Abstract
Background
Glioblastomas are highly resistant to therapy, and virtually all patients experience tumor recurrence after standard-of-care treatment. Surgical tumor resection is a cornerstone in glioblastoma therapy, but its impact on cellular phenotypes in the local postsurgical microenvironment has yet to be fully elucidated.
Methods
We developed a preclinical orthotopic xenograft tumor resection model in rats with integrated 18F-FET PET/CT imaging. Primary and recurrent tumors were subject to bulk and single-cell RNA sequencing. Differentially expressed genes and pathways were investigated and validated using tissue specimens from the xenograft model, 23 patients with matched primary/recurrent tumors, and a cohort including 190 glioblastoma patients. Functional investigations were performed in vitro with multiple patient-derived cell cultures.
Results
Tumor resection induced microglia/macrophage infiltration, angiogenesis as well as proliferation and upregulation of several stem cell-related genes in recurrent tumor cells. Expression changes of selected genes SOX2, POU3F2, OLIG2, and NOTCH1 were validated at the protein level in xenografts and early recurrent patient tumors. Single-cell transcriptomics revealed the presence of distinct phenotypic cell clusters in recurrent tumors which deviated from clusters found in primary tumors. Recurrent tumors expressed elevated levels of pleiotrophin (PTN), secreted by both tumor cells and tumor-associated microglia/macrophages. Mechanistically, PTN could induce tumor cell proliferation, self-renewal, and the stem cell program. In glioblastoma patients, high PTN expression was associated with poor overall survival and identified as an independent prognostic factor.
Conclusion
Surgical tumor resection is an iatrogenic driver of PTN-mediated self-renewal in glioblastoma tumor cells that promotes therapeutic resistance and tumor recurrence.
Keywords: glioblastoma, pleiotrophin, recurrence, self-renewal, tumor resection
Key Points.
Tumor resection induces iatrogenic glioblastoma stem cell propagation mediated, in part, by pleiotrophin.
Pleiotrophin expression increased in recurrent xenografts and patient glioblastomas and was an independent prognostic factor in glioblastoma patients.
Importance of the Study.
Surgical tumor resection is a cornerstone in glioblastoma therapy. By establishing and applying a preclinical glioblastoma resection model, we identified surgical resection as an inducer of the stem cell state in recurrent tumor cells mediated, in part, by pleiotrophin signaling. Glioblastoma stem cells have high intrinsic resistance toward radio- and chemotherapy, which is administered adjuvant in the postsurgical setting. As such, residual tumor cells become more prone to survive radio- and chemotherapy and facilitate tumor recurrence and ultimately patient death. Iatrogenic induction of the stem cell program in residual tumor cells left in the resection cavity may therefore contribute to the resistance mechanisms responsible for failures of both current standard-of-care treatment and novel clinical trials. Development and application of future therapeutic strategies should consider this stem cell enrichment in recurrent glioblastoma tumor cells to improve treatment efficacy and ultimately patient survival.
Glioblastoma (GBM) is the most common and malignant primary brain tumor. Current standard-of-care treatment includes surgical tumor resection and radio- and chemotherapy with concomitant and adjuvant temozolomide (TMZ). Despite therapeutic interventions, the median survival of GBM patients is approximately 15 months.1 Maximal surgical tumor resection is a cornerstone in GBM therapy, and complete resection is an independent prognostic factor for progression-free survival and overall survival.2 Microsurgical techniques and 5-aminolevulinic acid (5-ALA) aid the achievement of gross total resection, yet, approximately 90% of GBM patients experience tumor recurrence in the resection margin.3 The origin of recurrences can be ascribed to the diffusely infiltrative nature of GBM tumor cells,4 which makes complete resection nearly impossible. Although the resection margin is the region of recurrence, the margin has gained much less attention than central tumor areas, despite reports of intratumoral heterogeneity in these different regions.5 The few residual tumor cells left behind after surgery inhabit the reactive postsurgical microenvironment characterized by neuronal death, infiltration of microglia/macrophages, astrogliosis, angiogenesis, blood-brain barrier disruption, and edema,6 and eventually lead to tumor recurrence.
Surgical resection of breast-, lung-, and colorectal cancer has shown that surgical intervention can promote disease progression and metastasis.7 The central nervous system responds to damage through neuro-inflammation, mediated by many different intrinsic and extrinsic cell types, such as astrocytes, microglia, pericytes, endothelial cells, monocytes/macrophages, neutrophils, NK cells, and lymphocytes.8 The post-therapeutic inflammatory microenvironment secretes various cytokines, chemokines, and growth factors,9 which mediate tissue repair mechanisms, but simultaneously also support tumor cell proliferation and survival, thereby ultimately laying the foundation for tumor recurrence.10 In murine preclinical glioma resection models, it has been shown that reactive astrocytes can potentiate tumor aggressiveness following surgical resection alone,11 and that resection can suppress immune responses.12 However, the impact and potential phenotypic changes induced by tumor resection are largely overlooked, and its potential role in therapeutic resistance has yet to be fully uncovered.
Materials and Methods
Detailed methodological descriptions are available in the Supplementary material.
Cell Cultures
The patient-derived GBM spheroid cultures P3, T78, T86, T87, T110, T111, T123, T129, and T138 were cultured in serum-free neurobasal medium. Primary human microglia and immortalized SV-40 microglia were cultured in DMEM. Proliferation was measured with MTS assays. For limiting dilution assays, 100, 50, 10, 5, 1 cell(s)/well were seeded and analyzed with the ELDA software.13 For cell clot generation, tumor spheroids were fixed, washed, and two drops of plasma and one drop of thrombin were added.
In Vitro O-(2-[18F]Fluoroethyl)-l-Tyrosine (18F-FET) Uptake Assay
Tumor cells were exposed to 15 kBq GMP-grade 18F-FET for 60 min and washed. 18F-FET uptake was measured in the AtomLab 950 (Biodex Medical Systems).
GBM Xenograft Model and Tumor Resection Procedure
Orthotopic xenografts were generated in female nude rats (NIH-Foxn1rnu) aged 6-8 weeks. A craniotomy was centered around the implantation burr hole. The tumor was resected by gentle microdissection alternated with saline irrigation and suctioning. Hemostasis was achieved with Surgicel or electrocoagulation.
18F-FET PET/CT Scans and Data Analysis
Animals were fasted for 4 h and anesthetized. Through a tail-vein catheter, 50 MBq 18F-FET was injected. After 1 h, a 1-bed CT scan was performed followed by a 15-min static PET scan. Data were reconstructed using an OSEM3D/SP-MAP algorithm.
Tumor Dissociation and Sample Preparation
Single-cell suspensions were generated using the Brain Tumor Dissociation Kit and gentleMACS Dissociator (Miltenyi Biotec). Cellular debris and myelin were removed with debris removal solution, myelin removal beads II, and LS columns, before erythrocytes were lysed.
Cell Sorting
Fc receptors were blocked before the addition of CD11b PE, CD45 BB700, EAAT2 FITC, HLA-ABC Alexa Fluor 700, and live/dead fixable violet stain (Invitrogen). Cells were sorted with a FACSAria III sorter. The gating strategy is displayed in Supplementary Figure 1B. Sorting purity was validated by re-acquisition of sorted cells (Supplementary Figure 1C).
Total mRNA Sequencing
Total RNA was extracted and the NEBNext Poly(A) mRNA Magnetic Isolation module was used to isolate mRNA. cDNA synthesis was performed with the NEBNext Ultra II RNA Library Prep Kit. Sequencing was performed on an Illumina NovaSeq 6000.
Single-Cell Sequencing
Methanol-fixed single cells were rehydrated, passed through a 40 µm filter, and processed according to the 10× Genomics Chromium next GEM single-cell 3′ reagent kits v3.1 protocol. Library preparation was performed using the Chromium Next GEM Single Cell 3′ Library Kit v3.1.
Bioinformatics
Raw sequencing data were demultiplexed and converted into FASTQ files with bcl2Fastq. Data were aligned against the Homo sapiens genome and transcriptome using STAR.14 The edgeR R package15 was used for transforming raw counts into counts per million and for differential expression analysis using the glmFit and glmLRT functions. Detection of enriched BIOCARTA,16 KEGG,17 and REACTOME18 gene sets was based on pre-ranked gene set enrichment analysis (GSEA) and the fgsea R package.19
Single-cell RNA sequencing data were demultiplexed with the 10× Genomics CellRanger Software (V3.1.0) and analyzed with the Seurat R package (V4.0). After quality control, uniform manifold approximation and projection (UMAP) clustering was performed, and positive gene markers in each cluster were identified. GSEA was performed to identify enriched pathways. In silico protein interactions were investigated using STRING.20
Patient and Tissue Inclusion
Formaldehyde-fixed paraffin-embedded tissue sections from 11 patients with matching primary and early recurrent GBMs (≤6 months) were identified and compared to 12 matched primary and recurrent tumors, with recurrences 12-19 months after diagnosis. Patient survival was assessed with a well-annotated glioma cohort,21 including 190 GBM patients (184 IDH-wildtype). Patient characteristics are summarized in Supplementary Table 1.
Immunohistochemistry and Automated Quantitative Analysis
Tissue sections (3 μm) were subject to deparaffinization and heat-induced epitope retrieval. Sections were stained with H&E, vimentin, Iba-1, VEGF, SOX2, OLIG2, POU3F2, CD133, PTN, PTPRZ1, and Notch1. Stained slides were digitalized, and different classifiers were designed to quantify fractions of cells with positive staining, staining areas, and intensities using the Visiopharm software V2018.9.4.
Immunofluorescence Staining
Tissue sections were prepared as described for immunohistochemistry. Antibody detection was performed with DISCOVERY OmniMap anti-rabbit/mouse HRP coupled with the DISCOVERY Cy5 and FAM Kits. Nuclei were counterstained with DAPI. Images were acquired on a Leica DM6000B microscope with an Olympus DP72 camera.
ELISA
Tumor cells were resuspended in 20 mL medium, incubated for 48 h, and the supernatant was stored at −80°C. The human Pleiotrophin/PTN ELISA Kit (Invitrogen) was used according to the manufacturer’s instructions.
Statistical Analysis
Group comparisons were performed using Student’s t-test, Mann-Whitney U test, one-way ANOVA, and Tukey’s multiple comparison test or Kruskal-Wallis test with Dunn’s multiple comparison test. Correlations were assessed with Pearson R. Survival analysis was performed with Kaplan-Meier curves, log-rank tests, and Cox proportional hazard regression. Error bars represent mean ± SEM.
Ethics
Approvals were obtained from the Danish Data Inspection Authority (#16/11065), the Regional Scientific Ethical Committee of the Region of Southern Denmark (#S-20150148), and The Animal Experiments Inspectorate in Denmark (#2018-15-0201-01472).
Results
18F-FET Can Identify Preclinical GBM Growth
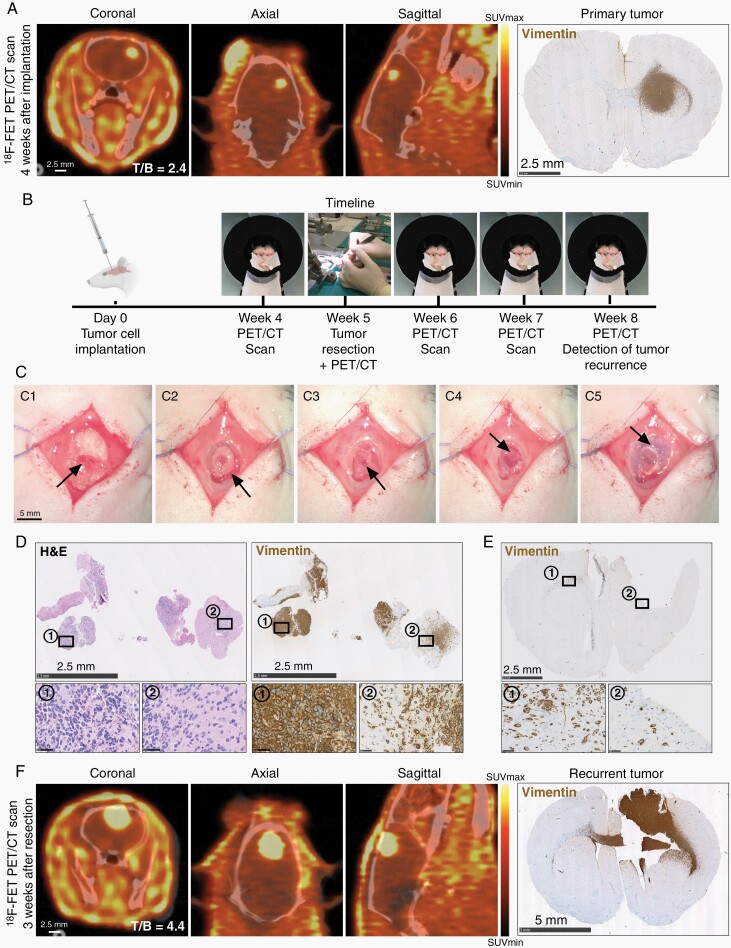
To establish PET/CT-based monitoring of tumor growth, we initially tested the 18F-FET uptake of 6 different patient-derived spheroid cultures in vitro (Supplementary Figure 2A). T78 cells had the highest uptake and were investigated in vivo along with the previously validated P3 cells.22 P3 xenografts could successfully be detected by 18F-FET PET 3-4 weeks after implantation and a combined compact/diffuse tumor growth pattern was histologically validated (Figure 1A). Surprisingly, T78 xenografts showed no 18F-FET uptake despite diffuse tumor growth (Supplementary Figure 2B), demonstrating poor correlation with in vitro 18F-FET uptake. Dynamic PET scans revealed that P3 xenografts showed >98% of maximum voxel intensity 60-min post-18F-FET injection (Supplementary Figure 2C). Subsequently, P3 xenografted animals were used, and PET/CT scanned 1-h post-18F-FET injection. A timeline indicating the different procedures is outlined in Figure 1B.
Fig. 1.
Establishment and validation of PET/CT-based tumor detection and surgical resection procedure. (A) Representative 18F-FET PET/CT and histological images from a P3 xenograft 4 weeks after tumor cell implantation. Scale = 2.5 mm. (B) Timeline depicting tumor cell implantation and subsequent procedures. (C) Tumor resection procedure; Exposure of the implantation burr hole (C1), craniotomy (C2), bone flap removal (C3), microsurgical resection (C4), and bone flap repositioning (C5). Scale = 5 mm. (D) H&E and tumor cell marker vimentin stains of resected tissue. Scale = 2.5 mm (overview) and 50 µm (inserts). (E) Histological section from a post-resection brain confirming gross total resection. Scale = 2.5 mm. (F) Representative 18F-FET PET/CT images and histological validation at recurrence, 3 weeks after resection. Scale = 2.5 mm (PET/CT) and 5 mm (histology). Abbreviations: CT, computed tomography; PET, positron emission tomography.
Resection of GBM Xenografts Closely Mimics Clinical Patient GBM Resection
Following 18F-FET PET/CT detection of primary tumors 3-4 weeks after implantation, tumor resection was performed (Figure 1C). As observed in H&E and tumor cell-specific vimentin staining, resected tissue fragments were comprised of both cell-dense bulk tumor (Figure 1D, region 1) and tumor-infiltrated brain parenchyma (Figure 1D, region 2), thereby closely resembling resected patient GBM specimens. Despite near gross total resection (Figure 1E), 72-h postsurgical 18F-FET PET/CT scans showed an increase in tracer uptake, presumably due to reactive changes induced by the resection (Supplementary Figure 2D). Therefore, the postsurgical scan was used as a new baseline scan. 18F-FET uptake remained stable for 2 weeks before further increasing (Figure 1F; Supplementary Figure 2D). As such, tumor recurrence was detectable approximately 2½ weeks after resection and was histologically validated (Figure 1F). Subsequently, all recurrent tumors were isolated 3 weeks after resection. Examples of 18F-FET tumor uptake at different time points are shown in Supplementary Figure 2E.
Surgical Tumor Resection Induces Tumor Cell Proliferation, Activation of Tumor-Promoting Signaling Pathways, Angiogenesis, and Infiltration With Microglia/Macrophages
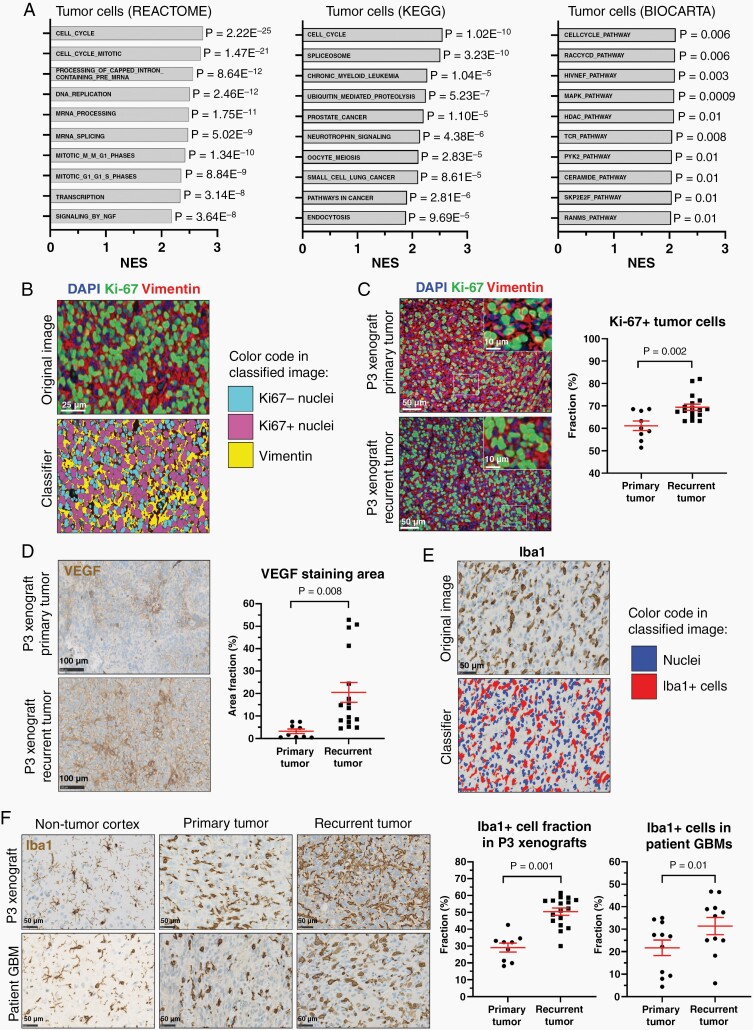
Next, tumor cells from primary and recurrent tumors were isolated based on human-specific MHC class 1 antigen HLA-ABC expression. Total RNA sequencing revealed 2266 genes with significant differential expression in recurrent tumor cells (1769 upregulated and 497 downregulated, Supplementary data 1). The top-10 positively enriched REACTOME, KEGG, and BIOCARTA pathways are represented in Figure 2A. Interestingly, cell cycle ranked among the most enriched pathways, and by quantifying the fraction of proliferating vimentin+/Ki-67+ tumor cells (Figure 2B), we found that tumor cells in recurrent tumors were more proliferative compared to primary tumors (69.4% vs 61.1%, P = .002, Figure 2C). Several pathways involved in cancer signaling were upregulated in recurrent tumor cells, exemplified by KEGG_PATHWAYS_IN_CANCER (NES = 1.9, P = 2.81E), KEGG_GLIOMA (NES = 1.91, P = .001), and 10 additional cancer annotated pathways (Supplementary data 1). Interestingly, a shift in the GBM subtype was observed after resection; primary tumors were classified as mesenchymal, while recurrent tumors matched with the proneural subtype.
Fig. 2.
Pathway enrichment and histological validation in recurrent tumors. (A) Top-10 enriched REACTOME, KEGG, and BIOCARTA pathways in recurrent tumors. (B) Representative example of a software-based classifier designed to quantify immunofluorescence double staining. Scale = 25 µm. (C) Quantification of proliferating tumor cells (vimentin+/Ki-67+) in primary (n = 9) vs recurrent (n = 17) xenografts. Scale = 50 µm (overview) and 10 µm (inserts). (D) Representative VEGF immunostaining and quantification in primary (n = 9) vs recurrent (n = 17) xenografts. Scale = 100 µm. (E) Software-based classifier designed for Iba1 quantification. Scale = 50 µm. (F) Iba1 staining quantification in primary (n = 9) vs recurrent (n = 17) xenografts and primary (n = 11) vs early recurrent (n = 11) patient GBMs. Scale = 50 µm. Abbreviations: GBMs, glioblastoma; VEGF, vascular endothelial growth factor.
Recurrent tumors upregulated the VEGF signaling pathway (Supplementary data 1), and VEGF protein upregulation could be validated in P3 xenografts, where recurrent tumors had a higher staining area fraction compared to primary tumors (20.5% vs 3.2%, P = .008, Figure 2D).
Additionally, a significantly higher proportion of microglia/macrophages was found in recurrent xenografts compared to primary tumors (50.4% vs 29.1%, P = .001, Figure 2E and F), and the same pattern was observed in recurrent patient GBMs (31.4% vs 21.7%, P = .01, Figure 2F). A complete list of upregulated pathways and gene sets in recurrent tumors is supplied in Supplementary data 1.
Surgical Tumor Resection Induces Self-Renewal in Recurrent Tumors
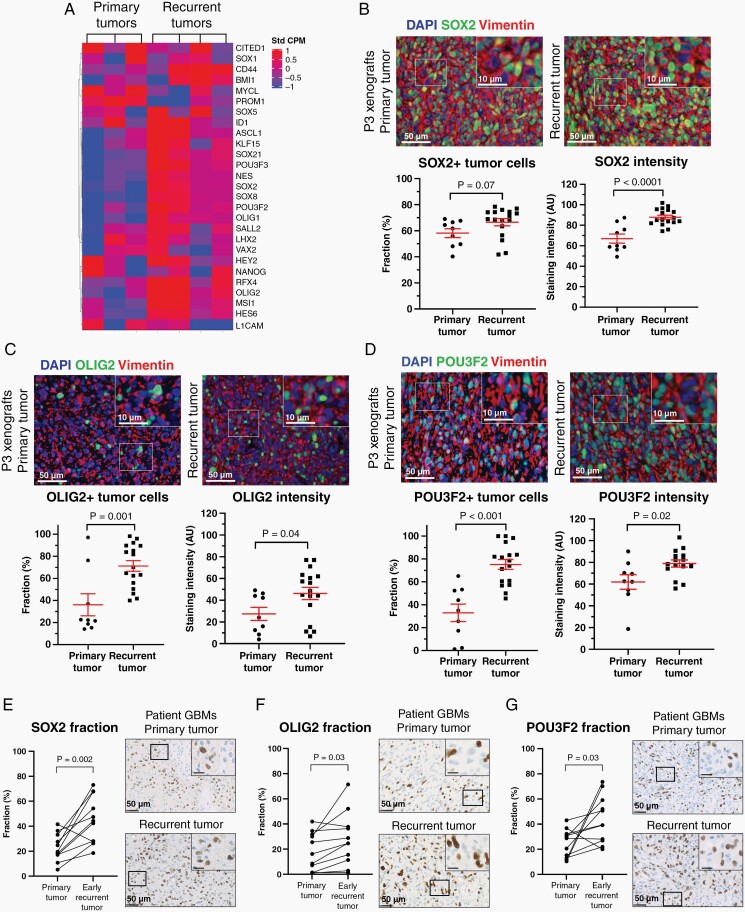
Among the upregulated single genes in recurrent tumor cells, a number of stem cell-related genes were identified (Supplementary Table 2 and Supplementary data 1), and therefore we hypothesized that recurrent tumors may be enriched for GBM stem cells (GSC). To address this, we compiled a 28-gene GSC signature based on pivotal GSC papers,23–25 and this signature was highly enriched in recurrent tumors (NES = 2.23, P = 8.55E, Figure 3A). Recurrent tumors further upregulated several GSEA annotated stem cell-related pathways, including BIOCARTA_NFKB_PATHWAY (NES = 1.84, P = .02), KEGG_WNT_SIGNALING_PATHWAY (NES = 1.67, P = .004), and REACTOME_SIGNALING_BY_NOTCH (NES = 2.27, P = 2.78E).
Fig. 3.
GSCs are enriched in recurrent xenografts and patient GBMs. (A) Relative mRNA expression levels of 28 GSC markers in primary (n = 3) vs recurrent (n = 4) pooled xenografts. (B–D) Representative images of SOX2, OLIG2, and POU3F2 immunofluorescence staining and software-based quantification in primary (n = 9) vs recurrent (n = 17) xenografts. Scale = 50 µm (overview) and 10 µm (inserts). (E–G) Validation of SOX2, OLIG2, and POU3F2 protein upregulation in primary (n = 11) vs early recurrent (n = 11) patient GBMs. Scale = 50 µm (overview) and 10 µm (inserts). AU = arbitrary units. Abbreviations: GBMs, glioblastomas; GSCs, GBM stem cells.
On the protein level, we confirmed upregulation of selected GSC markers including SOX2, OLIG2, POU3F2, Notch1, and CD133 in recurrent xenografts (Figure 3B–D; Supplementary Figure 3A–C). Interestingly, these findings could be validated in early recurrent patient GBMs, which had significantly higher fractions of SOX2+ cells (45.3% vs 22.9%, P = .002, Figure 3E), OLIG2+ cells (27.9% vs 18.1%, P = .03, Figure 3F), POU3F2+ cells (43.8% vs 24.8%, P = .03, Figure 3G), and Notch1+ staining area (34.7% vs 9.8%, P = .004, Supplementary Figure 3B) compared to the patient-matched primary tumors.
Recurrent Tumors Express Higher Levels of Pleiotrophin (PTN) Which Is Secreted Both by Tumor Cells and Tumor-Associated Microglia/Macrophages
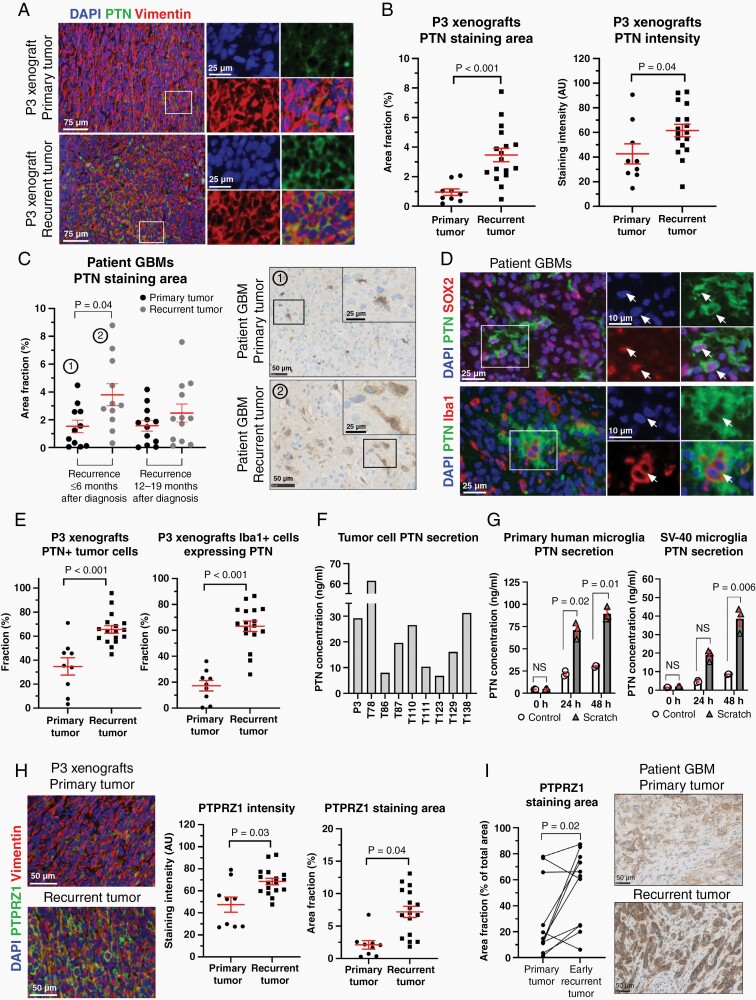
In recurrent tumor cells, the genes encoding the heparin-binding growth factor PTN and its primary receptor PTPRZ1 showed log2-fold changes of 2.99 and 2.86, respectively. PTN protein expression in P3 xenografts was upregulated in recurrent vs primary tumors (3.46% vs 0.95% positive staining area fraction, P < .001), as was PTN staining intensity (P = .04, Figure 4A and B). Early recurrent patient GBMs had significantly higher PTN expression compared to the patient-matched primary tumors (3.80% vs 1.53% positive staining area, P = .04, Figure 4C). Interestingly, PTN expression was found both in SOX2+ GSCs and Iba1+ tumor-associated microglia/macrophages in P3 xenografts (Figure 4D). In recurrent xenograft tumors, a significantly higher fraction of both tumor cells (65.5% vs 34.7%, P < .001, Figure 4E) and microglia/macrophages (63.2% vs 17.3%, P < .001, Figure 4E) showed co-localization with PTN compared to primary tumors. PTN secretion was validated in vitro using 9 different patient-derived GBM spheroid cultures (Figure 4F), as well as primary human microglia and SV-40 immortalized microglia cells (Figure 4G). Scratch assays, applied to mimic the resection procedure, demonstrated that mechanical stress augmented PTN secretion from microglia in a time-dependent manner (Figure 4G). PTN secretion from microglia/macrophages in the post-resection setting in vivo was validated in nontumor-bearing sham animals undergoing resection (Supplementary Figure 4). Expression of the primary PTN receptor, PTPRZ1, was significantly upregulated in recurrent tumors, both in the xenograft model (P = .04, Figure 4H) and in GBM patients (P = .02, Figure 4I).
Fig. 4.
PTN expression is found in both tumor cells and tumor-associated microglia/macrophages and increases in recurrent tumors. (A) Representative immunofluorescence images showing PTN expression in primary vs recurrent P3 xenografts. Scale = 75 µm (overview) and 25 µm (inserts). (B) Quantification of PTN expression in primary (n = 9) vs recurrent (n = 17) xenografts. (C) Quantification of PTN expression in patient GBMs validated PTN upregulation in early recurrent tumors. Scale = 50 µm (overview) and 25 µm (inserts). (D) PTN protein expression was seen both in SOX2+ GSCs and tumor-associated microglia/macrophages (white arrows). Scale = 25 µm (overview) and 10 µm (inserts). (E) Quantification of PTN-expressing tumor cells (vimentin+/PTN+) and microglia/macrophages (Iba1+/PTN+) in primary (n = 9) vs recurrent (n = 17) xenografts. (F, G) ELISA with 9 different patient-derived GBM spheroid cultures and primary human microglia as well as SV-40 microglia confirmed PTN secretion. (H, I) Quantification of PTPRZ1 expression in primary (n = 9) vs recurrent (n = 17) xenografts and primary (n = 11) vs early recurrent (n = 11) patient GBMs. Scale = 50 µm. AU = arbitrary units. Abbreviations: GBMs, glioblastomas; PTN, pleiotrophin.
PTN Induces Proliferation, Spheroid Growth, and Self-Renewal In Vitro
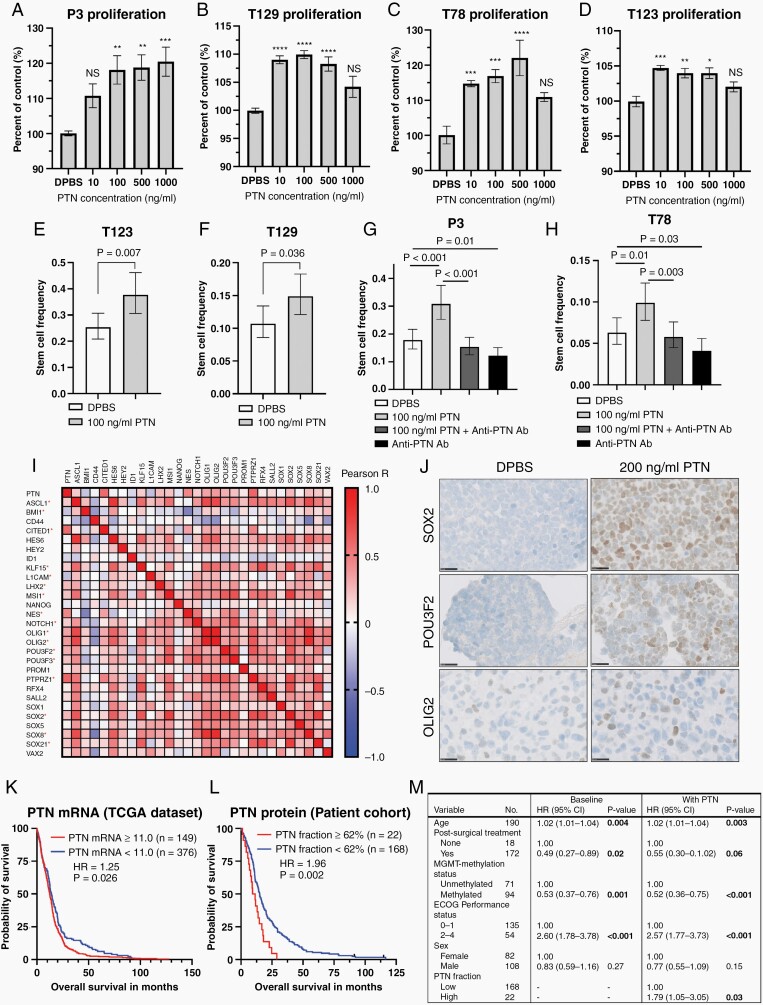
The addition of exogenous PTN to 4 different patient-derived GBM cultures induced significant tumor cell proliferation (at 10-100 ng/mL) and resulted in increased spheroid diameters (Figure 5A–D; Supplementary Figure 5). Limiting dilution assays showed that PTN significantly increased the tumor-initiating cell frequencies (P-values: T123 = 0.007; T129 = 0.036; P3 < 0.001; T78 = 0.01; Figure 5E–H). The addition of PTN along with anti-PTN antibody (0.5 µg/mL) abolished this effect, while the addition of anti-PTN antibody alone significantly reduced the fraction of tumor-initiating cells (P-values: P3 = 0.01; T78 = 0.03, Figure 5G and H).
Fig. 5.
PTN induces tumor cell proliferation, self-renewal, expression of GSC markers and is associated with poor patient survival. (A–D) Exogenous PTN induced proliferation after 5-day treatment of all 4 investigated spheroid cultures (n = 12 experiments). Scale = 200 µm (P3, T129, and T78) and 300 µm (T123). NS = non-significant. *P < .05, **P < .01, ***P < .001, ****P < .0001. (E–H) Limiting dilution assays with the addition of exogenous PTN and/or anti-PTN antibody (n = 3 experiments with 24 wells per condition/experiment). Error bars represent 95%CI. (I) Heatmap showing correlations between TCGA mRNA levels of PTN, PTPRZ1, NOTCH1, and the 28-gene GSC signature. Red asterisks indicate significant correlations with PTN. (J) Representative SOX2, POU3F2, and OLIG2 immunohistochemical staining performed on clotted T78 spheroids, cultured 4 days without EGF/FGF, and additional 48 h with/without exogenous PTN. Scale = 25 µm. (K, L) Survival analysis with TCGA PTN mRNA data and PTN protein levels in a clinical GBM patient cohort, stratified at optimized cutoff. Ticks indicate censored data. (M) Multivariate Cox regression performed on the patient cohort. Significant P-values are bold. Abbreviations: GBM, glioblastoma; GSC, GBM stem cell; PTN, pleiotrophin.
PTN Induces the GSC Program in GBM Tumor Cells
Based on PTN’s ability to promote GSCs, we hypothesized that PTN could have a functional role in the observed upregulation of tumor cell self-renewal. We investigated TCGA mRNA data and identified significant positive correlations between PTN mRNA levels and the majority of genes in the 28-gene GSC signature (Figure 5I). STRING analysis showed PTN being connected to the vast majority of GSC signature proteins through its receptor PTPRZ1, SOX2, and Notch1 (Supplementary Figure 6). Functional investigations in vitro confirmed that exogenous PTN could re-induce expression of SOX2, POU3F2, and to a lesser extent OLIG2 in tumor cells after culturing in the absence of growth factors EGF and FGF (Figure 5J).
High PTN Expression Is Associated With Poor Overall Patient Survival and Is an Independent Prognostic Factor
TCGA data showed that high PTN mRNA levels, at optimized dichotomization (PTN mRNA ≥ 11.0), were associated with significantly shorter overall survival compared to the PTN low group (P = .03, Figure 5K). These findings were reproduced in our clinical GBM patient cohort at optimized dichotomization (PTN fraction ≥ 62%), where patients with high PTN protein expression lived significantly shorter than patients with low expression (median survival: 10.4 vs 15.1 months, HR = 1.96, 95%CI 1.09-3.53, P = .002, Figure 5L). Cox regression confirmed high PTN protein expression as an independent prognostic factor (HR = 1.79, 95%CI 1.06-3.05, P = .03, Figure 5M).
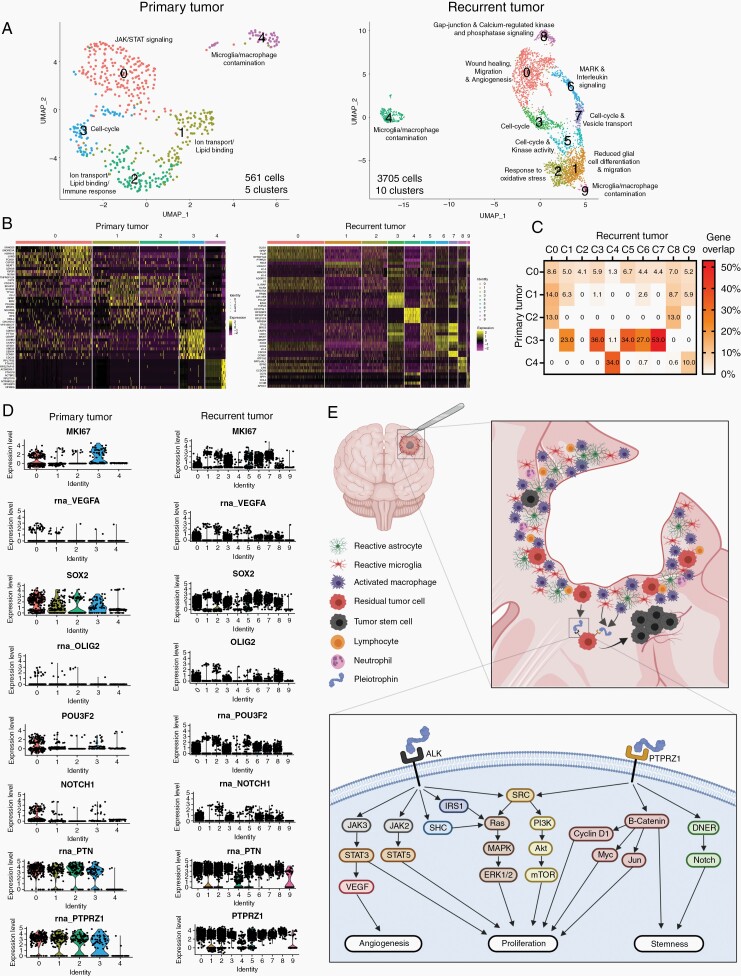
Single-Cell Transcriptomics Shows the Presence of Distinct Heterogeneous Cell Populations in Recurrent Tumors
Single-cell RNA sequencing of 561 cells from primary tumors resulted in segregation into 5 clusters. Clusters 0-3 represented tumor cells, while cluster 4 represented microglia/macrophage contamination (Figure 6A and B). The 3705 cells sequenced from recurrent tumors segregated into 10 clusters after data analysis. Clusters 4 and 9 represented monocyte and microglia/macrophage contamination, respectively, while the remaining clusters represented tumor cells (Figure 6A and B). Cluster-specific pathway analysis of tumor cells showed that the different clusters had enrichment of distinct pathways (Figure 6A). In primary tumors, cluster 0 was dominated by JAK/STAT signaling, clusters 1 and 2 were enriched for ion transport and lipid binding, while cluster 3 comprised cycling tumor cells. In recurrent tumors, cluster 0 was enriched for wound healing, migration, and angiogenesis pathways. Cluster 1 had no positive enriched pathways but was negatively enriched for glial differentiation and migration. Cluster 2 was characterized by oxidative stress response pathways, while clusters 3, 5, and 7 were enriched for cycling tumor cells. Cluster 6 showed accentuated MAPK and interleukin signaling, whereas cluster 8 was dominated by Gap junction- and calcium-regulated kinase and phosphatase signaling.
Fig. 6.
Single-cell transcriptomics of primary and recurrent tumors. (A, B) UMAP plots and heatmaps depicting clustering of tumor cells from primary and recurrent xenografts. (C) Gene expression overlap between clusters. (D) Single-cell expression levels of selected genes in primary vs recurrent tumors. (E) Graphical summary of the iatrogenic induction of PTN-mediated self-renewal and proliferation in glioblastoma recurrences. Abbreviations: PTN, pleiotrophin; UMAP, uniform manifold approximation and projection.
Next, we investigated whether gene expression signatures from primary tumors were recapitulated in recurrent tumors (Figure 6C). Gene expression overlap analysis revealed that genes from cluster 0 in primary tumors were found at low levels (1.3%-8.6%) in nearly all clusters in recurrent tumors, while genes expressed in clusters 1 and 2 in primary tumors were only found in a subset of clusters in recurrent tumors. Genes from cluster 3 in primary tumors were most commonly expressed in recurrent tumor cells with overlap ranging from 23% to 53%, and expression found in clusters 1, 3, 5, 6, and 7. Genes from the microglia/macrophage cluster 4 in primary tumors overlapped only with monocyte/microglia/macrophage clusters 4 and 9 in recurrent tumors. Single-cell RNA expression levels of selected genes accentuated in the bulk mRNA results are depicted in Figure 6D and validate mRNA upregulation at single-cell resolution. Interestingly, PTN RNA expression was very low in microglia/macrophages in primary tumors (cluster 4), while expression in monocytes/microglia/macrophages in recurrent tumors (clusters 4 and 9) was moderate/high, thereby also validating single-cell post-resection PTN RNA upregulation (Figure 6D). A graphical summary highlighting the PTN-mediated induction of proliferation and promotion of GSCs is illustrated in Figure 6E.
Discussion
In this study, we have established a 18F-FET PET/CT-based preclinical GBM resection model. 18F-FET could not detect the highly diffuse T78 xenografts, despite high tracer uptake in vitro, while P3 xenografts with moderate tracer uptake in vitro could be successfully visualized. P3 xenografts exhibit a combination of solid and infiltrative tumor growth, thereby closely resembling patient GBMs, while also having sufficient central tumor mass to be adequately visualized. 18F-FET PET has been used for tumor imaging in several preclinical GBM models with both murine- and human-derived cell cultures, and its sensitivity was found slightly superior to bioluminescent imaging approaches.26,27 This tracer therefore shows promise as an alternative imaging modality in preclinical brain tumor studies, as 18F-FET PET does not require preceding luciferase transfection of tumor cells necessary for bioluminescent imaging approaches.
We found a significant upregulation of several stem cell-related genes in recurrent tumor cells from the resection model and validated the upregulation at the protein level in xenografts and in patient GBMs. Interestingly, SOX2, POU3F2, and OLIG2, which were all upregulated in recurrent GBMs, have, along with SALL2, been described as essential neurodevelopmental transcription factors, which drive GSC propagation.24 It has been proposed that a subset of GSCs are in a plastic cellular state associated with an inflammatory wound response,28 and our demonstration of a shift from mesenchymal to proneural subtype fits with this hypothesis. Proneural GSCs predominantly express markers such as CD133, SOX2, and OLIG2 and have better capabilities of self-renewal and proliferation, while simultaneously being less invasive and treatment-resistant compared to mesenchymal GSCs.29 Since surgical tumor resection led to increased expression of these stem cell markers, the shift toward a proneural GSC phenotype seems plausible, and may facilitate better conditions for tumor regrowth. Intriguingly, these cells maintain GSC plasticity, and adjuvant intervention with TMZ can trigger a switch back toward more resistant phenotypes.30 Given the intrinsic resistance of GSCs toward conventional therapy with radio- and chemotherapy31 and with recurrent GSCs further augmenting this resistance,32 our results indicate that tumor resection and the subsequent enrichment of the GSC population contribute to therapeutic resistance. The presence of several distinct cell clusters in recurrent tumors, only partially overlapping with primary tumors, suggests the occurrence of phenotypic branching, facilitated by surgery alone. Recurrent tumors had cell clusters enriched for wound healing and decreased differentiation, respectively. This may imply the presence of different GSC populations in recurrent tumors, driven by injury response- and developmental programs, respectively, as described by Richards et al,28 and suggests that surgical GBM resection can contribute to GSC heterogeneity.
The increase of GSCs in recurrent tumors might be explained by an increase in PTN secretion. Our results showed that PTN could increase tumor cell self-renewal, while PTN inhibition decreased self-renewal. Furthermore, PTN induced expression of GSC markers in vitro, indicating that the increased PTN secretion in recurrent tumors can promote the stem cell program. Further supporting a role of PTN as a stem cell regulator, PTN expression has been linked to increases of cancer stem cells in lymphomas33 and chronic myeloid leukemia,34 and described as a regulator of hematopoietic stem cell maintenance.35 PTN further induced tumor cell proliferation, which is mediated by several downstream signaling pathways; The PTN-PTPRZ1 axis induces proliferation through β-catenin, PI3K-Akt-mTOR, and Ras-Raf-MEK-ERK signaling, while the PTN-ALK axis further promotes proliferation through the JAK/STAT signaling pathway.
Tumor-associated macrophages in GBMs have previously been shown to secrete PTN.36 Our finding of PTN secretion also occurring directly by tumor cells, supplement this knowledge to suggest that PTN acts through a combined autocrine and paracrine signaling mechanism where tumor cells and the tumor microenvironment synergize to influence tumor cell behavior and induce GSCs. Of major importance, we found high levels of PTN and PTPRZ1 present in all recurrent tumor cell clusters, which emphasizes these markers as promising targets in GBM.
As shown by Okolie et al,11 GBM tumor resection activates astrocytes to promote tumor cell proliferation and migration. Recently, tumor resection has been demonstrated to induce systemic immune suppression in a preclinical syngeneic GBM model,12 thereby further expanding the biological implications of GBM resection. Surgical tumor resection must therefore be considered as a double-edged sword in GBM therapy; resection is a necessity to relieve symptoms associated with tumor burden and to improve patient survival, but simultaneously tumor resection also induces tumor-promoting activation of the tumor microenvironment37 and directly promotes GSC propagation. Our resection model is an important step toward more advanced preclinical models of GBM. We have successfully recapitulated and investigated the isolated effect of surgical tumor resection. Parallels regarding the impact of resection on tumor biology may be drawn between our model and GBM patients, as conditions closely resemble each other during the time window from tumor resection until the onset of adjuvant therapy. Currently, the resection model does not fully resemble the entire treatment regimen offered to GBM patients. Future work should therefore aim to further refine the resection model to also include adjuvant therapies such as radio- and/or chemotherapy.
In conclusion, we showed that recurrent tumors from our resection model were enriched for GSCs, and importantly these findings could be validated in early recurrent patient GBMs. Mechanistically, PTN induced tumor cell proliferation, self-renewal, and expression of GSC markers, and PTN was a significant independent prognostic factor in a clinical GBM cohort. We have revealed novel insights into the biological implications of surgical GBM resection and propose that surgical intervention per se is an iatrogenic driver of GSC enrichment in GBMs.
Supplementary Material
Acknowledgments
The authors would like to thank technician Helle Wohlleben for assistance with immunohistochemical staining and M.D., Consultant in nuclear medicine, Peter Grupe, for input regarding radioisotopes and PET/CT scans.
Contributor Information
Arnon Møldrup Knudsen, Department of Clinical Research, University of Southern Denmark, Odense, Denmark; Department of Pathology, Odense University Hospital, Odense, Denmark.
Bo Halle, Department of Clinical Research, University of Southern Denmark, Odense, Denmark; Department of Neurosurgery, Odense University Hospital, Odense, Denmark.
Oriane Cédile, Hematology-Pathology Research Laboratory, Research Unit for Hematology and Research Unit for Pathology, University of Southern Denmark and Odense University Hospital, Odense, Denmark.
Mark Burton, Department of Clinical Genetics, Odense University Hospital, Odense, Denmark; Clinical Genome Center, University of Southern Denmark & Region of Southern Denmark, Odense, Denmark.
Christina Baun, Department of Nuclear Medicine, Odense University Hospital, Odense, Denmark; Danish Molecular Biomedical Imaging Center (DaMBIC), University of Southern Denmark, Odense, Denmark.
Helge Thisgaard, Department of Clinical Research, University of Southern Denmark, Odense, Denmark; Department of Nuclear Medicine, Odense University Hospital, Odense, Denmark.
Atul Anand, Department of Clinical Research, University of Southern Denmark, Odense, Denmark; Department of Pathology, Odense University Hospital, Odense, Denmark.
Christopher Hubert, Department of Biomedical Engineering, Cleveland Clinic Lerner Research Institute, Cleveland, Ohio, USA; Case Comprehensive Cancer Center, Case Western Reserve University, Cleveland, Ohio, USA; Cleveland Clinic Lerner College of Medicine of Case Western Reserve University, Cleveland, Ohio, USA.
Mads Thomassen, Department of Clinical Research, University of Southern Denmark, Odense, Denmark; Department of Clinical Genetics, Odense University Hospital, Odense, Denmark; Clinical Genome Center, University of Southern Denmark & Region of Southern Denmark, Odense, Denmark.
Signe Regner Michaelsen, Department of Pathology, Bartholin Institute, Copenhagen University Hospital, Copenhagen, Denmark.
Birgitte Brinkmann Olsen, Department of Clinical Research, University of Southern Denmark, Odense, Denmark; Department of Nuclear Medicine, Odense University Hospital, Odense, Denmark.
Rikke Hedegaard Dahlrot, Department of Clinical Research, University of Southern Denmark, Odense, Denmark; Department of Oncology, Odense University Hospital, Odense, Denmark.
Rolf Bjerkvig, Department of Biomedicine, University of Bergen, Bergen, Norway; NORLUX Neuro-Oncology Laboratory, Department of Oncology, Luxembourg Institute of Health, Strassen, Luxembourg.
Justin Durla Lathia, Case Comprehensive Cancer Center, Case Western Reserve University, Cleveland, Ohio, USA; Cleveland Clinic Lerner College of Medicine of Case Western Reserve University, Cleveland, Ohio, USA; Department of Molecular Medicine, Cleveland Clinic Lerner College of Medicine of Case Western Reserve University, Cleveland, Ohio, USA; Rose Ella Burkhardt Brain Tumor and Neuro-Oncology Center, Cleveland Clinic, Cleveland, Ohio, USA.
Bjarne Winther Kristensen, Department of Clinical Research, University of Southern Denmark, Odense, Denmark; Department of Pathology, Odense University Hospital, Odense, Denmark; Department of Pathology, Bartholin Institute, Copenhagen University Hospital, Copenhagen, Denmark; Department of Clinical Medicine and Biotech Research & Innovation Centre (BRIC), University of Copenhagen, Copenhagen, Denmark.
Funding
This study was supported by the Danish Cancer Society, University of Southern Denmark, and Odense University Hospital (grant Nos. A2152, A3015, and A3198).
Conflict of interest statement. The authors declare no conflicts of interest.
Authorship statement. Conceptualization and design: A.M.K., B.W.K., and R.B. Data acquisition: A.M.K., B.H., O.C., C.B., A.A., C.H., S.R.M., B.B.O., and R.H.D. Data analysis and interpretation: A.M.K., O.C., C.B., H.T., M.B., M.T., C.H., B.B.O., J.D.L., and B.W.K. Drafting of the manuscript: A.M.K. All authors have read and approved the final manuscript.
References
- 1. Stupp R, Hegi ME, Mason WP, et al. . Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 2. Brown TJ, Brennan MC, Li M, et al. . Association of the extent of resection with survival in glioblastoma: a systematic review and meta-analysis. JAMA Oncol. 2016;2(11):1460–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Petrecca K, Guiot MC, Panet-Raymond V, Souhami L. Failure pattern following complete resection plus radiotherapy and temozolomide is at the resection margin in patients with glioblastoma. J Neurooncol. 2013;111(1):19–23. [DOI] [PubMed] [Google Scholar]
- 4. Yool AJ, Ramesh S. Molecular targets for combined therapeutic strategies to limit glioblastoma cell migration and invasion. Front Pharmacol. 2020;11:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lemée JM, Clavreul A, Menei P. Intratumoral heterogeneity in glioblastoma: don’t forget the peritumoral brain zone. Neuro Oncol. 2015;17(10):1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jadhav V, Zhang JH. Surgical brain injury: prevention is better than cure. Front Biosci. 2008;13:3793–3797. [DOI] [PubMed] [Google Scholar]
- 7. Demicheli R, Retsky MW, Hrushesky WJ, Baum M, Gukas ID. The effects of surgery on tumor growth: a century of investigations. Ann Oncol. 2008;19(11):1821–1828. [DOI] [PubMed] [Google Scholar]
- 8. Burda JE, Sofroniew MV. Reactive gliosis and the multicellular response to CNS damage and disease. Neuron. 2014;81(2):229–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karve IP, Taylor JM, Crack PJ. The contribution of astrocytes and microglia to traumatic brain injury. Br J Pharmacol. 2016;173(4):692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hamard L, Ratel D, Selek L, Berger F, van der Sanden B, Wion D. The brain tissue response to surgical injury and its possible contribution to glioma recurrence. J Neurooncol. 2016;128(1):1–8. [DOI] [PubMed] [Google Scholar]
- 11. Okolie O, Bago JR, Schmid RS, et al. . Reactive astrocytes potentiate tumor aggressiveness in a murine glioma resection and recurrence model. Neuro Oncol. 2016;18(12):1622–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Otvos B, Alban TJ, Grabowski MM, et al. . Preclinical modeling of surgery and steroid therapy for glioblastoma reveals changes in immunophenotype that are associated with tumor growth and outcome. Clin Cancer Res. 2021;27(7):2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1–2):70–78. [DOI] [PubMed] [Google Scholar]
- 14. Dobin A, Davis CA, Schlesinger F, et al. . STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rouillard AD, Gundersen GW, Fernandez NF, et al. . The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database. 2016;2016(baw100). https://doi.org/10.1093/database/baw100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Croft D, O’Kelly G, Wu G, et al. . Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39(Database issue):D691–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Korotkevich G, Sukhov V, Sergushichev A. Fast gene set enrichment analysis. bioRxiv, https://doi.org/10.1101/060012, February 1, 2021, preprint: not peer reviewed. [Google Scholar]
- 20. Szklarczyk D, Gable AL, Lyon D, et al. . STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Knudsen AM, Eilertsen I, Kielland S, et al. . Expression and prognostic value of the transcription factors EGR1 and EGR3 in gliomas. Sci Rep. 2020;10(1):9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu H, Leiss L, Yang N, et al. . Surgical debulking promotes recruitment of macrophages and triggers glioblastoma phagocytosis in combination with CD47 blocking immunotherapy. Oncotarget. 2017;8(7):12145–12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suvà ML, Rheinbay E, Gillespie SM, et al. . Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157(3):580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gimple RC, Bhargava S, Dixit D, Rich JN. Glioblastoma stem cells: lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019;33(11–12):591–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nedergaard MK, Michaelsen SR, Urup T, et al. . 18F-FET microPET and microMRI for anti-VEGF and anti-PlGF response assessment in an orthotopic murine model of human glioblastoma. PLoS One. 2015;10(2):e0115315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim OS, Park JW, Lee ES, et al. . [18F]FET PET is a useful tool for treatment evaluation and prognosis prediction of anti-angiogenic drug in an orthotopic glioblastoma mouse model. Lab Anim Res. 2018;34(4):248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Richards LM, Whitley OKN, MacLeod G, et al. . Gradient of developmental and injury response transcriptional states defines functional vulnerabilities underpinning glioblastoma heterogeneity. Nat Cancer. 2021;2(2):157–173. [DOI] [PubMed] [Google Scholar]
- 29. Wang Z, Zhang H, Xu S, Liu Z, Cheng Q. The adaptive transition of glioblastoma stem cells and its implications on treatments. Signal Transduct Target Ther. 2021;6(1):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garnier D, Meehan B, Kislinger T, et al. . Divergent evolution of temozolomide resistance in glioblastoma stem cells is reflected in extracellular vesicles and coupled with radiosensitization. Neuro Oncol. 2018;20(2):236–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Osuka S, Van Meir EG. Overcoming therapeutic resistance in glioblastoma: the way forward. J Clin Invest. 2017;127(2):415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Orzan F, De Bacco F, Crisafulli G, et al. . Genetic evolution of glioblastoma stem-like cells from primary to recurrent tumor. Stem Cells. 2017;35(11):2218–2228. [DOI] [PubMed] [Google Scholar]
- 33. Wei X, Yang S, Pu X, et al. . Tumor-associated macrophages increase the proportion of cancer stem cells in lymphoma by secreting pleiotrophin. Am J Transl Res. 2019;11(10):6393–6402. [PMC free article] [PubMed] [Google Scholar]
- 34. Himburg HA, Roos M, Fang T, et al. . Chronic myeloid leukemia stem cells require cell-autonomous pleiotrophin signaling. J Clin Invest. 2020;130(1):315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Himburg HA, Termini CM, Schlussel L, et al. . Distinct bone marrow sources of pleiotrophin control hematopoietic stem cell maintenance and regeneration. Cell Stem Cell. 2018;23(3):370–381.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi Y, Ping YF, Zhou W, et al. . Tumour-associated macrophages secrete pleiotrophin to promote PTPRZ1 signalling in glioblastoma stem cells for tumour growth. Nat Commun. 2017;8:15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ratel D, van der Sanden B, Wion D. Glioma resection and tumor recurrence: back to Semmelweis. Neuro Oncol. 2016;18(12):1688–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.