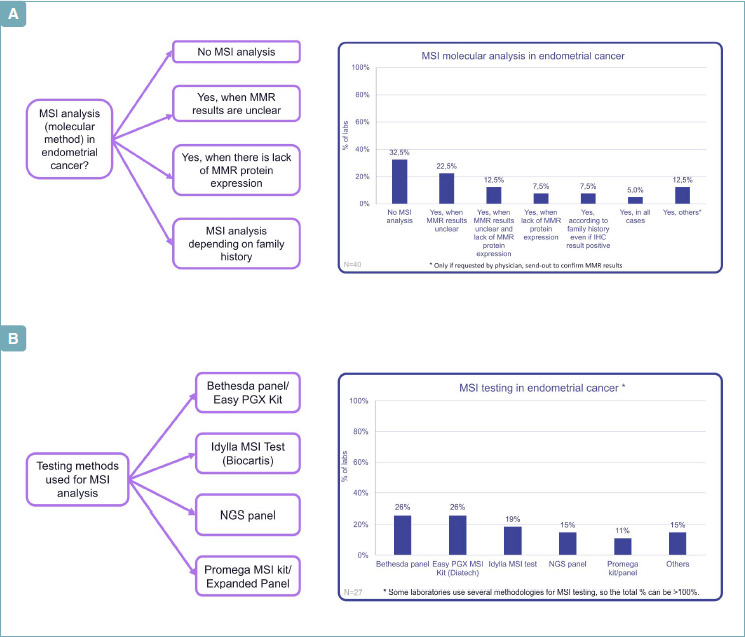
(A) MSI in endometrial carcinoma: when? (B) MSI in endometrial cancer: how?
MOLECULAR ANALYSIS: TECHNOLOGIES AND KIT EMPLOYED FOR MSI ASSESSMENT
In your lab what kind of method do you prefer for molecular analysis?
Regarding the testing methods for MSI analysis, there was no “clear predominant” methodology (Fig. 2B). Even if the Bethesda panel (26%) and Easy PGX MSI Kit (26%) from Diatech Pharmacogenetic were the most present, several methods emerged. Five labs used Idylla MSI test (19%), 4 used NGS (15%). The Promega kit was used in 3 cases.
MMR ANALYSIS ITALIAN STRATEGY
The most frequent strategy for MMR analysis in EC was IHC with four proteins (PMS2, MSH6, MSH2, MLH1), accounting for 58% of labs, followed by the MSI analysis alone (14%) or by the combination of IHC analysis and MSI testing (17%) (Fig. 3A).
Figure 3.
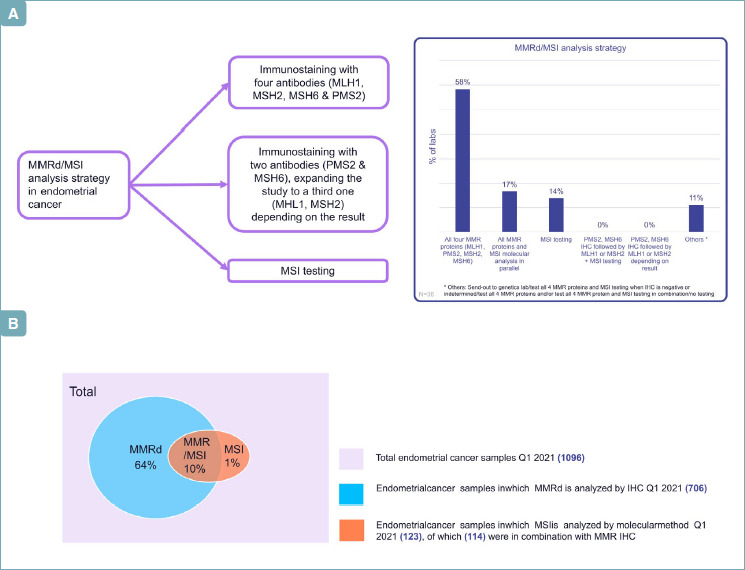
(A) MMRd/MSI analysis strategy. (B) MMR assessment in 1096 ECs from different Italian labs.
EC: SELECTION OF STUDY CASES
During the survey, 1096 ECs from the different labs were selected for the typology of MMR assessment (Fig. 3B): 706 cases were tested IHC (64%) and 123 by molecular analysis (11%), by confirming that in Italy IHC remains the ‘first choice test’. In 267 cases no integrative analysis was performed.
COMBINATION OF IHC AND MOLECULAR ANALYSIS
In case of negative MLH1/PMS2 do you perform further analyses?
50% of labs testing MMR IHC did not perform further analyses in case of negative MLH1/PMS2, whereas 53% of hubs performed further molecular investigations. Other strategies included analyses of MLH1 promoter hypermethylation (28%), MLH1 and BRAF p.V600E evaluation (6%) or BRAF p.V600/ MSI + BRAF p.V600 Pyrosequencing/ RT-PCR/ Genetic counseling.
MLH1 PROMOTER METHYLATION ANALYSIS: METHODS
How do you perform it?
The most frequent methods for the analysis of MLH1 promoter hypermethylation are methylation-specific PCR and pyrosequencing, respectively used in 38% and 31% of cases (Fig. 4A).
Figure 4.
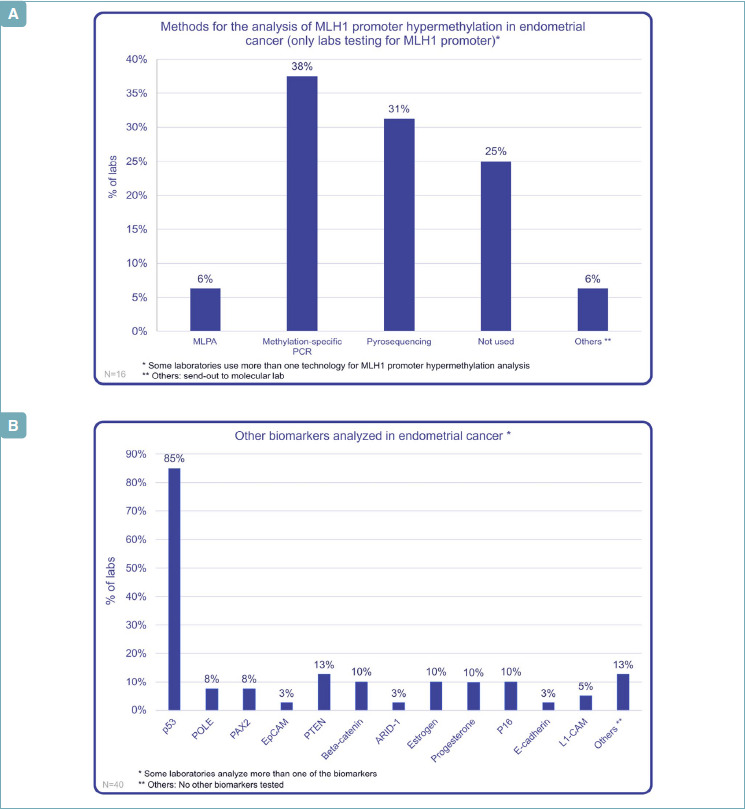
(A) MLH1 promoter hypermethylation in EC: which technology? (B) Other biomarkers in endometrial carcinoma.
OTHER BIOMARKERS
Do you also routinely analyze p53 antibody in EC?
In EC molecular characterization, numerous other prognostic and diagnostic molecules are employed. P53, together with MMR proteins, represents the most studied, with 85% of Italian Labs analyzing it by IHC: an interesting data, when compared to other molecules, such as ER and PR, analyzed only by 10% of Italian labs. A large number of laboratories analyzed more than one biomarker (Fig. 4B).
FOCUS ON POLE MUTATIONAL ANALYSIS
Do you perform POLE mutational analysis? If yes in which cases?
Only 3 Italian labs (8%) perform POLE mutational analysis, by a panel testing focused on the exonuclease domain of POLE exons 9, 11, 13, 14, in order to detect the POLE pathogenetic mutations variants (P286AR, V411L, S297F, A456P, S459F, F367S, L424I, M295R, P436R, M444K, D368Y).
In 1 of these 3 labs the molecular analysis was restricted to particularly circumstances, on request by the clinicians, especially in high grade, rare, ambiguous, and mixed histologies, in presence of abundance in TIL and/or peritumoral lymphocytes, in cases with ambiguous immunophenotype (possible multiple classifiers) or with subclonal p53 at IHC. In the other 2 labs, molecular analysis is performed in all cases. In 37 laboratories (90%), there was no availability of POLE molecular evaluation.
GUIDELINES
Which national/international guidelines do you refer to classify prognostic risk in endometrial cancer?
55% of Italian laboratories referred to national/international guidelines, analyzing biomarkers in EC (Fig. 5): 45% of labs refer to ESGO guidelines, 18% ASCO-CAP and 14% WHO. British association of gynecological pathologist guidelines, ESMO and NCCN were adopted in a lower percentage of cases.
Figure 5.
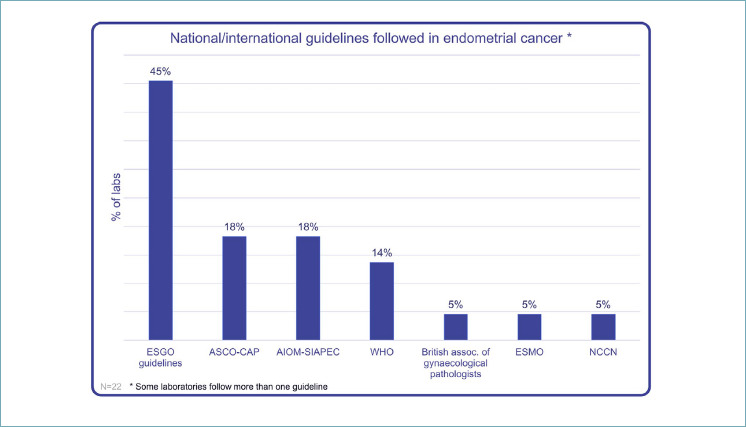
International/national guidelines in EC.
Discussion
SUMMARY OF MAIN RESULTS
The present study represents the first survey analysis addressed to 41 Italian labs, focusing on the most commonly adopted immunohistochemical and molecular biomarkers in daily clinical characterization of a diagnosed EC.
The first emerging data regards the IHC that still turns out as the standard ancillary tool in diagnostic pathology. The advantages of the IHC-based approaches:
the ready availability;
its inexpensive nature;
rapid turnaround time;
rapid identification of the defective protein;
correlation with morphology;
the need of only a limited amount of tissue;
feasibility for all types of FFPE specimens (small biopsies and/or surgical samples);
the propensity to IHC external quality assurance schemes.
The majority of Italian labs (47.5%) perform MMR IHC in all EC samples, while 32.5% perform it only in presence of morphological/clinical suspicion of Lynch Syndrome and in early-stage cancers.
MMR protein expression immunohistochemical testing offers advantages and challenges in identifying MMRd cases that potentially could benefit from immunotherapy and/or radiation therapy and in selected patients with Lynch Syndrome (LS), who should therefore be referred for genetic counseling and surveillance programs. 97% of Italian laboratories testing MMR use an immunoistochemical panel. Although a combination of only two antibodies MSH6 and PMS2 might reduce the cost without significantly decreasing the diagnostic accuracy, the immunohistochemical evaluation of 4 MMR proteins is confirmed as first choice test, followed by MSI analysis. The two methods have comparable sensitivity and show approximately 96% concordance, so that MMR proteins IHC can be considered the highly accurate surrogate of MSI molecular testing in EC.
MSI testing is restricted in the following selected events: unclear/indeterminate or equivocal IHC results; as confirmation, in case of instable immunophenotype; in presence of family history of LS.
MSI testing is generally based on PCR amplification of microsatellite markers, although in selected experienced centers (15%) NGS can represent an alternative novel test, especially in non-Lynch Syndrome-associated tumors, allowing MSI analysis, determination of tumor mutational burden (TMB) and identification of other possibly targetable alterations.
A discrete number of centers also performs the testing for MLH1 hypermethylation and BRAF p.V600 mutations, similarly to the colon cancer management in order to confirm a MLH1/PMS2 negative tumor as sporadic and to investigate the presence of somatic mutations.
Regarding other biomarkers, p53, together with MMR proteins, represents the most studied, with 85% of Italian labs analyzing it by IHC. An interesting data, when compared to other molecules, such as ER and PR, analyzed only by 10% of Italian Labs, and other such as Beta-catenin and L1-CAM that are integrated in pathological report only by 10% and 5% of Italian labs, respectively.
Pathogenic somatic mutations in the exonuclease domain of the replicative DNA polymerase epsilon (POLE) identify a subgroup of ultramutated ECs with enhanced immune response and better prognosis. According to the recent ESGO/ESTRO/ESP guidelines, all POLEmut carcinomas up to FIGO stage II, regardless of FIGO grade, histotype, or LVSI, are included in the low-risk group and could be managed by observation alone, with no need for adjuvant treatment, or as promising candidates (similar to MSI tumors) for checkpoint blockade immunotherapy.
The most common pathogenetic POLE mutations include P286R, V411L, S297F, A456P, and S459F; however the interpretation of novel and less frequent POLE mutations of uncertain pathogenicity remains challenging.
Moreover, the simultaneous presence of two or three molecular signatures has been identified in about 3% of EC cases, the so-called “multiple classifier”: MMRd/p53abn; POLEmut/p53abn; MMRd/POLEmut/p53abn; MMRd/POLEmut. In these latter subgroups, the clinical outcome is strictly related to the driver molecular subtype; in detail, pathogenetic POLE mutations prevail over the other signatures, conferring a good prognosis; on the other hand, the MMRd signature prevails over the p53abn signature.
From our survey data, complete molecular characterization of EC, including POLE panel testing, is restricted only to 3 labs, where POLE analysis is performed on physician demand or in histologically selected cases.
RESULTS IN THE CONTEXT OF PUBLISHED LITERATURE
EC is the sixth most frequent cancer in postmenopausal women worldwide and the most prevalent cause of mortality in the developed countries, with 382,096 new cases and 89,929 deaths reported in 2018 13-14. Despite recent advances in molecular pathology and in therapeutical regimens, the number of new cases is expected to further increase, owing to the increase in obesity rates and the aging population 15. To date, patients with stage III and IV cancers still experienced low 5-year OS rates, ranging from 57-66% and 20-26%, respectively, compared to early stage disease (stage I or II), with a 5-year overall survival ranging from 74-97% 16.
In this complex scenario, it is fundamental to improve risk stratification and open the way to targeted therapies. From the 2013 The Cancer Genome Atlas (TCGA) revolution in the molecular landscape of EC, pathological diagnosis and prognostic classification are continuously evolving, welcoming the possibility to incorporate molecular features into clinical practice and pathological definition.
Molecular classifiers are included in the new 5th edition of the WHO classification of tumors of the female genital tract, promoting an integrated histo-molecular diagnostic approach, for powerful prognostic information and therapeutical predictive purposes, with POLEmut ECs that have favorable outcomes and could be spared adjuvant therapy, MMRd ECs with intermediate prognosis that can benefit from radiotherapy and p53abn ECs with poor outcomes, standing to benefit from chemotherapy 17.
Ancillary molecular studies for POLE mutations, MMR/MSI definition, and p53 expression are highly encouraged to enrich and complement morphologic assessment of the histologic tumor type in the National Comprehensive Cancer Network guidelines 18, in order to define MMRd, p53 abn, POLEmut and NSMP subtypes of ECs.
To determine most of these profiles, IHC can be used as the first choice test, whereas POLE mutations does not have an IHC-based surrogate marker, still needing to be confirmed via DNA sequencing 19.
STRENGTHS AND WEAKNESSES
Our study represent the first survey analysis that focused on the most commonly adopted immunohistochemical and molecular analysis in daily clinical characterization of EC in Italian labs. This study has been replicated in Spain and UK in order to compare the practices between different countries. Although complete histo-molecular classification of EC is highly recommended by the International guidelines, in Italy the complete panel, including POLE analysis, is adopted only by a minority of labs, because this molecular test is not still reimbursed by the National Health Service. Thus, it may be possible to restrict POLE sequencing to low-risk EC showing abnormal or subclonal p53 staining, and omitted in advanced (stage III-IV) ECs since adjuvant therapy is always performed regardless of molecular classification.
IMPLICATIONS FOR PRACTICE AND FUTURE RESEARCH
Adoption of guidelines and standardization of pre-analytical and analytical procedure are effective tools for correct prognostic risk stratification. Therefore, it could be useful to build a network of certified centers for EC histo-molecular diagnosis and treatment, which can provide high quality standard of care, and offer patients the specific skills, experience, optimal levels of organization, and dedication.
Conclusions
From our Italian survey, the IHC based method for the biomarker analysis in EC emerges as the preferred and adopted diagnostic tool by the majority of investigated labs. It appears manageable in clinical practice, providing high diagnostic accuracy and high inter-observer concordance only when grounded on validated optimized laboratory protocols, together with correct pathological interpretation, according to the well established guidelines 20-22.
The most common prognostic assessment strategy in EC in Italy includes the analysis of MMR by IHC in all samples, followed by the analysis of MLH1 promoter hypermethylation and/or BRAF mutation and the analysis of other biomarkers such as p53.
In this way, we are able to identify MMR deficient or proficient ECs, p53 mutant or p53 wild-type ECs and multiple classifiers such as MMR deficient/p53 mutant ECs.
In the precision medicine era, optimizing and ensuring high histopathological diagnostic quality is essential to improve the management and outcomes of EC patients. The pathology report still remains a major component of patient multi-disciplinary management and the quality of the histo-molecular approach can contribute to avoid suboptimal treatment, guide genetic investigations and address women to adequate therapies.
Figures and tables
Appendix
Maria Maddalena Galante, Laboratorio Biologia Molecolare Oncologica, Ospedale di Lecce; Enrico M. Silini, Unità Patologia chirurgica, Azienda Ospedaliero-Universitaria di Parma; Vincenzo Canzonieri, CRO Aviano; Elena Rigoli, ASST Papa Giovanni XXIII, Bergamo; Giovanni Lanza, Anatomia Patologica Ferrara; Fabrizio Zanconati, SC Anatomia ed Istologia Patologica, Ospedale di Cattinara, Trieste; Sonia Nemolato, SC Anatomia Patologica P.O. Businco, Cagliari; Daniele Calistri, Istituto Romagnolo per lo Studio dei Tumori “Dino Amadori” - IRST IRCCS, Meldola; Laura Ardighieri, Servizio di Anatomia Patologica, ASST Spedali Civili di Brescia; Lorenzo Memeo, Istituto Oncologico del Mediterraneo, Viagrande; Laura Botticelli, Istituto Anatomia Patologica Policlinico di Modena; Mariantonia Carosi, Istituto Nazionale Tumori Regina Elena, Roma; Emanuela Filippi, Anatomia Patologica Ospedale Valduce Como; Claudio Di Cristofano, ICOT Latina, Università Sapienza di Roma; Maria Scatolini, Fondazione “Edo ed Elvo Tempia”, Laboratorio di Oncologia Molecolare, Biella; Laura Mariuzzi, Istituto di Patologia, Università di Udine; Anna Pesci, Dipartimento di Patologia, IRCCS Ospedale Sacro Cuore Don Calabria, Negrar-Verona; Gerardo Ferrara, Anatomia Patologica Ospedale Generale di Macerata; Antonio Cossu, Anatomia Patologica Università di Sassari; Alessandro D’Amuri, Anatomia Patologica Ospedale “A. Perrino” Brindisi; Angelina Pernazza, Anatomia e Istologia Patologica, Policlinico Umberto I, Roma; Daniela Fanni, Unità di Patologia, Dipartimento Scienze Mediche e salute pubblica, Università di Cagliari; Francesca Castiglione, SOD Istologia Patologica e Diagnostica Molecolare, AOU-Careggi; Andrea Palicelli, Azienda Unità Sanitaria Locale - IRCCS di Reggio Emilia; Gianluca Taccagni, Ospedale San Raffaele, Milano; Margherita Goia, Anatomia Patologica 1U Città della Salute e della Scienza Torino; Marta Jaconi, ASST Monza; Paola Pretelli, UOC Anatomia Patologica Carrara; Valerio Gaetano Vellone, Università di Genova; Renzo Boldorini, AOU “Maggiore della Carità” Novara; Luigi Insabato, Anatomia Patologica, Università degli Studi di Napoli Federico II ; Stefania Cesari, Ospedale San Matteo, Pavia; Renato Reitano, ASL Frosinone- UOC Anatomia Patologica; Alessandro Ubiali, UO Anatomia Patologica AUSL Piacenza; Maria Guido, Dipartimento di Patologia, Azienda ULSS 2 Marca Trevigiana, Treviso; Mattia Barbareschi, Ospedale S. Chiara Trento; Alfredo Santinelli, UOC Anatomia Patologica - AORMN Pesaro; Patrizia Falcone, Settore Biologia molecolare - SC Analisi cliniche - Ospedale Umberto Parini Aosta; Marianna Sciotino, Diaceutics, PLC, Belfast, United Kingdom; Antonio Capece, Diaceutics, PLC, Belfast, United Kingdom.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
FUNDING
This research received no external funding
ETHICAL CONSIDERATION
Our study was conducted in accordance with Good Clinical Practice guidelines and the Declaration of Helsinki (1975, revised in 2013). Patients’ initials or other personal identifiers did not appear in any image. Analysed data were collected as part of routine diagnosis.
AUTHOR CONTRIBUTIONS
Conceptualization: AS and GFZ; methodology: BB, FC, MF; software: BB, AP; validation: AS, GFZ; formal analysis: BB; investigation: AS, GA, NDA, GS, FI, EB; resources, Collaborators; writing/original draft preparation: AS, BB, NDA, GS; writing/review and editing: AS, GFZ; supervision: GFZ; project administration: GFZ.
All authors have read and agreed to the published version of the manuscript
References
- 1.Cancer Genome Atlas Research Network. Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497(7447):67-73. https://doi.org/10.1038/nature12113. 10.1038/nature12113 Erratum in: Nature 2013;500(7461):242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talhouk A, McConechy MK, Leung S, et al. Confirmation of ProMisE: a simple, genomics-based clinical classifier for endometrial cancer. Cancer 2017;123:802-813. https://doi.org/10.1002/cncr.30496. 10.1002/cncr.30496 Epub 2017 Jan 6. [DOI] [PubMed] [Google Scholar]
- 3.Kommoss S, McConechy MK, Kommoss F, et al. Final validation of the ProMisE molecular classifier for endometrial carcinoma in a large population-based case series. Ann Oncol 2018;29:1180-1188. https://doi.org/10.1093/annonc/mdy058 10.1093/annonc/mdy058 [DOI] [PubMed] [Google Scholar]
- 4.Stelloo E, Nout RA, Osse EM, et al. Improved Risk Assessment by Integrating Molecular and Clinicopathological Factors in Early-stage Endometrial Cancer – Combined Analysis of the PORTEC Cohorts. Clin Cancer Res 2016;22:4215-4224. https://doi.org/10.1158/1078-0432.CCR-15-2878 10.1158/1078-0432.CCR-15-2878 [DOI] [PubMed] [Google Scholar]
- 5.Concin N, Matias-Guiu X, Vergote I, et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. Int J Gynecol Cancer 2021;31:12-39. https://doi.org/10.1136/ijgc-2020-002230. 10.1136/ijgc-2020-002230 Epub 2020 Dec 18. [DOI] [PubMed] [Google Scholar]
- 6.Santoro A, Angelico G, Travaglino A, et al. New Pathological and Clinical Insights in Endometrial Cancer in View of the Updated ESGO/ESTRO/ESP Guidelines. Cancers (Basel) 2021;13:2623. https://doi.org/10.3390/cancers13112623 10.3390/cancers13112623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Travaglino A, Raffone A, Santoro A, et al. Clear cell endometrial carcinomas with mismatch repair deficiency have a favorable prognosis: a systematic review and meta-analysis. Gynecol Oncol 2021;162:804-808. https://doi.org/10.1016/j.ygyno.2021.07.007. 10.1016/j.ygyno.2021.07.007 Epub 2021 Jul 12. [DOI] [PubMed] [Google Scholar]
- 8.D’Alessandris N, Travaglino A, Santoro A, et al. TCGA molecular subgroups of endometrial carcinoma in ovarian endometrioid carcinoma: a quantitative systematic review. Gynecol Oncol 2021;163:427-432. https://doi.org/10.1016/j.ygyno.2021.08.011. 10.1016/j.ygyno.2021.08.011 Epub 2021 Aug 24. [DOI] [PubMed] [Google Scholar]
- 9.Jamieson A, Bosse T, McAlpine JN. The emerging role of molecular pathology in directing the systemic treatment of endometrial cancer. Ther Adv Med Oncol 2021;13:17588359211035959. https://doi.org/10.1177/17588359211035959 10.1177/17588359211035959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matias-Guiu X, Stanta G, Carneiro F, et al. European Society of Pathology (ESP) . The leading role of pathology in assessing the somatic molecular alterations of cancer: Position Paper of the European Society of Pathology. Virchows Arch 2020;476:491-497. https://doi.org/10.1007/s00428-020-02757-0. 10.1007/s00428-020-02757-0 Epub 2020 Mar 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fassan M. Molecular diagnostics in pathology: time for a next-generation pathologist? Arch Pathol Lab Med 2018;142:313-320. https://doi.org/10.5858/arpa.2017-0269-RA 10.5858/arpa.2017-0269-RA [DOI] [PubMed] [Google Scholar]
- 12.Angerilli V, Galuppini F, Pagni F, et al. The role of the pathologist in the next-generation era of tumor molecular characterization. Diagnostics (Basel) 2021;11:339. https://doi.org/10.3390/diagnostics11020339 10.3390/diagnostics11020339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. https://doi.org/10.3322/caac.21442 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 14.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. https://doi.org/10.3322/caac.21590 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 15.Pearson-Stuttard J, Zhou B, Kontis V, et al. Worldwide burden of cancer attributable to diabetes and high body-mass index: a comparative risk assessment. Lancet Diabetes Endocrinol 2018;6:e6-e15. https://doi.org/10.1016/S2213-8587(18)30150-5 10.1016/S2213-8587(18)30150-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mutch D. FIGO Update: Vancouver, Canada, October 2015. Gynecol Oncol 2016;140:6-7. https://doi.org/10.1016/j.ygyno.2015.12.002 10.1016/j.ygyno.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 17.León-Castillo A, de Boer SM, Powell ME, et al. Molecular classification of the. PORTEC-3 trial for high-risk endometrial cancer: impact on prognosis and benefit from adjuvant therapy. J Clin Oncol 2020;38:3388-3397. https://doi.org/10.1200/JCO.20.00549 10.1200/JCO.20.00549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.NCCN clinical practice guidelines in oncology (NCCN guidelines). Retrieved Dec 20, 2020. www.nccn.org [Google Scholar]
- 19.Imboden S, Nastic D, Ghaderi M, et al. Phenotype of POLE-mutated endometrial cancer. PLoS ONE 2019;14:e0214318. https://doi.org/10.1371/journal.pone.0214318 10.1371/journal.pone.0214318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Interpretation and Reporting Terminology for Mismatch Repair Protein Immunohistochemistry in Endometrial Cancer. BAGP Guidance Document: MMR Immunohistochemistry interpretation and terminology Retrieved Oct 21, 2020. [Google Scholar]
- 21.Köbel M, Ronnett BM, Singh N, et al. Interpretation of P53 immunohistochemistry in endometrial carcinomas: toward increased reproducibility. Int J Gynecol Pathol 2019;38;S123-S131. https://doi.org/10.1097/PGP.0000000000000488 10.1097/PGP.0000000000000488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh N, Piskorz AM, Bosse T, et al. p53 immunohistochemistry is an accurate surrogate for TP53 mutational analysis in endometrial carcinoma biopsies. J Pathol 2020;250:336-345. https://doi.org/10.1002/path.5375 10.1002/path.5375 [DOI] [PubMed] [Google Scholar]


