Abstract
Histones H2A, H2B, H3, H4 and H1 are highly conserved, positively charged proteins which form a disc-shaped protein core around which genomic DNA is wrapped to form a nucleosome. Immediately following DNA synthesis, replication-dependent canonical histones help package the DNA into nucleosomes to form compact chromatin fibers that can fit within the confines of the cell nucleus. Histone variants, which vary from the canonical histones in their primary amino acid sequence and expression patterns, replace their canonical counterparts throughout the cell cycle in important biological processes such as transcription, replication, DNA repair and heterochromatin formation. DNA damage is a continual threat to genomic stability and cell survival. Unrepaired DNA lesions are either lethal or can promote mutations if the damaged cells escape programmed cell death due to apoptosis. In order to repair DNA damage, cells use multiple DNA repair pathways, all of which require the recruitment of a multiple DNA damage signaling and repair factors. In order for these repair factors to be recruited efficiently and function properly at sites of DNA damage, the local chromatin environment surrounding the DNA lesion is often altered. Cells are able to regulate chromatin structure in the vicinity of DNA lesions through the addition of posttranslational modifications on histones and DNA, as well as through histone variant incorporation or removal. Recruitment or removal of histone variants at sites of DNA damage can alter the local chromatin structure by destabilizing it and making it more accessible to repair factors. Alternatively, some histone variants and their modifications may also provide specific binding sites for the recruitment of various DNA repair factors, thereby influencing repair pathway choice or repair efficiency, or both. This review seeks to provide an overview of our current understanding of the roles played by histone variants in DNA repair, especially in mammalian cells.
Keywords: DNA repair, DNA damage, genome stability, histone variant, chromatin, cancer
INTRODUCTION
Cells experience an estimated one hundred thousand DNA lesions per cell per day [1]. Such damage can result from the exposure to various external chemical, physical and biological agents, as well as due to by-products of normal internal cellular metabolism [2]. Left unresolved, these DNA lesions are detrimental to cell survival, as even a single unrepaired DNA double strand break (DSB) could result in cell death [3,4], or in mutations that could potentially drive genomic instability, which in turn is known to contribute strongly to cancer formation [5]. It is therefore crucial that cells have both highly efficient and effective methods to repair the DNA damage they incur daily.
The cell accomplishes efficient DNA repair through the use of multiple repair pathways which require the recruitment of a plethora of DNA repair factors to the lesion. The choice of the DNA repair pathway is determined based on the nature and location of the DNA damage, which directs the recruitment of specific DNA repair factors. Defects in a single DNA base pair (bp), such as a chemical alteration of the base via addition of a bulky alkyl group is repaired through the Base Excision Repair (BER) pathway; while the presence of a mismatched nucleotide is resolved by the Mismatch Repair (MMR) pathway (Figure 1) [6,7]. More extensive or complex damage, such as the formation of pyrimidine-pyrimidine dimers or intra-strand crosslinks, is repaired through the Nucleotide Excision Repair (NER) pathway (Figure 1) [8]. Additionally, Single-Strand Breaks (SSB) in DNA are repaired by the Single-Strand Break Repair (SSBR) pathway, while DSBs are repaired either by the Homologous Recombination (HR) or Nonhomologous End Joining (NHEJ) pathways (Figure 2). The HR pathway involves the copying of the missing DNA sequence in the vicinity of the DSB from the sister chromatid as an accurate method of DNA repair, while the NHEJ pathway uses the simple re-ligation of the two broken DNA ends to repair the damage, irrespective of whether any sequence information was altered or lost at the site of the break [9,10]. Throughout these DNA repair pathways, multiple chromatin components are removed and (re)deposited at the sites of the damage to both alter the chromosome architecture, as well as provide potential binding sites for the recruitment of DNA repair factors [2,11].
Figure 1. The role of histone variants in Nucleotide Excision Repair (NER) and Base Excision Repair (BER).
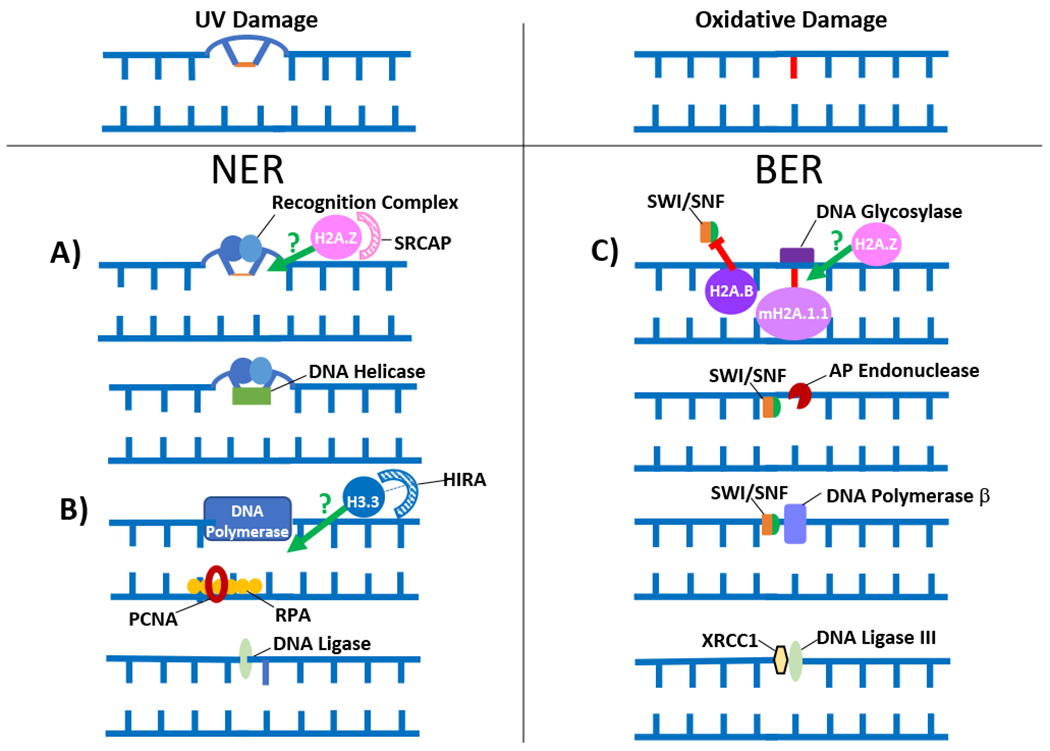
Red symbols (inhibitory) and green arrows (stimulatory) indicate where a histone variant may be potentially playing a role in the DNA repair pathway. Stripped arcs represent the most likely chaperones for the corresponding histones in the biological process under consideration. (A) H2A.Z (chaperoned by SRCAP) may assist the NER recognition complex to properly localize to the damage sites [111]. (B) H3.3 (with help from its chaperone HIRA) is potentially recruited to sites of UV damage where it may be required for replication fork progression [182,183]. (C) H2A.Z (along with its chaperone SRCAP) and mH2A.1.1 (chaperoned by LSH) have been shown to assist in promoting DNA glycosylase activity [110]. Additionally, nucleosomes containing the H2A.B are poorly remodeled by the SWI/SNF complex in vitro, suggesting that this variant either negatively affects the SWI/SNF remodeling function which appears to facilitate multiple steps of the BER pathway, or that H2A.B containing nucleosomes are inherently unstable and hence may not need to be remodeled, especially in vivo [138]. See text for more details. P = phosphorylation. Note: The shape, size and structure of the factors depicted are not drawn to scale.
Figure 2. The role of histone variants in HR and NHEJ pathways for repairing DSBs.
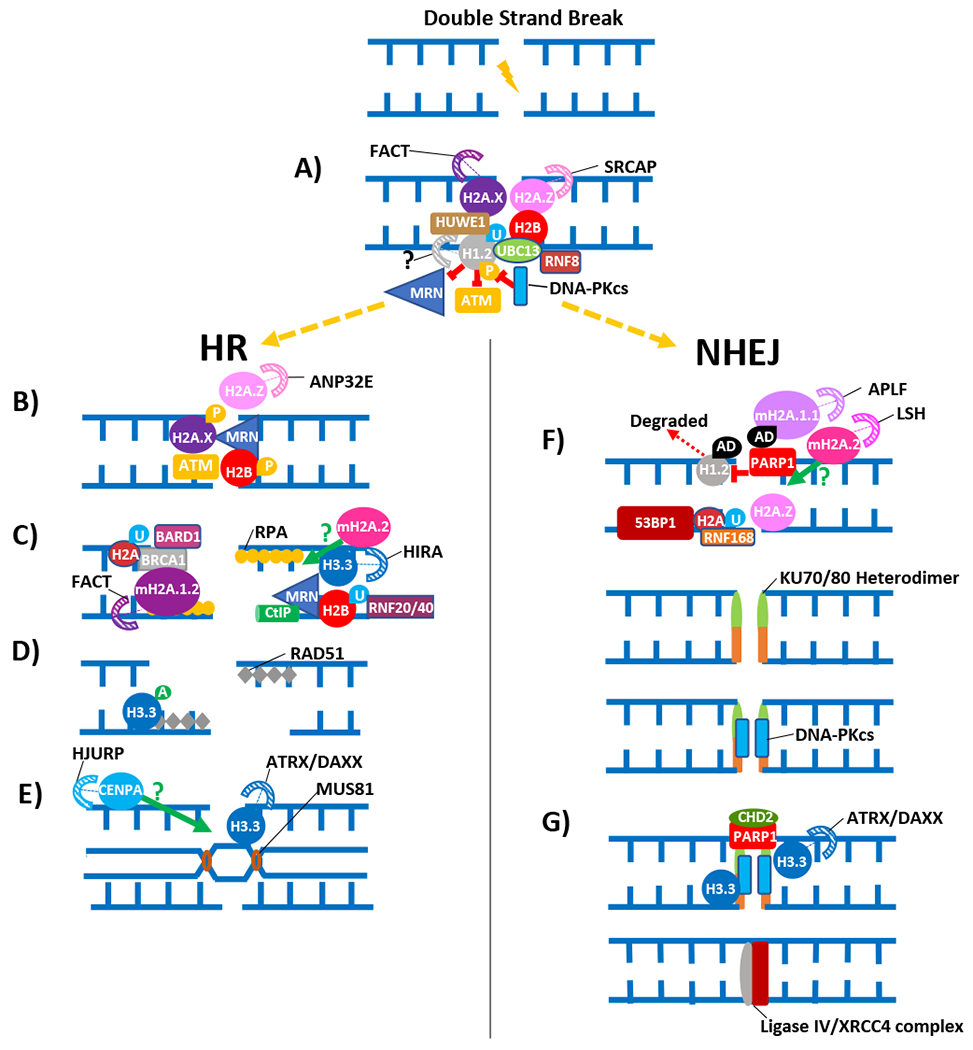
Red symbols and green arrows show the steps where the indicated histone variants may be potentially playing a negative or positive role respectively in the indicated DNA repair pathways. Stripped arcs represent the most likely chaperones for the corresponding histone variants. (A) At the DSB, histone H1 generally appears to inhibit the activities of the MRN complex and ATM in HR, as well as in c-NHEJ, although DNA-PK mediated phosphorylation H1 has been suggested to overcome this inhibition during c-NHEJ [41]. Histone variants H2A.X (chaperoned by FACT) and H2A.Z (chaperoned by SRCAP) are either already at the DSB sites, or are quickly recruited to these sites. The DSB can be repaired either by the HR pathway, or by the NHEJ pathways. (B) The DSB triggers a phosphorylation cascade, whereby H2A.X is phosphorylated at S139 almost as soon as a DSB occurs [60] by the ATM, ATR or DNA-PK kinases [67–69] and serves as a binding site for downstream DDR factors. Additionally, ATM also phosphorylates H2B at serine 14 which stimulates COMMD4 release [148,149]. H2A.Z may assist in the recruitment of various HR factors [97,98]. (C) Next, a ubiquitylation cascade follows, whereby H2BK120 is ubiquitylated by RNF20/RNF40 at DSBs, while H2A K125, K127 and K129 are ubiquitylated by the BRCA1/BARD1 complex to assist in proper end resection [58,150,151]. H3.3 may promote the coating of ssDNA by RPA at DSBs [186], while mH2A.1.2 (chaperoned by FACT) is recruited to DSBs to assist in BRCA1 localization [124–126]. Incorporation mH2A.2 (chaperoned by LSH) is crucial for proper Rad51 recruitment [130]. (D) H3.3 is acetylated by HAT1 to facilitate Rad51 recruitment [186]. (E) H3.3 (chaperoned by ATRX/DAXX) is believed to facilitate the recruitment of MUS81 to assist in double Holliday junction resolution [184,188]. cenH3 (deposited by HJURP) can also potentially assist in double Holliday junction formation or removal [160]. (F) Histone H1 is believed to dissociate from DSBs following poly-ADP ribosylation by PARP1 and is subsequently degraded to facilitate alt-NHEJ [49]. H2AK15 is ubiquitylated by RNF168 and promotes 53BP1 binding and c-NHEJ [58]. H2A.Z assists in Ku70/80 recruitment during repair by c-NHEJ [98]. mH2A.1.1 (chaperoned by APLF) binds to PAR chains on PARP1 at DSBs to promote the recruitment of KU70/80 and p53BP1 [120], while mH2A.2 (chaperoned by LSH) may also assist in proper p53BP1 localization [130]. (G) H3.3 (chaperoned by ATRX/DAXX) localizes to DSBs and may facilitate XRCC4 recruitment at the ligation step [185]. See text for more details. A = acetylation; AD = poly-ADP ribosylation (PARylation); P = phosphorylation; U = ubiquitylation. Note: The shape, size and structure of the factors depicted are not drawn to scale.
Histones are small positively charged proteins that help package the negatively charged DNA inside the nuclei of cells to form compact nucleoprotein filaments called chromatin in all eukaryotes. During DNA synthesis in S-phase, 147 base pairs of DNA wrap around an octameric structure known as the nucleosome core particle which is composed of 2 copies of each of the canonical core histone proteins H2A, H2B, H3 and H4 [12,13]. In most eukaryotes, a single linker histone or histone H1 molecule binds asymmetrically near the entry/exit site of the DNA around the octamer to complete the nucleosome [14,15]. The nucleosome forms the fundamental building blocks of chromatin and is repeated to package all the genomic DNA, thereby contributing to the first level of DNA compaction. Histone variants differ from their canonical counterparts in their primary amino acid sequence (Figure 3), and often in their timing/patterns of expression as well. Multiple histone variants have been shown to replace their canonical counterparts under specific circumstances and at certain genomic locations. Often, just a few amino acid alterations in their sequence allow for at least some of the variants to function in new ways that are distinct from their canonical counterparts. These histone variants play a role in various processes involving the DNA, such as replication, transcription, chromatin protection, chromatin compaction and DNA repair. Not surprisingly, histone mutations aberrant expression of histones have been increasingly linked to modulating carcinogenesis [16], leading to them being referred to as oncohistones [17]. Although in recent years a significant amount of research has been focused on the role that histone variants play in DNA repair, our knowledge of the specific functions of individual histone variants are still evolving and is far from being complete [18]. The aim of this review is to provide a broad overview of our current state of knowledge on the role of histone variants in DNA repair, especially in mammalian cells, with particular focus on resolving some of the controversies regarding the reported roles of specific histone variants in DNA repair.
Figure 3. Domain structure and sequence alignment of histone H1, H2A and H3 families.
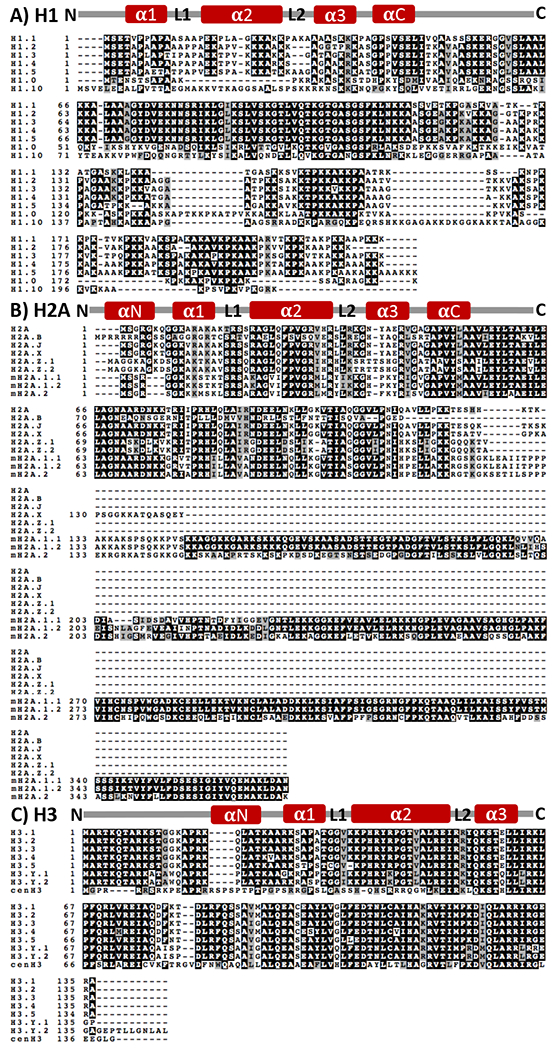
Specific members of these histone families have been implicated in different aspects of DNA repair. Amino acids highlighted in black represent identical residues in the protein sequences between the different histone variants, while those highlighted in grey represent conservative changes in amino acids that have similar characteristics/properties. Unshaded residues represent major non-conservative amino acid changes between the variants. Dashes represent amino acid residues that are absent in that particular histone variant. (A) Amino acid sequence alignment of the histone H1 family. (B) Amino acid sequence alignment of the histone H2A family. (C) Amino acid sequence alignment of the histone H3 family. Alignment was carried out with T-Coffee web [29]. The general domain structure of each family of histone variants is shown above the alignments and is not drawn to scale with the protein sequence.
HISTONE H1 FAMILY
Unlike the four core histones, members of the H1 family are not incorporated inside the core of the nucleosome, but rather sit asymmetrically on the outside of the nucleosome core particle [19–21]. Also known as the linker histone, H1 functions to asymmetrically protect the free linker DNA (~20bp) at either the entrance or the exit site of the DNA in the nucleosome. H1 is essential for the maintenance of proper chromatin structure [22], especially during higher order chromatin folding [23], and has been shown to play important roles in transcriptional regulation [24–26].
In comparison to other histone families, members of the H1 histone family are the most internally dissimilar and are consist of 11 members in mammals. The greatest variations in amino acid sequences are found in the C- and N-termini of these proteins, while most of the similarities occur within their central globular domains (Figure 3A) [27–29]. There are seven somatic H1 histones, including five (H1.1 through H1.5) that are replication-dependent and are upregulated during during DNA replication, plus the replacement variant H1.0 that accumulates in non-dividing cells and the ubiquituously expressed H1.10 [30]. Additionally, there are tissue specific variants comprising of the three testis specific H1.6, H1.7 and H1.9 variants, while H1.8 is an oocyte specific variant. Although significant progress over the past two and a half decades has shown H1 variant specific effects on gene expression [31,32], much of the research on the role of H1 in other chromatin activities such as DNA repair has been performed without distinguishing the specific variants involved in the process being studied [27]. Furthermore, compared to the core histones, currently relatively little is known about the specific physiological functions that H1 variants play in DNA repair, particularly in mammals [33]. Below, we discuss the H1 variants that have been implicated in aspects of DNA repair and/or cancer.
Somatic H1 Variants
H1.0
H1.0 behaves as a replacement variant as it accumulates in non-dividing and terminally differentiated cells [34]. It has been previously shown to be enriched within repetitive DNA elements in the nucleoli where it serves to stabilize these regions and may help prevent unregulated DNA synthesis [35,36]. H1.0 has also been found to be downregulated in various cancers [36,37] indicating a potential role as a tumor suppressor, though this needs to be investigated further. Given the known connections between tumorigenesis and DNA repair defects [5], it is possible that H1.0 may influence DNA repair, but this requires confirmation.
H1.10 (formerly known as H1.X)
H1.10 is found within euchromatic regions of the genome such as hypomethylated CpG islands and active genes, which suggests a role in transcriptional regulation for this variant [35]. Additionally, H1.10 has been shown to be upregulated in neuroendocrine tumors [38], suggesting a potential role as an oncogene as well. Along with several other linker histones (see below), H1.10 has also been reported to be polyubiquitylated via K63 ubiquitin chains at sites of DSBs, in a RNF8 (Ring Finger Protein 8), UBC13 (Ubiquitin conjugating E2 enzyme 13), and HUWE1 (HECT, UBA and WWE Domain Containing E3 Ubiquitin Protein Ligase 1) dependent manner. This modification provides a binding site crucial for the proper recruitment of RNF168 as well as p53-Binding Protein 1 (53BP1) protein that is a major DSB repair pathway choice factor and stimulates canonical or classic-NHEJ (c-NHEJ) [39,40].
H1.1-H1.5 histones
The bulk of the studies involving histone H1 use one or more of the relatively abundant replication dependent H1 histones, H1.1 through H1.5, often without distinguishing the exact subtype involved in the biological process being studied, with a few notable exceptions [31,32]. Previous research has found that the ligation step of c-NHEJ repair pathway is inhibited in vitro by the presence of histone H1 [41]. This inhibition is relieved by phosphorylation of H1 by the DNA-Dependent Protein Kinase (DNA-PK), a c-NHEJ factor (Figure 2A). Although this in vitro study presents a very plausible scenario, it utilized a mixture of bulk purified H1 histones rather than individual variants and needs further in vivo studies to determine any variant specific contributions.
Initial in vivo studies have found that cells deficient in the expression of replication dependent H1 histones exhibit resistance to DNA damage in both yeast [42] and mammalian cells [43]. In yeast cells, canonical H1 was postulated to play an inhibitory role within the HR repair pathway [42], while in mammalian cells the canonical H1 was postulated to restrict accessibility of DNA repair factors at DNA lesions, thereby preventing unregulated DNA repair specifically in the telomeric regions [43]. More importantly, mutations in the globular domain of a number of replication dependent H1 isoforms, especially H1.4 and H1.2, have been shown to be among the top driver mutations in B-cell lymphoma development, supporting their role as tumor suppressors. These mutations alter chromatin compaction and patterns of histone posttranslational modifications (PTMs) such as an increase in the activating methylation mark H3K36me2, and/or a loss of the repressive H3K27me3 mark, which result in chromatin decompaction and the upregulation of various stem cell renewal genes [44]. Although these results suggest important roles of the canonical H1 histones in DNA repair, they too do not clearly distinguish between the contributions of different H1 isoforms. Much of the available research on histone H1 in the context of DNA repair has been performed on the more abundant isoforms H1.2 and H1.4, which are discussed individually below.
H1.2
Linker histone variant H1.2 has been uniquely shown to play multiple roles in the context of DNA repair. H1.2 and H1.1 have a fairly dynamic binding affinity to nucleosomes when compared to H1.3-H1.5 [45]. H1.2 has been implicated in gene silencing by binding to the H3K27me3 repressive posttranslational methylation marks to increase chromatin compaction [46], as well as play an important role in proper cell cycle progression [47].
The condensation of chromatin by H1.2 has been shown to inhibit HR by preventing (Ataxia-Telangiectasia Mutated) ATM activity and proper localization of MRE11-RAD50-NBS1 (MRN), both of which are DSBR and HR factors [48]. H1.2 is believed to be displaced from DSB sites following ADP-ribosylation and subsequently degraded in a proteasome dependent manner to allow repair to continue. Furthermore, K63-linked polyubiquitin chains deposited on H1.2 through the action of the E3 ligase HUWE1 along with UBC13 and RNF8 help amplify the ubiquitylation cascade at the DSB site, while also providing a binding site for the c-NHEJ factor 53BP1 to regulate DSB repair (Figure 2A) [39,40]. In contrast to the inhibition of c-NHEJ by H1 [41], H1.2 has been shown to promote DSB repair via the alternative NHEJ (alt-NHEJ) pathway, although these results were obtained using cell extracts [49]. In addition to the reported roles of H1.2 in DNA repair, in the event of persistent unrepaired DSBs, H1.2 has been shown to be transported from the nucleus to the cytoplasm, where it stimulates the mitochondria to release cytochrome C, which in turn induces apoptosis (programed cell death) [50]. The involvement of any histone H1 chaperone in this transport (or in other DNA repair related activities of H1) is unclear, although H1 chaperones such as the proto oncoprotein SET (aka TAF1) that impact DNA damage sensitivity of mammalian cells have been reported to actively evict H1.2 in response to DNA damage [15,51].
H1.4
H1.4 has been shown to play a role in facilitating the condensation of chromatin by recruiting Heterochromatin Protein 1 (HP1) following the acetylation of its lysine 85 residue by P300/CBP-associated factor (PCAF) under normal conditions [52]. However, upon DNA damage, this lysine is rapidly deacetylated by Histone Deacetylase 1 (HDAC1), which is believed to result in the loss of HP1, leading to chromatin decondensation to allow efficient DNA repair. Since this lysine is highly conserved in all somatic H1 histones (Figure 3A), it is possible that other H1 proteins contribute to this regulation in a very similar way as well. As pointed out earlier, driver H1.4 mutations were found in 42.6% of Diffuse Large B-cell Lymphomas (DLBCL) [44] highlighting its importance in tumor suppression.
HISTONE H2A FAMILY
Histone H2A forms a dimer with H2B and, together with H3 and H4, makes up the nucleosome core particle (Figure 4). In comparison to the other core histones, H2A has the largest number of reported histone variants. Unlike other core histones that only have N-terminal tails, H2A also has a C-terminal tail which varies in length between variants and serves to regulate nucleosome formation and stability by directly binding to linker DNA. The majority of the differences found between the canonical H2A histone and its variants occurs on the C-terminus of the histone (Figure 3B) [53,54]. Additionally, fewer but structurally important differences also occur in the L1 loop between the first and second alpha helices of the histone fold domain in H2A variants (Figure 4), although how exactly these differences may be contributing to the functions of these variants in DNA repair is not fully understood [55–57]. Importantly, canonical histone H2A and several of its variants undergo many PTMs, including extensive ubiquitylation on multiple lysine residues to modulate important events in DSB repair [58]. Below, we review the known roles of H2A variants in the context of DNA repair.
Figure 4. Structural diversity of histone H2A variants within the nucleosome core particle.
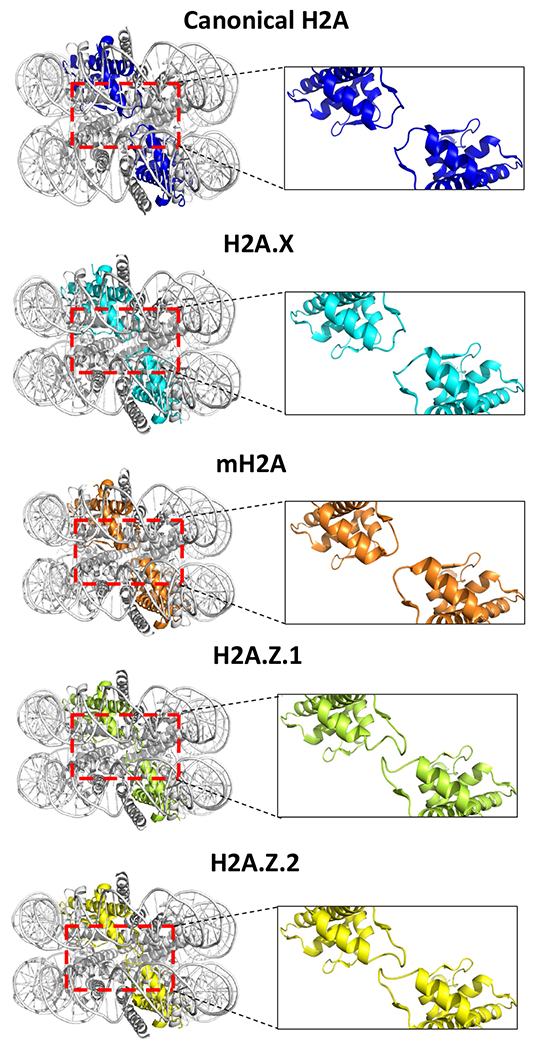
The side view of the crystal structures of human nucleosome core particles with either canonical or H2A variants is shown to highlight the location of the L1 loops of the H2A variants. A close-up view of the L1 loop region of the canonical and of each H2A variant is shown. Canonical H2A (PDB ID: 5Y0C) [16], H2A.X (PDB ID:6K1J) [63], mH2A (PDB ID:1U35) [57], H2A.Z.1 (PDB ID: 3WA9) [55] and H2A.Z.2 (PDB ID:3WAA)[55]. Nucleosome core particle figures were made using PyMol software.
H2A.X
The best studied histone variant in the context of DNA repair is H2A.X [59,60]. This variant shares a ~90% homology with the canonical H2A and only varies at a few amino acids (Figure 3B). H2A.X is estimated to make up 2-10% of all H2A in mammalian cells and is ubiquitously incorporated throughout the genome. However, if not already present, H2A.X is quickly recruited to sites of DNA damage in a Facilitates Chromatin Transcription (FACT) dependent manner [61,62]. The presence of H2A.X within the nucleosome increases the efficiency of recruitment of the DNA repair factor poly-ADP Ribose Polymerase 1 (PARP1) to sites of DNA damage [63].
In addition to the passive role of H2A.X in PARP1 recruitment during DNA repair, the H2A.X variant provides crucial PTM sites to initiate, amplify and regulate a cascade of DNA repair factor recruitment around DSB sites. Although H2A.X has many sites for potential PTMs, the most significant among these is the phosphorylation of S139 within it slightly extended C-terminal tail region, upon which it is referred to as γH2A.X (Figure 3B and Figure 4) [57]. γH2A.X is detected around DSBs immediately after their formation and can spread over a couple of megabases of chromatin [64,65]. Interestingly, recent evidence suggests that this spreading of γH2A.X from DSB sites is regulated by chromatin contacts in the context of topologically associating domains (TADs) and spreads bidirectionally within the TAD when the DSB is internal to the TAD, but asymmetrically into adjacent TADs when the DSB is located at the TAD border [66]. There are a number of kinases such as the phosphoinositide (PI) family of DNA Damage Response (DDR) kinases Ataxia Telangiectasia Mutated (ATM), ATM and Rad3-related (ATR) and DNA-PK that are responsible for this phosphorylation of H2A.X at sites of DSBs [67–69]. This modification assists both in the recruitment and retention of various DNA repair factors that not only help in the repair of the damage, but also stabilize the chromatin surrounding the break [70,71]. Specifically, γH2A.X foci formation provides a binding site for Mediator of DNA damage Checkpoint 1 protein (MDC1) [72] which then facilitates additional MRN complex recruitment [73] to prepare the DSB for repair as well as assist in the recruitment of a cascade of E3 ubiquitin ligases (RNF8, RNF168), that subsequently promote the recruitment of 53BP1 and Breast Cancer type 1 susceptibility protein (BRCA1), which are NHEJ and HR promoting factors respectively [74–77]. Almost immediately after the DSB is resolved, γH2A.X is removed either through histone exchange or through dephosphorylation [78]. Mice lacking H2A.X exhibit genomic instability and enhanced rates of cancer in the absence of p53 [79]. The importance of this H2A.X phosphorylation in the DDR can be further gauged by the fact that apart from mammals, other eukaryotic species also undergo this phosphorylation. In the budding yeast, the canonical H2A protein undergoes an analogous phosphorylation on S129 upon DNA damage and appears to perform similar functions [80]. In fruit flies, the histone variant H2Av, which appears to perform the functions of both H2A.X and H2A.Z variants (see below) in mammals, is phosphorylated on S137 upon DNA damage [81]. In contrast to the phosphorylation on S139, H2A.X can also be phosphorylated on Y142 in unstressed cells, apparently by the William-Beuren Syndrome Transcription Factor (WSTF) kinase. However, upon DNA damage, Y142 is dephosphorylated by EYA1 or EYA3 phosphatases which allows for the proper localization of DNA repair factors such as MDC1. If the phosphorylation of Y142 is not appropriately reversed upon DNA damage, it can result in the recruitment of factors to promote apoptosis [82,83]. A vast amount of literature is available on the role of H2A.X in DNA repair and cancer prevention, which cas been covered extensively in a number of excellent reviews and papers by the Bonner, Nussenzweig and other labs and we would like to point the readers to these [59,84–89]. Here, we will focus on the roles of other histone variants that have not be reviewed recently or in detail elsewhere.
H2A.Z
Histone H2A.Z is a highly conserved H2A variant and shares a near 60% homology (Figure 3B) with its canonical counterpart [90]. In vertebrates, H2A.Z is made up of 2 major isoforms (H2A.Z.1 and H2A.Z.2), which only differ by 3 amino acids [91,92]. Compared to the canonical H2A, the H2A.Z variant has an extended acidic patch as well as alterations in the L1 loop at the dimer interface within the nucleosome core particle (Figure 4), that can significantly alter the stability of the H2A.Z containing nucleosomes and hence their properties [55,90]. H2A.Z is found in actively transcribing genes as well as within chromosome centromeres [93]. Incorporation of H2A.Z into nucleosomes may render it less stable relative to nucleosomes containing canonical H2A [94]. H2A.Z is deposited into nucleosomes by p400 or Snf2 Related CREBBP Activator Protein (SRCAP) enzyme in human cells, or its homolog Swr1 in yeast cells, while it is evicted by the INO80 chromatin remodeling complex [95–97].
In the budding yeast, H2A.Z is evicted along with other core histones from DSBs in a Ino80 dependent manner [98], and cells lacking Ino80 show enhanced incorporation of H2A.Z at DSBs which blocks them from undergoing checkpoint adaptation in the presence of a persistent DSB [97]. Furthermore, the presence of Swr1, but not H2A.Z at DSBs appeared to promote the binding of Ku proteins and repair via c-NHEJ [98]. On the other hand, the Ino80 mediated eviction of H2A.Z promotes Mre11 recruitment and end-resection, as well as checkpoint signaling, although no effect on HR mediated repair was observed. It should be noted that these studies in yeast were performed using the galactose regulated HO endonuclease to generate a DSB at either the native MAT locus or ectopic sites, followed by Chromatin Immuno-Precipitation (ChIP) to detect the presence of specific proteins at the DSBs one or more hours after the induction of DSBs, which precludes the study of H2A.Z dynamics immediately after DSB formation.
In contrast to the general agreement on the role of H2A.Z in the context of DNA repair in yeast cells, results in mammalian cells were quite contradictory initially. In human cells, H2A.Z was shown to be recruited to sites of DSBs [99]. This study showed that H2A.Z functions to assist in the recruitment of a number of DNA repair factors for the HR pathway (MRE11, BRCA1 and RAD51), as well as in the c-NHEJ pathway (KU70/80 complex) [97–99]. However, subsequent findings directly contradict these early data and showed that H2A.Z did not recruit to sites of DNA damage and depletion of H2A.Z did not cause any alterations in the efficiency HR or NHEJ pathways [100]. One possible explanation for these discrepancies is that important differences in the reagents and methodologies used by the two groups could have affected their results [99,100]. A major difference was the method used for decreasing H2A.Z levels, with the Price lab using stable transfections of small hairpin RNA (shRNA) for H2A.Z knockdown [99], while the Canitrot lab used transient transfections of small interfering RNA (siRNA) [100]. Additionally, lasers of different wavelengths were used to induce DNA damage to observe H2A.Z recruitment, with the Price lab using 405 nm laser on cells that were pre-sensitized with Hoechst dye 33258 [99], while the Canitrot lab used a 532 nm laser to induce damage [100]. It should be noted that many such pre-sensitization drugs bind to or intercalate within DNA, thus potentially altering chromatin structure [101,102]. Although these differences may have potentially led to the different results obtained in these studies, they remained quite perplexing until it was shown more recently that H2A.Z is only temporarily needed at DSBs [103,104]. After being recruited to DSBs immediately after their formation, H2A.Z is then rapidly evicted within 10 minutes of incorporation from sites of DSBs in an Acidic Nuclear Phosphoprotein 32 Family Member E (ANP32E) dependent manner. This temporal specificity of H2A.Z was essential for proper C-terminal binding protein 1 interacting protein (CtIP) endonuclease function, as well as RAD51 localization to DSB sites for repair by the HR pathway [103–105]. Interestingly, additional research suggests that the DNA repair function of H2A.Z may be primarily due to the H2A.Z.2 isoform [105]. Hence, it is possible that some of the initial inconsistencies regarding the role of H2A.Z in DNA repair in the published data may also be the result of different isoforms being followed in the earlier studies.
So, what might be the function of the rapid deposition of H2A.Z initially at DSBs, followed by their subsequent removal a short time later? Evidence from yeast [98] and mammalian cells [99] both suggest that H2A.Z may contribute to NHEJ mediated repair and so one possibility is that this process may be facilitated by the initial deposition of H2A.Z at DSBs. Consistent with this, Swr1 mediated H2A.Z deposition has been suggested to facilitate the activity of Exonuclease 1 (Exo1) during MMR in yeast [106] and it is known that this nuclease also plays a role in end processing during DSB repair in yeast [107]. However, if NHEJ mediated repair does not fix the DSB quickly, then H2A.Z removal from the DSBs may be necessary to subsequently facilitate repair via the HR pathway. This two-step scenario would be consistent with much of the data on the role of H2A.Z in DNA repair available so far, as well as the reported sequential assembly of NHEJ factors prior to HR factors in both yeast [108] and mammalian cells [109].
In addition to its proposed role in HR repair pathways, H2A.Z has also been suggested to play a role in other DNA repair pathways. Nucleosomes with H2A.Z were shown to enhance BER activity in vitro in comparison to those composed of the canonical H2A [110]. Furthermore, H2A.Z incorporation has been shown to promote both MMR and NER pathways as well, although these results have only been demonstrated in yeast so far [106,111]. Overall, the information available so far suggests the possibility for H2A.Z to play roles in BER, MMR and NER pathways, in addition to its significant contributions to DSB repair.
mH2A (formerly known as macroH2A)
The histone variant mH2A shares a nearly 60% homology with the canonical H2A, but with an additional ~200 amino acid domain on the C-terminus of the protein (Figure 3B). Despite the addition of such a large domain, nucleosomes with mH2A retain the basic nucleosome core particle structure (Figure 4), although the mH2A nucleosomes appear to be more stable with the DNA more tightly bound than those with canonical H2A [57,112,113]. mH2A is specifically enriched within the inactive X-chromosome (Barr-body) in female cells. Not only does mH2A play an essential role in the maintenance and development of heterochromatin, but it is also involved in multiple molecular processes such as transcriptional repression and DNA repair [114]. mH2A has multiple isoforms (mH2A.1.1, mH2A.1.2 and mH2A.2) (Figure 3B), which adds further complexity to its potential roles [115,116].
The enzyme PARP1 is involved in sensing DNA damage, SSBR, recruiting multiple DNA repair factors, as well as stabilizing DNA replication forks [117]. PARP1 is the main enzyme which posttranslationally attaches poly(ADP-ribose) (PAR) chains to various protein targets. The attachment of PAR chains creates a binding site for various proteins that carry a conserved globular domain known as a macrodomain to trigger a veriety of downstream cellular signaling events.
Although the mH2A.1.1 and mH2A.1.2 isoforms share a nearly 70% homology (Figure 3B) with each other, a crucial difference is the presence of a macrodomain only in mH2A.1.1 [118,119]. Not surprisingly, mH2A.1.1, rather than mH2A.1.2, has been shown to be recruited to sites of DSB repair by binding to PAR chains on PARP1 after the latter localizes to DSBs, rather than being directly recruited to DSB sites as part of nucleosomes [120,121]. Once recruited, mH2A.1.1 assists in proper Checkpoint Kinase 2 (CHEK2) activation, as well as the recruitment of various DNA repair factors such as KU70/80 and 53BP1, both of which are NHEJ factors [120].
A reduction in the expression of mH2A.1.1 has been shown to increase the endogenous levels of 8-oxo-7,8-dihydroguanine (8-oxoG), a major type of oxidative DNA damage, as well as an enhanced vulnerability to alkylation damage by methyl methane sulfonate (MMS) that is repaired by BER [122]. Moreover, research also suggests the that the mH2A containing nucleosomes enhance the activity of BER in comparison to nucleosomes containing the canonical H2A [110,120]. These data suggest that mH2A.1.1 plays a role in BER repair. Further investigation suggested that the role of mH2A.1.1 in BER is fairly upstream and is mediated through its regulation of both PARP1 activity and degradation of PAR chains[110,122], but its precise role is not fully understood.
Despite mH2A.1.2 not having a macrodomain, it still appears to play an important role in DNA repair. This isoform has been shown to be enriched within chromosomal fragile sites and telomeres that have an increased incidence of DSBs as a result of replication stress [123]. mH2A.1.2 is also actively recruited to such DSBs in a FACT dependent manner [124,125]. At these sites, mH2A.1.2 enhances BRCA1 recruitment and retention to ensure that these DSBs are repaired through the HR and not the NHEJ pathway [126]. In contrast, as pointed out above, the mH2A.1.1 isoform promotes DSB repair via NHEJ [120,125].
The mH2A.2 isoform shares a 67% homology with mH2A.1 and also has a conserved globular domain. Despite these similarities, the macrodomain of mH2A.2 is unable to bind PAR chains, limiting its interaction with PARP1 [127]. mH2A.2 has been previously shown to play a prominent role in inhibiting somatic cell reprograming, as well as suppressing melanoma progression [128,129]. Additionally, a very recent study has shown that proper mH2A incorporation is crucial for RAD51 loading as well as regulating the balance of BRCA1 and 53BP1 binding, particularly at stalled replication forks, although this research did not distinguish between mH2A isoforms [130].
Based on the published data so far, mH2A undoubtedly plays important roles in DSB repair by both NHEJ and HR, as well as in the BER repair pathway. Due to the close connections between mH2A and PARP1, it is quite possible that mH2A isoforms may play additional roles in other DNA repair pathways, such as SSBR pathway. Further research is needed to determine the specific roles that mH2A isoforms may play within these pathways. However, it would be important to distinguish the roles of each isoform of mH2A in such studies, since they have been already shown to play significantly different roles in DNA repair pathways [120,125,126].
H2A.B (formerly known as H2A.Bbd)
H2A.B (H2A Barr-body deficient) is a rapidly evolving placental-mammal specific variant which only shares approximately 50% amino acid homology with its canonical H2A counterpart (Figure 3B). H2A.B has been found to be distributed throughout the genome, except for the inactive Barr body in human cells [131,132]. Interestingly, H2A.B containing nucleosomes only protect 118bp of DNA unlike the 147bp of DNA protected by canonical nucleosomes [132]. H2A.B incorporation has been shown to result in an open conformation of chromatin and is specifically enriched in the body of actively transcribed genes [133,134]. H2A.B is encoded by 3 genes and it was recently shown that loss of all three genes in mice results in aberrant post-meiotic male germ cell chromatin, but only very mild defects in fertility, whereas more significant effects were observed on post-implantation embryonic development in a pattern that is suggestive of H2A.B genes being strong parental-effect genes [135]. Recently, it has also been suggested that short H2A (sH2A) variants such as H2A.B may have oncogenic features and their aberrant expression may allow them to serve as oncohistones during carcinogenesis [17].
Generally, H2A.B is found to be transiently deposited at sites of DNA replication where it co-localizes with Proliferating Cell Nuclear Antigen (PCNA), a DNA synthesis factor, during early and late stages of replication [136,137]. H2A.B has also been shown to be recruited to sites of DNA damage, although this recruitment does not appear to play a direct role in any particular DNA repair pathway [136]. Instead, this study found that the exogenous expression of H2A.B enhances the sensitivity of cells to UV damage, suggesting a potential inhibitory effect of H2A.B on NER. Similarly, there is some indication that the presence of H2A.B in nucleosomes negatively affects the chromatin remodeling activity of SWI/SNF (SWItch/Sucrose Non-Fermentable) in facilitating the recruitment of various factors to properly remove 8-oxoG by the BER pathway, suggesting that H2A.B may need to be removed for efficient BER [138]. Interestingly, unlike most other histone variants, H2A.B has been shown to be able to replace its canonical counterpart without the use of a chaperone [134].
Currently, a specific role of H2A.B at sites of DNA damage remains unclear. However, based on the lack of the need of a chaperone for H2A.B and its ability to relax chromatin conformation, it is conceivable that H2A.B simply acts as a place holder for other H2A variants at DNA damage sites. Additionally, the transient incorporation of H2A.B at DNA damage sites could be enough to increase the accessibility of DNA to allow for the efficient recruitment for DNA repair factors. Although no significant defects in the repair of meiotic DSBs were observed in H2A.B knockout mice [135,139], it will be interesting to test the DNA damage sensitivity and the DDR of the H2A.B knockout mice, or somatic cells derived from them, to get a better insight into any role played by H2A.B in DNA repair.
H2A.J
H2A.J is a mammalian specific H2A variant which shares a near 94% homology to the canonical H2A and differs from it at just 5 residues [140]. Not surprisingly, due to this near identical amino acid sequence (Figure 3B), no significant structural differences were observed between nucleosome core particles containing H2A.J compared to those with canonical H2A (Figure 4) [141]. H2A.J has been shown to accumulate in senescent cells carrying DNA damage where it appears to promote the expression of inflammatory genes [141,142]. However, a specific role for H2A.J at DNA damage sites is currently not known.
HISTONE H2B
Of all the histone variants, H2B variants are the least studied, although well over a dozen H2B variant proteins have been identified and four variants (subH2B, H2B.E, H2B.W and hTSH2B) have been studied to some extent in mammalian cells [143–147]. To date, none of the H2B histone variants have been found a play a role in any of the known DNA repair pathways. However, the canonical H2B itself has been shown to serve as a major binding site for various DNA repair factors, especially after the deposition of specific PTMs. Under normal conditions the Copper Metabolism gene MURR1 Domain Containing 4 (COMMD4) protein has been shown to directly bind to unmodified H2B to protect it from unregulated PTMs, especially monoubiquitylation. However, soon after DSB occurrence, ATM phosphorylates H2BS14, stimulating the release of COMMD4 from H2B [148,149]. The removal of COMMD4 allows for the proper recruitment of RNF20-RNF40, which ubiquitylates H2BK120 within an hour of DSB formation. Furthermore, this ubiquitylated site serves as a binding site for, and promotes the recruitment of prominent HR repair factors, namely BRCA1, CtIP and NBS1 to assist in proper end resection [150,151]. Since these studies did not attempt to distinguish the H2B variants involved in these processes (which would be difficult, especially in the absence of reagents such as good H2B variant specific antibodies), it is formally possible that one or more H2B variants are involved in these processes, rather than the canonical H2B itself. Much more research is needed to understand the role of H2B variants in cell physiology and DNA repair, which is still in its infancy.
HISTONE H3 FAMILY
Histone H3 partners with H4 and is incorporated into nucleosomes immediately after DNA synthesis [152]. Histone H3 also has the largest number of known PTMs which can serve to modulate chromatin structure and function in a variety of ways [153]. Additionally, unlike the variants in other histone families, most H3 variants are nearly identical to the canonical H3, varying at just a few residues (Figure 3C).
cenH3 (CENPA)
cenH3 is a centromeric H3 variant that localizes specifically within the centromeres of chromosomes. cenH3 serves as an epigenetic marker for centromeres and has also been found to be essential for centromeric interactions within the kinetochore protein complex [154]. cenH3 shares only a ~45% homology with the canonical H3, which is the least among H3 variants by far (Figure 3C). cenH3 nucleosome core particle structure resembles its canonical counterpart, however, the DNA on either end of the nucleosome core particle is more flexible, with only 121bp of DNA wrapped around the core structure, presumably due to its longer unstructured N-terminal tail with a short α-helix (Figure 5) [155,156].
Figure 5. Structural diversity of histone H3 variants within the nuclesosome core particle.
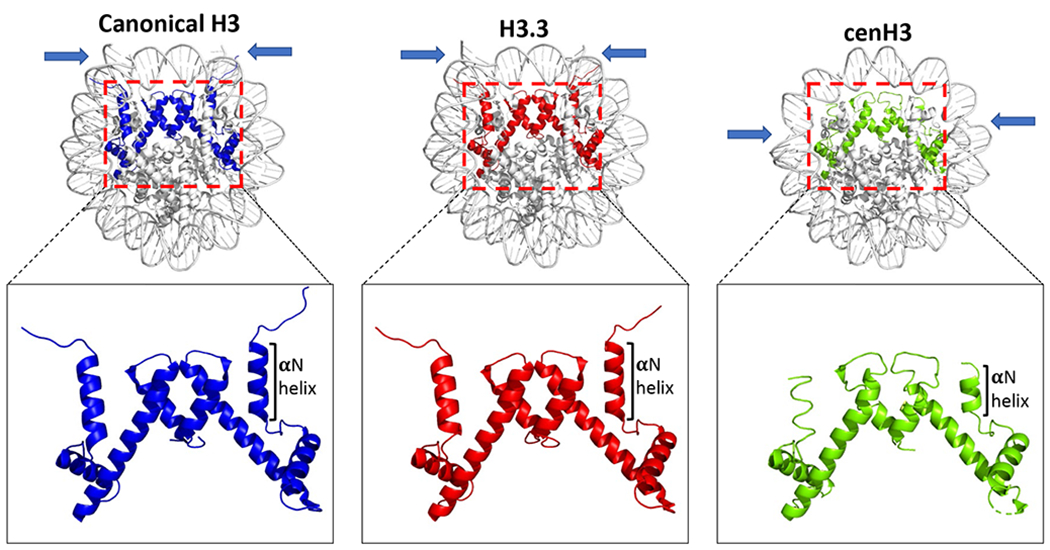
Crystal structures of human nucleosome core particles containing either canonical histone H3.1 (PDB ID: 5Y0C) [16], histone variant H3.3 (PDB ID:3AV2) [174] or histone variant cenH3 (PDB ID: 3AN2) [156] are shown. A close-up view of the histone H3 variants highlighting the α-N helix in cenH3 is shown. Note the shorter α-N helix in cenH3 compared to H3.1 and H3.3. The blue arrows point to the DNA ends visible in the crystal structures of the nucleosome core particles corresponding to the different H3 variants. Note the shorter tract of DNA associated with the histone octamer within the cenH3 nucleosome core particle. Nucleosome core particle figures were made using PyMol software.
cenH3 has been shown to reliably recruit to sites of DSBs [157]. Despite this observed recruitment, little is known about its role in the DSB repair pathway. Though not fully understood, evidence suggests that cenH3 may play a role in the proper recruitment or retention of ATM, which phosphorylates H2A.X immediately after DNA damage [154,157,158]. The Holliday Junction Recognition Protein (HJURP, a downstream Holliday junction binding factor in the HR pathway) has been shown to act as a primary chaperone for cenH3, incorporating it into new centromeric nucleosomes [159], suggesting that HJURP maybe depositing cenH3 at DSBs. A trivial possibility is that the enhanced expression of HJURP that is observed following DSBs, along with its interactions with proteins such as NBS1 [160], simply results in the deposition of cenH3 at DSBs due to the chaperone function of HJURP. However, this study demonstrated clear HJURP foci formation in response to radiation, in addition to a HR repair defect as well as genomic instability upon HJURP knockdown. Hence, it is formally possible that some of these phenotypes associated with HJURP knockdown may actually be associated with a defect in cenH3 deposition at the DNA damage sites.
Apart from its recruitment to DSBs [157], cenH3 has also been shown to undergo disassembly from centromeric repeats following their transcriptional activation in an ATM and p53 dependent manner upon DNA damage with the topoisomerase poison etoposide which causes DSBs [161]. This response may be important in initiating and/or maintaining a senescent state in cells that exhibit persistent DNA damage signaling. Overall, these data suggest that the normal homeostasis between cenH3 deposition and eviction from chromatin may be altered following DNA damage, raising the possibility that some of the cenH3 lost from centromeric regions under these conditions may then be redeposited at DSBs [157], perhaps in an attempt to form neocentromeres to avoid the potential loss of genetic material if DSB repair fails [162].
Interestingly, cenH3 and HJURP stabilize each other and both are overexpressed in cells lacking functional p53, which normally serves to repress these two genes [163,164]. p53 deficient tumor cells were also found to be dependent (or “addicted”) to high levels of HJURP and cenH3 for survival. Additionally, HJURP depletion in cells with wild type p53 resulted in activation of p53 and cell cycle arrest without concomitant DNA damage. On the other hand, cenH3 overexpression in cells with wild type p53 resulted in radiosensitivity, growth inhibition and senescence, although this was related to the suppression of several cell cycle related genes. Furthermore, in cells lacking p53, cenH3 overexpression resulted in epithelial to mesenchymal transition, which could contribute to carcinogenesis. Together, these data suggest a very significant and complex role of cenH3 in regulating aspects of DDR, cell cycle and carcinogenesis in a p53 dependent manner. However, more research is needed to explore any potential cross talk between these roles of cenH3 and its putative role in DNA repair.
In addition to its recruitment to DSB, cenH3 has also been shown to interact with a number of DNA repair factors such as Damage Specific DNA Binding Protein 1 (DDB1, a recognition protein in both NER and BER pathways) [165] and PARP1 [166]. However, these interactions are likely related to cenH3’s role in the centromere due to its simultaneous interaction with multiple proteins in the CEN complex. Finally, proper cenH3 assembly was also found to be dependent on Uracil-DNA glycosylase 2 (UNG2, a BER factor) activity in Xenopus [167,168]. These interactions suggest potential roles for cenH3 in HR and BER pathways, although the molecular details are yet to be investigated.
H3.3
H3.3 is nearly identical (96% homology) in amino acid sequence to the canonical H3 (H3.1 and H3.2) (Figure 3C) [169]. Previous research has shown that H3.3 is enriched in both the loosely packaged euchromatin, such as the promoters of actively transcribed genes, as well as the highly compact regions of genomes, such as the heterochromatin found at telomeres, pericentromeric regions, and rDNA. The deposition of H3.3 into these varying chromatin structures is dependent on two H3.3 specific chaperones, Histone Regulator (HIRA) and Alpha-Thallassemia Mental Retardation X-linked protein/Death domain Associated protein (ATRX/DAXX), which are responsible for its incorporation in euchromatin and heterochromatin respectively [170–172]. Interestingly, despite the near identical amino acid sequences of the major non-centromeric H3 variants (Figure 3C), nucleosomes with H3.3 have been found to be less stable than those containing canonical H3 (Figure 5), especially if the same nucleosomes also contain the H2A.Z variant (Figure 4) [173,174].
Specific point mutations in H3.3 have been shown to drive Diffuse Intrinsic Pontine Gliomas (DIPG, caused by the H3.3K27M mutation), high grade pediatric glioblastomas (H3.3G34R/V), giant cell tumors of the bone (H3.3G34W) and chondroblastomas (H3.3K36M) primarily in children [175–177]. Tumors associated with mutant H3.3 seem to have a generally early onset during development [177–179]. Pediatric high-grade gliomas (both H3.3K27M and H3.3G34R/V) make up about 11% of all reported pediatric central nervous system tumors, with approximately 400 new diagnosed cases in the United States of America every year [180]. Additionally, wild type H3.3 has been shown to be overexpressed in certain lung cancers where it promotes cancer metathesis and migration [181].
Defects in DNA repair are intimately linked to cancer formation [5]. Consistent with its putative role in cancer prevention, multiple studies, including our own unpublished studies, have found that H3.3 is recruited to sites of DNA damage, especially DSBs. Initially, H3.3 was shown to be recruited to sites of UV damage where it is required for replication fork progression in a HIRA dependent manner in mammalian [182] and chicken [183] cells. These findings suggest that H3.3 may play a role in the NER pathway.
The exact role of H3.3 in DNA repair is far from clear, in part due to the inconsistencies between the published studies [182–186]. H3.3 has been reported to be required for the efficient repair of DSBs via both NHEJ and HR pathways [184,185]. These studies suggested that H3.3 is deposited at DSBs in a Chromodomain Helicase DNA Binding Protein 2 (CHD2) and PARP1 dependent manner. Once deposited, H3.3 is believed to assist in the proper recruitment of X-Ray Repair Cross Complementing 4 (XRCC4) in a KU70/80 dependent manner to perform repair via the c-NHEJ pathway. However, data from these studies should be interpreted with caution since the U2OS cells used therein lack functional ATRX chaperone or p53, and relied on transient knockdown of H3.3 for assays that take weeks to complete [185]. Furthermore, these cancer cells also exhibit the Alternative Lengthening of Telomeres (ALT) phenotype and show highly aberrant H3.3S31 phosphorylation patterns [187]. Another study reported the deposition of H3.3 at sites of DSBs in a HIRA dependent manner, following which H3.3 was acetylated on lysine 12 by Histone Acetyltransferase 1 (HAT1) to facilitate proper RAD51 recruitment to Replication Protein A (RPA) coated single stranded DNA in the HR pathway [186]. Interestingly, subsequent research also suggests that H3.3 plays a crucial role in the recruitment of MUS81 and other downstream HR factors to assist in the proper formation of double Holliday junctions. Furthermore, even in this context, the recruitment the HR factors occurred in an ATRX/DAXX dependent manner [184,188].
So far, studies indicate that H3.3 does play important roles in DNA repair, specifically within pathways involved in DSB repair (HR & NHEJ), as well as possibly in the NER pathway. However, the data is still unclear on its specific functions within each pathway. Since nucleosomes containing H3.3 are less stable than those containing canonical H3 [173], it is possible that H3.3 incorporation may assist DNA repair by increasing the accessibility of the damage site to repair factors by opening up the chromatin. This would be particularly important for DSB repair via the HR pathway which involves long stretches of chromatin. H3.3 could also facilitate the proper recruitment of various DNA repair factors via direct interactions with them, or indirectly via interactions with its PTMs, including through its unique S31 residue in the N-terminus that can be reversibly phosphorylated. The S31 residue has been shown to be particularly important for the role of H3.3 in the NER pathway and for proper DNA synthesis after DNA repair [167]. Additionally, incorporation of H3.3 in chromatin has been shown to inhibit H1 binding in those regions [189]. Since histone H1 has been shown to promote DSB repair via NHEJ [41,49], it is possible that the presence of H3.3 in the vicinity of the DSBs serves as a novel repair pathway choice factor by destabilizing H1 binding, which in turn would suppress NHEJ, upon which HR may become the preferred pathway for DSB repair. Additional studies are needed to verify this possibility experimentally. Overall, although H3.3 appears to play a major role in multiple DNA repair pathways, further research is needed to clarify these roles at the molecular level. This would be particularly crucial for the development of therapeutics for cancers driven by H3.3 mutations.
Other H3 Variants
Apart from cenH3 and H3.3, additional H3 variants such as H3.4 (aka H3t), H3.5, H3.Y (aka H3.Y.1) and H3.Y.2 (aka H3.X) are known. However, it is currently unknown if they play a specific role in any DNA repair pathway. H3.4 is a testis specific H3 variant which shares 97% homology (Figure 3C) with the canonical H3 [190]. H3.4 incorporation results in nucleosomes with significantly less stability and is predicted to lead to more open chromatin [191]. H3.4 has also been be shown to play an essential role in spermatogenesis [192]. H3.5 is a primate specific variant of H3 which shares a ~93% homology with the canonical H3 and a ~96% homology with H3.3 (Figure 3C). Although H3.3 and H3.5 containing nucleosome core particles have nearly identical structures, the latter has been shown to have a decreased interaction with the neighboring H4 histone resulting in less stable nucleosomes [193]. H3.5 is specifically expressed in the seminiferous tubules of human testis and has been found to be deposited in the euchromatic regions of the genome. As a result of its similarity to H3.3 in amino acid sequence, H3.5 has been shown to compensate for the loss of H3.3 in cell culture [194,195]. This may endow H3.5 with a similar function to H3.3 in DNA repair within specific contexts, such as in HR during crossing-over in meiosis while forming gametes. However, further research needs to be done to verify this possibility. Finally, the primate specific H3 variants H3.Y.1 and H3.Y.2, both share a near 72% homology with the canonical H3 and have nearly identical amino acid sequence relative to each other (Figure 3C). Little is known about H3.Y.2, but H3.Y.1 has been shown to be incorporated around the transcriptional start sites of genes and affects the expression of genes involved in cell cycle progression. Nucleosomes with H3.Y.1 have also been found to form more relaxed and less tightly packaged chromatin than those containing either canonical H3 or H3.3 [196]. Additionally, just like H3.3 has been shown to counteract H1 deposition in chromatin [189], H3.Y.1 has also been shown to decrease the efficiency of linker histone H1 binding, and its incorporation would be predicted to further open up chromatin [195,196].
HISTONE H4
Until recently, there were no known H4 variants in mammalian cells [197–199]. However, two H4 variants H4.G and H4.I have been identified recently [200,201]. The H4G variant exhibits 85% identity to canonical human histone H4 and is missing the last 5 residues. It has been shown to be involved in regulating rDNA transcription and is overexpressed in breast cancer [200]. Currently, these newly identified histone H4 variants have not been studied in the context of DNA repair.
The relative lack of prominent histone H4 variants is more than compensated by the multiple PTMs on canonical histone H4 that have been shown to be important for DDR, specifically for regulating repair pathway choice between HR and NHEJ for repairing DSBs [202,203]. These PTMs have been shown to alter chromatin structure [204] to facilitate access to DNA damage sites, as well as provide a binding site for DNA repair factors [205]. For example, H4K20 has been shown to be methylated through much of the cell cycle [206]. Although this modification is fairly ubiquitous and found throughout the genome, both SET Domain Containing Lysine Methyltransferase 8 (SETD8) and Multiple Myeloma SET domain / Wold-Hirschhorn Syndrome Candidate 1 (MMSET/WHSC1) have been shown to sequentially methylate H4K20 at sites of DSBs [207–209]. This modification provides a binding site for the tandem Tudor domain of 53BP1, which in turn promotes c-NHEJ [205]. Paradoxically, the Nucleosome Acetyltransferase of histone H4 / Tat-Interactive Protein 60 (NuA4/TIP60) acetyltransferase complex has also been shown to bind to mono/di-methylated forms of H4K20 at sites of DSB, where it can potentially acetylate lysines at positions 5, 8, 12 and 16 on histone H4 to promote HR repair [210,211]. In addition to the competition between the NuA4/TIP60 complex and 53BP1 for binding to H4K20, recent research has found that the acetylation of H4K16 prevents 53BP1 binding at sites of DSBs [212]. Finally, the acetyltransferase HAT1 has been shown to facilitate the assembly of H3.1-H4 nucleosomes by preferentially acetylating H4K 5 and H4K12 in this complex over the analogous variant H3.3-H4 containing complexes [213].
CONCLUSIONS
Our knowledge of the roles that histone variants play in DNA repair is still evolving. While some histone variants have been studied to a considerable extent, much remains to be learned about the molecular roles that these variants play within specific DNA repair pathways, especially in mammals. In general, histone variants seem to play two specific roles within the DNA repair pathways. First, histone variants are actively recruited to sites of DNA damage to either assist/repel the binding of various DNA repair factors. This in turn could regulate the kinetics of DNA repair or influence the choice of the pathway used to repair the particular damage. Secondly, histone variants may also play of the role of a “place holder” for canonical nucleosomes during DNA repair, whereby variants are incorporated in nucleosomes to potentially stabilize chromatin, therefore preventing additional damage, while repair proceeds. This place holding function may be particularly important in repair pathways that require DNA synthesis (HR, BER, alt-NHEJ, NER), especially for repair that occurs outside of the S-phase of the cell cycle where the supply of the canonical histones is limited. Though we acknowledge the trivial possibility that some histone variants may simply be recruited to DNA damage sites in an opportunistic and non-consequential manner, it is very likely that several of the histone variants play very important roles in DNA repair under specific circumstances, as exemplified by the specific DNA repair defects observed in cells deficient in certain variants.
Table 1.
Histone variants with known or suspected roles in the DNA damage response or in specific DNA repair pathways. Percent homology indicates the percent similarity in the amino acid sequence of the variants compared to the primary amino acid sequence of canonical histones H2A type 1 for the H2A variants and H3.1 for the H3 variants.
| Variant Name | Histone Family | Percent Homology to canonical histone | Endogenous Localization | Involvement in DDR or DNA Repair Pathways | References |
|---|---|---|---|---|---|
| H1.2 | H1 | N/A | Enriched at silenced genes | HR, c-NHEJ, alt-NHEJ, apoptosis | [39,41,48–51] |
| H1.4 | H1 | N/A | Enriched at silenced genes | HR, c-NHEJ, alt-NHEJ | [52] |
| H1.0 | H1 | N/A | Repetitive DNA elements within the nucleoli; accumulates on silenced genes in non-dividing cells | N/A | [35–37] |
| H1.10 | H1 | N/A | Hypomethylated CpG islands and active genes | c-NHEJ | [35,39,51] |
| H2A.X | H2A | 90% | Ubiquitously throughout the genome | HR, c-NHEJ | [59,61,62,65,84–89]. |
| H2A.Z | H2A | 60% | Actively transcribing genes and centromeres | BER, HR, c-NHEJ | [90,93,96,98,99] |
| mH2A | H2A | 60% | Barr-body heterochromatin | BER, NER, HR | [110,122,123,125,126] |
| H2A.B | H2A | 50% | Ubiquitously (except in the Barr-body) | N/A | [131,132] |
| H2A.J | H2A | 94% | Enriched in senescent cells with DNA damage | N/A | [140,142] |
| cenH3 | H3 | 45% | Centromeres | BER, HR | [154,157,159,163,164,167,168] |
| H3.3 | H3 | 96% | Promoters of transcribing genes, telomeres, pericentromeric, rDNA heterochromatin | NER, HR, c-NHEJ | [169,182–186] |
| H3.4 | H3 | 97% | N/A | N/A | [190] |
| H3.5 | H3 | 93% | Euchromatic regions (seminiferous tubules) | N/A | [194,195] |
| H3.Y.1 | H3 | 72% | Transcription start sites | N/A | [195,196] |
N/A = Not applicable.
Highlights:
Specific histone variants play important roles in DNA repair following their recruitment to sites of DNA damage.
Histone variants regulate chromatin structure at sites of DNA damage to facilitate the recruitment and/or retention of specific DNA repair factors, often following posttranslational modifications.
In doing so, they affect the efficiency of DNA repair, or influence DNA repair pathway choice, or both.
Acknowledgements
We would like to thank our colleagues in the Biomedical Sciences Department at Florida State University for helpful and stimulating discussions. We specially thank Dr. Robert Tomko for help with navigating the nucleosome crystal structures using PyMol. Research in the Gunjan lab is supported by funding from the National Institutes of Health (CA206051-01A1), the National Science Foundation (MCB-1818026) and the and Florida Department of Health Live Like Bella Pediatric Cancer Research Initiative (20L01).
Abbreviations:
- 53BP1
p53 Binding Protein 1
- 8-oxoG
8-oxo-7,8-dihydroguanine
- alt-NHEJ
Alternative (or microhomology mediated) Nonhomologous End Joining
- ANP32E
Acidic Nuclear Phosphoprotein 32 Family Member E
- APLF
Aprataxin and PNK-like factor
- ALT
Alternative Lengthening of Telomeres
- ATM
Ataxia-Telangiectasia Mutated
- ATR
ATM and Rad3-related
- ATRX
Alpha-Thallassemia Mental Retardation X-linked protein
- BARD1
BRCA1 Associated RING Domain 1
- BER
Base Excision Repair
- bp
base pair
- BRCA1
Breast Cancer type 1
- BRCA2
Breast Cancer type 2
- c-NHEJ
Classic (or Canonical) Nonhomologous End Joining
- CHD2
Chromodomain Helicase DNA Binding Protein 2
- CHEK2
Checkpoint Kinase 2
- ChIP
Chromatin Immuno-Precipitation
- COMMD4
Copper Metabolism gene MURR1 Domain Containing 4
- CtIP
C-terminal binding protein 1 interacting protein
- DAXX
Death domain Associated
- DDB1
Damage Specific DNA Binding Protein 1
- DDR
DNA Damage Response
- DIPG
Diffuse Intrinsic Pontine Glioma
- DLBCL
Diffuse Large B-cell Lymphoma
- DNA-PK
DNA-Dependent Protein Kinase
- DSB
Double Strand Break
- DSBR
Double Strand Break Repair
- Exo1
Exonuclease 1
- FACT
Facilitates Chromatin Transcription
- HAT1
Histone Acetyltransferase 1
- HDAC1
Histone Deacetylase 1
- HIRA
Histone Regulator
- HJURP
Holliday Junction Recognition Protein
- HP1
Heterochromatin Protein 1
- HR
Homologous Recombination
- HUWE1
HECT, UBA And WWE Domain Containing E3 Ubiquitin Protein Ligase 1
- LSH
Lymphoid Specific Helicase
- MDC1
Mediator of DNA damage Checkpoint 1
- MMR
Mismatch Repair
- MMS
Methyl Methane Sulfonate
- MMSET
Multiple Myeloma SET domain
- MRN
MRE11-RAD50-NBS1 complex
- NER
Nucleotide Excision Repair
- NuA4
Nucleosome Acetyltransferase of histone H4
- PAR
poly(ADP-ribose)
- PARP1
poly-ADP Ribose Polymerase 1
- PCAF
P300/CBP-associated factor
- PCNA
Proliferating Cell Nuclear Antigen
- PTM
Posttranslational Modifications
- RPA
Replication Protein A
- RNF8
Ring Finger Protein 8
- RNF20
Ring Finger Protein 20
- RNF40
Ring Finger Protein 40
- RNF168
Ring Finger Protein 168
- SETD8
SET Domain Containing Lysine Methyltransferase 8
- shRNA
small hairpin RNA
- siRNA
small interfering RNA
- SRCAP
Snf2 Related CREBBP Activator Protein
- SSB
Single-Strand Break
- SSBR
Single-Strand Break Repair
- SWI/SNF
SWItch/Sucrose Non-Fermentable
- TAD
Topologically associating domain
- TAF1
Template Activating Factor 1
- TIP60
Tat-Interactive Protein 60
- UBC13
Ubiquitin-conjugating E2 enzyme 13
- UNG2
Uracil-DNA glycosylase 2
- WHSC1
Wold-Hirschhorn Syndrome Candidate 1
- WSTF
William-Beuren Syndrome Transcription Factor
- XRCC1
X-Ray Repair Cross Complementing 1
- XRCC4
X-Ray Repair Cross Complementing 4
References
- [1].Hoeijmakers J, DNA Damage, Aging, and Cancer, The New England Journal Medicine. 361 (2009) 1475–1485. 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- [2].Ciccia A, Elledge SJ, The DNA Damage Response: Making it safe to play with knives, Molecular Cell. 40 (2010) 179–204. 10.1016/J.MOLCEL.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Frankenberg-Schwager M, Frankenberg D, DNA double-strand breaks: their repair and relationship to cell killing in yeast, International Journal of Radiation Biology. 58 (1990) 569–575. 10.1080/09553009014551931. [DOI] [PubMed] [Google Scholar]
- [4].Bennett C, Lewis A, Baldwin K, Resnick M, Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid, Proceedings of the National Academy of Sciences of the United States of America. 90 (1993) 5613–5517. 10.1073/PNAS.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Alhmoud JF, Woolley JF, al Moustafa A-E, Malki MI, DNA Damage/Repair Management in Cancers, Cancers. 12 (2020) 1–22. 10.3390/CANCERS12041050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li G, Mechanisms and functions of DNA mismatch repair, Cell Research 2008 18:1. 18 (2007) 85–98. 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- [7].Schermerhorn K, Delaney S, A chemical and kinetic perspective on base excision repair of DNA, Accounts of Chemical Research. 47 (2014) 1238–1246. 10.1021/AR400275A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Vaughn CM, Sancar A, Mechanisms and Maps of Nucleotide Excision Repair, in: DNA Damage, DNA Repair and Disease, 2017: pp. 1–23. 10.1039/9781839162541-00001. [DOI] [Google Scholar]
- [9].Chang H, Pannunzio NR, Adachi N, Lieber MR, Non-homologous DNA end joining and alternative pathways to double-strand break repair, Nature Reviews Molecular Cell Biology 2017 18:8. 18 (2017) 495–506. 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li X, Heyer W, Homologous recombination in DNA repair and DNA damage tolerance, Cell Research 2008 18:1. 18 (2008) 99–113. 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Downs JA, Nussenzweig MC, Nussenzweig A, Chromatin dynamics and the preservation of genetic information, Nature 2007 447:7147. 447 (2007) 951–958. 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- [12].Luger K, Mäder A, Richmond R, Sargent D, Richmond T, Crystal structure of the nucleosome core particle at 2.8 A resolution, Nature. 389 (1997) 251–260. 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- [13].Zhou Y, Gerchman S, Ramakrishnan V, Travers A, Muyldermans S, Position and orientation of the globular domain of linker histone H5 on the nucleosome, Nature. 395 (1998) 402–405. 10.1038/26521. [DOI] [PubMed] [Google Scholar]
- [14].Harshman S, Young N, Parthun M, Freitas M, H1 histones: current perspectives and challenges, Nucleic Acids Research. 41 (2013) 9593–9609. 10.1093/NAR/GKT700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Flanagan TW, Brown DT, Molecular dynamics of histone H1, Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 1859 (2016) 468–475. 10.1016/J.BBAGRM.2015.10.005. [DOI] [PubMed] [Google Scholar]
- [16].Arimura Y, Ikura M, Fujita R, Noda M, Kobayashi W, Horikoshi N, Sun J, Shi L, Kusakabe M, Harata M, Ohkawa Y, Tashiro S, Kimura H, Ikura T, Kurumizaka H, Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome, Nucleic Acids Research. 46 (2018) 10007–10018. 10.1093/NAR/GKY661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chew GL, Bleakley M, Bradley RK, Malik HS, Henikoff S, Molaro A, Sarthy J, Short H2A histone variants are expressed in cancer, Nature Communications 2021 12:1. 12 (2021) 1–9. 10.1038/s41467-020-20707-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ferrand J, Rondinelli B, Polo SE, Histone Variants: Guardians of Genome Integrity, Cells. 9(11) (2020) 1–31. 10.3390/CELLS9112424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].White AE, Hieb AR, Luger K, A quantitative investigation of linker histone interactions with nucleosomes and chromatin, Scientific Reports. 6 (2016) 1–14. 10.1038/srep19122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang S, Vogirala VK, Soman A, Berezhnoy N. v., Liu ZB, Wong ASW, Korolev N, Su C-J, Sandin S, Nordenskiöld L, Linker histone defines structure and self-association behaviour of the 177 bp human chromatosome, Scientific Reports. 11 (2021) 1–16. 10.1038/s41598-020-79654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Brown D, Izard T, Misteli T, Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo, Nature Structural & Molecular Biology. 13 (2006) 250–255. 10.1038/NSMB1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Robinson P, Rhodes D, Structure of the “30 nm” chromatin fibre: a key role for the linker histone, Current Opinion in Structural Biology. 16 (2006) 336–343. 10.1016/J.SBI.2006.05.007. [DOI] [PubMed] [Google Scholar]
- [23].Fan Y, Nikitina T, Zhao J, Fleury T, Bhattacharyya R, Bouhassira E, Stein A, Woodcock C, Skoultchi A, Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation, Cell. 123 (2005) 1199–1212. 10.1016/J.CELL.2005.10.028. [DOI] [PubMed] [Google Scholar]
- [24].Vignali M, Workman J, Location and function of linker histones, Nature Structural Biology. 5 (1998) 1025–1028. 10.1038/4133. [DOI] [PubMed] [Google Scholar]
- [25].Thomas J, Histone H1: location and role, Current Opinion in Cell Biology. 11 (1999) 312–317. 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- [26].Hergeth SP, Schneider R, The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle, EMBO Reports. 16 (2015) 1439–1453. 10.15252/EMBR.201540749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Brockers K, Schneider R, Histone H1, the forgotten histone, Epigenomics. 11 (2019) 363–366. 10.2217/epi-2019-0018. [DOI] [PubMed] [Google Scholar]
- [28].Ponte I, Vila R, Suau P, Sequence complexity of histone H1 subtypes, Molecular Biology and Evolution. 20 (2003) 371–380. 10.1093/MOLBEV/MSG041. [DOI] [PubMed] [Google Scholar]
- [29].Notredame C, Higgins D, Heringa J, T-Coffee: A novel method for fast and accurate multiple sequence alignment, Journal of Molecular Biology. 302 (2000) 205–217. 10.1006/JMBI.2000.4042. [DOI] [PubMed] [Google Scholar]
- [30].Happel N, Doenecke D, Histone H1 and its isoforms: contribution to chromatin structure and function, Gene. 431 (2009) 1–12. 10.1016/J.GENE.2008.11.003. [DOI] [PubMed] [Google Scholar]
- [31].Brown DT, Alexander BT, Sittman DB, Differential effect of H1 variant overexpression on cell cycle progression and gene expression., Nucleic Acids Research. 24 (1996) 486–493. 10.1093/NAR/24.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Brown DT, Gunjan A, Alexander BT, Sittman DB, Differential effect of H1 variant overproduction on gene expression is due to differences in the central globular domain., Nucleic Acids Research. 25 (1997) 5003–5009. 10.1093/NAR/25.24.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Harvey AC, Downs JA, What functions do linker histones provide?, Molecular Microbiology. 53 (2004) 771–775. 10.1111/J.1365-2958.2004.04195.X. [DOI] [PubMed] [Google Scholar]
- [34].Zlatanova J, Doenecke D, Histone H1 zero: a major player in cell differentiation?, FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology. 8 (1994) 1260–1268. 10.1096/FASEBJ.8.15.8001738. [DOI] [PubMed] [Google Scholar]
- [35].Mayor R, Izquierdo-Bouldstridge A, Millán-Ariño L, Bustillos A, Sampaio C, Luque N, Jordan A, Genome distribution of replication-independent histone H1 variants shows H1.0 associated with nucleolar domains and H1X associated with RNA polymerase II-enriched regions, The Journal of Biological Chemistry. 290 (2015) 7474–7491. 10.1074/JBC.M114.617324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].De S, Brown D, Lu Z, Leno G, Wellman S, Sittman D, Histone H1 variants differentially inhibit DNA replication through an affinity for chromatin mediated by their carboxyl-terminal domains, Gene. 292 (2002) 173–181. 10.1016/S0378-1119(02)00675-3. [DOI] [PubMed] [Google Scholar]
- [37].Medrzycki M, Zhang Y, McDonald J, Fan Y, Profiling of linker histone variants in ovarian cancer, Frontiers in Bioscience (Landmark Edition). 17 (2012) 396–406. 10.2741/3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Warneboldt J, Haller F, Horstmann O, Danner BC, Füzesi L, Doenecke D, Happel N, Histone H1x is highly expressed in human neuroendocrine cells and tumours, BMC Cancer. 8 (2008) 1–9. 10.1186/1471-2407-8-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B, Narita T, Sixma TK, Choudhary C, Bekker-Jensen S, Mailand N, Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage, Nature 2015 527:7578. 527 (2015) 389–393. 10.1038/nature15401. [DOI] [PubMed] [Google Scholar]
- [40].Mandemaker IK, van Cuijk L, Janssens RC, Lans H, Bezstarosti K, Hoeijmakers JH, Demmers JA, Vermeulen W, Marteijn JA, DNA damage-induced histone H1 ubiquitylation is mediated by HUWE1 and stimulates the RNF8-RNF168 pathway, Scientific Reports 2017 7:1. 7 (2017) 1–11. 10.1038/s41598-017-15194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kysela B, Chovanec M, Jeggo PA, Phosphorylation of linker histones by DNA-dependent protein kinase is required for DNA ligase IV-dependent ligation in the presence of histone H1, Proceedings of the National Academy of Sciences. 102 (2005) 1877–1882. 10.1073/PNAS.0401179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Downs JA, Kosmidou E, Morgan A, Jackson SP, Suppression of Homologous Recombination by the Saccharomyces cerevisiae Linker Histone, Molecular Cell. 11 (2003) 1685–1692. 10.1016/S1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- [43].Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang S, Blasco MA, Skoultchi AI, Fernandez-Capetillo O, Global chromatin compaction limits the strength of the DNA damage response, Journal of Cell Biology. 178 (2007) 1101–1108. 10.1083/JCB.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yusufova N, Kloetgen A, Teater M, Osunsade A, Camarillo JM, Chin CR, Doane AS, Venters BJ, Portillo-Ledesma S, Conway J, Phillip JM, Elemento O, Scott DW, Béguelin W, Licht JD, Kelleher NL, Staudt LM, Skoultchi AI, Keogh M-C, Apostolou E, Mason CE, Imielinski M, Schlick T, David Y, Tsirigos A, Allis CD, Soshnev AA, Cesarman E, Melnick AM, Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture, Nature 2020 589:7841. 589 (2020) 299–305. 10.1038/s41586-020-3017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Th’ng J, Sung R, Ye M, Hendzel M, H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain, The Journal of Biological Chemistry. 280 (2005) 27809–27814. 10.1074/JBC.M501627200. [DOI] [PubMed] [Google Scholar]
- [46].Kim J, Kim K, Punj V, Liang G, Ulmer TS, Lu W, An W, Linker histone H1.2 establishes chromatin compaction and gene silencing through recognition of H3K27me3, Scientific Reports 2015 5:1. 5 (2015) 1–16. 10.1038/srep16714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sancho M, Diani E, Beato M, Jordan A, Depletion of Human Histone H1 Variants Uncovers Specific Roles in Gene Expression and Cell Growth, PLOS Genetics. 4 (2008) 1–17. 10.1371/JOURNAL.PGEN.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Li Z, Li Y, Tang M, Peng B, Lu X, Yang Q, Zhu Q, Hou T, Li M, Liu C, Wang L, Xu X, Zhao Y, Wang H, Yang Y, Zhu WG, Destabilization of linker histone H1.2 is essential for ATM activation and DNA damage repair, Cell Research. 28 (2018) 756–770. 10.1038/s41422-018-0048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rosidi B, Wang M, Wu W, Sharma A, Wang H, Iliakis G, Histone H1 functions as a stimulatory factor in backup pathways of NHEJ, Nucleic Acids Research. 36 (2008) 1610–1623. 10.1093/NAR/GKN013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Konishi A, Shimizu S, Hirota J, Takao T, Fan Y, Matsuoka Y, Zhang L, Yoneda Y, Fujii Y, Skoultchi A, Tsujimoto Y, Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks, Cell. 114 (2003) 673–688. 10.1016/S0092-8674(03)00719-0. [DOI] [PubMed] [Google Scholar]
- [51].Mandemaker I, Zhou D, Bruens S, Dekkers D, Verschure P, Edupuganti R, Meshorer E, Demmers J, Marteijn J, Histone H1 eviction by the histone chaperone SET reduces cell survival following DNA damage, Journal of Cell Science. 133 (2020) 1–11. 10.1242/JCS.235473. [DOI] [PubMed] [Google Scholar]
- [52].Li Y, Li Z, Dong L, Tang M, Zhang P, Zhang C, Cao Z, Zhu Q, Chen Y, Wang H, Wang T, Lv D, Wang L, Zhao Y, Yang Y, Wang H, Zhang H, Roeder R, Zhu W, Histone H1 acetylation at lysine 85 regulates chromatin condensation and genome stability upon DNA damage, Nucleic Acids Research. 46 (2018) 7716–7730. 10.1093/NAR/GKY568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Li Z, Kono H, Distinct Roles of Histone H3 and H2A Tails in Nucleosome Stability, Scientific Reports 2016 6:1. 6 (2016) 1–12. 10.1038/srep31437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Vogler C, Huber C, Waldmann T, Ettig R, Braun L, Izzo A, Daujat S, Chassignet I, Lopez-Contreras AJ, Fernandez-Capetillo O, Dundr M, Rippe K, Längst G, Schneider R, Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding, PLOS Genetics. 6 (2010) 1–12. 10.1371/JOURNAL.PGEN.1001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Horikoshi N, Sato K, Shimada K, Arimura Y, Osakabe A, Tachiwana H, Hayashi-Takanaka Y, Iwasaki W, Kagawa W, Harata M, Kimura H, Kurumizaka H, Structural polymorphism in the L1 loop regions of human H2A.Z.1 and H2A.Z.2, Acta Crystallographica. Section D, Biological Crystallography. 69 (2013) 2431–2439. 10.1107/S090744491302252X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kurumizaka H, Kujirai T, Takizawa Y, Contributions of Histone Variants in Nucleosome Structure and Function, Journal of Molecular Biology. 433 (2021). 10.1016/JJMB.2020.10.012. [DOI] [PubMed] [Google Scholar]
- [57].Chakravarthy S, Gundimella SKY, Caron C, Perche P-Y, Pehrson JR, Khochbin S, Luger K, Structural characterization of the histone variant macroH2A, Molecular and Cellular Biology. 25 (2005) 7616–7624. 10.1128/MCB.25.17.7616-7624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Uckelmann M, Sixma TK, Histone ubiquitination in the DNA damage response, DNA Repair. 56 (2017) 92–101. 10.1016/J.DNAREP.2017.06.011. [DOI] [PubMed] [Google Scholar]
- [59].Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y, γH2AX and cancer, Nature Reviews. Cancer 8 (2008) 957. 10.1038/NRC2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM, DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139, Journal of Biological Chemistry. 273 (1998) 5858–5868. 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- [61].Piquet S, Parc FL, Bai SK, Chevallier O, Adam S, Polo SE, The Histone Chaperone FACT Coordinates H2A.X-Dependent Signaling and Repair of DNA Damage, Molecular Cell. 72 (2018) 888–901. 10.1016/J.MOLCEL.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Heo K, Kim K, Choi S, Choi J, Kim K, Gu J, Lieber M, Yang A, An W, FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16, Molecular Cell. 30 (2008) 86–97. 10.1016/J.MOLCEL.2008.02.029. [DOI] [PubMed] [Google Scholar]
- [63].Sharma D, de Falco L, Padavattan S, Rao C, Geifman-Shochat S, Liu CF, Davey CA, PARP1 exhibits enhanced association and catalytic efficiency with γH2A.X-nucleosome, Nature Communications 2019 10:1. 10 (2019) 1–12. 10.1038/s41467-019-13641-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bewersdorf J, Bennett B, Knight K, H2AX chromatin structures and their response to DNA damage revealed by 4Pi microscopy, Proceedings of the National Academy of Sciences of the United States of America. 103 (2006) 18137–18142. 10.1073/PNAS.0608709103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Rogakou EP, Boon C, Redon C, Bonner WM, Megabase Chromatin Domains Involved in DNA Double-Strand Breaks In Vivo, The Journal of Cell Biology. 146 (1999) 905–915. http://www.jcb.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Collins PL, Purman C, Porter SI, Nganga V, Saini A, Hayer KE, Gurewitz GL, Sleckman BP, Bednarski JJ, Bassing CH, Oltz EM, DNA double-strand breaks induce H2Ax phosphorylation domains in a contact-dependent manner, Nature Communications 2020 11:1. 11 (2020) 1–9. 10.1038/s41467-020-16926-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Löbrich M, Jeggo P, ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation, Cancer Research. 64 (2004) 2390–2396. 10.1158/0008-5472.CAN-03-3207. [DOI] [PubMed] [Google Scholar]
- [68].Ward I, Chen J, Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress, The Journal of Biological Chemistry. 276 (2001) 47759–47762. 10.1074/JBC.C100569200. [DOI] [PubMed] [Google Scholar]
- [69].Burma S, Chen B, Murphy M, Kurimasa A, Chen D, ATM phosphorylates Histone H2AX in Response to DNA Double-strand Breaks, The Journal of Biological Chemistry. 276 (2001) 42462–42467. 10.1074/JBC.C100466200. [DOI] [PubMed] [Google Scholar]
- [70].Dickey JS, Redon CE, Nakamura AJ, Baird BJ, Sedelnikova OA, Bonner WM, H2AX: Functional roles and potential applications, Chromosoma. 118 (2009) 683–692. 10.1007/s00412-009-0234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Foster ER, Downs JA, Histone H2A phosphorylation in DNA double-strand break repair, The FEBS Journal. 272 (2005) 3231–3240. 10.1111/J.1742-4658.2005.04741.X. [DOI] [PubMed] [Google Scholar]
- [72].Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP, MDC1 Directly Binds Phosphorylated Histone H2AX to Regulate Cellular Responses to DNA Double-Strand Breaks, Cell. 123 (2005) 1213–1226. 10.1016/J.CELL.2005.09.038. [DOI] [PubMed] [Google Scholar]
- [73].Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J, Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention, The EMBO Journal. 23 (2004) 2674–2683. 10.1038/SJ.EMBOJ.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J, RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins, Cell. 131 (2007) 887–900. 10.1016/J.CELL.2007.09.040. [DOI] [PubMed] [Google Scholar]
- [75].Attikum H, Gasser S, Crosstalk between histone modifications during the DNA damage response, Trends in Cell Biology. 19 (2009) 207–217. 10.1016/J.TCB.2009.03.001. [DOI] [PubMed] [Google Scholar]
- [76].Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C, RNF168 Binds and Amplifies Ubiquitin Conjugates on Damaged Chromosomes to Allow Accumulation of Repair Proteins, Cell. 136 (2009) 435–446. 10.1016/J.CELL.2008.12.041. [DOI] [PubMed] [Google Scholar]
- [77].Stewart G, Panier S, Townsend K, Al-Hakim A, Kolas N, Miller E, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd P, Stankovic T, Taylor A, Durocher D, The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage, Cell. 136 (2009) 420–434. 10.1016/J.CELL.2008.12.042. [DOI] [PubMed] [Google Scholar]
- [78].Cha H, Lowe JM, Li H, Lee JS, Belova GI, Bulavin D. v., Fornace AJ, Wip1 directly dephosphorylates γ-H2AX and attenuates the DNA damage response, Cancer Research. 70 (2010) 4112–4122. 10.1158/0008-5472.CAN-09-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Celeste A, Petersen S, Romanienko P, Fernandez-Capetillo O, Chen H, Sedelnikova O, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio M, Redon C, Pilch D, Olaru A, Eckhaus M, Camerini-Otero R, Tessarollo L, Livak F, Manova K, Bonner W, Nussenzweig M, Nussenzweig A, Genomic Instability in Mice Lacking Histone H2AX, Science (New York, N.Y.). 296 (2002) 922–927. 10.1126/SCIENCE.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Downs J, Lowndes N, Jackson S, A role for Saccharomyces cerevisiae histone H2A in DNA repair, Nature. 408 (2000) 1001–1004. 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- [81].Madigan JP, Chotkowski HL, Glaser RL, DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis, Nucleic Acids Research. 30 (2002) 3698–3705. 10.1093/NAR/GKF496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, Patel DJ, Elledge SJ, Allis CD, WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity, Nature. 457 (2008) 57–62. 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG, Tyrosine Dephosphorylation of H2AX Modulates Apoptosis and Survival Decisions, Nature. 458 (2009) 591. 10.1038/NATURE07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Podhorecka M, Skladanowski A, Bozko P, H2AX phosphorylation: Its role in DNA damage response and cancer therapy, Journal of Nucleic Acids. 2010 (2010) 1–9. 10.4061/2010/920161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Turinetto V, Giachino C, Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions, Nucleic Acids Research. 43 (2015) 2489–2498. 10.1093/NAR/GKV061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A, H2AX: the histone guardian of the genome, DNA Repair. 3 (2004) 959–967. 10.1016/J.DNAREP.2004.03.024. [DOI] [PubMed] [Google Scholar]
- [87].Celeste A, Fernandez-Capetillo O, Kruhlak M, Pilch D, Staudt D, Lee A, Bonner R, Bonner W, Nussenzweig A, Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks, Nature Cell Biology. 5 (2003) 675–679. 10.1038/NCB1004. [DOI] [PubMed] [Google Scholar]
- [88].Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, Kimura H, Ikura M, Nishikubo S, Ito T, Muto A, Miyagawa K, Takeda S, Fishel R, Igarashi K, Kamiya K, DNA Damage-Dependent Acetylation and Ubiquitination of H2AX Enhances Chromatin Dynamics, Molecular and Cellular Biology. 27 (2007) 7028–7040. 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Li A, Eirín-López JM, Ausió J, H2AX: tailoring histone H2A for chromatin-dependent genomic integrity, Biochemistry and Cell Biology = Biochimie et Biologie Cellulaire. 83 (2005) 505–515. 10.1139/005-114. [DOI] [PubMed] [Google Scholar]
- [90].Suto RK, Clarkson MJ, Tremethick DJ, Luger K, Crystal structure of a nucleosome core particle containing the variant histone H2A.Z, Nature Structural Biology. 7 (2000) 1121–1124. http://structbio.nature.com. [DOI] [PubMed] [Google Scholar]
- [91].Coon JJ, Ueberheide B, Syka JEP, Dryhurst DD, Ausio J, Shabanowitz J, Hunt DF, Protein identification using sequential ion/ion reactions and tandem mass spectrometry, Proceedings of the National Academy of Sciences. 102 (2005) 9463–9468. 10.1073/PNAS.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Dryhurst D, Ishibashi T, Rose KL, Eirín-López JM, McDonald D, Silva-Moreno B, Veldhoen N, Helbing CC, Hendzel MJ, Shabanowitz J, Hunt DF, Ausió J, Characterization of the histone H2A.Z-1 and H2A.Z-2 isoforms in vertebrates, BMC Biology. 7 (2009). 10.1186/1741-7007-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Greaves IK, Rangasamy D, Ridgway P, Tremethick DJ, H2A.Z contributes to the unique 3D structure of the centromere, Proceedings of the National Academy of Sciences. 104 (2007) 525–530. 10.1073/PNAS.0607870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausió J, Characterization of the Stability and Folding of H2A.Z Chromatin Particles, Journal of Biological Chemistry. 276 (2001) 41945–41949. 10.1074/JBC.M108217200. [DOI] [PubMed] [Google Scholar]
- [95].Stargell L, Bowen J, Dadd C, Dedon P, Davis M, Cook R, Allis C, Gorovsky M, Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila, Genes & Development. 7 (1993) 2641–2651. 10.1101/GAD.7.12B.2641. [DOI] [PubMed] [Google Scholar]
- [96].Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL, Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity, Cell. 144 (2011) 200–213. 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Papamichos-Chronakis M, Krebs JE, Peterson CL, Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage, Genes and Development. 20 (2006) 2437–2449. 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Attikum H. v., Fritsch O, Gasser SM, Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks, EMBO Journal. 26 (2007) 4113–4125. 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD, Histone H2A.Z Controls a Critical Chromatin Remodeling Step Required for DNA Double-Strand Break Repair, Molecular Cell. 48 (2012) 723–733. 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Taty-Taty GC, Courilleau C, Quaranta M, Carayon A, Chailleux C, Aymard F, Trouche D, Canitrot Y, H2A.Z depletion impairs proliferation and viability but not DNA double-strand breaks repair in human immortalized and tumoral cell lines, Cell Cycle. 13 (2014) 399–407. 10.4161/cc.27143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Wlodkowic D, Darzynkiewicz Z, Please do not disturb: destruction of chromatin structure by supravital nucleic acid probes revealed by a novel assay of DNA-histone interaction, Cytometry. Part A : The Journal of the International Society for Analytical Cytology. 73 (2008) 877–879. 10.1002/CYTO.A.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Kumar A, Brown D, Leno G, DNA intercalators differentially affect chromatin structure and DNA replication in Xenopus egg extract, Anti-Cancer Drugs. 15 (2004) 633–639. 10.1097/01.CAD.0000131686.14013.4F. [DOI] [PubMed] [Google Scholar]
- [103].Gursoy-Yuzugullu O, Ayrapetov MK, Price BD, Histone chaperone Anp32e removes H2A.Z from DNA double-strand breaks and promotes nucleosome reorganization and DNA repair, Proceedings of the National Academy of Sciences. 112 (2015) 7507–7512. 10.1073/PNAS.1504868112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Alatwi HE, Downs JA, Removal of H2A.Z by INO80 promotes homologous recombination, EMBO Reports. 16 (2015) 986–994. 10.15252/EMBR.201540330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Nishibuchi I, Suzuki H, Kinomura A, Sun J, Liu N, Horikoshi Y, Shima H, Kusakabe M, Harata M, Fukagawa T, Ikura T, Ishida T, Nagata Y, Tashiro S, Reorganization of damaged chromatin by the exchange of histone variant H2A.Z-2, International Journal of Radiation Oncology, Biology, Physics. 89 (2014) 736–744. 10.1016/J.IJROBP.2014.03.031. [DOI] [PubMed] [Google Scholar]
- [106].Van C, Williams JS, Kunkel TA, Peterson CL, Deposition of histone H2A.Z by the SWR-C remodeling enzyme prevents genome instability, DNA Repair. 25 (2015) 9–14. 10.1016/j.dnarep.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Mimitou EP, Symington LS, Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing, Nature. 455 (2008) 770–774. 10.1038/NATURE07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Frank-Vaillant M, Marcand S, Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination, Molecular Cell. 10 (2002) 1189–1199. 10.1016/S1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- [109].Kim J, Krasieva T, Kurumizaka H, Chen D, Taylor A, Yokomori K, Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells, The Journal of Cell Biology. 170 (2005) 341–347. 10.1083/JCB.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Li C, Delaney S, Histone H2A Variants Enhance the Initiation of Base Excision Repair in Nucleosomes, ACS Chemical Biology. 14 (2019) 1041–1050. 10.1021/acschembio.9b00229. [DOI] [PubMed] [Google Scholar]
- [111].Yu Y, Deng Y, Reed SH, Millar CB, Waters R, Histone variant Htz1 promotes histone H3 acetylation to enhance nucleotide excision repair in Htz1 nucleosomes, Nucleic Acids Research. 41 (2013) 9006–9019. 10.1093/NAR/GKT688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Bowerman S, Hickok RJ, Wereszczynski J, Unique Dynamics in Asymmetric macroH2A-H2A Hybrid Nucleosomes Result in Increased Complex Stability, Journal of Physical Chemistry B. 123 (2019) 419–427. 10.1021/ACS.JPCB.8B10668/SUPPL_FILE/JP8B10668_SI_001.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Changolkar LN, Pehrson JR, Reconstitution of nucleosomes with histone macroH2A1.2, Biochemistry. 41 (2002) 179–184. 10.1021/BI0157417. [DOI] [PubMed] [Google Scholar]
- [114].Gamble MJ, Kraus WL, Multiple facets of the unique histone variant macroH2A: From genomics to cell biology, Cell Cycle. 9 (2010) 2568–2574. 10.4161/cc.9.13.12144. [DOI] [PubMed] [Google Scholar]
- [115].Chadwick BP, Willard HF, Histone H2A variants and the inactive X chromosome: identification of a second macroH2A variant, Human Molecular Genetics. 10 (2001) 1101–1113. [DOI] [PubMed] [Google Scholar]
- [116].Pehrson JR, Fuji RN, Evolutionary conservation of histone macroH2A subtypes and domains., Nucleic Acids Research. 26 (1998) 2837. 10.1093/NAR/26.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Chaudhuri AR, Nussenzweig A, The multifaceted roles of PARP1 in DNA repair and chromatin remodelling, Nature Reviews Molecular Cell Biology. 18 (2017) 610–621. 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG, The macro domain is an ADP-ribose binding module, EMBO Journal. 24 (2005) 1911–1920. 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Posavec M, Timinszky G, Buschbeck M, Macro domains as metabolite sensors on chromatin, Cell. Mol. Life Sci 70 (2013) 1509–1524. 10.1007/s00018-013-1294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Xu C, Xu Y, Gursoy-Yuzugullu O, Price BD, The histone variant macroH2A1.1 is recruited to DSBs through a mechanism involving PARP1, FEBS Letters. 586 (2012) 3920–3925. 10.1016/j.febslet.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Timinszky G, Till S, Hassa P, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer E, Scheffzek K, Hottiger M, Ladurner A, A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation, Nature Structural & Molecular Biology. 16 (2009) 923–929. 10.1038/NSMB.1664. [DOI] [PubMed] [Google Scholar]
- [122].Ruiz PD, Hamilton GA, Park JW, Gamble MJ, MacroH2A1 Regulation of Poly(ADP-Ribose) Synthesis and Stability Prevents Necrosis and Promotes DNA Repair, Molecular and Cellular Biology. 40 (2020) 1–17. 10.1128/MCB.00230-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Kim J, Sun C, Tran AD, Chin P-J, Ruiz PD, Wang K, Gibbons RJ, Gamble MJ, Liu Y, Oberdoerffer P, The macroH2A1.2 histone variant links ATRX loss to alternative telomere lengthening, Nature Structural & Molecular Biology. 26 (2019) 213–219. 10.1038/s41594-019-0192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Kim J, Sturgill D, Sebastian R, Khurana S, Tran AD, Edwards GB, Kruswick A, Burkett S, Hosogane EK, Hannon WW, Weyemi U, Bonner W, Luger K, Oberdoerffer P, Replication Stress Shapes a Protective Chromatin Environment across Fragile Genomic Regions, Molecular Cell. 69 (2018) 36–47. 10.1016/J.MOLCEL.2017.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Sebastian R, Hosogane EK, Sun EG, Tran AD, Reinhold WC, Burkett S, Sturgill DM, Gudla PR, Pommier Y, Aladjem MI, Oberdoerffer P, Epigenetic Regulation of DNA Repair Pathway Choice by MacroH2A1 Splice Variants Ensures Genome Stability, Molecular Cell. 79 (2020) 836–845.e7. 10.1016/j.molcel.2020.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Khurana S, Kruhlak MJ, Kim J, Tran AD, Liu J, Nyswaner K, Shi L, Jailwala P, Sung M-H, Hakim O, Oberdoerffer P, A Macrohistone Variant Links Dynamic Chromatin Compaction to BRCA1-Dependent Genome Maintenance, Cell Reports. 8 (2014) 1049–1062. 10.1016/J.CELREP.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Kozlowski M, Corujo D, Hothorn M, Guberovic I, Mandemaker IK, Blessing C, Sporn J, Gutierrez-Triana A, Smith R, Portmann T, Treier M, Scheffzek K, Huet S, Timinszky G, Buschbeck M, Ladurner AG, MacroH2A histone variants limit chromatin plasticity through two distinct mechanisms, EMBO Reports. 19 (2018) 1–13. 10.15252/embr.201744445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO, Menendez S, Vardabasso C, LeRoy G, Vidal CI, Polsky D, Osman I, Garcia BA, Hernando E, Bernstein E, The histone variant macroH2A suppresses melanoma progression through regulation of CDK8, Nature 2010 468:7327. 468 (2010) 1105–1109. 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Gaspar-Maia A, Qadeer ZA, Hasson D, Ratnakumar K, Leu NA, Leroy G, Liu S, Costanzi C, Valle-Garcia D, Schaniel C, Lemischka I, Garcia B, Pehrson JR, Bernstein E, MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency, Nature Communications. 4 (2013) 1–12. 10.1038/NCOMMS2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Xu X, Ni K, He Y, Ren J, Sun C, Liu Y, Aladjem MI, Burkett S, Finney R, Ding X, Sharan SK, Muegge K, The epigenetic regulator LSH maintains fork protection and genomic stability via MacroH2A deposition and RAD51 filament formation, Nature Communications. 12 (2021) 1–16. 10.1038/s41467-021-23809-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Chadwick BP, Willard HF, A Novel Chromatin Protein, Distantly Related to Histone H2A, Is Largely Excluded from the Inactive X Chromosome, The Journal of Cell Biology. 152 (2001) 375–384. http://www.jcb.org/cgi/content/full/152/2/375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Bao Y, Konesky K, Park YJ, Rosu S, Dyer PN, Rangasamy D, Tremethick DJ, Laybourn PJ, Luger K, Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA, EMBO Journal. 23 (2004) 3314–3324. 10.1038/sj.emboj.7600316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Tolstorukov MY, Goldman JA, Gilbert C, Ogryzko V, Kingston RE, Park PJ, Histone Variant H2A.Bbd Is Associated with Active Transcription and mRNA Processing in Human Cells, Molecular Cell. 47 (2012) 596–607. 10.1016/j.molcel.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Hirano R, Arimura Y, Kujirai T, Shibata M, Okuda A, Morishima K, Inoue R, Sugiyama M, Kurumizaka H, Histone variant H2A.B-H2B dimers are spontaneously exchanged with canonical H2A-H2B in the nucleosome, Communications Biology. 4 (2021) 1–11. 10.1038/s42003-021-01707-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Molaro A, Wood AJ, Janssens D, Kindelay SM, Eickbush MT, Wu S, Singh P, Muller CH, Henikoff S, Malik HS, Biparental contributions of the H2A.B histone variant control embryonic development in mice, PLOS Biology. 18 (2020) 1–28. 10.1371/JOURNAL.PBIO.3001001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Sansoni V, Casas-Delucchi CS, Rajan M, Schmidt A, Bönisch C, Thomae AW, Staege MS, Hake SB, Cardoso MC, Imhof A, The histone variant H2A.Bbd is enriched at sites of DNA synthesis, Nucleic Acids Research. 42 (2014) 6405–6420. 10.1093/nar/gku303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Arimura Y, Kimura H, Oda T, Sato K, Osakabe A, Tachiwana H, Sato Y, Kinugasa Y, Ikura T, Sugiyama M, Sato M, Kurumizaka H, Structural basis of a nucleosome containing histone H2A.B/H2A.Bbd that transiently associates with reorganized chromatin, Scientific Reports. 3 (2013) 1–9. 10.1038/srep03510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Menoni H, Gasparutto D, Hamiche A, Cadet J, Dimitrov S, Bouvet P, Angelov D, ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes, Molecular and Cellular Biology. 27 (2007) 5949–5956. 10.1128/MCB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Anuar ND, Kurscheid S, Field M, Zhang L, Rebar E, Gregory P, Buchou T, Bowles J, Koopman P, Tremethick DJ, Soboleva TA, Gene editing of the multi-copy H2A.B gene and its importance for fertility, Genome Biology. 20 (2019) 1–16. 10.1186/S13059-019-1633-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Nishida H, Suzuki T, Tomaru Y, Hayashizaki Y, A novel replication-independent histone H2a gene in mouse, BMC Genetics. 6 (2005) 1–7. 10.1186/1471-2156-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Tanaka H, Sato S, Koyama M, Kujirai T, Kurumizaka H, Biochemical and structural analyses of the nucleosome containing human histone H2A.J, Journal of Biochemistry. 167 (2020) 419–427. 10.1093/JB/MVZ109. [DOI] [PubMed] [Google Scholar]
- [142].Contrepois K, Coudereau C, Benayoun BA, Schuler N, Roux P, Bischof O, Courbeyrette R, Carvalho C, Thuret J, Ma Z, Derbois C, Nevers M, Volland H, Redon CE, Bonner WM, Deleuze J, Wiel C, Bernard D, Snyder MP, Rübe CE, Olaso R, Fenaille F, Mann C, Histone variant H2A.J accumulates in senescent cells and promotes inflammatory gene expression, Nature Communications. 8 (2017) 1–18. 10.1038/NCOMMS14995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Santoro SW, Dulac C, The activity-dependent histone variant H2BE modulates the life span of olfactory neurons, ELife. 1 (2012) 1–32. 10.7554/ELIFE.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Govin J, Escoffier E, Rousseaux S, Kuhn L, Ferro M, Thévenon J, Catena R, Davidson I, Garin J, Khochbin S, Caron C, Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis, The Journal of Cell Biology. 176 (2007) 283–294. 10.1083/JCB.200604141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Boulard M, Gautier T, Mbele GO, Gerson V, Hamiche A, Angelov D, Bouvet P, Dimitrov S, The NH2 Tail of the Novel Histone Variant H2BFWT Exhibits Properties Distinct from Conventional H2B with Respect to the Assembly of Mitotic Chromosomes, Molecular and Cellular Biology. 26 (2006) 1518–1526. 10.1128/MCB.26.4.1518-1526.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Zalensky AO, Siino JS, Gineitis AA, Zalenskaya IA, Tomilin NV, Yau P, Morton Bradbury E, Human Testis/Sperm-specific Histone H2B (hTSH2B), Journal of Biological Chemistry. 277 (2002) 43474–43480. 10.1074/jbc.M206065200. [DOI] [PubMed] [Google Scholar]
- [147].Aul RB, Oko RJ, The major subacrosomal occupant of bull spermatozoa is a novel histone H2B variant associated with the forming acrosome during spermiogenesis, Developmental Biology. 239 (2001) 376–387. 10.1006/DBIO.2001.0427. [DOI] [PubMed] [Google Scholar]
- [148].Suraweera A, Gandhi NS, Beard S, Burgess JT, Croft L. v., Bolderson E, Naqi A, Ashton NW, Adams MN, Savage KI, Zhang S-D, O’Byrne KJ, Richard DJ, COMMD4 functions with the histone H2A-H2B dimer for the timely repair of DNA double-strand breaks, Communications Biology. 4 (2021) 1–11. 10.1038/s42003-021-01998-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Fernandez-Capetillo O, Allis CD, Nussenzweig A, Phosphorylation of histone H2B at DNA double-strand breaks, Journal of Experimental Medicine. 199 (2004) 1671–1677. 10.1084/jem.20032247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, Shkedy D, Smorodinsky NI, van Vliet N, Kuster B, Mann M, Ciechanover A, Dahm-Daphi J, Kanaar R, Hu MCT, Chen DJ, Oren M, Shiloh Y, Requirement of ATM-Dependent Monoubiquitylation of Histone H2B for Timely Repair of DNA Double-Strand Breaks, Molecular Cell. 41 (2011) 529–542. 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira D, Shimada M, Tauchi H, Suzuki H, Tashiro S, Zou L, Komatsu K, Regulation of homologous recombination by RNF20-dependent H2B ubiquitination, Molecular Cell. 41 (2011) 515–528. 10.1016/J.MOLCEL.2011.02.002. [DOI] [PubMed] [Google Scholar]
- [152].Verreault A, Kaufman P, Kobayashi R, Stillman B, Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4, Cell. 87 (1996) 95–104. 10.1016/S0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- [153].Bannister AJ, Kouzarides T, Regulation of chromatin by histone modifications, Cell Research. 21 (2011) 381–395. 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Quénet D, Dalal Y, The CENP-A nucleosome: A dynamic structure and role at the centromere, Chromosome Research. 20 (2012) 465–479. 10.1007/sl0577-012-9301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Tachiwana H, Kagawa W, Kurumizaka H, Comparison between the CENP-A and histone H3 structures in nucleosomes, Nucleus. 3 (2012) 6–11. 10.4161/nucl.18372. [DOI] [PubMed] [Google Scholar]
- [156].Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park S-Y, Kimura H, Kurumizaka H, Crystal structure of the human centromeric nucleosome containing CENP-A, Nature. 476 (2011) 232–236. 10.1038/naturel0258. [DOI] [PubMed] [Google Scholar]
- [157].Zeitlin SG, Baker NM, Chapados BR, Soutoglou E, Wang JYJ, Berns MW, Cleveland DW, Double-strand DNA breaks recruit the centromeric histone CENP-A, Proceedings of the National Academy of Sciences of the United States of America. 106 (2009) 15762–15767. 10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Pessina F, Lowndes NF, The RSF1 Histone-Remodelling Factor Facilitates DNA Double-Strand Break Repair by Recruiting Centromeric and Fanconi Anaemia Proteins, PLOS Biology. 12 (2014) 1–17. 10.1371/JOURNAL.PBIO.1001856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Dunleavy E, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G, HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres, Cell. 137 (2009) 485–497. 10.1016/J.CELL.2009.02.040. [DOI] [PubMed] [Google Scholar]
- [160].Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y, Daigo Y, Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells, Cancer Research. 67 (2007) 8544–8553. 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- [161].Hédouin S, Grillo G, Ivkovic I, Velasco G, Francastel C, CENP-A chromatin disassembly in stressed and senescent murine cells, Scientific Reports . 7 (2017) 1–14. 10.1038/srep42520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Barra V, Fachinetti D, The dark side of centromeres: types, causes and consequences of structural abnormalities implicating centromeric DNA, Nature Communications. 9 (2018) 1–17. 10.1038/S41467-018-06545-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Filipescu D, Naughtin M, Podsypanina K, Lejour V, Wilson L, Gurard-Levin ZA, Orsi GA, Simeonova I, Toufektchan E, Attardi LD, Toledo F, Almouzni G, Essential role for centromeric factors following p53 loss and oncogenic transformation, Genes & Development. 31 (2017) 463–480. 10.1101/GAD.290924.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Jeffery D, Gatto A, Podsypanina K, Renaud-Pageot C, Landete RP, Bonneville L, Dumont M, Fachinetti D, Almouzni G, CENP-A overexpression promotes distinct fates in human cells, depending on p53 status, Communicatios Biology. 4 (2021) 1–18. 10.1038/s42003-021-01941-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K, Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase, Genes to Cells : Devoted to Molecular & Cellular Mechanisms. 9 (2004) 105–120. 10.llll/J.1365-2443.2004.00705.X. [DOI] [PubMed] [Google Scholar]
- [166].Saxena A, Saffery R, Wong LH, Kalitsis P, Choo KHA, Centromere Proteins Cenpa, Cenpb, and Bub3 Interact with Poly(ADP-ribose) Polymerase-1 Protein and Are Poly(ADP-ribosyl)ated *, Journal of Biological Chemistry. 277 (2002) 26921–26926. 10.1074/JBC.M200620200. [DOI] [PubMed] [Google Scholar]
- [167].Zeitlin SG, Chapados BR, Baker NM, Tai C, Slupphaug G, Uracil DNA N-Glycosylase Promotes Assembly of Human Centromere Protein A, PLoS ONE. 6 (2011) 17151–17151. 10.1371/journal.pone.0017151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Zeitlin SG, Patel S, Kavli B, Slupphaug G, Xenopus CENP-A assembly into chromatin requires base excision repair proteins, DNA Repair. 4 (2005) 760–772. 10.1016/j.dnarep.2005.02.007. [DOI] [PubMed] [Google Scholar]
- [169].Szenker E, Ray-Gallet D, Almouzni G, The double face of the histone variant H3.3, Cell Research. 21 (2011) 421–434. 10.1038/cr.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Elsässer SJ, Noh K-M, Diaz N, David Allis C, Banaszynski LA, Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells, Nature. 522 (2015) 240–244. 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD, Distinct Factors Control Histone Variant H3.3 Localization at Specific Genomic Regions, Cell. 140 (2010) 678–691. 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y, Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis, Cell. 116 (2004) 51–61. 10.1016/S0092-8674(03)01064-X. [DOI] [PubMed] [Google Scholar]
- [173].Jin C, Felsenfeld G, Nucleosome stability mediated by histone variants H3.3 and H2A.Z, Genes & Development. 21 (2007) 1519–1529. 10.1101/GAD.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Tachiwana H, Osakabe A, Shiga T, Miya Y, Kimura H, Kagawa W, Kurumizaka H, Structures of human nucleosomes containing major histone H3 variants, Acta Crystallographica. Section D, Biological Crystallography. 67 (2011) 578–583. 10.1107/S0907444911014818. [DOI] [PubMed] [Google Scholar]
- [175].Bjerke L, Mackay A, Nandhabalan M, Burford A, Jury A, Popov S, Bax D, Carvalho D, Taylor K, Vinci M, Bajrami I, McGonnell I, Lord C, Reis R, Hargrave D, Ashworth A, Workman P, Jones C, Histone H3.3. mutations drive pediatric glioblastoma through upregulation of MYCN, Cancer Discovery. 3 (2013) 512–519. 10.1158/2159-8290.CD-12-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Fang D, Gan H, Lee J, Han J, Wang Z, Riester SM, Jin L, Chen J, Zhou H, Wang J, Zhang H, Yang N, Bradley EW, Ho TH, Rubin BP, Bridge JA, Thibodeau SN, Ordog T, Chen Y, Wijnen AJ, Oliveira AM, Xu R, Westendorf JJ, Zhang Z, The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas, Science. 352 (2016) 1344–1348. 10.1126/SCIENCE.AAE0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Lulla RR, Saratsis AM, Hashizume R, Mutations in chromatin machinery and pediatric high-grade glioma, Science Advances. 2 (2016) 1–9. 10.1126/SCIADV.1501354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Chen W, DiFrancesco L, Chondroblastoma: An Update, Archives of Pathology & Laboratory Medicine. 141 (2017) 867–871. 10.5858/ARPA.2016-0281-RS. [DOI] [PubMed] [Google Scholar]
- [179].Sobti A, Agrawal P, Agarwala S, Agarwal M, Giant Cell Tumor of Bone - An Overview, Archives of Bone and Joint Surgery. 4 (2016) 2–9. /pmc/articles/PMC4733230/ (accessed July 24, 2021). [PMC free article] [PubMed] [Google Scholar]
- [180].Bavle A, Chintagumpala M, Pediatric high-grade glioma: a review of biology, prognosis, and treatment, Journal of Radiation Oncology . 7 (2018) 7–15. 10.1007/S13566-018-0344-9. [DOI] [Google Scholar]
- [181].Park S, Choi E, Bae M, Kim S, Park JB, Yoo H, Choi JK, Kim Y, Lee S, Kim I, Histone variant H3F3A promotes lung cancer cell migration through intronic regulation, Nature Communications . 7 (2016) 1–14. 10.1038/ncomms12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Adam S, Polo SE, Almouzni G, Transcription Recovery after DNA Damage Requires Chromatin Priming by the H3.3 Histone Chaperone HIRA, Cell. 155 (2013) 94–106. 10.1016/J.CELL.2013.08.029. [DOI] [PubMed] [Google Scholar]
- [183].Frey A, Listovsky T, Guilbaud G, Sarkies P, Sale J, Histone H3.3 is required to maintain replication fork progression after UV damage, Current Biology. 24 (2014) 2195–2201. 10.1016/J.CUB.2014.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Juhász S, Elbakry A, Mathes A, Löbrich M, ATRX Promotes DNA Repair Synthesis and Sister Chromatid Exchange during Homologous Recombination, Molecular Cell. 71 (2018) 11–24. 10.1016/J.MOLCEL.2018.05.014. [DOI] [PubMed] [Google Scholar]
- [185].Luijsterburg M, de Krijger I, Wiegant W, Shah R, Smeenk G, de Groot A, Pines A, Vertegaal A, Jacobs J, Shah G, van Attikum H, PARP1 Links CHD2-Mediated Chromatin Expansion and H3.3 Deposition to DNA Repair by Non-homologous End-Joining, Molecular Cell. 61 (2016) 547–562. 10.1016/J.MOLCEL.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Yang X, Li L, Liang J, Shi L, Yang J, Yi X, Zhang D, Han X, Yu N, Shang Y, Histone Acetyltransferase 1 Promotes Homologous Recombination in DNA Repair by Facilitating Histone Turnover, The Journal of Biological Chemistry. 288 (2013) 18271–18282. 10.1074/JBC.M113.473199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Chang FTM, Chan FL, McGhie JDR, Udugama M, Mayne L, Collas P, Mann JR, Wong LH, CHK1-driven histone H3.3 serine 31 phosphorylation is important for chromatin maintenance and cell survival in human ALT cancer cells, Nucleic Acids Research. 43 (2015) 2603–2614. 10.1093/NAR/GKV104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Elbakry A, Juhász S, Mathes A, Löbrich M, DNA repair synthesis and histone deposition partner during homologous recombination, Molecular & Cellular Oncology. 5 (2018). 10.1080/23723556.2018.1511210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Braunschweig U, Hogan GJ, Pagie L, van Steensel B, Histone H1 binding is inhibited by histone variant H3.3, The EMBO Journal. 28 (2009) 3635–3645. 10.1038/EMB0J.2009.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Albig W, Ebentheuer J, Klobeck G, Kunz J, Doenecke D, A solitary human H3 histone gene on chromosome 1, Human Genetics. 97 (1996) 486–491. 10.1007/BF02267072. [DOI] [PubMed] [Google Scholar]
- [191].Tachiwana H, Kagawa W, Osakabe A, Kawaguchi K, Shiga T, Hayashi-Takanaka Y, Kimura H, Kurumizaka H, Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T, Proceedings of the National Academy of Sciences of the United States of America. 107 (2010) 10454–10459. 10.1073/PNAS.1003064107/-/DCSUPPLEMENTAL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Jun Ueda A, Harada A, Urahama T, Ohkawa Y, Kurumizaka H, Yamagata K Correspondence Testis-Specific Histone Variant H3t Gene Is Essential for Entry into Spermatogenesis, Cell Reports. 18 (2017) 593–600. 10.1016/j.celrep.2016.12.065. [DOI] [PubMed] [Google Scholar]
- [193].Urahama T, Harada A, Maehara K, Horikoshi N, Sato K, Sato Y, Shiraishi K, Sugino N, Osakabe A, Tachiwana H, Kagawa W, Kimura H, Ohkawa Y, Kurumizaka H, Histone H3.5 forms an unstable nucleosome and accumulates around transcription start sites in human testis, Epigenetics & Chromatin. 9 (2016). 10.1186/S13072-016-0051-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Schenk R, Jenke A, Zilbauer M, Wirth S, Postberg J, H3.5 is a novel hominid-specific histone H3 variant that is specifically expressed in the seminiferous tubules of human testes, Chromosoma. 120 (2011) 275–285. 10.1007/S00412-011-0310-4. [DOI] [PubMed] [Google Scholar]
- [195].Wiedemann S, Mildner S, Bönisch C, Israel L, Maiser A, Matheisl S, Straub T, Merkl R, Leonhardt H, Kremmer E, Schermelleh L, Hake S, Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y, The Journal of Cell Biology. 190 (2010) 777–791. 10.1083/JCB.201002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Kujirai T, Horikoshi N, Sato K, Maehara K, Machida S, Osakabe A, Kimura H, Ohkawa Y, Kurumizaka H, Structure and function of human histone H3.Y nucleosome, Nucleic Acids Research. 44 (2016) 6127–6141. 10.1093/NAR/GKW202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Akhmanova A, Miedema K, Hennig W, Identification and characterization of the Drosophila histone H4 replacement gene, FEBS Letters. 388 (1996) 219–222. 10.1016/0014-5793(96)00551-0. [DOI] [PubMed] [Google Scholar]
- [198].Kamakaka RT, Biggins S, Histone variants: deviants?, Genes & Development. 19 (2005) 295–316. 10.1101/GAD.1272805. [DOI] [PubMed] [Google Scholar]
- [199].Talbert PB, Ahmad K, Almouzni G, Ausió J, Berger F, Bhalla PL, Bonner WM, Cande WZ, Chadwick BP, Chan SWL, Cross GAM, Cui L, Dimitrov SI, Doenecke D, Eirin-López JM, Gorovsky MA, Hake SB, Hamkalo BA, Holec S, Jacobsen SE, Kamieniarz K, Khochbin S, Ladurner AG, Landsman D, Latham JA, Loppin B, Malik HS, Marzluff WF, Pehrson JR, Postberg J, Schneider R, Singh MB, Smith MM, Thompson E, Torres-Padilla M-E, Tremethick DJ, Turner BM, Waterborg JH, Wollmann H, Yelagandula R, Zhu B, Henikoff S, A unified phylogeny-based nomenclature for histone variants, Epigenetics & Chromatin. 5 (2012) 1–19. 10.1186/1756-8935-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Long M, Sun X, Shi W, Yanru A, Leung STC, Ding D, Cheema MS, MacPherson N, Nelson CJ, Ausio J, Yan Y, Ishibashi T, A novel histone H4 variant H4G regulates rDNA transcription in breast cancer, Nucleic Acids Research. 47 (2019) 8399–8409. 10.1093/NAR/GKZ547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Singh R, Bassett E, Chakravarti A, Parthun M, Replication-dependent histone isoforms: a new source of complexity in chromatin structure and function, Nucleic Acids Research. 46 (2018) 8665–8678. 10.1093/NAR/GKY768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Ge Z, Nair D, Guan X, Rastogi N, Freitas M, Parthun M, Sites of acetylation on newly synthesized histone H4 are required for chromatin assembly and DNA damage response signaling, Molecular and Cellular Biology. 33 (2013) 3286–3298. 10.1128/MCB.00460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Dhar S, Gursoy-Yuzugullu O, Parasuram R, Price BD, The tale of a tail: histone H4 acetylation and the repair of DNA breaks, Philosophical Transactions of the Royal Society B: Biological Sciences. 372 (2017). 10.1098/RSTB.2016.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL, Histone H4-K16 acetylation controls chromatin structure and protein interactions, Science (New York, N.Y.). 311 (2006) 844–847. 10.1126/SCIENCE.1124000. [DOI] [PubMed] [Google Scholar]
- [205].Sanders SL, Portoso M, Mata J, Bähler J, Allshire RC, Kouzarides T, Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage, Cell. 119 (2004) 603–614. 10.1016/J.CELL.2004.ll.009/ATTACHMENT/69DD6368-9817-4462-91A4-C9669D651A1D/MMC4.PDF. [DOI] [PubMed] [Google Scholar]
- [206].Pesavento JJ, Yang H, Kelleher NL, Mizzen CA, Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle, Molecular and Cellular Biology. 28 (2008) 468–486. 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Oda H, Hübner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D, Regulation of the Histone H4 Monomethylase PR-Set7 by CRL4Cdt2 Mediated PCNA Dependent Degradation during DNA Damage, Molecular Cell. 40 (2010) 364. 10.1016/J.MOLCEL.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z, MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites, Nature. 470 (2011) 124–129. 10.1038/NATURE09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Hajdu I, Ciccia A, Lewis SM, Elledge SJ, Wolf–Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage, Proceedings of the National Academy of Sciences. 108 (2011) 13130–13134. 10.1073/PNAS.1110081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Kimura A, Horikoshi M, Tip60 acetylates six lysines of a specific class in core histones in vitro, Genes to Cells : Devoted to Molecular & Cellular Mechanisms. 3 (1998) 789–800. 10.1046/J.1365-2443.1998.00229.X. [DOI] [PubMed] [Google Scholar]
- [211].Jacquet K, Fradet-Turcotte A, Avvakumov N, Lambert JP, Roques C, Pandita RK, Paquet E, Herst P, Gingras AC, Pandita TK, Legube G, Doyon Y, Durocher D, Côté J, The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation, Molecular Cell. 62 (2016) 409–421. 10.1016/J.MOLCEL.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Hsiao K-Y, Mizzen CA, Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair, Journal of Molecular Cell Biology. 5 (2013) 157–165. 10.1093/jmcb/mjs066. [DOI] [PubMed] [Google Scholar]
- [213].Zhang H, Han J, Kang B, Burgess R, Zhang Z, Human Histone Acetyltransferase 1 Protein Preferentially Acetylates H4 Histone Molecules in H3.1-H4 over H3.3-H4, The Journal of Biological Chemistry. 287 (2012) 6573. 10.1074/JBC.M111.312637. [DOI] [PMC free article] [PubMed] [Google Scholar]


