Abstract
The zebrafish caudal fin has become a popular model to study cellular and molecular mechanisms of regeneration due to its high regenerative capacity, accessibility for experimental manipulations, and relatively simple anatomy. The formation of a regenerative epidermis and blastema are crucial initial events and tightly regulated. Both the regenerative epidermis and the blastema are highly organized structures containing distinct domains, and several signaling pathways regulate the formation and interaction of these domains. Bone is the major tissue regenerated from the progenitor cells of the blastema. Several cellular mechanisms can provide source cells for blastemal (pre-)osteoblasts, including dedifferentiation of differentiated osteoblasts and de novo formation from other cell types, providing intriguing examples of cellular plasticity. In recent years, omics analyses and single-cell approaches have elucidated genetic and epigenetic regulation, increasing our knowledge of the surprisingly complex coordination of various mechanisms to achieve successful restoration of a seemingly simple structure.
The ability for regeneration of adult form and function after injury is highly variable among animal species (Ricci and Srivastava 2018). Some vertebrates like the ray-finned zebrafish Danio rerio can regenerate, among other tissues, the heart, central nervous system, and appendages (González-Rosa et al. 2017; Becker and Becker 2020; Daponte et al. 2021). In contrast, regeneration capacity in many adult mammalian species, including humans, is rather low, as they cannot restore lost appendages (Daponte et al. 2021), myocardial infarction results in a repair process with extremely low cardiomyocyte proliferation (Cui et al. 2018), and axons and neurons are not replaced after injury to the central nervous system (Tran et al. 2018).
For decades, appendage regeneration in adult vertebrates has largely been studied in salamanders. Only since the early 2000s, zebrafish fins have become widely used models to study regeneration. Major advantages of the salamander model include the similarity of its anatomy to that of mammalian limbs, its relatively large size, which makes techniques like cell transplantations and electroporation feasible, and the complexity of pattern along all three spatial axes, in particular the proximodistal axis. The latter provides a large space of analysis for studies into the regeneration of form, most importantly into mechanisms of positional memory. Zebrafish have become popular models for regeneration studies largely due to their genetic tractability. Salamanders have very big genomes and all species commonly used for regeneration studies have a long generation time, putting some constraints on genetic manipulations (Joven et al. 2019). Nevertheless, recent breakthroughs in full genome sequencing and the development of transgenesis and CRISPR-Cas9-based mutagenesis have massively expanded the experimental toolbox in salamanders. Advantages of the zebrafish model include their longer history as a genetic model organism, which results in the availability of a large number of transgenic and mutant lines, and the relative ease of establishing new lines, which is also due to their high fecundity and small size. The zebrafish fin regeneration model features highly robust and rapid regeneration, and partial amputation, in particular of the relatively large caudal fin, is a simple surgical procedure that is well tolerated by the fish. Caudal fins can also be fairly easily imaged due to their flat and thin anatomy; thus, several features of regeneration can be monitored in vivo using straightforward stereomicroscopy techniques. Thus, fin regeneration is even amenable to high content approaches like chemical and genetic screens (Oppedal and Goldsmith 2010; Chen et al. 2015; Mishra et al. 2020).
Remarkable advancements have been made in elucidation of cellular and molecular mechanisms underlying and regulating fin regeneration. These include, among others, the identification of a variety of signaling pathways regulating the sequential steps of regeneration and the mechanisms underlying the attainment of preinjury fin size and shape (Gemberling et al. 2013; Pfefferli and Jaźwińska 2015; Wehner and Weidinger 2015). Also, much progress has been made in identification of source cells for regeneration and to which extent cellular plasticity is involved. Not all of these topics can be covered in this review, and we will concentrate on certain aspects.
THE ZEBRAFISH CAUDAL FIN MODEL
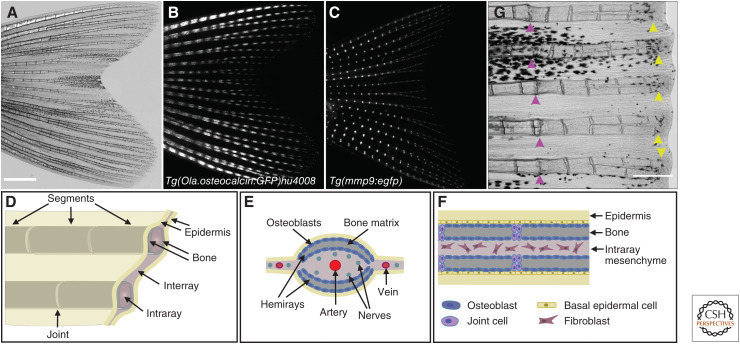
Although all zebrafish fins—the paired pelvic and pectoral fins and the unpaired anal, dorsal and caudal fins—regenerate, most regeneration studies use the bilobed caudal fin as it is the largest one and the easiest to access for injury and imaging. The skeletal elements of the fin can be divided into endochondral bones at the base, which are muscularized, and exoskeletal rays, the lepidotrichia, which run from proximal to distal through the fin (Bird and Mabee 2003). This exoskeletal part of the fin does not contain muscles. The fin rays are segmented and occasionally bifurcated, and the segments are connected by collagenous ligaments, the joints (Fig. 1A–D). Zebrafish grow throughout their life span, and the accompanying growth of the fin is achieved by the distal addition of new segments. Each segment consists of two opposed, concave hemirays (Fig. 1E). The bone of the rays is formed via direct ossification (without a cartilage template) by a monolayer of osteoblasts that lines the inner and outer surface of the hemirays (Fig. 1E,F). Rays are separated by soft interray tissue (fibroblasts) that contains venous capillaries (Becerra et al. 1983). At the tip of each ray, fish-specific nonmineralized skeletal elements, the so-called actinotrichia, complete each ray (König et al. 2018).
Figure 1.
Anatomy of a zebrafish fin. (A) Brightfield image of an adult zebrafish fin. (B) In the transgenic line Tg(Ola.osteocalcin:GFP)hu4008 (Knopf et al. 2011), GFP is expressed in mature osteoblasts lining the segments and is absent in the joints. (C) The transgenic line Tg(mmp9:egfp) (Ando et al. 2017) displays specific EGFP expression at fin ray joints. (D) Schematic representation of the fin with segmented bony rays surrounded by connective tissue. (E) Schematic cross-section highlighting the two concave bony hemirays of each segment, and the localization of blood vessels. Osteoblasts line the hemirays in a single layer. (F) Schematic longitudinal section showing the basal epidermal cell layer abutting the outer layer of osteoblasts. Segments are separated by joint cells. (G) Fins can be repeatedly amputated and form new bone. Pink arrowheads, first amputation side 9 d before imaging. New bone has already been formed in the regenerate. Yellow arrowheads, second amputation side 1 d before imaging. Atop of each ray, the histological distinctive blastema can be recognized. Scale bars, 1000 µm (A–C); 500 µm (G).
Because the endochondral base of zebrafish fins is quite reduced and hardly protrudes from the body, the bulk of the “visible” part of the fins is formed from the rays, and partial fin amputations, as they are routinely used in zebrafish fin regeneration studies, only remove this exoskeletal portion of the fin. Of note, amputations through the endochondral base of the pectoral fin are met with a highly incomplete regenerative response (Pápai et al. 2019). Interestingly, the ability to regenerate the endochondral skeleton of appendages is quite variable among teleosts; while several species can—like zebrafish—only regenerate the exoskeleton of the fins, Polypterus (the most basal living ray-finned fish), lungfishes (a sister group to tetrapods), and knifefish (gymnotiform electric fishes) can regenerate their endoskeleton (Kirschbaum and Meunier 1981; Cuervo et al. 2012; Nogueira et al. 2016). In the developing zebrafish pectoral fin, endoskeletal structures do regenerate, but this ability declines during maturation (Yoshida et al. 2020). Future studies will hopefully provide mechanistic insight into the differential capacity to regenerate the appendage endoskeleton in teleosts.
In contrast to the endochondral bones at the pectoral fin base, the exoskeletal elements of the teleost fin rays are not homologous to bones in amniote limbs. Thus, while the functional part of the appendages that extends from the body regenerates very efficiently both in zebrafish and salamanders, interesting differences exist in the regenerative ability of endochondral skeletal elements between these models.
The regeneration of fins is epimorphic, that is, lost tissue is restored by proliferation of source cells, and these cells form a morphologically identifiable structure called a blastema at the amputation site (Poss et al. 2003; Pfefferli and Jaźwińska 2015). Amputated fins are restored in their size, shape, and anatomical pattern within 3 weeks. Regeneration capacity is independent of fish age (Shao et al. 2011), and fins can be repeatedly amputated (Fig. 1G; Azevedo et al. 2011). Regeneration of the fin can be divided into defined phases: (1) wound healing and formation of a regenerative epidermis (RE), (2) blastema formation, and (3) regenerative outgrowth (Poss et al. 2003).
WOUND HEALING AND THE REGENERATIVE EPIDERMIS
Before the regeneration program starts with blastema formation, a general injury response can be observed, with neutrophils migrating toward the injury site (Petrie et al. 2014). Within few hours after injury, the amputation plane is covered by a thin epidermal layer. This wound epithelium is formed through migration of nonproliferative epithelial cells over the wound (Poleo et al. 2001; Santos-Ruiz et al. 2002). Over the following hours to days, the epithelium becomes multilayered and matures into the RE. In zebrafish, the fin epidermis consists of basal cells, suprabasal epithelial cells, and superficial epithelial cells (SECs). In the multicolor Cre-lox-based skinbow transgenic line, SECs are barcoded with distinguishable tags, which allows for simultaneous tracking of individual cells (Chen et al. 2016). The combination with a panepithelial reporter line showed that the first epidermal cells recruited to the amputation site are basal and suprabasal cells, quickly followed by SECs. SECs of interray regions display a higher motility than SECs overlying bony rays, and SECs are recruited over long distances into regenerating tissue. Additionally, de novo created SECs contribute to the surface epithelium. Concordantly, lineage tracing using fibronectin 1b (fn1b) as a marker expressed in all strata of the epidermis, showed that RE cells predominantly originate from interray regions (Shibata et al. 2018). Importantly, epidermal cells face different fates depending on their time of recruitment. Whereas epidermal cells recruited from 1 day postamputation (dpa) onward contribute to the RE, cells recruited earlier transiently cover the wound, but later undergo apoptosis (Shibata et al. 2018). Intriguingly, many epidermal cells of the RE in the distal fin area are expelled within 15 dpa and replenished by widespread proliferation of both basal and nonbasal epidermal cells. Conversion of suprabasal or surface epidermal cells into basal cells to replenish basal cells does not occur. In the larval fin fold, which has become a popular model for mechanisms of wound healing, the epithelium consists of only a basal and a suprabasal layer. Here, cell migration only occurs in the basal layer, while suprabasal cells are potentially dragged by mechanical coupling, assisted by purse string contractions (Gault et al. 2014).
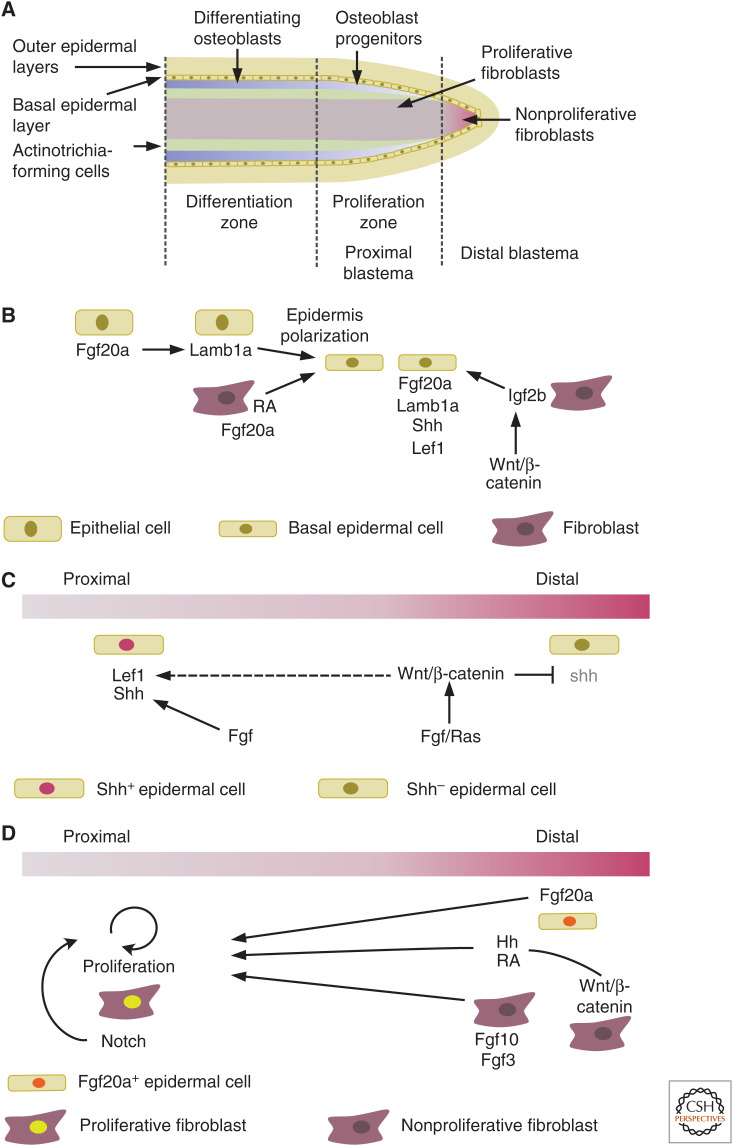
The blastema is a highly organized structure with distinct domains for proliferation and differentiation (Fig. 2A), which we will discuss in detail in the next section. The RE not only acts as a physical barrier to protect the regenerating tissue, but also plays an important role in the formation and organization of the blastema through paracrine signaling to the adjacent mesenchymal cells. Particularly, the basal layer of the RE, a single layer of cells aligned atop of blastemal cells, is a central signaling hub. In the basal layer of the RE, type 1 insulin-like growth factor receptor (igfr1) (Chablais and Jaźwińska 2010), laminin β1a (lamb1a) (Chen et al. 2015), fibroblast growth factor 20a (fgf20a) (Shibata et al. 2016), and Hh ligand sonic hedgehog (shh) (Armstrong et al. 2017) are expressed in a distinct pattern. Fgf20a expression is activated early in the wound epidermis, and inhibition of Fgf signaling by overexpression of a dominant-negative Fgf receptor 1 (dnfgfr1) impairs blastema formation (Shibata et al. 2016), demonstrating a functional role of the RE in fin regeneration. Epidermis formation is also modulated by Fgf20a emerging form the underlying mesenchymal cells (Whitehead et al. 2005), as does retinoic acid (RA) (Blum et al. 2012). Dnfgfr1 overexpression also reduces lamb1a expression, indicating that Fgf20a induces the expression of lamb1a in adjacent epithelial cells (Fig. 2B; Chen et al. 2015). Mutant analyses show that lamb1a is instrumental for the polarization of the basal cell layer of the RE along the apicobasal axis, and for the formation of the basement membrane. This polarization is crucial for the directed activation and position of various factors, including phosphorylated Igf1r and components of the Hh and Wnt/β-catenin signaling pathways, and subsequently for osteoblast patterning and bone formation (Wehner et al. 2014; Chen et al. 2015). Igfr1 in the epidermis is activated by insulin-like growth factor 2b (igf2b) emanating from the blastema (Chablais and Jaźwińska 2010). Transcription of igf2b in blastemal cells is in turn regulated by Wnt/β-catenin signaling, as induction of the Wnt signaling pathway inhibitor dickkopf (dkk1) reduces its expression, thus indirectly linking this pathway with IGF pathway activation (Fig. 2B; Wehner et al. 2014). As mentioned, expression of the various factors in the epidermis is partly nonoverlapping, defining several epidermal domains. These domains are regulated by Fgfs, as shown with in situ analysis on dnfgfr1-expressing fins. Fgfs maintain shh expression in proximal regions, where the transcription factor lymphoid enhancer-binding factor 1 (lef1) is also expressed (Fig. 2C; Lee et al. 2009). In distal regions, Fgf together with Ras signaling induces wingless-type MMTV integration site family, member 5b (wnt5b) expression, resulting in the confinement of the shh domain to the proximal region. Lef1 is best known as a mediator and feedback target gene of Wnt/β-catenin signaling and its expression does depend on active Wnt/β-catenin signaling also in the regenerating fin, as overexpression of dkk1 results in loss of expression (Stoick-Cooper et al. 2007). However, active β-catenin signaling could not be demonstrated in the epidermis (Poss et al. 2000a; Stewart et al. 2014; Wehner et al. 2014). Thus, it appears that activation of Wnt/β-catenin signaling in the blastema indirectly regulates epidermal lef1 expression and epidermal patterning via secondary signals (Wehner and Weidinger 2015).
Figure 2.
Blastema organization and regulation. (A) Schematic longitudinal section of the distal tip of a regenerating ray. Progenitors of distinct cell fates are found in different regions of the blastema, cells at the very tip are largely nonproliferative. (B) Regulation of epidermis formation. (C) Regulation of epidermal patterning. (D) Regulation of blastema proliferation.
Additional studies employing tissue-specific pathway manipulation will be helpful to further elucidate the complex regulation patterns between the different pathways and cell types.
BLASTEMA FORMATION AND ORGANIZATION
From 12 h postamputation onward, a blastema forms distally to the amputation site in each bony ray (Sehring and Weidinger 2020). The blastema is not a homogenous mass of cells; rather molecular markers and lineage-tracing approaches have shown that it is composed of a collection of progenitor cells that are predominantly lineage-restricted and spatially segregated, with their position within the blastema reflecting their site of origin in the stump (Fig. 2A). Intraray fibroblasts give rise to the mesenchymal cells at the core of the blastema, while dedifferentiated osteoblasts maintain their location to the lateral sides, beneath the epidermis. In addition, clonal analysis of regenerating fibroblasts revealed that they contribute to distinct proximodistal blastema regions depending on their proximodistal location in the stump (Tornini et al. 2016). Early fgf20a signaling in the wound epidermis is required for the recruitment of fibroblasts to the blastema (Shibata et al. 2016). Several pathways regulate the proliferation of blastemal cells, such as FGF (Poss et al. 2000b; Lee et al. 2005), RA (Blum et al. 2012; Wehner et al. 2014), and Shh signaling (Quint et al. 2002; Lee et al. 2009). Importantly, proliferation and thus growth of the blastema has to be tightly coupled with mechanisms regulating blastema organization, thereby ensuring directed outgrowth and subsequent differentiation and tissue formation in the growing regenerate. The distalmost blastema contains largely nonproliferative fibroblasts, while further proximally cells are highly proliferative (Fig. 2A; Nechiporuk and Keating 2002). Wnt/β-catenin signaling is active in the nonproliferative distal blastema and regulates blastemal cell proliferation. Systemic overexpression of the negative Wnt regulator axin1 suppresses proliferation, but can be rescued by both RA and Smoothened agonist (SAG) treatment, indicating that Wnt/β-catenin signaling acts via RA and hh signaling (Fig. 2D; Wehner et al. 2014). Furthermore, fibroblast growth factor3 (fgf3) and fibroblast growth factor 10a (fgf10a) are expressed in the distal blastema, and have been suggested to be important for cell proliferation (Shibata et al. 2016). In the proximal blastema, cells are kept in a proliferative progenitor state by Notch signaling (Grotek et al. 2013; Münch et al. 2013). Fibroblasts proliferate and express extracellular matrix proteins, such as tenascin C (Jaźwińska et al. 2007).
A recent study showed that in mutants for midkine-a, a cytokine growth factor, the initial blastemal proliferation burst is reduced (Ang et al. 2020). The authors observed similar effects in extraocular muscle and retinal neuron regeneration, suggesting a general role for midkine-a as a potential generic wound signal that regulates the early onset of proliferation. A fundamental question in regeneration research has been why some organs are completely restored by structural regeneration after injury, while others only display wound healing. Injuries of such differently responding organs might either induce different signals, or the different outcomes are due to deviating responses to a common set of wounding-induced signals. The potency of generic wound signals to induce regeneration was demonstrated in a missing-tissue context (Owlarn et al. 2017). Transient blockage of either Wnt/β-catenin or FGF signaling generated dormant fins, where wound healing and formation of the RE happen, but blastema formation and regenerative outgrowth do not occur. In these fins, injuries that under normal conditions do not induce regeneration but only wound healing (e.g., skin incisions in the interray) stimulate regenerative growth beyond the primary amputation plane. Together, these studies imply that generic wounding-induced signals also trigger regeneration. Further studies into the mechanisms underlying the differential response of tissues to generic wound signals, depending on the need to regenerate structure or to merely heal the wound, will be of high interest.
In summary, in the last decade a great number of studies has revealed molecular mechanisms and signaling pathways regulating blastema formation and regenerative growth. Yet, many questions about the epistasis and interaction of pathways, downstream effectors and cross talk between tissues remain. Single-cell approaches have started to provide additional insights into the cellular mechanisms of regeneration and are expected to also reveal candidate molecular regulators for further analysis. scRNASeq identified two distinct transcriptional states within the mesenchyme of noninjured fins (likely representing osteoblasts and other cells), while seven distinct clusters arose in the mesenchyme of the regenerating fin (Hou et al. 2020). These could be assigned to one putative lineage for osteoblasts and three nonosteoblastic cellular states. Comparing the top differentially expressed genes in these lineages allowed for allocation of specific roles for the lineages (for example, only one lineage showed up-regulation of the RA-degrading enzyme cytochrome P450, family 26, subfamily A, polypeptide 1 cyp26b1). Intriguingly, these data suggest that tryptophan hydroxylase 1b (tph1b), aldehyde dehydrogenase 1 family, member A2 (aldh1a2), and actinodin1 (and1), which have been proposed as markers for joint fibroblasts (Tornini et al. 2017), fibroblasts adjacent to osteoblasts (Blum and Begemann 2015), and actinotrichia-forming cells, respectively (König et al. 2018), rather represent different states of the same nonosteoblastic cell population during early regeneration stages instead. Lineage-tracing approaches will be required to confirm this, but it is likely that future single-cell studies will refine our current concepts of cell fates in the regenerating fin, and might also reveal unexpected cellular heterogeneity, in particular in the lineage that has so far been studied in greatest detail, the bone-forming cells.
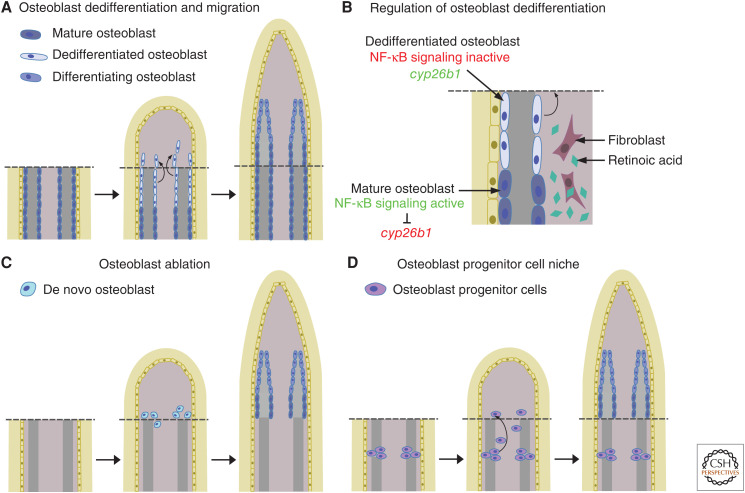
OSTEOBLAST FATE
Osteoblasts constitute an important part of each fin ray blastema. Whereas current data indicate that stem cells, and not differentiated osteoblasts, provide the source cells for bone repair in mammalian fracture healing (Park et al. 2012), differentiated osteoblasts appear to represent an important source for bone-forming cells during zebrafish fin regeneration (Knopf et al. 2011). Mature osteoblasts in the stump close to the amputation plane dedifferentiate, that is, they lose characteristics of the differentiation state, in particular expression of the differentiation marker bone γ-carboxyglutamate (gla) protein (bglap, previous name osteocalcin), and they gain progenitor characteristics (e.g., expression of the progenitor marker RUNX family transcription factor 2 runx2) (Knopf et al. 2011; Sousa et al. 2011; Stewart and Stankunas 2012). These cells remain lineage restricted and only redifferentiate into osteoblasts in the regenerate (Knopf et al. 2011; Tu and Johnson 2011). Such lineage restriction was also proposed for other cell types of the fin, such as epidermal cells, dermal fibroblasts, and endothelial cells (Tu and Johnson 2011; Stewart and Stankunas 2012). Dedifferentiated osteoblasts migrate distally toward the amputation plane and integrate into the forming blastema (Knopf et al. 2011; Sousa et al. 2011), providing a cellular source for bone regeneration (Fig. 3A). Identifying signals and genes that promote (or inhibit) osteoblast dedifferentiation is of critical importance to understand this mechanism. So far, two pathways have been shown to be involved in regulating osteoblast dedifferentiation (Fig. 3B). Intriguingly, both are negative regulators. Osteoblast dedifferentiation is inhibited by RA, which is synthesized after amputation, as it is an important signaling pathway necessary for blastema formation and function (Blum et al. 2012; Blum and Begemann 2015). To counteract this, osteoblasts up-regulate expression of the RA-degrading enzyme cyp26b1 and thus enable their dedifferentiation (Blum and Begemann 2015). In addition to RA signaling, NF-κB signaling has also been shown to modulate osteoblast dedifferentiation (Mishra et al. 2020). In mature osteoblasts, NF-κB signaling is active and prevents their dedifferentiation in a cell-autonomous manner. Upon amputation, NF-κB signaling is down-regulated in osteoblasts, thus allowing for their dedifferentiation. Combination of RA and NF-κB signaling interference using small molecules suggests that NF-κB signaling acts upstream of RA signaling and that cyp26b1 expression is negatively regulated by NF-κB signaling (Fig. 3B). NF-κB signaling emerged as modulator of osteoblast dedifferentiation from a high content in vivo chemical screen for regulators of fin regenerative growth and osteoblast dedifferentiation. Noteworthy, NF-κB signaling does not affect regenerative growth, and the majority of target compounds identified in this screen affected either dedifferentiation or regenerative growth. Thus, osteoblasts dedifferentiate even if subsequent regenerative growth is impaired, and vice versa regenerative growth occurs albeit osteoblast dedifferentiation is inhibited (potential other sources for osteoblasts are discussed below). This suggests that these two processes are at least partly independent from each other and are individually regulated. Further studies on other potential pathways emerging from this screen will elucidate this hypothesis. Such studies should also be able to test whether osteoblastic dedifferentiation is inherently negatively regulated, that is that dedifferentiation occurs through the loss of signals maintaining the differentiated state and not through the activation of signals inducing dedifferentiation.
Figure 3.
Cellular sources of osteoblasts. (A) Differentiated osteoblasts close to the amputation plane dedifferentiate, migrate into the blastema and redifferentiate. (B) Both retinoic acid and NF-κB signaling negatively regulate osteoblast dedifferentiation. (C) After osteoblast ablation, osteoblasts are formed de novo in the regenerate. (D) Osteoblast progenitor cells in the joints contribute to the osteoblast population in the regenerate.
During the outgrowth phase of regeneration, when new tissue is generated, dedifferentiated osteoblasts have to redifferentiate to produce bone matrix. Redifferentiation is tightly regulated in space and time to ensure that preosteoblasts in the distal blastema remain proliferative to contribute to progenitor expansion, while osteoblasts in the proximal blastema redifferentiate to build new bone. In the regenerating fin, runx2+ preosteoblasts are responsive to canonical Wnt signaling generated by distal blastema cells, which keeps them in the proliferative state (Stewart et al. 2014). More proximally, autocrine BMP signaling is required for the commitment of runx2+ cells to Sp7 transcription factor (sp7, previous name osterix) positive cells. RA signaling, which negatively regulates osteoblast dedifferentiation, also inhibits osteoblast redifferentiation (Blum and Begemann 2015). RA treatment down-regulates expression of bone morphogenetic protein 2b (bmp2b), the ligand presumed to be responsible for activation of Bmp signaling in preosteoblasts, while also inhibiting expression of the Wnt signaling pathway inhibitor dickkopf 1b (dkk1b). These data suggest that RA signaling acts through modulating both BMP and Wnt/β-catenin signaling. Both Bmp and Wnt signaling pathways are also regulated by the Hippo-Yap pathway. In mesenchymal cells of the proximal blastema, Yes1-associated transcriptional regulator (Yap) promotes expression of bmp2a, thus regulating osteoblast differentiation in a paracrine fashion (Brandão et al. 2019). Additionally, overexpression of a dominant-negative form of Yap reduces expression of dkk1a and bmp2a. These data indicate that Yap negatively modulates Wnt signaling by regulating dkk1a expression in a potential synergistic manner with Bmp signaling, ensuring the confinement of active Wnt signaling to the distal region of the blastema.
Dedifferentiated osteoblasts are not the sole origin for osteoblasts during regeneration. When fins are depleted of virtually all osteoblasts via genetic ablation (using nitroreductase driven by sp7 regulatory elements), regeneration of bone is not perturbed (Singh et al. 2012). This indicates that de novo differentiation of cells into osteoblasts can occur (Fig. 3C). If such nonosteoblastic cells contribute to bone regeneration by default, or if their activation is a back-up mechanism when dedifferentiation from mature osteoblasts fails, is an intriguing question. Cell-lineage analysis has identified a population of cells expressing matrix metalloproteinase 9 (mmp9) as potential osteoblast progenitor cells (OPCs) (Ando et al. 2017). These cells are located in the joints between fin ray segments, and in the noninjured fin they do not express osteoblast markers such as osterix or bglap. Upon amputation, however, these cells migrate into the regenerate and contribute to the osteoblast population (Fig. 3D). Genetic ablation of mmp9+ cells using the nitroreductase system resulted in reduction of osteoblast numbers and of calcified fin ray tissue, indicating that both osteoblast dedifferentiation and differentiation from OPCs are deployed during unperturbed fin regeneration. Joint cells come from the same lineage as osteoblasts (Sousa et al. 2011; Tu and Johnson 2011), and a lineage tracing study in medaka showed that collagen, type X, α1a (col10a1)+ cells contribute to osteoblasts and joint cells during regeneration (Dasyani et al. 2019). Further studies about the fate of various bone-associated cell populations during regeneration using lineage tracing will be necessary to ascertain the contribution of these different pools to bone regeneration. As mentioned above, regenerating fibroblasts locate to distinct proximodistal blastema regions depending on their original position in the stump (Tornini et al. 2016). It would be intriguing to analyze whether such heterogeneity can also be observed for osteoblasts.
GENETIC AND EPIGENETIC REGULATORS OF FIN REGENERATION
The highly variable regeneration capacity between species could result from regeneration-specific genes existing only in species with high regeneration capacity. While one such gene, Prod1, a cell-surface molecule implicated in mediating positional identity, is described to exist only in highly regenerative salamanders (Da Silva et al. 2002; see also Otsuki and Tanaka 2021), the prevalent hypothesis is that the predominant mechanism underlying differential regenerative outcomes between species is the differential regulation of conserved genes. Transcriptional profiling and proteomic analyses have shown that the activity of hundreds of genes is modified during zebrafish fin regeneration (Schebesta et al. 2006; Saxena et al. 2012; Kang et al. 2016); thus gene regulation is dynamic (early blastema formation and dedifferentiation phase, later redifferentiation phase) and customized to different cell types (epidermis, different blastema compartments).
DNA methylome analysis of fin regenerates of different timepoints showed neither global changes in DNA methylation nor differentially methylated genomic regions during regeneration (Lee et al. 2020). However, immunohistochemical analysis revealed a transient DNA demethylation during the early phase of fin regeneration in blastema cells, together with up-regulation of growth arrest and DNA-damage-inducible, a (gadd45), an important mediator of DNA methylation (Hirose et al. 2013). Possibly, pooled tissue analysis in the first study obscured changes occurring only in a subset of cells. Indeed, the authors show that DNA methylation, while globally comparable between sp7+ and sp7− cells, varies in local genomic regions, and they identified over 2000 regions that are differentially methylated in sp7+ compared to sp7− cells. Yet, DNA methylation analysis of sp7+ cells at different timepoints suggests that these osteoblast-specific DNA methylation regions change very little, if at all, during regeneration (Lee et al. 2020). Another important epigenetic feature controlling gene expression is chromatin accessibility. Indeed, genomic regions classified as cis-regulatory regions gained chromatin accessibility during regeneration (Lee et al. 2020). Interestingly, these putative enhancers contained low DNA methylation in the noninjured state, potentially allowing a fast gene activation after amputation. Chromatin accessibility is regulated by factors remodeling the configuration of chromatin in response to intrinsic and extrinsic signals. One of four major ATP-dependent chromatin remodeling complexes is the nucleosome remodeling and deacetylase (NuRD) complex. During the redifferentiation phase, the transcription of several components of NuRD was shown to be up-regulated in the proliferative compartment of the blastema (Pfefferli et al. 2014). Furthermore, inhibition of the histone deacetylase 1 (Hdac1) blocked redifferentiation of osteoblast precursors but not blastema formation and osteoblast dedifferentiation, stressing the independent regulation of these processes. Taken together, chromatin accessibility, but not DNA methylation, might be involved in regulating regeneration-specific gene transcription.
In recent years, several studies have analyzed which genetic elements are involved in the differential regulation of gene activation during regeneration. A pioneering study coined the term “tissue regeneration enhancer element” (TREE) for regulatory elements involved in activating expression of regeneration-associated genes (Kang et al. 2016). The authors showed that a leptinb-linked enhancer (LEN) can regulate regeneration-activated gene expression. Interestingly, within this element, different modules are present for tissue-specific regulation (fin vs. heart), although neither heart nor fin regeneration was impaired in leptinb mutants. In a follow-up study, the authors identified further TREES and demonstrated that these regulatory elements are located near genes known to be important for regeneration, such as fgf20a, midkine a (mdka), and connexin 43 (cx43) (Thompson et al. 2020). Consistent with the methylome study (Lee et al. 2020), no variation in the methylation state within these enhancer regions could be observed between different timepoints of regeneration. Another regulatory component is the ctgfa reporter in regeneration (careg) element, an upstream genomic region of the connective tissue growth factor a (ctgfa) gene (Pfefferli and Jaźwińska 2017). During regeneration, careg is activated in mesenchymal cells through TGF-β/activin-β signaling. Intriguingly, careg is also active during heart regeneration; this together with the involvement of LEN in both fin and heart regeneration suggests common regulatory mechanisms for regeneration. However, it is not clear yet whether these regulatory elements respond to generic wounding-induced signals, or are only activated in contexts where structural regeneration occurs. Comparative analysis between killifish and zebrafish fin regeneration revealed conserved regeneration-responsive enhancers (RREs) with binding sites for the AP-1 transcription factor complex as a shared characteristic (Wang et al. 2020). Genes regulated by RREs are mainly expressed in blastema cells. Coinciding, comparison between zebrafish regenerates at different timepoints revealed regulatory elements with predicted binding sites for the AP-1 complex to be most differentially active (Thompson et al. 2020). One gene under the control of an RRE is the TGF-β-related ligand inhba, which is known to be required for fin regeneration (Jaźwińska et al. 2007), and minimal sequences from the enhancer can direct gene expression in response to injury (Wang et al. 2020).
Altogether, these studies indicate that common regenerative-positive elements are involved in activation of altered gene expression during regeneration. Fast gene expression responses might be facilitated by specific chromatin remodeling.
CONCLUDING REMARKS
The zebrafish fin provides a powerful model to study regeneration in vivo. Although fin regeneration is a complex process, the understanding of molecular regulators involved in fin regeneration and their interactions has substantially advanced in the past decade. Prior studies often relied on systemic interference in the whole fish. In the future, the combination of conditional genetic and single-cell approaches will allow for precise tissue-specific manipulation and cell-type-specific analysis, yielding deeper understanding of the origin and recipients of signals and how these mechanisms contribute to the regeneration of the various tissues.
ACKNOWLEDGMENTS
Research in the Weidinger laboratory is funded by the Deutsche Forschungsgemeinschaft (SFB 1149, project No. 251293561; SFB 1279, project No. 316249678; WE 4223/6-1, project No. 414077062; WE 4223/8-1, project No. 433187294).
Footnotes
Editors: Kenneth D. Poss and Donald T. Fox
Additional Perspectives on Regeneration available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Ando K, Shibata E, Hans S, Brand M, Kawakami A. 2017. Osteoblast production by reserved progenitor cells in zebrafish bone regeneration and maintenance. Dev Cell 43: 643–650.e3. 10.1016/j.devcel.2017.10.015 [DOI] [PubMed] [Google Scholar]
- Ang NB, Saera-Vila A, Walsh C, Hitchcock PF, Kahana A, Thummel R, Nagashima M. 2020. Midkine-a functions as a universal regulator of proliferation during epimorphic regeneration in adult zebrafish. PLoS ONE 15: e0232308. 10.1371/journal.pone.0232308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong BE, Henner A, Stewart S, Stankunas K. 2017. Shh promotes direct interactions between epidermal cells and osteoblast progenitors to shape regenerated zebrafish bone. Development 144: 1165–1176. 10.1242/dev.143792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo AS, Grotek B, Jacinto A, Weidinger G, Saúde L. 2011. The regenerative capacity of the zebrafish caudal fin is not affected by repeated amputations. PLoS ONE 6: e22820. 10.1371/journal.pone.0022820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra J, Montes GS, Bexiga SR, Junqueira LC. 1983. Structure of the tail fin in teleosts. Cell Tissue Res 230: 127–137. 10.1007/BF00216033 [DOI] [PubMed] [Google Scholar]
- Becker T, Becker CG. 2020. Dynamic cell interactions allow spinal cord regeneration in zebrafish. Curr Opin Physiol 14: 64–69. 10.1016/j.cophys.2020.01.009 [DOI] [Google Scholar]
- Bird NC, Mabee PM. 2003. Developmental morphology of the axial skeleton of the zebrafish, Danio rerio (Ostariophysi: Cyprinidae). Dev Dyn 228: 337–357. 10.1002/dvdy.10387 [DOI] [PubMed] [Google Scholar]
- Blum N, Begemann G. 2012. Retinoic acid signaling controls the formation, proliferation and survival of the blastema during adult zebrafish fin regeneration. Development 139: 107–116. 10.1242/dev.065391 [DOI] [PubMed] [Google Scholar]
- Blum N, Begemann G. 2015. Osteoblast de- and redifferentiation are controlled by a dynamic response to retinoic acid during zebrafish fin regeneration. Development 142: 2894–2903. [DOI] [PubMed] [Google Scholar]
- Brandão AS, Bensimon-Brito A, Lourenço R, Borbinha J, Soares AR, Mateus R, Jacinto A. 2019. Yap induces osteoblast differentiation by modulating Bmp signalling during zebrafish caudal fin regeneration. J Cell Sci 132: jcs231993. 10.1242/jcs.231993 [DOI] [PubMed] [Google Scholar]
- Chablais F, Jaźwińska A. 2010. IGF signaling between blastema and wound epidermis is required for fin regeneration. Development 137: 871–879. 10.1242/dev.043885 [DOI] [PubMed] [Google Scholar]
- Chen CH, Merriman AF, Savage J, Willer J, Wahlig T, Katsanis N, Yin VP, Poss KD. 2015. Transient laminin β 1a induction defines the wound epidermis during zebrafish fin regeneration. PLoS Genet 11: e1005437. 10.1371/journal.pgen.1005437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Puliafito A, Cox BD, Primo L, Fang Y, Di Talia S, Poss KD. 2016. Multicolor cell barcoding technology for long-term surveillance of epithelial regeneration in zebrafish. Dev Cell 36: 668–680. 10.1016/j.devcel.2016.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo R, Hernández-Martínez R, Chimal-Monroy J, Merchant-Larios H, Covarrubias L. 2012. Full regeneration of the tribasal Polypterus fin. Proc Natl Acad Sci 109: 3838–3843. 10.1073/pnas.1006619109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B, Zheng Y, Sun L, Shi T, Shi Z, Wang L, Huang G, Sun N. 2018. Heart regeneration in adult mammals after myocardial damage. Acta Cardiol Sin 34: 115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daponte V, Tylzanowski P, Forlino A. 2021. Appendage regeneration in vertebrates: what makes this possible? Cells 10: 242. 10.3390/cells10020242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva SM, Gates PB, Brockes JP. 2002. The newt ortholog of CD59 is implicated in proximodistal identity during amphibian limb regeneration. Dev Cell 3: 547–555. 10.1016/S1534-5807(02)00288-5 [DOI] [PubMed] [Google Scholar]
- Dasyani M, Tan WH, Sundaram S, Imangali N, Centanin L, Wittbrodt J, Winkler C. 2019. Lineage tracing of col10a1 cells identifies distinct progenitor populations for osteoblasts and joint cells in the regenerating fin of medaka (Oryzias latipes). Dev Biol 455: 85–99. 10.1016/j.ydbio.2019.07.012 [DOI] [PubMed] [Google Scholar]
- Gault WJ, Enyedi B, Niethammer P. 2014. Osmotic surveillance mediates rapid wound closure through nucleotide release. J Cell Biol 207: 767–782. 10.1083/jcb.201408049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemberling M, Bailey TJ, Hyde DR, Poss KD. 2013. The zebrafish as a model for complex tissue regeneration. Trends Genet 29: 611–620. 10.1016/j.tig.2013.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Rosa JM, Burns CE, Burns CG. 2017. Zebrafish heart regeneration: 15 years of discoveries. Regeneration 4: 105–123. 10.1002/reg2.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotek B, Wehner D, Weidinger G. 2013. Notch signaling coordinates cellular proliferation with differentiation during zebrafish fin regeneration. Development 140: 1412–1423. 10.1242/dev.087452 [DOI] [PubMed] [Google Scholar]
- Hirose K, Shimoda N, Kikuchi Y. 2013. Transient reduction of 5-methylcytosine and 5-hydroxymethylcytosine is associated with active DNA demethylation during regeneration of zebrafish fin. Epigenetics 8: 899–906. 10.4161/epi.25653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Lee HJ, Chen Y, Ge J, Osman FOI, McAdow AR, Mokalled MH, Johnson SL, Zhao G, Wang T. 2020. Cellular diversity of the regenerating caudal fin. Sci Adv 6: eaba2084 10.1126/sciadv.aba2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaźwińska A, Badakov R, Keating MT. 2007. Activin-βA signaling is required for zebrafish fin regeneration. Curr Biol 17: 1390–1395. 10.1016/j.cub.2007.07.019 [DOI] [PubMed] [Google Scholar]
- Joven A, Elewa A, Simon A. 2019. Model systems for regeneration: salamanders. Development 146: dev167700. 10.1242/dev.167700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Hu J, Karra R, Dickson AL, Tornini VA, Nachtrab G, Gemberling M, Goldman JA, Black BL, Poss KD. 2016. Modulation of tissue repair by regeneration enhancer elements. Nature 532: 201–206. 10.1038/nature17644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschbaum F, Meunier FJ. 1981. Experimental regeneration of the caudal skeleton of the glass knifefish, Eigenmannia virescens (Rhamphichthydae, Gymnotoidei). J Morphol 168: 121–135. 10.1002/jmor.1051680202 [DOI] [PubMed] [Google Scholar]
- Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, Mahatma G, Fisher S, Brand M, Schulte-Merker S, et al. 2011. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell 20: 713–724. 10.1016/j.devcel.2011.04.014 [DOI] [PubMed] [Google Scholar]
- König D, Page L, Chassot B, Jaźwińska A. 2018. Dynamics of actinotrichia regeneration in the adult zebrafish fin. Dev Biol 433: 416–432. 10.1016/j.ydbio.2017.07.024 [DOI] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. 2005. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development 132: 5173–5183. 10.1242/dev.02101 [DOI] [PubMed] [Google Scholar]
- Lee Y, Hami D, De Val S, Kagermeier-Schenk B, Wills AA, Black BL, Weidinger G, Poss KD. 2009. Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev Biol 331: 270–280. 10.1016/j.ydbio.2009.05.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Hou Y, Chen Y, Dailey ZZ, Riddihough A, Jang HS, Wang T, Johnson SL. 2020. Regenerating zebrafish fin epigenome is characterized by stable lineage-specific DNA methylation and dynamic chromatin accessibility. Genome Biol 21: 52. 10.1186/s13059-020-1948-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R, Sehring I, Cederlund M, Mulaw M, Weidinger G. 2020. NF-κB signaling negatively regulates osteoblast dedifferentiation during zebrafish bone regeneration. Dev Cell 52: 167–182.e7. 10.1016/j.devcel.2019.11.016 [DOI] [PubMed] [Google Scholar]
- Münch J, González-Rajal A, de la Pompa JL. 2013. Notch regulates blastema proliferation and prevents differentiation during adult zebrafish fin regeneration. Development 140: 1402–1411. 10.1242/dev.087346 [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Keating MT. 2002. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development 129: 2607–2617. 10.1242/dev.129.11.2607 [DOI] [PubMed] [Google Scholar]
- Nogueira AF, Costa CM, Lorena J, Moreira RN, Frota-Lima GN, Furtado C, Robinson M, Amemiya CT, Darnet S, Schneider I. 2016. Tetrapod limb and sarcopterygian fin regeneration share a core genetic programme. Nat Commun 7: 13364. 10.1038/ncomms13364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppedal D, Goldsmith MI. 2010. A chemical screen to identify novel inhibitors of Fin regeneration in zebrafish. Zebrafish 7: 53–60. 10.1089/zeb.2009.0633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Otsuki L, Tanaka EM. 2021. Positional memory in vertebrate regeneration: a century's insights from the salamander limb. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a040899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owlarn S, Klenner F, Schmidt D, Rabert F, Tomasso A, Reuter H, Mulaw MA, Moritz S, Gentile L, Weidinger G, et al. 2017. Generic wound signals initiate regeneration in missing-tissue contexts. Nat Commun 8: 2282. 10.1038/s41467-017-02338-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pápai K, Csikós K, Vellai V. 2019. No correlation between endo- and exoskeletal regenerative capacities in teleost species. Fishes 4: 51. 10.3390/fishes4040051 [DOI] [Google Scholar]
- Park D, Spencer JA, Koh BI, Kobayashi T, Fujisaki J, Clemens TL, Lin CP, Kronenberg HM, Scadden DT. 2012. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell 10: 259–272. 10.1016/j.stem.2012.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie TA, Strand NS, Tsung-Yang C, Rabinowitz JS, Moon RT. 2014. Macrophages modulate adult zebrafish tail fin regeneration. Development 141: 2581–2591. 10.1242/dev.098459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferli C, Jaźwińska A. 2015. The art of fin regeneration in zebrafish. Regen 2: 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferli C, Jaźwińska A. 2017. The careg element reveals a common regulation of regeneration in the zebrafish myocardium and fin. Nat Commun 8: 15151. 10.1038/ncomms15151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferli C, Müller F, Jaźwińska A, Wicky C. 2014. Specific NuRD components are required for fin regeneration in zebrafish. BMC Biol 12: 30. 10.1186/1741-7007-12-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poleo G, Brown CW, Laforest L, Akimenko MA. 2001. Cell proliferation and movement during early fin regeneration in zebrafish. Dev Dyn 221: 380–390. 10.1002/dvdy.1152 [DOI] [PubMed] [Google Scholar]
- Poss KD, Shen J, Keating MT. 2000a. Induction of lef1 during zebrafish fin regeneration. Dev Dyn 219: 282–286. [DOI] [PubMed] [Google Scholar]
- Poss KD, Shen J, Nechiporuk A, McMahon G, Thisse B, Thisse C, Keating MT. 2000b. Roles for Fgf signaling during zebrafish fin regeneration. Dev Biol 222: 347–358. 10.1006/dbio.2000.9722 [DOI] [PubMed] [Google Scholar]
- Poss KD, Keating MT, Nechiporuk A. 2003. Tales of regeneration in zebrafish. Dev Dyn 226: 202–210. 10.1002/dvdy.10220 [DOI] [PubMed] [Google Scholar]
- Quint E, Smith A, Avaron F, Laforest L, Miles J, Gaffield W, Akimenko MA. 2002. Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc Natl Acad Sci 99: 8713–8718. 10.1073/pnas.122571799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci L, Srivastava M. 2018. Wound-induced cell proliferation during animal regeneration. Wiley Interdiscip Rev Dev Biol 7: e321. 10.1002/wdev.321 [DOI] [PubMed] [Google Scholar]
- Santos-Ruiz L, Santamaría JA, Ruiz-Sánchez J, Becerra J. 2002. Cell proliferation during blastema formation in the regenerating teleost fin. Dev Dyn 223: 262–272. 10.1002/dvdy.10055 [DOI] [PubMed] [Google Scholar]
- Saxena S, Singh SK, Meena Lakshmi MG, Meghah V, Bhatti B, Brahmendra Swamy CV, Sundaram CS, Idris MM. 2012. Proteomic analysis of zebrafish caudal fin regeneration. Mol Cell Proteomics 11: M111.014118. 10.1074/mcp.M111.014118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schebesta M, Lien CL, Engel FB, Keating MT. 2006. Transcriptional profiling of caudal fin regeneration in zebrafish. ScientificWorldJournal 6: 38–54. 10.1100/tsw.2006.326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehring IM, Weidinger G. 2020. Recent advancements in understanding fin regeneration in zebrafish. Wiley Interdiscip Rev Dev Biol 9: e367. 10.1002/wdev.367 [DOI] [PubMed] [Google Scholar]
- Shao J, Chen D, Ye Q, Cui J, Li Y, Li L. 2011. Tissue regeneration after injury in adult zebrafish: the regenerative potential of the caudal fin. Dev Dyn 240: 1271–1277. 10.1002/dvdy.22603 [DOI] [PubMed] [Google Scholar]
- Shibata E, Yokota Y, Horita N, Kudo A, Abe G, Kawakami K, Kawakami A. 2016. Fgf signalling controls diverse aspects of fin regeneration. Development 143: 2920–2929. [DOI] [PubMed] [Google Scholar]
- Shibata E, Ando K, Murase E, Kawakami A. 2018. Heterogeneous fates and dynamic rearrangement of regenerative epidermis-derived cells during zebrafish fin regeneration. Development 145: dev162016. 10.1242/dev.162016 [DOI] [PubMed] [Google Scholar]
- Singh SP, Holdway JE, Poss KD. 2012. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell 22: 879–886. 10.1016/j.devcel.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa S, Afonso N, Bensimon-Brito A, Fonseca M, Simões M, Leon J, Roehl H, Cancela ML, Jacinto A. 2011. Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development 138: 3897–3905. 10.1242/dev.064717 [DOI] [PubMed] [Google Scholar]
- Stewart S, Stankunas K. 2012. Limited dedifferentiation provides replacement tissue during zebrafish fin regeneration. Dev Biol 365: 339–349. 10.1016/j.ydbio.2012.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S, Gomez AW, Armstrong BE, Henner A, Stankunas K. 2014. Sequential and opposing activities of Wnt and BMP coordinate zebrafish bone regeneration. Cell Rep 6: 482–498. 10.1016/j.celrep.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. 2007. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 134: 479–489. 10.1242/dev.001123 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Ou J, Lee N, Shin K, Cigliola V, Song L, Crawford GE, Kang J, Poss KD. 2020. Identification and requirements of enhancers that direct gene expression during zebrafish fin regeneration. Dev 147: dev191262. 10.1242/dev.191262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornini VA, Puliafito A, Slota LA, Thompson JD, Nachtrab G, Kaushik AL, Kapsimali M, Primo L, Di Talia S, Poss KD. 2016. Live monitoring of blastemal cell contributions during appendage regeneration. Curr Biol 26: 2981–2991. 10.1016/j.cub.2016.08.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornini VA, Thompson JD, Allen RL, Poss KD. 2017. Live fate-mapping of joint-associated fibroblasts visualizes expansion of cell contributions during zebrafish fin regeneration. Dev 144: 2889–2895. 10.1242/dev.155655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran AP, Warren PM, Silver J. 2018. The biology of regeneration failure and success after spinal cord injury. Physiol Rev 98: 881–917. 10.1152/physrev.00017.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, Johnson SL. 2011. Fate restriction in the growing and regenerating zebrafish fin. Dev Cell 20: 725–732. 10.1016/j.devcel.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hu CK, Zeng A, Alegre D, Hu D, Gotting K, Granillo AO, Wang Y, Robb S, Schnittker R, et al. 2020. Changes in regeneration-responsive enhancers shape regenerative capacities in vertebrates. Science 369: eaaz3090. 10.1126/science.aaz3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner D, Weidinger G. 2015. Signaling networks organizing regenerative growth of the zebrafish fin. Trends Genet 31: 336–343. 10.1016/j.tig.2015.03.012 [DOI] [PubMed] [Google Scholar]
- Wehner D, Cizelsky W, Vasudevaro MD, Özhan G, Haase C, Kagermeier-Schenk B, Röder A, Dorsky RI, Moro E, Argenton F, et al. 2014. Wnt/β-catenin signaling defines organizing centers that orchestrate growth and differentiation of the regenerating zebrafish caudal fin. Cell Rep 6: 467–481. 10.1016/j.celrep.2013.12.036 [DOI] [PubMed] [Google Scholar]
- Whitehead GG, Makino S, Lien CL, Keating MT. 2005. Developmental biology: fgf20 is essential for initiating zebrafish fin regeneration. Science 310: 1957–1960. 10.1126/science.1117637 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Kawakami K, Abe G, Tamura K. 2020. Zebrafish can regenerate endoskeleton in larval pectoral fin but the regenerative ability declines. Dev Biol 463: 110–123. 10.1016/j.ydbio.2020.04.010 [DOI] [PubMed] [Google Scholar]