Abstract
A rapid protocol for the extraction of total nucleic acids from environmental samples is described. The method facilitates concomitant assessment of microbial 16S rRNA diversity by PCR and reverse transcription-PCR amplification from a single extraction. Denaturing gradient gel electrophoresis microbial community analysis differentiated the active component (rRNA derived) from the total bacterial diversity (ribosomal DNA derived) down the horizons of an established grassland soil.
The molecular analysis of 16S rRNA is now central to studies examining the diversity of microorganisms in the environment. Traditional methods based upon cultivation underestimate diversity considerably, whereas modern molecular methods (PCR, cloning, and sequencing) have provided a greater insight into the extent of prokaryotic diversity (for a review see reference 6). Methodologies for the analyses of a DNA-based phylogeny (using the 16S rRNA gene) are now well established but the direct targeting of 16S rRNA, as a potential indicator of activity (4), has received comparatively less attention, due primarily to the lack of suitable protocols for extraction from natural environments.
Methods currently employed for DNA extraction vary widely, from direct methods of in situ lysis to indirect methods of initial cell extraction prior to lysis. In both cases, the methods used often include various combinations of bead beating, detergents, enzymatic lysis, and solvent extractions to obtain a crude preparation of nucleic acid (see, e.g., references 5 and 8). The utility of the published methods varies, particularly in soil systems, since inhibitory compounds such as humic acids and clay minerals are often coextracted. Therefore, additional purification procedures are required for successful PCR amplification. These additional steps can prevent the simultaneous extraction of the labile RNA (3) and reduce DNA yield. Reliable extraction methods have been reported for isolation of RNA from soils (2, 3, 11) and other environments (10), but they typically involve multiple steps for purification, rendering them impractical for processing large numbers of samples. Here we describe the first direct method for the rapid coextraction of RNA and DNA from soil for the comparison of bacterial diversity by 16S rRNA reverse transcription-PCR (RT-PCR) and 16S ribosomal DNA (rDNA)-PCR. To demonstrate the efficacy and reproducibility of the method, we present the denaturing gradient gel electrophoresis (DGGE) analysis of the diversity of bacterial populations in a humified upland soil based on 16S rDNA and 16S rRNA templates.
Sampling and extraction protocol.
Replicate cores of a brown forest soil (pH 4.5 to 5.0) were collected from the Sourhope Field Experiment Site in the Scottish Borders (United Kingdom) to a depth of 20 to 25 cm. Each replicate core was divided into four horizons characterized by standard nomenclature (Fh, H, Ah upper, and Ah lower). Prior to nucleic acid extraction, all solutions and glassware were rendered RNase free by diethyl pyrocarbonate treatment (1), and only certified RNase- and DNase-free plasticware was used. Nucleic acids were extracted from 0.5 g (wet weight) of soil using Bio-101 Multimix 2 Matrix tubes in combination with the FastPrep FP120 bead beating system (Bio-101, Vista, Calif.). Extractions were performed by the addition of 0.5 ml of hexadecyltrimethylammonium bromide (CTAB) extraction buffer and 0.5 ml of phenol-chloroform-isoamyl alcohol (25:24:1) (pH 8.0). CTAB extraction buffer, modified from the method of Kowalchuk et al. (7), was prepared by mixing equal volumes of 10% (wt/vol) CTAB (Sigma, Poole, United Kingdom) in 0.7 M NaCl with 240 mM potassium phosphate buffer, pH 8.0 (14). Samples were lysed for 30 s at a machine speed setting of 5.5 m/s, and the aqueous phase containing nucleic acids were separated by centrifugation (16,000 × g) for 5 min at 4°C. The aqueous phase was then extracted, and phenol was removed by mixing with an equal volume of chloroform-isoamyl alcohol (24:1) followed by repeated centrifugation (16,000 × g) for 5 min at 4°C. Total nucleic acids were subsequently precipitated from the extracted aqueous layer with 2 volumes of 30% (wt/vol) polyethelene glycol 6000 (Fluka BioChemika)–1.6 M NaCl for 2 h at room temperature, followed by centrifugation (18,000 × g) at 4°C for 10 min. Pelleted nucleic acids were then washed in ice cold 70% (vol/vol) ethanol and air dried prior to resuspension in 50 μl of RNase free Tris-EDTA buffer (pH 7.4) (Severn Biotech, Kidderminster, United Kingdom).
In related studies, the method has been used to extract total nucleic acids from environments such as activated sludge and limestone quarried rock faces (data not shown). Nucleic acids from these samples were precipitated directly using standard salt and alcohol methods (14), as no darkly colored contaminants were coextracted.
Agarose gel analysis of nucleic acid extracts.
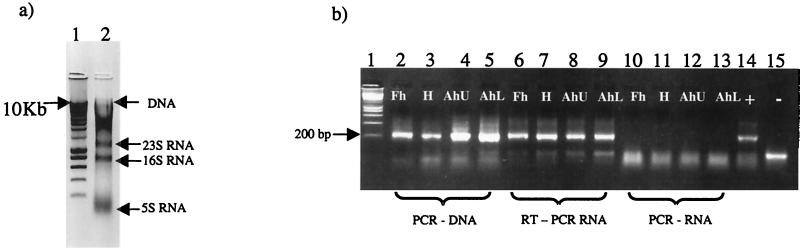
Extraction of DNA and RNA was confirmed and quantified by gel electrophoresis (Fig. 1a). Typical yields of approximately 10 to 20 μg of DNA (∼10 kb) and 2 to 5 μg of 16S rRNA per g (dry weight) of soil were observed in the total extracts from the uppermost horizons (Fh and H), which is consistent with results of other extraction methods (3, 8). Yields of nucleic acid typically decreased with depth. This may indicate a reduction in biological activity or, more likely, reflect the decreasing biomass relative to the soil matrix down the soil profile. Plate count estimates of the relative density of bacteria on R2A (Difco-Oxoid) supported this conclusion: Fh = 7.9 × 107, H = 4.6 × 107, AhU = 4.7 × 107, and Ahl = 3.7 × 107 CFU/g (wet weight) of soil.
FIG. 1.
(a) Negative image of a 1% ethidium bromide-stained agarose gel of the total nucleic extract. Lane 1, HyperLadder I (Bioline, London, United Kingdom); lane 2, total nucleic extract from 0.5 g of Sourhope soil. (b) Ethidium bromide (1.5%)-stained agarose gel showing PCR and RT-PCR amplification products from each soil horizon (Fh, H, Ah upper [AhU], and Ah lower [AhL]). Lane 1, HyperLadder I; lanes 2 to 5 amplification products from 16S rDNA for each soil horizon; lanes 6 to 9, amplification products from reverse-transcribed 16S rRNA; lanes 10 to 13, amplified 16S rRNA (controls without reverse transcriptase) for each horizon; lane 14, amplified bacterial 16S rDNA (positive control); lane 15, no-template negative control.
PCR-based analyses of DNA and RNA extracts.
To demonstrate the efficacy of the method, extracted nucleic acids were divided into two aliquots for the preparation of pure DNA or RNA templates. To obtain pure DNA, half of the sample was incubated at 37°C with RNase A (Sigma) at a final concentration of 100 μg ml−1 for 10 min. RNA for RT-PCR analysis was obtained by treating the other 25 μl of the sample with 3 U of RQ1 RNase-free DNase (Promega Corp.) according to the manufacturer's instructions. Prior to reverse transcription, the template secondary structure was melted by incubating 0.5 μl of the universal 16S rRNA V3 region primer 530R (100 pmol/μl) (12) with 2 μl of RNA and 12.75 μl of nuclease-free water at 70°C for 5 min. Samples of annealed primer-template were then immediately placed on ice, and a reaction mixture was added, containing for each reaction 1.5 μl of MgSO4 (25 mM), 5 μl of 5× reaction buffer (supplied with the Access RT-PCR System [Promega Corp.]), 1.25 μl of deoxynucleoside triphosphate mix (10 mM) (Promega Corp.), and 2 μl of avian myeloblastosis virus reverse transcriptase (8 U/μl) (Promega Corp.) (Note that the buffer supplied with the avian myeloblastosis virus reverse transcriptase is not suitable for use in RT-PCR, as it contains spermidine.) Reverse transcription was carried out at 48°C for 45 min, and the enzyme was subsequently heat inactivated for 5 min at 99°C. PCR amplification of both the DNA and cDNA templates was performed in 100-μl volumes using 1 μl of DNA or cDNA template with universal bacterial primers spanning the V3 region of the 16S rRNA and incorporating a GC-clamped primer, as documented previously (15). Efficient amplification of the 16S rDNA from the DNA extract was occasionally variable when using 1 μl of undiluted template. However, 1/10 dilutions proved reliable and produced high yields of PCR product (Fig. 1b). In contrast, RT-PCR consistently produced strong amplification from undiluted RNA samples. Particular attention was paid to ensure that DNA did not contaminate RNA preparations by always performing RT-PCR on RNA samples in the absence of the reverse transcriptase enzyme.
DGGE fingerprinting of soil horizon communities by DNA and RNA analyses.
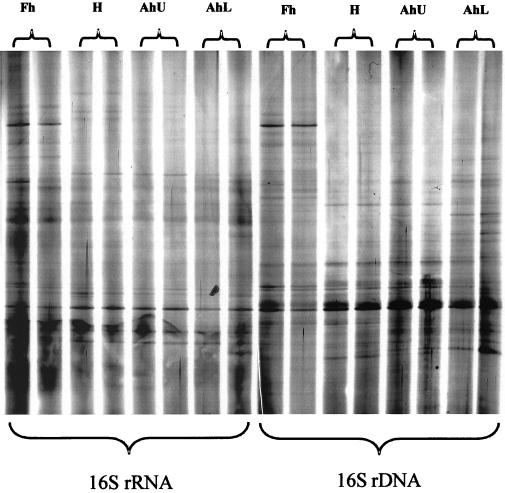
DGGE analyses of PCR amplified products was performed as described elsewhere (15). The DGGE profiles resulting from either RNA or DNA templates confirmed the complex microbial diversity present in soils (Fig. 2). Silver staining (SilverSequence; Promega Corp.) coupled with digital scanning provided greater resolution of faint bands than Sybr Gold staining (Molecular Probes, Eugene, Oreg.) and enabled more representative profiling using Phoretix (Newcastle Upon Tyne, United Kingdom) one-dimensional gel analysis software.
FIG. 2.
Scanned image of a silver-stained DGGE gel (10% acrylamide, 30 to 60% denaturant) profiling the microbial communities by soil horizon and nucleic acid template. For each horizon, two replicate profiles from two independently extracted soil cores are displayed.
Community composition and stratification in soil horizons.
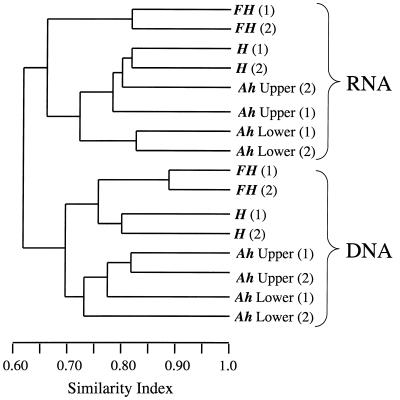
The presence or absence of bands in each community profile was analyzed by the unweighted pairwise grouping method with mathematical averages (utilizing the Dice coefficient of similarity). Analyses indicated strong delineation of the profiles into clusters corresponding to the nucleic acid template (DNA or RNA) (Fig. 3). This analysis clearly demonstrated that differences existed between the profiles, presumably due to the active or total diversity assessed by rRNA or rDNA, respectively. Further, replicate Fh samples (top 5 cm) grouped separately from the other horizons (for both the RNA- and DNA-derived profiles), indicative of a different community structure in the near-rhizosphere environment compared to soil depth profiles. Replicate homogeneity was also demonstrated by the clustering of separately extracted and analyzed replicate cores, indicating the consistency of the extraction and analysis methods.
FIG. 3.
Dendrogram showing clustering analyses of the digitized profiles from Fig. 2, using the unweighted pairwise grouping method with mathematical averages (Dice coefficient of similarity). The analyses takes into account the presence or absence of bands at certain positions in each lane, standardized across the gel using Rf values.
Community composition determined by sequence analyses.
In order to assess any significant bias inherent to this extraction method, full-length 16S rDNAs were cloned in pGEM-T Easy (Promega Corp.). Sequence analyses of the gene library and DGGE bands confirmed the clone dominance of the alpha-Proteobacteria and the presence of the Acidobacterium-Holophaga group, including the recently described Sourhope 3 cluster (9). Therefore, the sequences detected with the protocol described above were consistent with those reported from an adjacent field site at Sourhope (9) or a low-pH soil environment (13), which used alternative primer sets to amplify cesium chloride-purified DNA.
Conclusions.
The study of 16S rRNA genes has provided a greater knowledge of the diversity of bacteria in the environment and has also revolutionized bacterial systematics. However, in order to detect specific functional groups of microorganisms, different techniques are required to differentiate the active components within a sample. While the benefits of using an RNA directed approach are still to be fully realized, the use of the methodology described here will allow examination of the correlation between an RNA-based phylogeny and the activities of specific taxa. These data, when integrated with measurements of biogeochemical processes, should permit a greater understanding of microbial community structure and functionality in the environment.
Acknowledgments
This work was supported as part of the Soil Biodiversity NERC thematic program through grant GST/32/2136 to M.J.B., A.S.W., and A.G.O. and an associated studentship (to R.I.G.).
We thank Sarah Buckland for help with sample collection.
REFERENCES
- 1.Blumberg D D. Creating a ribonuclease-free environment. Methods Enzymol. 1987;152:20–24. doi: 10.1016/0076-6879(87)52005-5. [DOI] [PubMed] [Google Scholar]
- 2.Duarte G F, Rosado A S, Seldin L, KeijzerWolters A C, van Elsas J D. Extraction of ribosomal RNA and genomic DNA from soil for studying the diversity of the indigenous bacterial community. J Microb Methods. 1998;32:21–29. [Google Scholar]
- 3.Felske A, Engelen B, Nubel U, Backhaus H. Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl Environ Microbiol. 1996;62:4162–4167. doi: 10.1128/aem.62.11.4162-4167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felske A, Wolterink A, Van Lis R, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frostegard A, Courtois S, Ramisse V, Clerc S, Bernillon D, LeGall F, Jeannin P, Nesme X, Simonet P. Quantification of bias related to the extraction of DNA directly from soils. Appl Environ Microbiol. 1999;65:5409–5420. doi: 10.1128/aem.65.12.5409-5420.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated micro-organisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 7.Kowalchuk G, A, Bodelier P L E, Heilig G H J, Stephen J R, Laanbroek H J. Community analysis of ammonia-oxidising bacteria, in relation to oxygen availability in soils and root-oxygenated sediments, using PCR, DGGE and oligonucleotide probe hybridisation. FEMS Microbiol Ecol. 1998;27:339–350. [Google Scholar]
- 8.Krsek M, Wellington E M H. Comparison of different methods for the isolation and purification of total community DNA from soil. J Microbiol Methods. 1999;39:1–16. doi: 10.1016/s0167-7012(99)00093-7. [DOI] [PubMed] [Google Scholar]
- 9.McCaig A E, Glover L A, Prosser J I. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl Environ Microbiol. 1999;65:1721–1730. doi: 10.1128/aem.65.4.1721-1730.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miskin I P, Farrimond P, Head I M. Identification of novel bacterial lineages as active members of microbial populations in a freshwater sediment using a rapid RNA extraction procedure and RT-PCR. Microbiology. 1999;145:1977–1987. doi: 10.1099/13500872-145-8-1977. [DOI] [PubMed] [Google Scholar]
- 11.Moran M A, Torsvik V L, Torsvik T, Hodson R E. Direct extraction and purification of rRNA for ecological studies. Appl Environ Microbiol. 1993;59:915–918. doi: 10.1128/aem.59.3.915-918.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radajewski S, Ineson P, Parekh N R, Murrell J C. Stable-isotope probing as a tool in microbial ecology. Nature. 2000;403:646–649. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 15.Whiteley A S, Bailey M J. Bacterial community structure and physiological state within an industrial phenol bioremediation system. Appl Environ Microbiol. 2000;66:2400–2407. doi: 10.1128/aem.66.6.2400-2407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]