Abstract
A multiplex fluorogenic PCR assay for simultaneous detection of pathogenic Salmonella strains and Escherichia coli O157:H7 was developed and evaluated for use in detecting very low levels of these pathogens in meat and feces. Two sets of primers were used to amplify a junctional segment of virulence genes sipB and sipC of Salmonella and an intragenic segment of gene eae of E. coli O157:H7. Fluorogenic reporter probes were included in the PCR assay for automated and specific detection of amplified products. The assay could detect <10 CFU of Salmonella enterica serovar Typhimurium or E. coli O157:H7 per g of meat or feces artificially inoculated with these pathogens and cultured for 6 to 18 h in a single enrichment broth. Detection of amplification products could be completed in ≤4 h after enrichment.
Salmonella strains and enterohemorrhagic Escherichia coli O157:H7 are among the most important foodborne bacterial pathogens (9, 12). Cattle are important reservoirs of E. coli O157:H7 strains, and the majority of human infections due to this organism are associated with ingestion of undercooked, contaminated beef, water, or raw milk (6, 7). Salmonella, on the other hand, exhibits a broad host spectrum, which comprises most animal species, including mammals, birds, and cold-blooded animals (1). A variety of food products, especially contaminated poultry, beef, pork, and cheese, are the most important sources of human salmonellosis (3).
The objective of the present study was to assess the fidelity and utility of two sets of primers and two fluorogenic probes in a multiplex PCR format for simultaneous and semiautomated detection of pathogenic Salmonella strains and E. coli O157:H7. We also describe the use of a single nonselective broth for enrichment of meats and feces harboring very low numbers of these two pathogens. This PCR assay was optimized to obtain a strong and reproducible fluorescence signal from probes labeled with two reporter dyes that allowed immediate and specific detection of Salmonella and E. coli O157:H7.
Bacterial strains, culture media, growth conditions, and sample preparation.
The bacterial strains that were used are listed in Table 1. The strains of E. coli and Salmonella were obtained from the National Animal Disease Center, Ames, Iowa. Thomas Whittam (The Pennsylvania State University, University Park) kindly provided some of the E. coli strains. Bacterial strains were propagated and maintained on Trypticase soy agar (TSA) plates. Liquid cultures were obtained by growing bacteria in GNTSB (prepared by mixing equal volumes of gram-negative broth and Trypticase soy broth) for 18 h at 37°C with continuous agitation (160 rpm) in a circulating-air incubator (New Brunswick Scientific, Edison, N.J.). TSA and MacConkey agar were used to enumerate bacteria. TSA, gram-negative broth, Trypticase soy broth, and MacConkey agar were purchased from BBL (Becton Dickson Microbiology Systems, Cockeysville, Md.). Meat and feces were tested, as described previously (11), for the presence of endogenous bacterial flora and for Salmonella and E. coli O157:H7 contamination. Meat and feces (1- or 25-g portions) found to be free of Salmonella and E. coli O157:H7 contamination by PCR (2, 11) were artificially inoculated with 0.1-ml aliquots of 10-fold serial dilutions (prepared from 1:1 mixture of an 18-h culture of S. typhimurium and E. coli O157:H7). Inoculated samples were cultured in GNTSB (9 ml of GNTSB was added to 1 g of meat or feces) for 6 to 18 h at 37°C. Cultures were centrifuged at 1,000 × g for 2 min to remove large particles. A washing step was performed by centrifuging (12,000 × g for 3 min) 0.05 ml of supernatant mixed with 0.95 ml of GNTSB. The bacterial pellet was processed for DNA isolation as described previously (11).
TABLE 1.
Specificity of fluorogenic PCR to detect Salmonella and E. coli O157:H7
| Bacterial strainsa | Serotype, species, or groupb | eae gene presentc | sipB-sipC genes presentc | Fluorogenic PCR detection
|
|
|---|---|---|---|---|---|
| E. coli O157:H7 specific | Salmonella specific | ||||
| Nontoxigenic E. coli | O111:H12 (2) | − | − | − | − |
| H21 (3) | − | − | − | − | |
| O149:HN | − | − | − | − | |
| O157:H43 | − | − | − | − | |
| O128:H7 (2) | − | − | − | − | |
| H21 | − | − | − | − | |
| H47 | − | − | − | − | |
| ETEC | O101 | − | − | − | − |
| O149:H10 | − | − | − | − | |
| RDEC | O132 | + | − | − | − |
| O15:HN | + | − | − | − | |
| EPEC | O55:NM (2) | + | − | − | − |
| H6 (2) | + | − | − | − | |
| O26:NM (3) | + | − | − | − | |
| O111:H2 (3) | + | − | − | − | |
| O128:H2 (4) | + | − | − | − | |
| O45:H2 | + | − | − | − | |
| O55:H7 (2) | + | − | + | − | |
| STEC | O111:NM (2) | + | − | − | − |
| O111:H8 | + | − | − | − | |
| O26:H11 | + | − | − | − | |
| O45:H2 | − | − | − | − | |
| O113 | − | − | − | − | |
| OX3:H2 | − | − | − | − | |
| EHEC | O26:H11 (3) | + | − | − | − |
| O111:NM | + | − | − | − | |
| O157:H7 (5) | + | − | + | − | |
| O157:NM (1) | + | − | + | − | |
| Salmonella | Typhimurium | − | + | − | + |
| Enteriditis | − | + | − | + | |
| Samftenberg | − | + | − | + | |
| Litchfield | − | + | − | + | |
| DT104 (2) | − | + | − | + | |
| B1RC8 | − | + | − | + | |
| B1RC9 | − | + | − | + | |
| EILC9 | − | + | − | + | |
| E1LC10 | − | + | − | + | |
| C1CC117 | − | + | − | + | |
| Enterobacter | E. cloacae | − | − | − | − |
| Hafnia | H. alvie | + | − | − | − |
| Klebsiella | K. pneumoniae | − | − | − | − |
| Proteus | P. vulgaris | − | − | − | − |
| Pseudomonas | P. aeruginosa | − | − | − | − |
| Staphylococcus | S. aureus | − | − | − | − |
ETEC, enterotoxigenic E. coli; RDEC, rabbit diarrheagenic E. coli; EPEC, enteropathogenic E. coli; STEC, Shiga toxin-producing E. coli; EHEC, enterohemorrhagic E. coli.
Parentheses indicate the number of isolates of a serotype tested.
The presence of the eae gene in E. coli and that of sipBC in Salmonella was determined by colony blot hybridization using probes specific for these genes (2, 11; http://www.bio.psu.edu/people/faculty/whittam/lab/deca/).
Design of primers and fluorogenic probes.
The nucleotide sequences of primers and fluorogenic probes used in amplification and detection of genes sipB-sipC and eae have been reported previously (2, 11). The reporter dye FAM (6-carboxyfluorescein) or HEX (6-carboxyhexafluorescein) was conjugated at the 5′ ends of these probes, and the quencher dye TAMRA (6-carboxytetramethylrhodamine) was conjugated at the 3′ ends. The FAM-labeled probe was used for detecting the 250-bp sipB-sipC gene fragment of Salmonella, and the HEX-labeled probe facilitated the detection of the 150-bp eae gene fragment of E. coli O157:H7. Primers and probes were synthesized by Integrated DNA Technologies (Coralville, Iowa).
PCR amplification.
Template DNA (5 μl) was added to 45 μl of a master mixture (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 10 mM Na2 EDTA, 3 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 450 nM each sipB-sipC primer, 300 nM each eae primer, 100 nM each fluorogenic probe, and 2.5 Units of AmpliTaq Gold DNA polymerase [PE Applied Biosystems, Foster City, Calif.]) and amplified under conditions described previously (11).
Fluorogenic detection of PCR products.
The specific detection of PCR-amplified products was achieved by reading a 96-well plate in a computer-controlled dual-scanning microplate spectrofluorometer (SPECTRAmax GEMINI; Molecular Devices, Sunnyvale, Calif.). A series of six no-DNA template controls (NTCs) were included in each reaction plate to establish the background fluorescence and to calculate the detection threshold (DT). The excitation (ex) and emission (em) wavelengths used for reporter (FAM and HEX) and quencher (TAMRA) dyes were as follows: FAM, ex at 490 nm and em at 515 nm; HEX, ex at 535 nm and em at 560 nm; and TAMRA, ex at 490 nm and em at 585. The fluorescence data were collected and analyzed by using the fluorescence data management program SOFTmax PRO. Samples exhibiting reporter fluorescence, expressed as relative fluorescence units (RFU), higher than the DT were assigned a plus score, indicating the presence of the target gene(s). Samples exhibiting fluorescence equal to or less than the DT were assigned a minus score, indicating the absence of the target genes and thus no detectable amplification.
Calculation of DT.
The DT was computed as [mean reporter fluorescence + confidence interval (ς/√n)] × correction coefficient. The confidence interval was computed at a significance level of 99.9%, where ς represents the standard deviation of the mean and n is the number of NTCs. To calculate the correction coefficient, DNA from a 10-fold serial dilution (prepared from a 1:1 mixture of overnight-grown cultures of S. enterica serovar Typhimurium and E. coli O157:H7) was subjected to PCR amplification. The correction coefficient was calculated by dividing the mean fluorescence value of the highest 10-fold dilution producing detectable 250- and 150-bp amplicons on an agarose gel by the mean fluorescence value of the NTCs.
Amplification of 250- and 150-bp fragments of target genes.
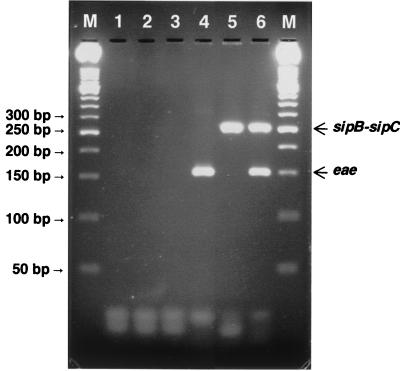
As shown in Fig. 1, the sipB-sipC and eae primer pair generated the predicted 250- and 150-bp DNA bands from S. enterica serovar Typhimurium and E. coli O157:H7 strain 2409, respectively. Amplified products were not detected from strains lacking these genes.
FIG. 1.
Agarose gel electrophoresis of PCR products obtained from different strains of Salmonella and E. coli using primers designed to amplify the 250-bp junctional fragment of sipB-sipC and the 150-bp intragenic fragment of eae. Lanes: M, 50-bp molecular size standards; 1, NTC; 2, E. coli strain DEC A6 (eae) template DNA; 3, Salmonella strain A 1cc217 (sipB-sipC) template DNA; 4, E. coli O157:H7 (eae+) template DNA; 5, S. enterica serovar Typhimurium (sipB+-sipC+) template DNA; 6, E. coli O157:H7 and serovar Typhimurium template DNA. Arrows indicate the relative positions of amplified fragments.
Specificity of FAM- and HEX-labeled fluorogenic probes.
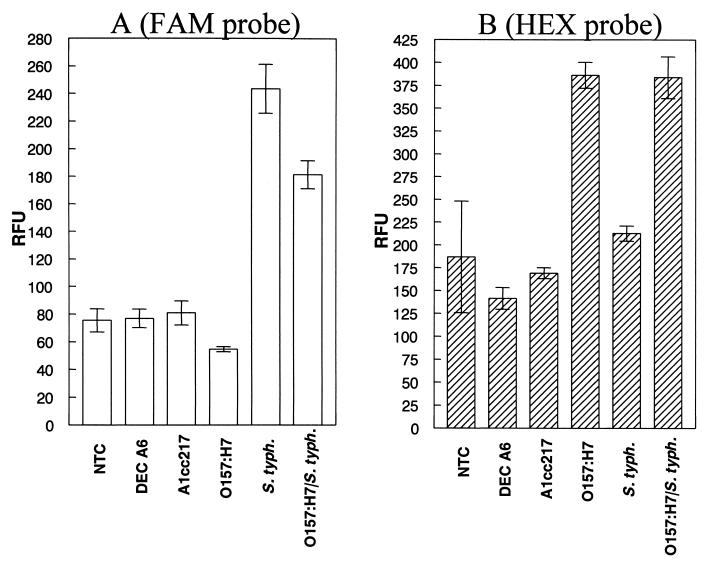
The ability of FAM- and HEX-labeled probes to specifically detect the sipB-sipC and eae genes was determined by measuring the RFU of each PCR sample. As shown in Fig. 2A, a FAM-specific fluorescence signal was generated by PCR samples containing DNA from S. enterica serovar Typhimurium harboring the sipB and sipC genes. Similarly, PCR samples containing DNA from E. coli O157:H7 produced a HEX-specific signal (Fig. 2B). PCR samples that received the template DNA from serovar Typhimurium and E. coli O157:H7 produced a positive fluorescence signal for both reporter dyes.
FIG. 2.
The specificity of FAM- and HEX-conjugated probes for detection of the 250-bp sipB-sipC fragment of S. enterica serovar Typhimurium and the 150-bp E. coli O157:H7-specific eae fragment, respectively, was examined using DNA templates from different bacterial strains (indicated on the x axis). Following PCR, the RFU of each sample were determined (plotted on the y axis). A DT of 97.8 RFU was obtained for the Salmonella-specific FAM-labeled probe (A), and a DT of 314.0 RFU was obtained for the E. coli O157:H7-specific HEX-labeled probe (B). A PCR sample was scored positive if the RFU value was greater than the DT. Error bars indicated standard deviations of the mean (n = 3).
Specificity of fluorogenic PCR to detect pathogenic Salmonella and E. coli O157:H7.
As shown in Table 1, only those E. coli strains previously shown to harbor the E. coli O157:H7-specific eae gene (11) were scored positive for amplification by the fluorogenic detection system. Among the E. coli strains tested, all E. coli O157:H7 and O157:NM isolates were detected positive. These two serotypes cannot be distinguished from each other in eae-based PCR assays (4, 10). However, detection of E. coli O157:NM is an advantage considering the frequent isolation of this serotype from patients with hemolytic uremic syndrome (14). Recent studies have shown that many E. coli O157:NM isolates contain fliC, the gene encoding the H7 flagellar antigen (5). Based on this finding, fliC-harboring E. coli O157:NM isolates are considered the nonmotile variants of E. coli O157:H7. The E. coli O157:H7-specific eae probe also facilitated the detection of E. coli O55:H7, an enteropathogenic E. coli strain. All Salmonella isolates tested positive in this assay, and these isolates have been shown to harbor sipB-sipC (2). All 47 strains lacking eae of E. coli O157:H7 or the sipB-sipC genes of Salmonella resulted in RFU less than or equal to the DT and were scored negative for amplification by the detection system (Table 1).
Detection limits of fluorogenic PCR in beef and feces.
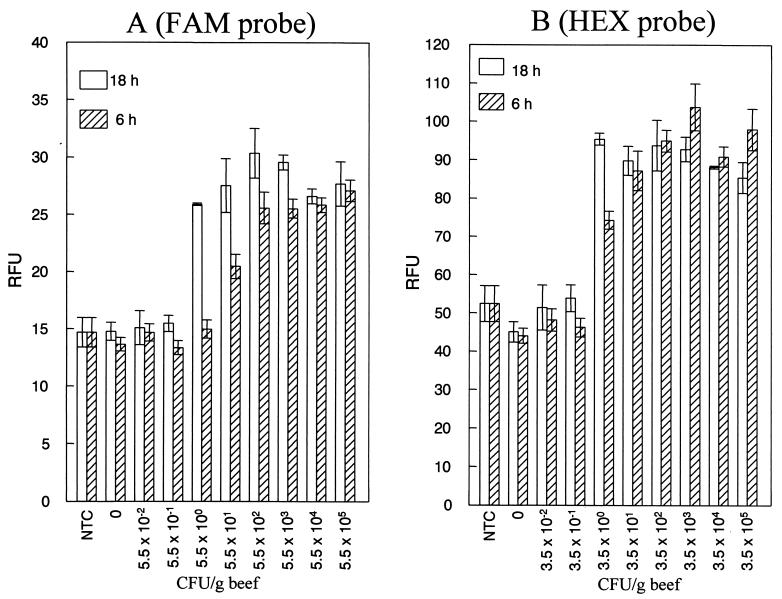
The detection sensitivity of fluorogenic PCR using DNA isolated from a 10-fold serial dilution (prepared from a 1:1 mixture of S. enterica serovar Typhimurium and E. coli O157:H7 overnight cultures) was determined to be 2.5 × 103 CFU per ml or 10 CFU per PCR (data not shown). However, most bacterial pathogens, including E. coli O157:H7 and Salmonella, are present in very low numbers in foods and feces. Foods and feces also contain substances that are inhibitory to the process of PCR amplification. Detection of very low levels of bacterial contamination in foods and feces necessitates that these samples be cultured for a few hours in an appropriate enrichment broth. This enrichment serves two purposes. First, it dilutes out substances inhibitory to the PCR process, and second, it provides conditions conducive for growth and multiplication of bacterial pathogens to a detectable number. The detection sensitivity of this PCR assay for S. enterica serovar Typhimurium ranged from 55 to 5.5 CFU per g of beef after 6 to 18 h of enrichment (Fig. 3A). For E. coli O157:H7, the detection sensitivity was 3.5 CFU per g of beef after 6 or 18 h of enrichment (Fig. 3B). In ground chicken, the detection sensitivity ranged from 47 to 0.47 CFU per g of chicken for S. enterica serovar Typhimurium (data not shown) and from 3.1 to 0.31 CFU per g of chicken for E. coli O157:H7 after 6 to 18 of enrichment (data not shown). The detection sensitivity in feces ranged from 5.8 × 104 to 5.8 CFU per g of feces for Salmonella (data not shown) and from 55 to 5.5 CFU per g of feces for E. coli O157:H7 (data not shown) after 6 to 18 h of enrichment. Thus, by using a single enrichment broth, the fluorogenic PCR assay could detect between 1 and 10 CFU of Salmonella and E. coli O157:H7 after 18 h enrichment of meat and feces artificially inoculated with these two pathogens. Neither strain interfered with the detection of the other strain in enriched meat or fecal samples. The presence of endogenous bacterial flora (determined as total aerobic plate counts) to the level of 106 CFU per g of beef and 105 CFU per g of feces has no effect on the detection sensitivity of this assay. This observation is in agreement with a previously reported study (11) showing that endogenous bacteria when present at or below 108 CFU per g of beef or feces do not interfere with detection of low numbers of target bacterial cells using our PCR assay. The lower detection sensitivity observed for Salmonella in feces enriched for only 6 h is probably due to the slow growth of Salmonella in these types of samples. However, once the samples (meat or feces) were enriched for 18 h, the detection sensitivity for Salmonella reached the same limits as for E. coli O157:H7. The detection sensitivity obtained in our assay is better than or comparable to those of protocols that rely on laborious and time-consuming methods to prepare DNA from meat and fecal samples (8, 10, 13). Immunomagnetic capture of target bacterial cells, which is employed in some of these methods to increase detection sensitivity, is not required in our assay to detect very low levels of contamination.
FIG. 3.
Sensitivity of fluorogenic PCR in beef. DNA isolated from beef inoculated with various dilutions (plotted on the x axis) of a mixed culture of Salmonella and E. coli O157:H7 and enriched for 6 to 18 h was subjected to PCR amplification. The RFU of each sample were determined (plotted on the y axis). The DTs for the Salmonella-specific FAM-labeled probe and for the E. coli O157:H7-specific HEX-labeled probe were 18.4 and 66.0 RFU, respectively. A PCR sample was scored positive if the RFU value was greater than the DT. Error bars indicate the standard deviations of the mean (n = 3).
In conclusion, the fluorogenic PCR assay described in the present study provides a highly sensitive and specific closed-tube system for simultaneous detection of pathogenic Salmonella and E. coli O157:H7 in meats and feces. In addition, the automated PCR amplification and detection of target gene amplicons are conducive for screening large number of samples in a single assay. This method, therefore, should be a significant tool in monitoring foods of animal origin and the environment where these animals are produced and processed into foods for the presence of potential human pathogens. The enrichment, sample preparation, DNA isolation, PCR amplification, and fluorogenic detection procedures described here can also be used for detection of other foodborne pathogens by combining appropriate primers and fluorogenic probes. The availability of additional reporter dyes and detection systems capable of detecting fluorescent signals from several reporters present in a single PCR tube will allow simultaneous detection of several pathogens in a single PCR assay.
Acknowledgments
We thank Robert Morgan for technical assistance and assistance in preparation of the manuscript, Sandy Johnson for secretarial assistance, and Irene Wesley and Tom Casey for critical review of the manuscript.
REFERENCES
- 1.Baird-Parker A C. Foodborne salmonellosis. Lancet. 1990;336:1231–1235. doi: 10.1016/0140-6736(90)92844-8. [DOI] [PubMed] [Google Scholar]
- 2.Carlson S A, Bolton L F, Briggs C E, Hurd H S, Sharma V K, Fedorka-Cray P J, Jones B D. Detection of multiresistant Salmonella typhimurium DT104 using multiplex and fluorogenic PCR. Mol Cell Probes. 1999;13:213–222. doi: 10.1006/mcpr.1999.0240. [DOI] [PubMed] [Google Scholar]
- 3.Ekperigin H E, Nagaraja K V. Microbial food borne pathogens. Salmonella. Vet Clin N Am Food Anim Pract. 1998;14:17–29. [PubMed] [Google Scholar]
- 4.Fratamico P M, Sackitey S K, Wiedmann M, Deng M Y. Detection of Escherichia coli O157:H7 by multiplex PCR. J Clin Microbiol. 1995;33:2188–2191. doi: 10.1128/jcm.33.8.2188-2191.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gannon V P, S. D S, Graham T, King R K, Rahn K, Rend S. Use of the flagellar H7 gene as a target in multiplex PCR assays and improved specificity in identification of enterohemorrhagic Escherichia coli strains. J Clin Microbiol. 1997;35:656–662. doi: 10.1128/jcm.35.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffin P M, Tauxe R V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 7.Hancock D D, Besser T E, Kinsel M L, Tarr P I, Rice D H, Paros M G. The prevalence of Escherichia coli O157.H7 in dairy and beef cattle in Washington State. Epidemiol Infect. 1994;113:199–207. doi: 10.1017/s0950268800051633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGraw E A, Li J, Selander R K, Whittam T S. Molecular evolution and mosaic structure of alpha, beta, and gamma intimins of pathogenic Escherichia coli. Mol Biol Evol. 1999;16:12–22. doi: 10.1093/oxfordjournals.molbev.a026032. [DOI] [PubMed] [Google Scholar]
- 9.Mead P S, Slutsker L, Dietz V, McCaig L F, Bresee J S, Shapiro C, Griffin P M, Tauxe R V. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oberst R D, Hays M P, Bohra L K, Phebus R K, Yamashiro C T, Paszko-Kolva C, Flood S J, Sargeant J M, Gillespie J R. PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with an internal fluorogenic probe and the 5′ nuclease (TaqMan) assay. Appl Environ Microbiol. 1998;64:3389–3396. doi: 10.1128/aem.64.9.3389-3396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma V K, Dean-Nystrom E A, Casey T A. Semi-automated fluorogenic PCR assays (TaqMan) for rapid detection of Escherichia coli O157:H7 and other Shiga toxigenic E. coli. Mol Cell Probes. 1999;13:291–302. doi: 10.1006/mcpr.1999.0251. [DOI] [PubMed] [Google Scholar]
- 12.Todd E C. Costs of acute bacterial foodborne disease in Canada and the United States. Int J Food Microbiol. 1989;9:313–326. doi: 10.1016/0168-1605(89)90099-8. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Slavik M F. Rapid detection of Salmonella in chicken washes by immunomagnetic separation and flow cytometry. J Food Prot. 1999;62:717–723. doi: 10.4315/0362-028x-62.7.717. [DOI] [PubMed] [Google Scholar]
- 14.Willshaw G A, Thirlwell J, Jones A P, Parry S, Salmon R L, Hickey M. Vero cytotoxin-producing Escherichia coli O157 in beefburgers linked to an outbreak of diarrhoea, haemorrhagic colitis and haemolytic uraemic syndrome in Britain. Lett Appl Microbiol. 1994;19:304–307. doi: 10.1111/j.1472-765x.1994.tb00461.x. [DOI] [PubMed] [Google Scholar]