Abstract
Background
Targeted gene NGS testing is available through many academic institutions and commercial entities and is increasingly incorporated in practice guidelines for glioblastoma (GBM). This single-center retrospective study aimed to evaluate the clinical utility of incorporating NGS results in the management of GBM patients at a clinical trials-focused academic center.
Methods
We identified 1011 consecutive adult patients with pathologically confirmed GBM (IDHwt or IDHmut) who had somatic tumor sequencing (Oncopanel, ~500 cancer gene panel) at DFCI from 2013–2019. Clinical records of all IDHwt GBM patients were reviewed to capture clinical trial enrollment and off-label targeted therapy use based on NGS results.
Results
Of the 557 IDHwt GBM patients with sequencing, 182 entered clinical trials at diagnosis (32.7%) and 213 (38.2%) entered after recurrence. Sequencing results for 130 patients (23.3%) were utilized for clinical trial enrollment for either targeted therapy indications (6.9 % upfront and 27.7% at recurrent clinical trials and 3.1% for off-label targeted therapy) or exploratory studies (55.4% upfront and 6.9% recurrent clinical trials). Median overall survival was 20.1 months with no survival difference seen between patients enrolled in clinical trials compared to those who were not, in a posthoc analysis.
Conclusions
While NGS testing has become essential for improved molecular diagnostics, our study illustrates that targeted gene panels remain underutilized for selecting therapy in GBM-IDHwt. Targeted therapy and clinical trial design remain to be improved to help leverage the potential of NGS in clinical care.
Keywords: GBM, next-generation sequencing, targeted therapy
Key Points.
Beyond diagnostic confirmation, use of NGS to select therapies in GBM remains constrained by lack of effective targeted therapy options.
Development of more effective targeted therapies could help leverage the potential of NGS in GBM.
Importance of the Study.
With the increased use of targeted gene NGS panels in clinical care for the diagnosis of glioblastoma, the therapeutic and clinical trial utility of these panels for patient management has not been well studied. This single-center retrospective study evaluates the practice patterns and utility of incorporating targeted gene sequencing results into therapeutic decision-making for GBM patients at a clinical trials-focused academic center.
In 2016, a revision to the WHO classification incorporated molecular diagnostics to complement histological diagnosis and grading.1 Molecular testing is now integral to practice guidelines,2 endorsed by the National Comprehensive Cancer Network (NCCN) and expounded upon by the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy (cIMPACT-NOW). The 2021 WHO classification (5th Edition) reflects updates from cIMPACT-NOW3,4—seven of which have now been published—and relies on testing for molecular alterations for the diagnosis of gliomas.5–11 Furthermore, in glioma, molecular signatures acquired longitudinally can help broaden our understanding of each tumor’s natural history, vulnerabilities, response to treatment, and progression pattern, potentially paving the way to improved precision medicine.12,13
The concept of targeted therapy based on individualized tumor genotyping has long been attractive in glioma. However, only certain targeted therapies are currently FDA approved for use in gliomas, such as mammalian target of rapamycin (mTOR) inhibitors in tuberous sclerosis associated subependymal giant cell astrocytoma, neurotrophic tyrosine receptor kinase (NTRK)-inhibitors for NTRK fusions, and programmed cell death protein 1 (PD-1) inhibitors in mismatch repair deficient-/high tumor mutational burden/high microsatellite instability tumors. Many other targets are attractive due to their identification as key oncogenic drivers in gliomas such as IDH mutations in astrocytoma and oligodendroglioma. In IDH-wildtype (IDHwt) gliomas, specifically, glioblastoma (GBM), a number of different targets have been identified including epidermal growth factor receptor (EGFR) amplification and mutations, cyclin dependent kinase inhibitor 2A and 2B (CDKN2A/B), and phosphoinositide 3-kinase (PI3K) pathways, the v-Raf murine sarcoma viral oncogene homolog V1 (BRAF) V600E mutations, and fusion events such as in fibroblast growth factor receptor (FGFR) and neurotrophic tyrosine receptor kinase (NTRK).12 However, efforts to find effective therapies have so far fallen short when it comes to life-prolonging treatments in GBM. There are a number of possible explanations for this including the redundant nature of several oncogenic pathways, the lack of CNS penetrance from tested therapies due to the blood-brain barrier, and substantial intratumoral and intertumoral heterogeneity.14 Outside of these challenges, it is also important to consider that the targetable mutations can be rare, for example, only about 2% of adult GBM harbor a BRAF V600E mutation and better characterization of such subtypes may be helpful in screening patients for such mutations.15 In the broader world of oncology, and across tumor sites, precision oncology has seen some success stories, but widespread success has been elusive, particularly in brain tumors.16
In recent years, molecular diagnostics in the form of next-generation sequencing (NGS)-based targeted gene assays have become commercially available and has been incorporated into routine clinical care for a number of cancer types at many institutions. However, given the specialized laboratory infrastructure associated with NGS, as well as its limited availability and accessibility, the current impact of widespread targeted NGS assays for IDHwt GBM patients in treatment remains unclear. In this study, we describe our single-center experience of incorporating targeted gene NGS testing into therapeutic decision-making for patients with IDHwt GBM.
Materials and Methods
This study was conducted with approval from the Dana-Farber/Harvard Cancer Center Institutional Review Board. A waiver was granted for informed consent due to the retrospective nature of the study.
Cohort Selection
This retrospective study was carried out with methodology consistent with the STROBE (Strengthening the Reporting of Observational studies in Epidemiology) guidelines.17
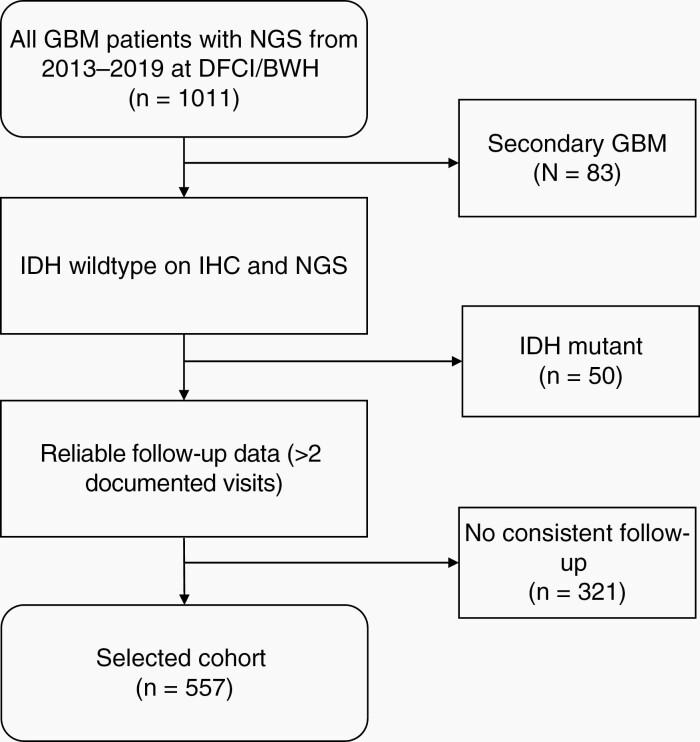
Adult patients (≥18-years-old) with a pathological diagnosis of glioblastoma WHO grade 4, treated at Dana-Farber Cancer Institute/Brigham and Women’s Hospital from January 1, 2013 to July 31, 2019, and had molecular information through the institutional NGS targeted gene panel (ie, OncoPanel, ~500 cancer-causing genes) were screened for this study. Exclusion criteria were: i) progression of previous low-grade glioma (secondary glioblastoma), ii) IDHmut high-grade glioma, and iii) lack of consistent (ie, >2) clinical visits with documented clinical notes after the first consultation (Figure 1).
Fig. 1.
Patient selection: 1011 patients with glioblastoma at Dana-Farber Cancer Institute who had in-house targeted panel sequencing between 2013 and 2019 were retrospectively identified. All secondary GBMs, IDH-mutant tumors and patients without consistent follow-up were excluded. This algorithm resulted in 557 patients which were included in the final cohort.
Data Collection
A review of the medical records and clinical charts of each patient was performed (MA, ML, SW, GY), and clinical data including sex, age, histomolecular diagnosis, all tumor-directed therapy, progression dates based on clinical notes, progression-free survival, and overall survival were collected retrospectively. Specifically, all therapeutic clinical trial enrollment information was captured, and required biomarkers for enrollment purposes including O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation status, or specific mutations were annotated. Clinical trials not investigating tumor-directed treatments were not included.
Pathology and Molecular Data
Histomolecular diagnosis in accordance with the 2016 WHO classification1 was collected. Isocitrate dehydrogenase (IDH) mutations by either immunohistochemistry for IDH1 R132H or NGS were assessed and all IDH1 or IDH2-mutant tumors were excluded. MGMT promoter methylation status was recorded where available. All patients included in this retrospective cohort had their tumor sequenced using the previously validated OncoPanel18,19 at the Center for Cancer Genome Discovery (Dana-Farber Cancer Institute) and the Center for Advanced Molecular Diagnosis (Brigham and Women’s Hospital). This institutional targeted gene NGS assay surveys cancer-associated genes for single nucleotide variants, insertions, and deletions, and copy number variants; as well as intronic regions for rearrangement detection and is based on the Illumina HiSeq 2500 platform (targeted genes are listed in Supplementary Appendix 1).
Statistical Analysis
Statistical analyses were performed using Stata/SE Version 16. Kaplan-Meier estimator and log-rank test were performed to assess for difference in survival data in a posthoc fashion due to the retrospective nature of the study. Time-to-event analyses for overall survival (OS) were calculated using the Kaplan-Meier method from time of glioblastoma diagnosis to date of death.
Results
Patient Characteristics
From 1011 patients with newly diagnosed GBM and available NGS OncoPanel data, 878 were newly diagnosed IDHwt patients (Figure 1). 321 patients were excluded due to lack of reliable follow-up at our institution; 557 patients were included for the final cohort. Baseline and relevant characteristics of the whole cohort are summarized in Table 1. 508 patients (91.2%) had documented MGMT promoter methylation data, including 198 (39.0%) with methylated tumors, 30 (5.9%) with partially methylated tumors, and 280 (55.1%) with unmethylated tumors.
Table 1.
Patient Demographic for Cohort of 557 Glioblastoma Patients
| Total | Upfront Clinical Trial | Any Clinical Trial | |||
|---|---|---|---|---|---|
| (N = 557) | |||||
| Yes (N = 182) | No | Yes | No | ||
| (N = 375) | (N = 314) | (N = 243) | |||
| Median age at diagnosis (95% CI) | 60.6 | 58.5 | 61.5 | 58.7 | 62.9 |
| (59.1–61.7) | (57.4–60.6) | (60.2–62.9) | (57.3–60.3) | (61.2–64.6) | |
| P = .01 | P < .01 | ||||
|
Sex (%)
Male |
227 (40.8) | 108 | 222 | 193 | 137 |
| (59.3) | (59.2) | (61.4) | (56.3) | ||
| P = .98 | P = .23 | ||||
| KPS at baseline (%) | N = 462 | 150 (82.4) | 241 (64.2) | 229 (72.9) | 162 (66.7) |
| ≥ 80 | 396 (85.7) | P < .01 | P = .11 | ||
| Extent of resection (%) | N = 549 | 83 (46.4) | 146 (39.2) | 153 (49.5) | 76 (31.4) |
| GTR | 229 (41.6) | ||||
| P = .11 | P < .01 | ||||
| MGMT methylation status (%) | N = 508 | 63 (38.7) | 135 (39.1) | 115 (40.8) | 83 (36.7) |
| Methylated | 198 (39.0) | ||||
| Partially methylated | 30 (5.9) | ||||
| Unmethylated | 280 (55.1) | 11 (6.7) | 19 (5.5) | 18 (6.4) | 12 (5.3) |
| 89 (54.6) | 191(55.4) | 149 (52.8) | 131 (58.0) | ||
| P = .86 | P = .50 | ||||
| Concurrent chemoradiation with TMZ | N = 553 | 169 (92.9) | 361 (97.3) | 299 (95.2) | 231 (96.7) |
| 530 (95.2) | |||||
| P = .01 | P = .40 | ||||
| Adjuvant treatment | 417 (74.8) | 112 (61.9) | 305 (83.6) | 233 (74.9) | 184 (78.2) |
| TMZ | |||||
| P < .01 | P = .36 | ||||
| Median number of TMZ cycles (95% CI) | 6 (6–6) | 6 (6–6) | 6 (5–6) | 6 (6–6) | 5 (5–6) |
| P = .41 | P < .01 |
Reported Molecular Alterations of Clinical Significance
Our analysis included the clinically significant Tier 1 and Tier 2 single nucleotide variants (SNV) and estimated copy number variants (CNV) based on the tiering system of our institutional panel.20 Relevant genes required for eligibility in GBM clinical trials at our institution (EGFR, PTEN, MET, FGFR1, FGFR3, H3F3A, PDGFRA, CDKN2A/B, RB1, TP53) and the frequency of patients with each molecular alteration are shown in Table 2. The most common alterations included mutations in TERT promoter (n = 180, 63.6%), CDKN2A (n = 254, 45.6%), CDKN2B (n = 229, 41.1%), PTEN (n = 196, 35.2%) and EGFR amplification (n = 186, 33.4%). Results directly relevant for FDA approved therapies included high tumor mutational burden (TMB high defined as >17 mut/Mb)21 (n = 11, 2%), BRAF V600E (n = 12, 2.2%) and FGFR3-TACC fusions. (n = 4; ~1%).
Table 2.
Frequency of Molecular Alteration in Select Genes in Cohort
| Gene | Total (%) |
|---|---|
| TERT promoter (N = 283) | 180 (63.6) |
| CDKN2A | 254 (45.6) |
| CDKN2B | 229 (41.1) |
| PTEN | 196 (35.2) |
| EGFR amplification | 186 (33.4) |
| TP53 | 161 (28.9) |
| MTAP (N = 283) | 108 (38.2) |
| CDK4 | 61 (11.0) |
| NF1 | 54 (9.7) |
| PIK3CA | 51 (9.2) |
| PDGFRA | 43 (7.7) |
| RB1 | 41 (7.4) |
| MDM2 | 36 (6.5) |
| MDM4 | 31 (5.6) |
| PIK3C2B | 31 (5.6) |
| PIK3R1 | 21 (3.8) |
| STAG2 | 16 (2.9) |
| ATRX | 14 (2.5) |
| BRAF V600E | 12 (2.2) |
| PTPN11 | 7 (1.3) |
| NOTCH1 | 3 (0.5) |
| POLE (N = 283) | 2 (0.7) |
| APC | 2 (0.4) |
| SETD2 | 1 (0.2) |
Standard of Care vs Clinical Trial Enrollment
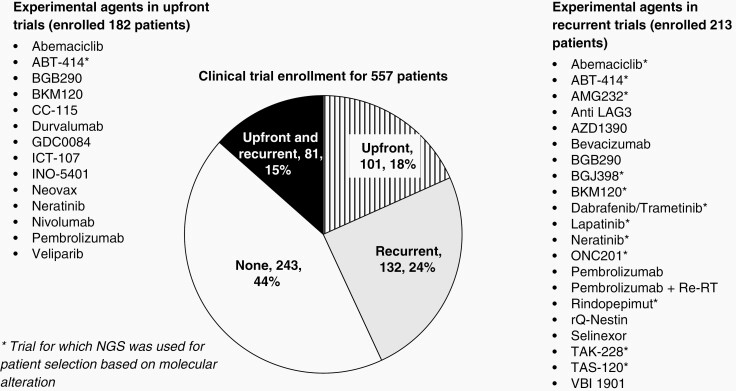
The vast majority of our patients received concurrent chemoradiation with temozolomide as upfront treatment (n = 530, 95.2%; Table 1). While 417 (74.8%) of patients received adjuvant therapy with temozolomide, 182 (32.7%) were enrolled in clinical trial immediately after first diagnosis (“upfront setting”), of which 112 (61.5%) received temozolomide as part of the trial (with or without another agent). Of these 182 patients, 75 (41.2%) also enrolled in subsequent clinical trials at recurrence. In total, 213 (38.2%) of patients were enrolled in a clinical trial at first or subsequent recurrence. Altogether, 314 (56.4%) patients enrolled in 77 trials upfront or at recurrence (Appendix 2).
Figure 2 shows the different investigational drug agents used in the context of a clinical trial in this cohort. The demographics of patients enrolled in therapeutic clinical trials in the upfront setting or at any point during their diagnosis were also assessed and compared (Table 1). Patients in clinical trials (both in the upfront setting and at any timepoint) were younger and exhibited better Karnofsky Performance Scale (KPS) scores at baseline (P < .05). Patients enrolled in clinical trials also were more likely to have had a gross total resection (P < .01) and were more likely to receive more cycles of temozolomide compared to those who did not participate in a clinical trial (P < .01).
Fig. 2.
Clinical trial enrollment for the 557 patients included in our study and the experimental agents used in the clinical trial setting.
Utilization of Molecular Data for Tumor-Directed Therapy or Trial Enrollment
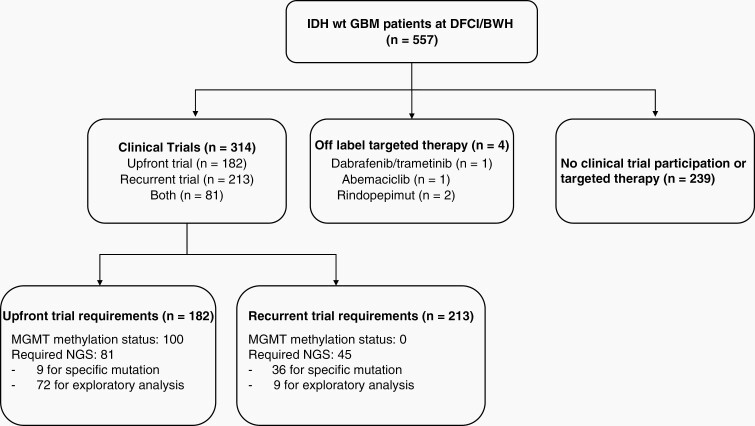
Among the 182 patients who were enrolled in clinical trials in the upfront setting, 100 (54.9%) required MGMT methylation status information and 81 (44.5%) required sequencing information for enrollment (Figure 3). Twelve of the 77 trials required molecular testing for eligibility based on 10 genes: EGFR, PTEN, MET, FGFR1, FGFR3, H3F3A, PDGFRA, CDKN2A/B, RB1, TP53. Of these 81 patients requiring sequencing information, only 9 patients (11.1%) required a specific mutation to meet inclusion criteria. Of the 4 molecularly selective upfront trials that selected patients based on the identification of specific genetic aberrations, several were trials with EGFR mutation/amplification-directed treatments (NCT01800695, NCT01480479, NCT02573324, NCT03107780) and a phase 0/1 study of AMG 232, an MDM2 inhibitor for patients with p53 wild type glioblastoma (NCT03107780). The remaining 65 patients (85.5%) required sequencing information for exploratory/biomarker analyses and this information did not guide therapy selection. Thirty-four (52.3%) of these patients were enrolled in the Individualized Screening Trial of Innovative Glioblastoma Therapy (INSIGhT) trial22 (NCT02977780), a Bayesian adaptive platform trial for newly diagnosed glioblastoma patients with unmethylated MGMT promoter. In this design, randomization at diagnosis occurs to multiple experimental arms or one control arm, and patient subtypes including biomarkers are identified, after which adaptive randomization occurs based on accumulating trial results.
Fig. 3.
Flow chart showing utilization of NGS and MGMT methylation data for patient enrollment in clinical trial or for off-label therapy.
In the recurrent setting, only 45 patients out of the 213 (21.1%) who were enrolled in clinical trials required sequencing data for enrollment eligibility. Of these, 36 (80%) required specific mutations as inclusion criteria for biologically driven therapies (Figure 2). All trials and their requirements with regards to MGMT methylation status or molecular screening are summarized in Appendix 2.
Out of the 11 patients identified to have hypermutated tumors, 3 (27.3%) received immune checkpoint inhibition. Only four patients received compassionate use targeted therapy: one with dabrafenib/trametinib (BRAF V600E mutant), one with abemaciclib (CDK4/6 amplification), and two with rindopepimut (EGFRvIII amplification).
Survival Times and Follow-Up
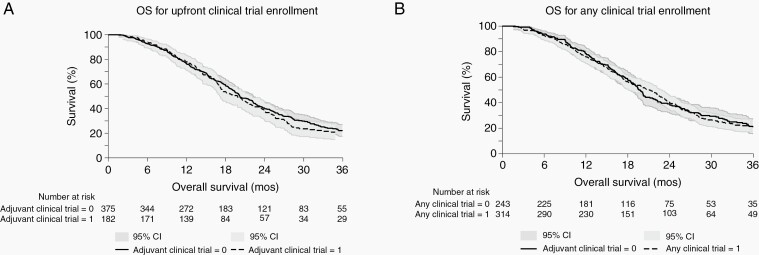
Given that patients without consistent follow-up were excluded for this analysis, the median follow-up time could be estimated by the survival time until death or last censored date. Median overall survival for this cohort was 20.1 months (95% confidence interval [CI]: 18.9–22.0); specifically, 17.1 months for the MGMT unmethylated patients (n = 280; 95% CI:15.5–18.0); 20.4 months for the MGMT partially methylated patients (n = 30, 95% CI: 10.3–22.3) and 28.3 months for the MGMT methylated patients (n = 198, 95% CI: 25.7–34.2). Median progression-free survival was 9.1 months (95%CI: 8.5–9.7) for the cohort. There was no difference in overall survival in patients who were enrolled in clinical trials in the upfront setting compared to those in the recurrent setting (19.0 months [95% CI: 17.1–22.2] vs 20.2 months [95% CI: 19.0–22.3]), P = .60) in our posthoc survival analysis. In addition, there was no difference in overall survival for patients who were enrolled in a clinical trial (either in the upfront or recurrent settings) compared to those who were not part of any therapeutic clinical trial (20.4 months [95% CI: 17.3–23.3] vs 20.2 months [95% CI: 18.9–22.3]), P = .13) (Figure 4).
Fig. 4.
Survival curves of patients enrolled in upfront clinical trials vs not (A) and patients enrolled in any clinical trial vs not (B). There was no difference in overall survival in patients who were enrolled in clinical trials upfront compared than those who did not (19.0 months [95% CI: 17.1–22.2] vs 20.2 months [95% CI: 19.0–22.3]), P = .60). In addition, there was no difference in overall survival in patients who were enrolled in a clinical trial (at any point—upfront or at the recurrent stage) compared to those who were not part of any clinical trial (20.4 months [95% CI: 17.3–23.3] vs 20.2 months [95% CI: 18.9–22.3]), P = .13).
Discussion
With the implementation of the 2021 WHO Classification of Central Nervous Tumours 5th edition, molecular testing is an increasingly crucial component of the standard-of-care diagnostic work-up of an infiltrative glioma. National Comprehensive Cancer Network guidelines (CNS cancers v1.2021) additionally encourages the molecular testing of glioblastomas to help identify driver mutations that may be targetable in the context of a clinical trial or on a compassionate use basis. Targeted NGS gene panel assays can help simultaneously test for the growing number of diagnostically important—and potentially targetable—molecular alterations in infiltrative gliomas in a straightforward and cost-effective manner. However, although access to targeted gene sequencing panels for gliomas has increased in the past decade, beyond their marked value in diagnosis, the utility and clinical benefit for guiding therapy based on these tests remain unclear. Molecular testing in brain tumors has become increasingly feasible in the timeline of real-world best clinical practice, with some groups reporting an integrated workflow of 5 days from tissue biopsy to finalized report of a 130 gene NGS panel.23 Given the increase in the number of trials for newly diagnosed glioblastoma, molecular information, including MGMT status, is typically helpful at the first consultation with the neuro-oncologist prior to radiation planning; however, this is not always feasible and can present a logistical challenge.
A retrospective cross-sectional study using publicly available data across cancer types found that about 4.5% of patients derived benefit from “genome-targeted” drugs (where the drug targeted the specific aberration identified by the genomic test) and 6% benefited from “genome-informed” therapy (all genome-targeted drugs or drugs given after genomic test, regardless of whether drug targeted aberration or acted via an alternate mechanism of action).24 In our study of IDHwt GBM, we found that the targeted gene sequencing results were utilized for clinical decision-making in 23% of patients—mostly as a prerequisite for clinical trial enrollment. In a subset of these patients (8% overall), specific mutations or molecular results were required for enrollment due to mechanism of action of the drugs. Most of these trials were studies of recurrent gliomas and the rate was much lower in the upfront clinical trial setting.
In our analysis, there was no survival difference between patients who enrolled in clinical trials versus those that did not. This finding is worth highlighting as previous studies, with smaller patient numbers have indicated that patients benefit from enrollment in clinical trials compared to standard therapy.25,26 It is possible that this survival difference may be less pronounced in centers where routine care of patients receiving standard-of-care therapy does not differ significantly from those on clinical trials. In addition, a more relevant survival outcome would be a comparison of survival for patients that used molecular selection for enrollment, although this was not feasible in our study due to the small sample sizes within these groups. This analysis would be a helpful addition to the understanding of the utility of NGS in clinical care and could be performed through multi-institutional review of such trials.
Most of the trials involved sequencing of patients’ tumors for exploratory biomarker purposes, and while such efforts may eventually further our understanding of potential targets and inform future patient selection, the enrolled individual generally does not undergo mutation-derived targeted therapy in those cases. A unique example of the use of molecular biomarkers in an adaptive setting is the Individualized Screening Trial of Innovative Glioblastoma Therapy (INSIGhT) trial22 (ClinicalTrials.gov Identifier: NCT02977780), which is a Bayesian adaptive platform trial for newly diagnosed GBM patients with a two-phase approach. In this trial, biomarkers are identified, after which adaptive randomization occurs based on accumulating trial results. In this trial, and in the larger similarly designed GBM-AGILE study, while the biomarker-specific probability of treatment impact on progression-free survival is used to drop arms that would have a low probability of treatment impact on overall survival, molecular biomarkers are not used at the time of accrual for specific arm assignment until specific criteria have been met. Therefore, in this particular trial, “targeted therapy” is not the basis of the first phase of the trial for each individual patient.27
Only four patients in our cohort had their molecular information used for off-label targeted therapies (which are FDA-approved for other cancer types but not for GBM as a specific indication) during their follow-up time at DFCI. The accessibility of off-label therapies to individual patients is often dependent on third-party coverage and FDA approval. This landscape has been evolving in recent years and there are now several tumor type-agnostic therapies approved by the FDA, such as NTRK inhibitors (eg, larotrectinib and entrectinib) for NTRK fusions and immune checkpoint inhibitors (eg, pembrolizumab) for mismatch repair deficient/microsatellite instability-high solid tumors. Initiating off-label treatments outside of clinical trials in the upfront setting remains controversial if it is done in lieu of or in addition to standard-of-care therapy. Therefore, anecdotally, off-label targeted therapies are often initiated by the provider if there is available sequencing data to guide this decision at the time when salvage treatment is justified. This window may be very short for patients with an aggressive tumor type such as GBM, therefore obtaining targeted gene sequencing results “for future use” at the time of initial surgery has been an emerging phenomenon in clinical practice, catalyzed in part by growing access to NGS testing. This approach may be feasible for molecular targets such as EGFR amplification and CDKN2A/B deletion, both of which are usually unchanged at the time of recurrence in GBM but may be less effective in tumors where targets such as EGFR mutations may be lost in recurrent tumors.28,29 The overall clinical utility of this “for future use” sequencing strategy—given GBM’s increasingly recognized proclivity for continued tumor evolution and acquiring mutations due to standard-of-care therapy21,28—as well as the strategy’s cost-effectiveness, remain to be determined.
Utilization of MGMT methylation status is now well established both as a predictive marker of response to alkylating therapy and a prognostic biomarker. MGMT methylation status can be determined by quantitative or semi-quantitative PCR methods and is mandated by the NCCN guidelines, although uniform and routine implementation can remain challenging.30,31 While methylation status has anecdotally been more helpful in treatment decisions for elderly or frail patients with glioblastoma,32,33 there is an impetus to rethink the role of standard-of-care concurrent and adjuvant chemoradiation in MGMT unmethylated patients with glioblastoma, especially in the post-COVID19 era.34 Most upfront GBM trials with investigational drug therapies stratify enrollment based on MGMT methylation status, due to an appreciation of the unique risks and benefits posed by standard therapy vs unproven therapies to different MGMT methylation statuses. While we did not specifically evaluate the utility of MGMT methylation status in guiding treatment decisions in this study, most upfront clinical trials at our institution utilized this information for initial enrollment, with MGMT status representing perhaps the most commonly utilized biomarker at that juncture.
Many institutional sequencing panels and commercial panels are now available, each of which can examine several hundreds of selected GBM relevant genes.35 While our results show that targeted NGS may be currently underutilized for selecting targeted therapies, this study does not argue against the benefits of sequencing in the clinical care of patients. The diagnostic and prognostic value to patients are clear and targeted sequencing panels can be a cost-effective solution compared to individual selected gene panels addressing specific alterations that define gliomas within the WHO 2021 classification3 (eg, IDH1, IDH2, ATRX, T53, CDKN2A/B, 1p/19q, TERT promoter, CIC, FUBP1, NOTCH1, chromosome 7 gain or 10 loss, EGFR, MYB, MYBL1, BRAF, FGFR, H3K27, H3G34). Additionally, in the face of a growing number of molecular alterations/signatures that are potentially targetable (eg, mismatch repair deficiency/microsatellite instability-high, NTRK, PIK3CA, CDK4/6, PDGFRA, etc.) in glioblastoma, NGS gene panel assays can provide a more scalable solution for neuropathology laboratories than the continuous validation and implementation of new single-gene or single-purpose molecular assays. The answer in better leveraging the potential of multi-gene NGS assay data may ultimately lie in creating computational platforms that automatically match a patient’s identified molecular alterations with relevant clinical trials and – at a fundamental level –in improving the drug development pipeline for targeted agents in GBM. Certain subgroups within IDHwt GBM may in particular benefit from multi-gene NGS testing, for example, younger patients may be more likely to have targetable mutations such as those in BRAF or FGFR, and the clinical yield of sequencing patients with pediatric type IDHwt gliomas would be more pronounced.
Designing precision medicine-focused clinical trials for GBM to find more effective agents remains an area of pressing need, and while accrual to molecular selective trials is often a challenge due to the low frequency of certain driver mutations in high-grade gliomas (eg, NTRK and BRAF), multi-institutional trials with novel trial designs including basket studies are approaches that can be effective. Novel adjunctive technologies to overcome the blood-brain-barrier that are currently under investigation such as focused ultrasound (FUS)36 may also represent effective combinatorial strategies to be used with targeted agents. Beyond clinical trials, molecularly annotated and clinically annotated “real-life” cohorts offer a unique chance to study precision medicine in GBM patients, guided by NGS data, whether or not patients receive targeted therapy or standard-of-care therapy. Lastly, this type of data may be particularly valuable to study patterns of response, identify and understand biomarkers of long-term response and resistance and sequencing of serum or CSF37,38 may also represent an emerging application that will help in these efforts.
Limitations
This single-institution experience at an NCI-Designated Cancer Center does not necessarily reflect the current practice patterns at other institutions, specifically in terms of availability of targeted gene NGS testing in a clinical setting, access to clinical trials, and use of off-label therapies. However, the Center for Neuro-Oncology at Dana-Farber Cancer Institute is a comprehensive brain tumor program that sees large patient volumes, and at which next-generation sequencing is performed on most glioblastoma specimens, multiple clinical trials are available both in the upfront and recurrent setting, and off-label therapies are also often discussed with patients. While we estimate that other large comprehensive brain tumor programs may have similar opportunities for NGS testing in IDH-wt GBM, our results may in fact be an overestimate the use of NGS testing for access to therapies outside of specialized centers. Evaluation of potential survival benefits will also require assessment in the context of each individual trial as more results from such studies emerge over time.
Conclusions
With the rise of more cost-effective and efficient sequencing technology, comprehensive repositories of molecular data combined with clinical data could provide further insight into the therapeutic impact of molecular typing in glioblastoma patients. However, until effective targeted therapies against the more common molecular targets such as EGFR, CDKN2A/B, and PI3kinase can be developed, the therapeutic utility of targeted gene NGS testing in GBM patients will not be leveraged to its full potential. Patients need to be included in a discussion with regards to the expectations and to emphasize that these tests are highly valuable for diagnostic purposes and for consideration of clinical trials.
Supplementary Material
Acknowledgments
J.B.I. gratefully acknowledges support from the NIH/NCI (K12CA090354) and the Conquer Cancer Foundation/Sontag Foundation.
Conflict of interest statement. M.T. reports consulting or advisory role from Agios Pharmaceutical, Integragen, and Taiho Oncology, outside the submitted work; research funding from Sanofi, outside of submitted work.
Authorship statement. All authors contributed to the experimental design, data interpretation, draft writing, and revision, and have approved the final version.
Contributor Information
Mary Jane Lim-Fat, Division of Neurology, Department of Medicine, Sunnybrook Health Sciences Centre, University of Toronto, Toronto, Ontario, Canada.
Gilbert C Youssef, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Mehdi Touat, Sorbonne Université, Inserm, CNRS, UMR S 1127, Institut du Cerveau et de la Moelle épinière, ICM, AP-HP, Hôpitaux Universitaires La Pitié Salpêtrière - Charles Foix, Service de Neurologie 2-Mazarin, Paris, France.
J Bryan Iorgulescu, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA; Department of Pathology, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Sydney Whorral, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Marie Allen, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Rifaquat Rahman, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Ugonma Chukwueke, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
J Ricardo McFaline-Figueroa, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Lakshmi Nayak, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Eudocia Q Lee, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Tracy T Batchelor, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Omar Arnaout, Department of Neurosurgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Pier Paolo Peruzzi, Department of Neurosurgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
E Antonio Chiocca, Department of Neurosurgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
David A Reardon, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
David Meredith, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA; Department of Pathology, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Sandro Santagata, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA; Department of Pathology, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Rameen Beroukhim, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Wenya Linda Bi, Department of Neurosurgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Keith L Ligon, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA; Department of Pathology, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA; Department of Oncologic Pathology, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
Patrick Y Wen, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Funding
The authors received no financial support for the research, authorship and/or publication of this article.
References
- 1. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 2. Horbinski C, Ligon KL, Brastianos P, et al. . The medical necessity of advanced molecular testing in the diagnosis and treatment of brain tumor patients. Neuro Oncol. 2019;21(12):1498–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Louis DN, Perry A, Wesseling P, et al. . The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–1251. doi: 10.1093/neuonc/noab106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wen PY, Packer RJ. The 2021 WHO classification of tumors of the central nervous system: clinical implications. Neuro Oncol. 2021;23(8):1215–1217. doi: 10.1093/neuonc/noab120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Louis DN, Wesseling P, Paulus W, et al. . cIMPACT-NOW update 1: not otherwise specified (NOS) and not elsewhere classified (NEC). Acta Neuropathol. 2018;135(3):481–484. [DOI] [PubMed] [Google Scholar]
- 6. Louis DN, Giannini C, Capper D, et al. . cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol. 2018;135(4):639–642. [DOI] [PubMed] [Google Scholar]
- 7. Brat DJ, Aldape K, Colman H, et al. . cIMPACT-NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 2018;136(5):805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ellison DW, Hawkins C, Jones DTW, et al. . cIMPACT-NOW update 4: diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAFV600E mutation. Acta Neuropathol. 2019;137(4):683–687. [DOI] [PubMed] [Google Scholar]
- 9. Brat DJ, Aldape K, Colman H, et al. . cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020;139(3):603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Louis DN, Wesseling P, Aldape K, et al. . cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol. 2020;30(4):844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ellison DW, Aldape KD, Capper D, et al. . cIMPACT-NOW update 7: advancing the molecular classification of ependymal tumors. Brain Pathol. 2020;30(5):863–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wen PY, Weller M, Lee EQ, et al. . Glioblastoma in adults: a Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro Oncol. 2020;22(8):1073–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lim-Fat MJ, Nayak L, Meredith DM. Genomic biomarker assessment in gliomas: impacts of molecular testing on clinical practice and trial design. Surg Pathol Clin. 2020;13(2):209–215. [DOI] [PubMed] [Google Scholar]
- 14. Woodworth GF, Dunn GP, Nance EA, Hanes J, Brem H. Emerging insights into barriers to effective brain tumor therapeutics. Front Oncol. 2014;4:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lim-Fat MJ, Song KW, Iorgulescu JB, et al. . Clinical, radiological and genomic features and targeted therapy in BRAF V600E mutant adult glioblastoma. J Neurooncol. 2021;152(3):515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prasad V, Fojo T, Brada M. Precision oncology: origins, optimism, and potential. Lancet Oncol. 2016;17(2):e81–e86. [DOI] [PubMed] [Google Scholar]
- 17. Von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP.. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: Guidelines for Reporting Observational Studies. Vol 370.; 2007. www.plosmedicine.org. Accessed August 26, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sholl LM, Do K, Shivdasani P, et al. . Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1(19):e87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garcia EP, Minkovsky A, Jia Y, et al. . Validation of oncopanel: a targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med. 2017;141(6):751–758. [DOI] [PubMed] [Google Scholar]
- 20. Li MM, Datto M, Duncavage EJ, et al. . Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the association for molecular pathology, American society of clinical oncology, and college of American pathologists. J Mol Diagn. 2017;19(1):4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Touat M, Li YY, Boynton AN, et al. . Mechanisms and therapeutic implications of hypermutation in gliomas. Nature. 2020;580(7804):517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alexander BM, Trippa L, Gaffey S, et al. . Individualized screening trial of innovative glioblastoma therapy (INSIGhT): a bayesian adaptive platform trial to develop precision medicines for patients with glioblastoma. JCO Precis Oncol. 2019;(3):1–13. doi: 10.1200/po.18.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sahm F, Schrimpf D, Jones DT, et al. . Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016;131(6):903–910. [DOI] [PubMed] [Google Scholar]
- 24. Marquart J, Chen EY, Prasad V. Estimation of the percentage of US patients with cancer who benefit from genome-driven oncology. JAMA Oncol. 2018;4(8):1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shahar T, Nossek E, Steinberg DM, et al. . The impact of enrollment in clinical trials on survival of patients with glioblastoma. J Clin Neurosci. 2012;19(11):1530–1534. [DOI] [PubMed] [Google Scholar]
- 26. Field KM, Drummond KJ, Yilmaz M, et al. . Clinical trial participation and outcome for patients with glioblastoma: multivariate analysis from a comprehensive dataset. J Clin Neurosci. 2013;20(6):783–789. [DOI] [PubMed] [Google Scholar]
- 27. Alexander BM, Ba S, Berger MS, et al. ; GBM AGILE Network. Adaptive global innovative learning environment for glioblastoma: GBM AGILE. Clin Cancer Res. 2018;24(4):737–743. [DOI] [PubMed] [Google Scholar]
- 28. Barthel FP, Johnson KC, Varn FS, et al. ; GLASS Consortium. Longitudinal molecular trajectories of diffuse glioma in adults. Nature. 2019;576(7785):112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Draaisma K, Chatzipli A, Taphoorn M, et al. . Molecular evolution of IDH wild-type glioblastomas treated with standard of care affects survival and design of precision medicine trials: a report from the EORTC 1542 study. J Clin Oncol. 2020;38(1):81–99. [DOI] [PubMed] [Google Scholar]
- 30. Mansouri A, Hachem LD, Mansouri S, et al. . MGMT promoter methylation status testing to guide therapy for glioblastoma: refining the approach based on emerging evidence and current challenges. Neuro Oncol. 2019;21(2):167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lamba N, Chukwueke UN, Smith TR, et al. . Socioeconomic disparities associated with MGMT promoter methylation testing for patients with glioblastoma. JAMA Oncol. 2020;6(12):1972–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wick W, Platten M, Meisner C, et al. ; NOA-08 Study Group of Neuro-oncology Working Group (NOA) of German Cancer Society. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13(7):707–715. [DOI] [PubMed] [Google Scholar]
- 33. Perry JR, Laperriere N, O’Callaghan CJ, et al. ; Trial Investigators. Short-course radiation plus temozolomide in elderly patients with glioblastoma. N Engl J Med. 2017;376(11):1027–1037. [DOI] [PubMed] [Google Scholar]
- 34. Kamson DO, Grossman SA. The role of temozolomide in patients with newly diagnosed wild-type IDH, Unmethylated MGMTp Glioblastoma during the COVID-19 pandemic. JAMA Oncol. 2021;7(5):675–676. [DOI] [PubMed] [Google Scholar]
- 35. Kotelnikova EA, Pyatnitskiy M, Paleeva A, Kremenetskaya O, Vinogradov D. Practical aspects of NGS-based pathways analysis for personalized cancer science and medicine. Oncotarget. 2016;7(32):52493–52516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meng Y, Hynynen K, Lipsman N. Applications of focused ultrasound in the brain: from thermoablation to drug delivery. Nat Rev Neurol. 2021;17(1):7–22. [DOI] [PubMed] [Google Scholar]
- 37. Miller AM, Shah RH, Pentsova EI, et al. . Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565(7741):654–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meng Y, Pople CB, Suppiah S, et al. . MR-guided focused ultrasound liquid biopsy enriches circulating biomarkers in patients with brain tumors. Neuro Oncol. 2021;23(10):1789–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.