Abstract
Salamanders, such as axolotls and newts, can regenerate complex tissues including entire limbs. But what mechanisms ensure that an amputated limb regenerates a limb, and not a tail or unpatterned tissue? An important concept in regeneration is positional memory—the notion that adult cells “remember” spatial identities assigned to them during embryogenesis (e.g., “head” or “hand”) and use this information to restore the correct body parts after injury. Although positional memory is well documented at a phenomenological level, the underlying cellular and molecular bases are just beginning to be decoded. Herein, we review how major principles in positional memory were established in the salamander limb model, enabling the discovery of positional memory-encoding molecules, and advancing insights into their pattern-forming logic during regeneration. We explore findings in other amphibians, fish, reptiles, and mammals and speculate on conserved aspects of positional memory. We consider the possibility that manipulating positional memory in human cells could represent one route toward improved tissue repair or engineering of patterned tissues for therapeutic purposes.
Observing organisms as they rebuild and pattern tissue after injury instills a sense of wonder. In adult humans, perhaps the most dramatic exemplar is the liver, which can recover in mass and function from a 70% resection. However, the recovered liver is an unfaithful reproduction as it does not regain its original shape. Organ shape and function are originally acquired during embryonic development through the actions of patterning molecules such as morphogens. Wolpert proposed that patterning molecules infuse cells with “positional information” conferring spatial identities such as “liver” or “limb,” “dorsal” or “ventral” relative to anatomical landmarks in the embryo (Wolpert 1969). These identities enable cells to engage one another and to contribute toward correct assembly of the body plan. An intriguing possibility is that human cells retain remnants of positional information beyond embryogenesis, as “positional memory.” If this is true, could positional memory somehow be harnessed to repair or regenerate adult tissues, additional to the liver?
Salamanders are interesting cases that have revealed the existence of positional memory in vertebrates and its necessity for pattern formation during regeneration. Salamanders are amphibians able to regenerate diverse tissues including entire limbs, large parts of their central nervous system, visual system, gills, and tails (for a primer, see Joven et al. 2019). Limbs are surgically accessible for investigating positional memory. Although salamanders regenerate amputated limbs with high fidelity, surgically excising defined regions of the limb (e.g., posterior) before amputation results in a regenerated limb with incorrect patterning. Conversely, juxtaposing defined combinations of tissues with distinct positional memories can generate additional limbs even in uninjured salamanders. An important possibility is that adult humans harbor positional memories that could be leveraged to create a pattern, as in salamanders. Salamanders are informative as they possess similar tissue types to humans and regenerate at relevant size scales using evolutionarily conserved molecules.
We begin this review by introducing the salamander limb model. We discuss the requisite properties of a positional memory molecule and highlight known players satisfying these criteria. We summarize the historic insights that led to the current positional memory framework in the form of “lessons from the salamander limb.” Finally, we integrate findings from humans and other vertebrates to propose some potential avenues toward a more global understanding of positional memory.
THE SALAMANDER LIMB MODEL FOR ELUCIDATING RULES OF POSITIONAL MEMORY
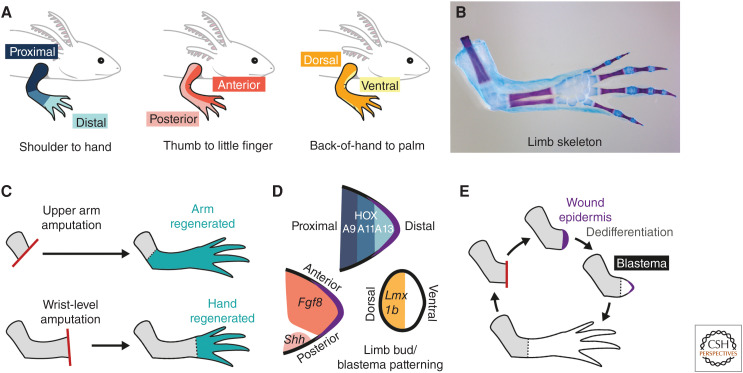
Salamander limbs are structurally homologous to those of humans across their three axes: proximodistal, anteroposterior, and dorsoventral (Fig. 1A). Limb skeletal elements act as convenient assays for patterning without the need for molecular markers, as they have a characteristic morphology, arrangement, and number—for example, 24 skeletal elements in the arm of an axolotl (Ambystoma mexicanum) (Fig. 1B). Amputating a salamander limb at any place between the body wall and hand/foot regenerates only the missing structures: An upper arm amputation regenerates upper arm, lower arm, and hand, whereas a wrist-level amputation regenerates only hand (Fig. 1C). Thus, salamander cells interpret spatial location to direct position-specific regeneration (positional memory).
Figure 1.
Salamander limb patterning and regeneration. (A) The three primary limb axes in the salamander limb (top), and their equivalent axes in the human limb (see text below schematics). (B) Skeletal elements in the salamander limb have a characteristic patterning and arrangement. Image depicts the right forelimb of an axolotl stained with Alcian blue (cartilage) and Alizarin red (bone). The axolotl forelimb has four digits, and a total of 24 skeletal elements. (Image credit: Yuka Taniguchi-Sugiura.) (C) Position-dependent regeneration after amputation. (Top) Following amputation at the upper arm level, the rest of the upper arm, the lower arm, and the hand are regenerated. (Bottom) Following a wrist-level amputation, only the missing hand is regenerated. (D) Similar patterning mechanisms in the limb bud and cone-stage blastema. Proximodistal axis: HOX proteins are expressed in nested domains corresponding to the future proximodistal axis (HOXA13-expressing cells give rise to the most distal elements). Anteroposterior axis: Fgf8 is expressed in anterior cells, Shh in posterior cells. Dorsoventral axis: Lmx1b is expressed in dorsal, but not ventral, cells. Black outline indicates epidermis. Purple indicates wound epidermis. (E) Major steps in salamander limb regeneration. Following amputation (red line), a wound epidermis (purple) is established at the amputation plane. Cells in the limb stump (gray) are recruited toward the wound epidermis. Connective tissue cells dedifferentiate and, together with other lineage-restricted progenitors, form a blastema. Blastema cells proliferate and redifferentiate in lineage-restricted manners.
One strength of salamander models is their tolerance to transplantation, with relatively little immune rejection compared with mammals or fish. This opens up creative possibilities to test positional memory. For example, posterior limb regions can be “deleted” by surgically excising the posterior half of a limb and replacing it with the anterior half taken from a separate donor. Limb parts can be removed, rotated, and regrafted to invert tissue axes. One weakness has been the difficulty in interrogating genome-wide regulation and epigenetic mechanisms owing to the large genome sizes of salamanders (e.g., 32 Gb for an axolotl—more than 10 times as large as that of a human). However, recent genome assemblies for the axolotl and the Iberian ribbed newt (Pleurodeles waltl), plus the generation of genetic toolkits has now established them as genetically tractable models (Elewa et al. 2017; Nowoshilow et al. 2018; Schloissnig et al. 2021).
LIMB DEVELOPMENT AND REGENERATION
As in other vertebrates, the salamander limb originates as a limb bud—a proliferative, mesodermal outgrowth from the lateral flank of the embryo that is encapsulated by an overlying epidermis. The future limb axes are established by spatially restricted patterns of gene expression within the limb bud (Fig. 1D). The homeodomain-containing transcription factors HoxA9 and HoxA13 are expressed in nested domains along the future proximodistal axis, with HoxA13 more distal (Gardiner et al. 1995). Anterior limb bud cells express the secreted factor Fgf8 (Fibroblast growth factor 8) (Han et al. 2001), posterior cells express the secreted factor Shh (Sonic hedgehog) (Imokawa and Yoshizato 1997; Torok et al. 1999), and dorsal (but not ventral) cells express the LIM homeobox transcription factor Lmx1b (Shimokawa et al. 2013). These genes belong to an evolutionarily conserved limb patterning circuitry and, apart from Fgf8, are expressed in comparable spatial domains in the limb bud mesoderm as their orthologs in other vertebrate limb buds. The segments of the limb—upper arm/leg, lower arm/leg, and hand/foot—are laid down in a largely sequential manner and differentiate over the course of several weeks. There are some differences in limb bud patterning in salamanders compared with well-studied amniotes such as mice (Mus musculus) and chicks (Gallus gallus). For example, it is unclear to what extent a signaling apical ectodermal ridge is present in salamanders and several genes expressed in the ectoderm in amniotes, such as the secreted factors Fgf8 and Wnt7a, are instead expressed in the mesenchyme in salamanders (Christensen et al. 2002; Shimokawa et al. 2013; Schloissnig et al. 2021).
Amputation of the salamander limb triggers regeneration (Fig. 1E). The first steps are wound healing and formation of a wound epidermis necessary to sustain regeneration. Within days, a proliferative cell mass called a blastema forms under the wound epidermis. The blastema regenerates almost all limb tissues. In axolotls, up to 78% of blastema cells originate as connective tissue in the limb stump dermis, estimated by grafting triploid-labeled dermal tissue to diploid host arms followed by amputation and quantification of triploid cells in the resulting blastema (Muneoka et al. 1986). Connective tissue cells migrate toward the wound epidermis and “dedifferentiate” to give rise to blastema cells. Blastema cells have been proposed to carry out regeneration by “returning” to an embryonic state and recapitulating aspects of limb bud patterning—hence, the “dedifferentiation” moniker. Strong support for this concept came from comparing single blastema cell transcriptomes isolated at sequential time points during axolotl limb regeneration to the transcriptomes of limb bud cells. This comparison revealed an increase in similarity (correlation score) between the global transcriptomes of these cell types during the first days of regeneration (Gerber et al. 2018). To validate this in a more targeted manner, these investigators identified a set of ∼100 genes whose expression was deemed specific either to axolotl stage 44 embryonic limb bud or to uninjured adult connective tissue (Gerber et al. 2018; T Gerber, pers. comm.). Using these restricted gene lists as an input for quadratic programming analysis enabled quantification of the similarity of single blastema cells to embryonic limb bud identity or uninjured adult cell identity. As in the global transcriptome comparisons, an increase in similarity between blastema cells and stage 44 limb bud cells could be observed during regeneration, peaking at 11 days postamputation and beginning to decrease by 18 days postamputation (Gerber et al. 2018).
The similarity between regenerative and developmental patterning is evident by in situ hybridization after several days of proliferation, when blastema cells express genes including Hox, Fgf8, Shh, and Lmx1b in similar domains as in the limb bud (Fig. 1D). Proximal and distal blastema cells also differ in their surface properties. In hanging drop cocultures, proximal blastemas engulf distal blastemas preferentially (Nardi and Stocum 1984). Proximal or distal blastemas grafted to the side of an upper arm stump displace to their corresponding axial positions during regeneration (affinophoresis) (Crawford and Stocum 1988).
Connective tissue-derived blastema cells redifferentiate into connective tissue, tendon cells, and skeletal and periskeletal elements (Gerber et al. 2018). Separate lineage-committed progenitors regenerate other cell types, such as epidermis, muscle, and Schwann cells (Kragl et al. 2009). Proliferation and differentiation of blastema cells over several weeks regenerates the limb, at least partially by recapitulating growth and patterning mechanisms used during limb development, as discussed above.
WHAT IS POSITIONAL MEMORY?
Blastema cells possess position-specific gene expression and surface properties. But, as they only exist transiently during regeneration, blastema cells cannot be considered reservoirs of positional memory. Positional memory is a property of steady-state cells (e.g., connective tissue cells), with the potential to maintain information over a lifetime. Positional memory is decoded during regeneration and the spatial information transferred to blastema cells to direct patterning.
Patterning genes including Fgf8 and Shh are not expressed in the steady-state limb, precluding them from encoding memory transcriptionally. However, there is no requirement for positional memory to be transcribed. Other possibilities include epigenetic “priming” at patterning gene loci, or stable molecules in the extracellular environment. The requisite properties of positional memory were defined to include such possibilities, based on experiments performed in salamanders and other models.
Positional memory must:
Differ spatially in steady-state tissue;
Be instructive for patterning; and
Be interpretable externally (i.e., by neighboring, or more distant, cells).
Deletion of a positional memory gene from a steady-state cell should preclude that cell from exerting its usual contribution toward regenerative patterning. Misexpression of a positional memory gene, followed by transplantation of the cell to an ectopic site, should induce predictable alterations in tissue patterning. However, current understanding of the “rules” of positional memory is not sufficient to predict all experimental outcomes. For example, it is not clear whether deletion of a “distal” memory gene would automatically render a cell “proximal.” It is also unclear what result would arise if competing memories, such as “anterior” and “posterior,” are coinduced experimentally within the same cell.
WHICH POSITIONAL MEMORY MOLECULES HAVE BEEN IDENTIFIED?
Brockes’ group discovered and named the first positional memory-encoding gene, Prod1, in the Eastern red-spotted newt (Notophthalmus viridescens) (da Silva et al. 2002). Prod1 imparts proximal identity and encodes a salamander-only cell surface protein with similarity to mammalian CD59 (Garza-Garcia et al. 2009). Upper arm cells transcribe 1.7× more Prod1 than hand cells, and the protein is present in Schwann cells. After amputation, proximal blastema cells express 1.7× more Prod1 compared with distal blastema cells (da Silva et al. 2002; Kumar et al. 2007a). Notophthalmus PROD1 is linked to the cell surface via a GPI anchor and contributes to the distinct surface properties of proximal and distal blastema cells. Yeast two-hybrid screening identified a ligand for Prod1 called newt anterior gradient (nAG)—a secreted thioredoxin fold-containing protein whose expression is up-regulated after amputation (Kumar et al. 2007b). Consistent with a role in proximal identity, retinoid treatment of distal blastemas (which proximalizes cells—see below) increased Prod1 transcription, whereas Prod1 misexpression in distal axolotl blastema cells forced them to contribute to proximal structures after transplantation (Echeverri and Tanaka 2005). Intriguingly, Prod1 loss-of-function resulted in loss of lower arm and digit patterning during development, which appears to conflict with the proximal function observed during regeneration (Kumar et al. 2015).
Almost 20 years later, Yun's group identified a proximal memory gene in axolotls: the evolutionarily conserved but poorly characterized Tarzarotene-induced gene 1 (Tig1) (Oliveira et al. 2021). Like Prod1, axolotl Tig1 encodes a cell membrane protein transcribed in a declining manner from upper arm to hand at steady state. Interestingly, single-cell RNA-sequencing data appears to indicate that only a subset of connective tissue cells transcribes Tig1 in any given arm segment (in a relatively binary “on/off” manner) and that the abundance of this cell population determines the proximal-high, distal-low distribution of Tig1 expression, rather than a smooth gradient of Tig1 expression in all connective tissue cells. However, this interpretation is dependent on the sensitivity of the single-cell RNA-sequencing data. After amputation, connective tissue-derived blastema cells express Tig1 in a proximally enriched gradient (Gerber et al. 2018; Oliveira et al. 2021). Tig1’s proximalizing properties were elucidated similarly to Prod1: it is up-regulated by retinoid treatment and its misexpression interferes with distal patterning. Tig1 misexpression in distal blastema up-regulated Meis1, Prod1, and endogenous Tig1 and, conversely, down-regulated the distal gene HoxA13. Meis transcription factors are expressed proximally during mouse, chick, and axolotl limb bud development, confer proximal cell identity and surface affinities, and are up-regulated transcriptionally by retinoic acid (Capdevila et al. 1999; Mercader et al. 1999, 2000, 2005). Meis proteins are also retinoic acid effectors in axolotl blastema cells, confer proximal properties, and, moreover, can bind Prod1 regulatory elements (Mercader et al. 2005; Shaikh et al. 2011). Thus, Tig1 likely proximalizes cells by targeting an evolutionarily conserved circuitry that additionally incorporates Prod1 in salamanders. The loss-of-function phenotype of Tig1 is not known.
The discoveries of Prod1 and Tig1, the two strongest candidates to encode positional memory in salamanders, were made possible by a rich understanding of the properties of positional memory. We now summarize important salamander limb literature that enabled this understanding in the form of five “lessons from the salamander limb.”
LESSON 1: POSITIONAL MEMORIES ENCODE PATTERNING INFORMATION
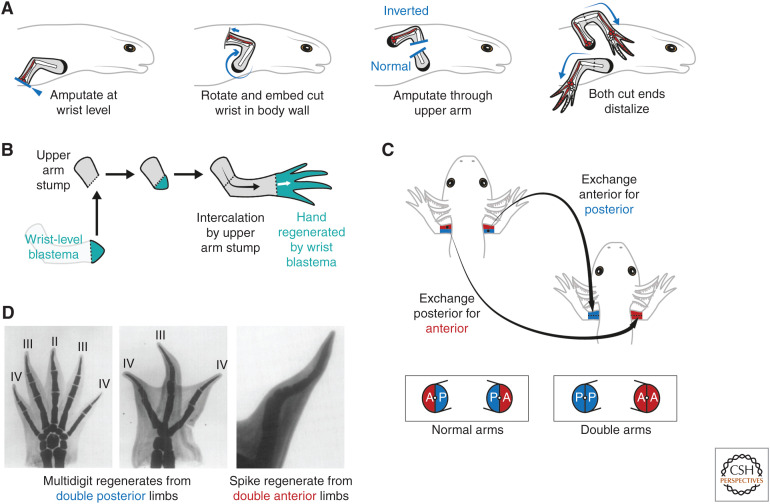
The amputated salamander limb regenerates only missing parts (Fig. 1C). This implied a cellular property that specifies proximodistal position at the amputation plane and directs production of more distal cells. Butler (1955) used circularization surgery to invert axolotl limbs about their proximodistal axis and found that inverted limbs still regenerated distal structures (Fig. 2A). This indicated that salamander limb cells are confined to generating distal, and not proximal, cells during regeneration (“rule of distal transformation”). Salamander cells adhere to this rule even when juxtaposed ectopically. When a wrist-level blastema was grafted to an upper arm stump, the wrist blastema regenerated only the hand, whereas the stump gave rise to the intervening upper arm and lower arm by intercalation, thereby forming a normally patterned limb (Fig. 2B; Iten and Bryant 1975; Stocum 1975; Maden 1980; Pescitelli and Stocum 1980).
Figure 2.
Properties of positional memory in the salamander limb. (A) Butler's circularization surgery to generate distoproximal inverted limbs. The limb is amputated at the wrist level, then the exposed surface embedded into a pocket made in the body wall muscles posterior to the shoulder. After healing and innervation, an amputation through the upper limb releases two arm stumps: one with normal proximodistal polarity (bottom arm) and the other with inverted, distoproximal polarity (top arm). Both arm stumps distalize and regenerate upper arm, lower arm, and hand elements in sequence. This experiment showed that salamander cells are constrained to generating distal, but not proximal, cells during regeneration (rule of distal transformation). The ulna (posterior lower arm bone) and posterior wrist and digit elements are shaded in red to denote the anteroposterior axis. Note that the inverted arm regenerates a mirror image limb compared with the normal arm (compare red shading). (Schematics after Dent 1954.) (B) The rule of distal transformation as shown through an intercalation assay. Transplantation of a distal, wrist-level blastema (turquoise) to a proximal, upper arm stump (gray) results in a normally patterned regenerate. The wrist-level blastema regenerates only wrist and hand, whereas the upper arm stump intercalates the intervening arm elements. (C) Generation of surgical “double” arms in which specific positional memories are deleted. Double anterior arms are generated by removing the posterior half (blue) of a recipient arm and grafting in its place the anterior half (red) from a donor arm. Fusion along the midline generates an arm lacking posterior identity. Similar operations can be used to generate double posterior arms, and also double dorsal or double ventral arms (not shown). Skeletal elements were not transplanted in this assay. (Schematic after Tank 1978.) (D) Double posterior limbs amputated immediately after construction in axolotls regenerated multidigit limbs (left and center). Note that these regenerates are often symmetrical about the midline and can harbor more digits than normal (four-digit) limbs. In contrast, double anterior limbs often failed to regenerate, or regenerated hypomorphic “spike” structures (right). (Images in Panel D from Holder et al. 1980; reprinted, with permission, from Elsevier © 1980.)
Dent noticed that proximodistal inverted arms in Notophthalmus regenerated in mirror-image fashion—what had previously been right arms regenerated left hands (Fig. 2A; Dent 1954). Circularization surgery inverts not only the proximodistal axis but also the anteroposterior axis, a determinant of handedness. Dent realized that mirror image limbs could arise if anteroposterior patterning in the regenerate is determined by fixed anteroposterior properties (i.e., memories) in the limb stump. If anteroposterior and dorsoventral memories indeed exist, removing part of the limb before amputation should alter patterning. Surgically swapping the posterior half of a limb with the anterior half taken from a donor limb creates a chimeric “double anterior” limb lacking posterior tissue (Fig. 2C). After a 30-day healing period, amputation of all combinations of double upper arms (anterior, posterior, dorsal, or ventral) led to failed regeneration or reduced skeletal patterning (Bryant 1976; Bryant and Baca 1978; Tank 1978; Tank and Holder 1978). This indicated that anterior, posterior, dorsal, and ventral cells harbor distinct memories. After removal, memories cannot be compensated by other cells, resulting in loss of pattern during regeneration.
Further experiments in axolotls revealed that double arms amputated immediately after construction regenerated more pattern than those amputated after a healing period (Tank and Holder 1978). When amputated immediately, double posterior limbs regenerated more digits than double anterior limbs, whereas double dorsal and double ventral limbs regenerated similar numbers (Fig. 2D; Tank and Holder 1978; Holder et al. 1980; Burton et al. 1986). This indicated that posterior memory could encode more patterning information than other memories in the axolotl upper arm. Similar conclusions were reached from experiments in which only skin was replaced (Slack 1980b), as well as amputations of “half limbs” (half of the limb is removed, and not replaced) (Wigmore and Holder 1985), “half irradiated limbs” (half of the limb is x-irradiated, permanently blocking cell proliferation) (Maden and Wallace 1976; Maden 1979a,b) and double hindlimbs (Stocum 1978). It is not known how posterior tissue encodes more information: perhaps graded expression of a memory molecule (Slack 1980b), an uneven density of positional values, or simply more cells (Tank and Holder 1979). Intriguingly, anterior and ventral regions regenerated the most pattern in Notophthalmus limbs, suggesting a different organization of positional memory compared to axolotls (Bryant 1976; Bryant and Baca 1978). However, the results of transplantation-based experiments can be challenging to interpret because of variations in the age, size, and species of animals used and differences in grafting precision between investigators.
LESSON 2: CONFRONTATION BETWEEN DISPARATE POSITIONAL MEMORIES CAN PROVOKE PATTERN
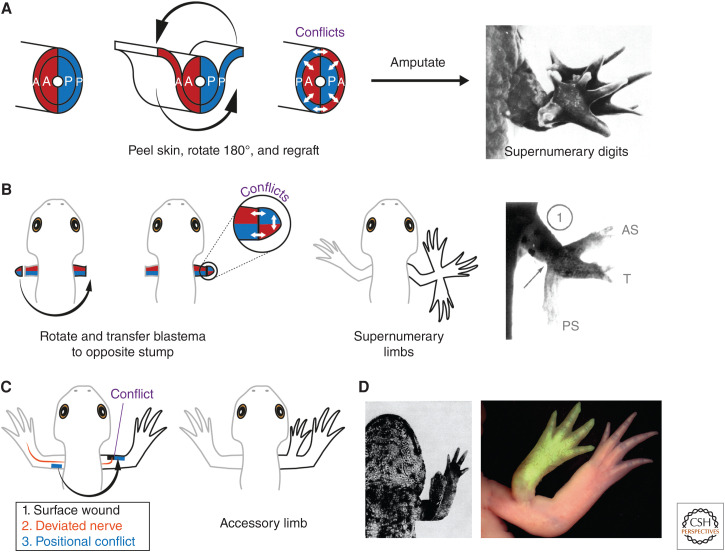
When a skin cuff was removed from the limb, rotated 180°, grafted back into position then amputated, mispatterned limbs bearing up to 13 digits (supernumerary digits) regenerated in the axolotl (Fig. 3A; Carlson 1974). Based on similar results in other salamanders (Glade 1957; Droin 1959; Rahmani 1960; Settles 1967; Lheureux 1972; Carlson 1974), Carlson proposed that skin rotation generates an ectopic interface at which the positional memories of skin cells and internal tissue are brought into conflict. This “morphogenetic conflict” drives pattern formation after amputation (Carlson 1975b).
Figure 3.
Confrontations in positional information give rise to pattern. (A) Skin rotation assay to induce morphogenetic conflict and mispatterning. A skin cuff is removed from the arm, rotated 180° then regrafted in place. Ectopic confrontations are generated between the skin and underlying limb tissue, as shown by the white arrows. Schematic depicts conflicts between anterior tissue (red) and posterior tissue (blue). However, conflicts in the dorsoventral and proximodistal axes are generated similarly. Following amputation, these limbs regenerate with altered pattern, such as too many digits (supernumerary digits; image depicts a limb with 13 digits). (Panel A from Carlson 1974; reprinted, with permission, from Elsevier © 1974.) (B) Generation of supernumerary limbs by blastema transplantation. An arm blastema is removed from the left arm, rotated, and grafted to the amputated stump of the right arm in a manner that the anteroposterior axes are opposed between stump and blastema. This generates morphogenetic conflicts (white arrows) that give rise to supernumerary limbs. The supernumerary limbs have the same handedness as the stump (i.e., right-handed), whereas the “main” regenerate has graft handedness (i.e., left-handed)—see Fig. 5 for details. Image depicts supernumerary limbs generated in Notophthalmus. The main regenerate generated from the transplant (T) is in the center; antero- and posterior supernumerary limbs (AS and PS) have formed at the host–graft interface. (Panel B from Iten and Bryant 1975; reprinted, with permission, from Elsevier © 1975.) (C) The Lheureux/accessory limb model to generate supernumerary limbs in the absence of amputation. The requirements are a surface-level wound (black), a nerve supply (orange), usually obtained by deviating a brachial nerve, and provision of positional conflict (blue). Provisional conflict can be reliably induced by grafting a patch of skin from the opposite side of the limb to the wound site. When all three conditions are satisfied, a patterned limb is generated. Note that accessory limbs usually harbor skeletal elements distal to the elbow, but not the upper arm. Assay according to Lheureux (1977) and Endo et al. (2004), respectively. (D) Examples of accessory limbs generated in Pleurodeles (left, Lheureux 1977) and axolotl (right, Vieira et al. 2020). The accessory limb in the axolotl was labeled with transgenic, GFP-expressing tissue (green). (Panel D, left side, from Lheureux 1977; reprinted, with permission, from Company of Biologists © 1977. Panel D, right side, from Vieira et al. 2020; reprinted, with permission, from S. Karger AG © 2020.)
The most dramatic morphogenetic conflicts generate entire ectopic limbs (supernumerary limbs). Iten and Bryant transplanted a limb blastema to an amputated stump in a manner that their dorsoventral axes coincided, but their anteroposterior axes conflicted. Forty percent of transplants generated supernumerary limbs at the anterior and/or posterior surfaces close to the graft boundary (Fig. 3B; Iten and Bryant 1975). Interestingly, this phenotype arose when late bud blastemas were transplanted, but not early bud blastemas. It has been proposed that positional information in early blastemas is malleable and can be “corrected” by the stump, only later becoming fixed and able to provoke morphogenetic conflict (McCusker and Gardiner 2013; McCusker et al. 2015).
In specific circumstances, positional conflict can generate a supernumerary limb without amputation (Balinsky 1925). Bodemer (1958) generated supernumerary limbs in Notophthalmus by inducing a surface-level wound in the chest/limb region, “extensively traumatizing” the underlying tissue using scissors, then deviating a nerve. Lheureux (1977) realized that tissue trauma could be substituted by grafting a skin patch from the opposite side of the limb to the wound site (i.e., inducing positional confrontation). Thus, positional confrontation in the context of a wound and nerve supply is sufficient to form a limb: formalized as the accessory limb model in axolotls (Fig. 3C,D; Endo et al. 2004). The accessory limb model enables discovery of positional information-encoding molecules. For example, the secreted molecules FGF8 and SHH could substitute for anterior or posterior skin grafts, respectively (Nacu et al. 2016). A decellularized extracellular matrix from posterior skin could induce some skeletal pattern at anterior wounds with a deviated nerve (Phan et al. 2015). However, when these accessory limbs were reamputated, they did not regenerate (Nacu et al. 2016; Vieira et al. 2021), indicating that a heritable diversity of positional identities had not been established. Thus, SHH signaling and extracellular matrix alter positional information in blastema cells, but are not sufficient to induce positional memory.
In contrast to the well-defined patterns arising from anteroposterior and dorsoventral confrontations, confrontation between proximal and distal cells results in less overt outcomes. Grafting distal blastema to a proximal stump results in intercalation and a normally patterned limb (Fig. 2B). Grafting a late-stage upper arm blastema to a wrist stump results in a duplicated upper arm and lower arm (Stocum 1975). In both cases, the transplanted blastema cells obey the rule of distal transformation and behave as they would have without transplantation.
LESSON 3: POSITIONAL INFORMATION CAN BE MANIPULATED
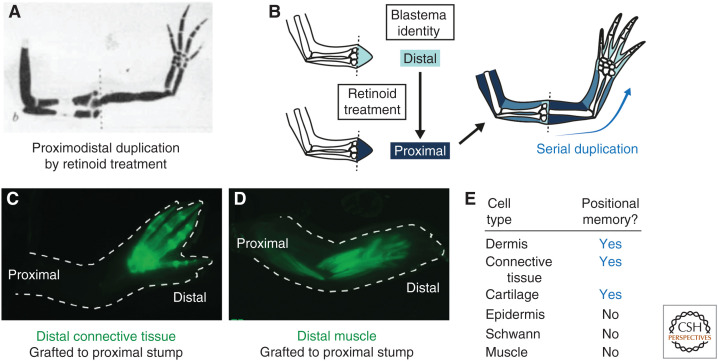
Experiments on tadpoles suggested that vitamin A and its derivatives (retinoids) can alter patterning during limb regeneration (Niazi and Saxena 1978). Maden bathed axolotls in retinoids, which caused limbs to regenerate with proximodistal duplications. The highest doses of retinol palmitate triggered serial duplication of a complete arm from a wrist-level amputation (Fig. 4A; Maden 1982). This indicated that retinoids proximalize positional information. The retinoid-sensitive window was during dedifferentiation stages in axolotls and between early bud and medium bud blastema stages in Pleurodeles (Maden 1983; Niazi et al. 1985). As retinoid sensitivity is transient during regeneration (i.e., treatment before or after the sensitive window does not induce skeletal duplications), retinoids do not appear to alter positional memory in steady-state cells, but probably act on blastema cells (Thoms and Stocum 1984). About 1 ng of retinoic acid per axolotl cell was estimated to respecify it to the most proximal identity (Fig. 4B; Maden et al. 1985). Retinoids also posteriorize and ventralize identity (Thoms and Stocum 1984; Kim and Stocum 1986; Ludolph et al. 1990; McCusker et al. 2014). Retinoids likely activate a proximalizing transcriptional network involving Meis, Prod1, and Tig1 (Mercader et al. 2005; Shaikh et al. 2011; Oliveira et al. 2021). Although positional information is thought to be primarily a hallmark of lateral plate mesoderm derivatives (see following section), a retinoic signaling reporter axolotl revealed activity primarily in the epidermis during limb regeneration (Monaghan and Maden 2012). This reporter comprises eight tandem retinoic acid response elements (RAREs) isolated from upstream of the mouse rarb (retinoic acid receptor β) gene, theoretically acting as a transcriptional readout for retinoic acid signaling. In the future, it will be important to repeat this analysis using cognate axolotl RARE sequences, as well as more direct assays such as those that detect retinoic acid binding to its receptors using fluorescence resonance energy transfer (FRET) (Shimozono et al. 2013).
Figure 4.
Positional information can be altered pharmacologically and is harbored by specific cell types. (A) A regenerated limb from an axolotl that had been bathed in retinol palmitate for 15 days after amputation. A serial duplication of the upper arm, lower arm, and hand has arisen from a wrist-level amputation. (Panel A from Maden 1982; reprinted, with permission, from Springer © 1982.) (B) Schematic depicting the interpretation of the experiment in A. Retinoids proximalize blastema cells, but not positional memory in steady-state cells. Proximalized wrist blastema cells initiate regeneration from the upper arm level, appearing to violate the rule of distal transformation. (C) Connective tissue-derived blastema cells were labeled transgenically with GFP (green). When distal GFP-labeled cells were transplanted to an unlabeled, upper arm stump, the GFP-labeled descendants contributed to hand elements, but not more proximal limb elements. Thus, connective tissue lineages harbor positional memory. (D) Similar experiment as C but performed with GFP-labeled muscle lineages. Distal muscle transplanted to an upper arm stump contributes to all arm segments. Thus, muscle does not harbor positional memory. (Panels C and D from Nacu et al. 2013; reprinted, with permission, from Company of Biologists © 2013.) (E) Positional memory-encoding cell types in the axolotl limb. (Summarized from Carlson 1974, Kragl et al. 2009, and Nacu et al. 2013.)
To date, genetic mutants that alter positional memory have not been described in salamanders. In contrast, a temperature-sensitive allele of the pola2 (DNA polymerase α subunit 2) gene—called pola2mem—was proposed to alter positional memory in zebrafish (Danio rerio) fins (Wang et al. 2019). pola2mem mutant tailfin blastemas fail to grow out from the amputation plane at the restrictive temperature (33°C) as a result of reduced blastema cell proliferation—consistent with the requirement of DNA polymerase α for DNA replication. A proximal amputation induces fourfold more pola2 expression compared with a distal amputation, suggesting a spatial, position-dependent role for this gene. Consistent with an early role for pola2 during regeneration, a 24-h restriction of pola2 activity during blastema formation, followed by recovery at the permissive temperature (26°C), resulted in regenerated tailfins that were 16%–17% reduced in size compared with heterozygous siblings. Strikingly, these smaller tailfins regenerated small fins when amputated a second time despite being grown at the permissive temperature, indicating a heritable change consistent with an altered cellular memory. Thus, heritable information concerning position and size of the regenerate appears to be programmed into zebrafish tailfin blastema cells during early regeneration—and these memories can be altered through pola2 inactivation (Wang et al. 2019). The connection between amputation plane-dependent proliferation and proximodistal cellular identity remains to be defined rigorously in this experimental paradigm. However, an interesting comparison can be made with zebrafish treated with FK506, a pharmacological inhibitor of the phosphatase calcineurin, which similarly regenerated fins with inappropriate size. FK506-treated zebrafish had increased retinoic acid signaling and regenerated larger-than-normal caudal fins after amputation (Kujawski et al. 2014). As in salamanders, increased retinoic acid signaling was accompanied by phenotypes consistent with proximalization, including increased shh expression domain size and a distal shift in distal bone bifurcation in the regenerated fin rays (Kujawski et al. 2014). However, in contrast to pola2mem mutants, the larger fins regenerated after FK506 treatment did not regenerate large fins following a second round of amputation and instead “reverted” to their pretreatment size unless continually treated with FK506 (Daane et al. 2018). This result suggests that FK506 treatment/increased retinoic acid signaling does not heritably proximalize positional memory but instead modifies positional information in blastema cells, similar to the conclusions reached through retinoic acid treatment of the salamander limb (Fig. 4B).
LESSON 4: POSITIONAL MEMORY IS ENCODED IN SPECIFIC CELL TYPES
Carlson's rotation assays (see Lesson 2) revealed that dermis and muscle could provoke morphogenetic conflict, whereas epidermis and bone could not (Carlson 1975a,b). This suggested that only some cell types harbor positional memory. Tanaka's group revisited this question using molecular markers and transgenic axolotls. One assay for positional memory is to test whether a cell obeys the rule of distal transformation in an intercalation setup (Stocum 1975; Maden 1980). When GFP-labeled distal fingertip cartilage was transplanted into the upper arm of an unlabeled host, then amputated, GFP-labeled descendants contributed to the regenerated hand but not to the proximal arm (Kragl et al. 2009). A similar result was obtained for connective tissue-derived blastema cells (Fig. 4C; Nacu et al. 2013). Moreover, both lineages showed nuclear localization of MEIS (proximal marker) after upper arm, but not lower arm, amputation (Kragl et al. 2009; Nacu et al. 2013). Thus, cartilage and connective tissue cells harbor proximodistal memory. Additional experiments showed that these cells also harbor anteroposterior and dorsoventral memories (Lheureux 1972, 1975; Carlson 1974; Gardiner and Bryant 1989; Endo et al. 2004). In contrast, muscle fibers, muscle stem cells, and Schwann cells did not obey the rule of distal transformation (Fig. 4D). Carlson's conclusion that muscle harbors positional memory was likely a result of connective tissue cells residing within muscle bundles. Thus, dermal cells in the skin, internal connective tissue, and cartilage cells harbor positional memory, whereas epidermis, Schwann cells, and muscle cells do not (Fig. 4E). The positional memory-encoding cell types are derivatives of the embryonic lateral plate mesoderm, thus sharing a common developmental source. It is possible that the ability to harbor positional memory becomes restricted to this cellular lineage sometime during the determination of the lateral plate. An important unknown is how memory-encoding and non-memory-encoding cells interact physically and functionally in the blastema.
LESSON 5: THEORETICAL MODELS CONNECT MEMORY TO PATTERNING
Insights from influential patterning models, including the salamander limb, were synthesized into theoretical models aimed at explaining pattern formation during regeneration.
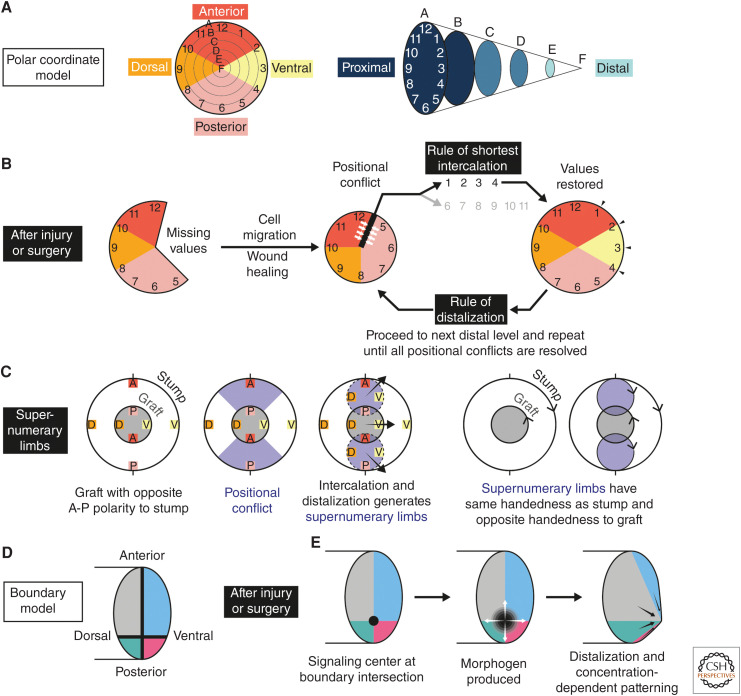
French, Bryant, and Bryant proposed the “polar coordinate model” (see Fig. 5A–C for discussion; French et al. 1976; Bryant et al. 1981). According to this model, when two cells with disparate positional identities become juxtaposed after injury, they respond by proliferating and generating a cell with an intermediate positional value (rule of shortest intercalation). If that intermediate value already exists nearby, the new cell instead distalizes (rule of distalization). Maden (1977) implicated the wound epidermis in distalization. Repeated rounds of intercalation and distalization lead to regeneration.
Figure 5.
The polar coordinate model and the boundary model for pattern formation. (A–C) The polar coordinate model proposed by French, Bryant, and Bryant in 1976, with the modified rule of distalization in 1981. (A, left) Cells are assigned positional values based on their location on the circumference and radius of a circle. As applied to the salamander limb, the circumferential values 1 through 12 correspond to transverse identities (anterior, posterior, dorsal, or ventral), whereas radial values A through F correspond to proximal-to-distal positions. The number of values and their distributions are arbitrary. Although circumferential values imply that positional memory is present only at the surface of the limb, later iterations of this model implicitly assume that corresponding identities are present in the deep tissues. (Right) A three-dimensional depiction of the polar coordinates to emphasize the proximodistal values. (B) The main features of the model are that (1) cells intercalate missing positional values, and (2) it is driven by local cell–cell interactions. After loss of tissue, wound healing and influx of regenerative cells result in normally nonadjacent positional values becoming juxtaposed. This generates positional conflicts (white arrows), which are resolved according to the rule of shortest intercalation. First, cell proliferation is stimulated, then newly generated cells adopt positional values intermediate between the values of the conflicting neighbors. Intercalation of missing values occurs by the shortest arc—in the schematic, the confrontation between 12 and 5 is intercalated via the shorter arc of 1-2-3-4 and not by the longer arc of 6-7-8-9-10-11. If intercalation would result in a value that is already represented by a nearby cell, the value of the new cell is instead distalized (e.g., from A to B) according to the rule of distalization. Intercalation and distalization are repeated until no more intercalation can take place, thus terminating regeneration. (C) The polar coordinate model can explain experimental phenotypes like the supernumerary limbs generated by Iten and Bryant. For simplicity, the circumferential values 1 through 12 have been replaced with A (anterior), P (posterior), D (dorsal), and V (ventral). The schematic represents an “end-on” view of the limb. When a blastema of opposite handedness is grafted to an amputated stump, positional conflicts are stimulated between graft and stump (purple regions). These conflicts are resolved through intercalation and distalization, as in B, resulting in the formation of up to three limbs that grow out in the directions indicated by the black arrows. Importantly, the model correctly assigns the handedness of all three limbs (indicated with arrowheads on the schematic). To determine handedness, compare the relative positions of A, P, D, and V in each outgrowing limb. The supernumerary limbs inherit stump handedness, whereas the main regenerate has graft handedness. (D–E) Salient features of Meinhardt's boundary model (1983). (D) Positional values are distributed in quadrants separating the anteroposterior and dorsoventral axes of the limb. Larger anterodorsal and anteroventral domains abut smaller posterodorsal and posteroventral domains at boundaries (thick black lines). (E) After injury or surgery, a distalizing morphogen is secreted by wound epidermis cells (also referred to as apical ectodermal ridge or cap) and induces proliferation and patterning of underlying blastema cells. This acknowledges the role of the wound epidermis in sustaining blastema outgrowth during normal regeneration. The morphogen-secreting signaling center (black circle) is established at the intersection between the A-P and D-V boundaries. Blastema cells at the intersection secrete a morphogen maintenance factor that promotes secretion of distalizing morphogen from the overlying epidermis. More distal blastema cells secrete higher levels of morphogen maintenance factor, in turn leading to higher concentrations of distalizing morphogen. This positive feedback induces progressive distalization. Like the polar coordinate model, positional confrontations are sensed locally—however, this is coupled to long-range morphogen production. The boundary model can explain supernumerary limb number and handedness, similar to the polar coordinate model, and suggests a molecular mechanism. Some experimental evidence challenges aspects of the boundary model—for example, only one boundary (e.g., anterior vs. posterior) is sufficient in the accessory limb model to generate a patterned limb, in contrast to the two boundaries predicted by the model. However, the notions of signaling centers and secreted morphogens fit well with understanding of limb patterning during embryogenesis. Aspects of the polar coordinate model and boundary model rely on common principles, for example, local sensing of positional conflicts and tight coupling between transverse conflicts and distalization. The models are not mutually exclusive.
The polar coordinate model is driven by local cell–cell interactions. Meinhardt's “boundary model” additionally incorporated secreted morphogens that induce proliferation and patterning over a longer range (see Fig. 5D,E for discussion) (Meinhardt 1983b). The boundary model subdivides the limb into transverse “zones” corresponding to anterior, posterior, dorsal, and ventral identities. Confrontation between zones postinjury induces production of a distalizing morphogen at the boundary. To explain limb handedness, the model requires three or four zones (two boundaries) to be confronted and morphogen to be produced from the intersection point. Distal cells induce higher concentrations of morphogen, leading to progressive distalization and regeneration (Meinhardt 1983a).
Both models couple confrontations in the transverse axes to distalization. Slack's “serial threshold model” instead uncoupled the axes and assigned them different intercalation rates (dorsoventral fastest; proximodistal slowest) (Slack 1980a). This model incorporated additional concepts such as posterior dominance and progressive stabilization of positional identity.
These models and others, account for limb regeneration and successfully handle many experimental phenotypes. Although they do not specify molecular mechanisms, they act as frameworks to understand adult tissue patterning.
GLOBAL PRINCIPLES OF POSITIONAL MEMORY IN VERTEBRATES
We now speculate on wider themes in positional memory by incorporating insights from other tissues and vertebrate models.
How Is Embryonic Positional Information Inherited into Adults?
Whereas this issue has not been addressed explicitly in the salamander limb, it is easy to imagine that keeping embryonic patterning genes “switched on” would enable adult cells to retain positional memory from development. Lmx1b, which determines dorsal identity in limb buds, remains dorsally enriched in adult axolotl limbs, although functional perturbation is lacking (Satoh and Makanae 2014). The axolotl spinal cord, which regenerates after amputation, expresses patterning genes in similar domains to those during embryogenesis, including the transcription factors Pax7 and Msx1 in dorsal cells and the transcription factor FoxA2+ secreted Shh in ventral cells (Schnapp et al. 2005; Sun et al. 2018). Zebrafish fins retain embryo-like expression of transcription factors including alx4a/lhx9 (anterior), hand2 (posterior), and meis1a/dlx5a (proximal) (Nachtrab et al. 2013; Rabinowitz et al. 2017). It would be interesting to determine whether Tig1 is a proximal patterning gene in axolotl limb buds, similar to its expression in adult limb, although Prod1 is not proximally enriched in the Notophthalmus limb bud, thus differing from its adult expression pattern (Kumar et al. 2015). It is important to note that patterning gene expression is not in itself sufficient for regeneration, as adult human spinal cord expresses dorsoventral patterning genes yet regenerates poorly (Ghazale et al. 2019).
How Do Adult Cells Stably Maintain Positional Memory?
Tig1 misexpression in axolotl blastema cells up-regulates proximal genes including itself (Oliveira et al. 2021). If similar self-reinforcing loops exist at steady state, they could maintain positional memory. Mutually repressive transcriptional circuits could prevent cells from acquiring alternative identities. Tig1 and Prod1 are not DNA-binding proteins, but spatially expressed transcription factors (see above) could contribute to such circuitry. Epigenetic regulation is likely to play a role. In adult mouse muscle stem cells, the enhancers of positionally expressed genes, including Hox, show differential DNA methylation (Evano et al. 2020; Yoshioka et al. 2021). DNA methylation, selective chromatin accessibility, and histone marks could stabilize memory gene expression and/or contribute to position-specific activation after injury. An intriguing possibility is that a unique epigenetic signature is responsible for positional memory and that this signature is restricted to regenerative tissues. This could be tested using epigenetic profiling techniques recently adapted to axolotls (Schloissnig et al. 2021; Wei et al. 2021).
Small Differences Could Be Sufficient to Specify Position
It is striking that a ∼1.5–1.7× expression difference could be sufficient for Prod1 and Tig1 to establish proximal versus distal identity in the salamander limb (da Silva et al. 2002; Oliveira et al. 2021). This is particularly interesting when thinking about different regions of the same limb segment (for example, proximal upper arm vs. distal upper arm). Could such subtle expression differences account for intrasegment patterning, or is positional information within a segment encoded differently? Lmx1b, although lacking functional characterization, shows a 2× expression difference between dorsal and ventral axolotl skin (Satoh and Makanae 2014). In contrast, zebrafish hand2 is expressed ∼200× more in posterior pectoral fin compared with anterior (Nachtrab et al. 2013). These quantifications were performed on bulk tissues. In the future, it will be important to purify and measure relevant expression differences in positional memory-encoding cells.
How Is Positional Memory Decoded into Patterning Gene Expression?
Prod1 and Tig1 are proximally enriched at steady state and in the blastema, suggesting that they contribute to both memory and patterning. If this is true, some other difference must be invoked to explain memory “activation” postinjury. The Prod1 ligand nAG is up-regulated after amputation (Kumar et al. 2007b), indicating that interactor availability could “decode” memory. Another possibility is a change in expression level: Lmx1b expression increases following amputation (Satoh and Makanae 2014). How is positional memory transferred from steady-state to blastema cells? A simple explanation is that there is a direct lineage relationship between steady-state and blastema cells expressing the same genes. Information transfer would occur through lineage-intrinsic “handover.” However, this does not explain how Prod1 is expressed in Schwann cells at steady state but in putative connective tissue-derived blastema cells after amputation (Kumar et al. 2007a). An intriguing unknown is whether positional memory genes perform additional functions during homeostasis, or whether they are “poised” for function solely after injury.
Links between Positional Memory and Regeneration Termination?
A fascinating question is how regenerating appendages “know” when to stop growing. Prod1 represents a potential link between positional memory and termination of regeneration. nAG binding to Prod1 increased blastema cell proliferation in vitro (Kumar et al. 2007b; Grassme et al. 2016). As Prod1 is more highly expressed proximally than distally, an interesting possibility is that a gradual reduction in nAG-Prod1-induced proliferation during regeneration connects distal identity to cell-cycle termination. This could be interrogated using recently established cell-cycle-reporting transgenic axolotls (Cura Costa et al. 2021; Duerr et al. 2021).
Body-Wide Compatibility of Positional Codes
An interesting consideration is whether patterning codes around the body are compatible with one another. Forelimb–hindlimb transplantations in salamanders indicate that limbs use similar codes. Diverse zebrafish fins are likely to share codes with one another, as they express similar patterning transcription factors (Nachtrab et al. 2013). But what about more distinct tissues? The accessory limb model reveals conflicts in positional values between salamander cells—indeed, the earliest accessory limbs were induced almost 100 years ago by grafting ear vesicle or nose placode to the lateral body wall of salamander embryos (Balinsky 1925; Glick 1931). Bodemer implanted various organs into a nerve-deviated chest/limb wound in Notophthalmus. Grafts of liver and lung induced accessory limbs, whereas spleen and spinal cord did not (Bodemer 1959). Thus, certain organs can provoke morphogenetic conflict with the limb field. This could be a reflection of similar signaling pathways—for example, Shh or Fgf signaling—being used in certain organs, making some organs better patterning “donors” than others. However, the situation is likely to be more complex as Shh is a patterning molecule in both limb and spinal cord, yet spinal cord implants were not able to provoke accessory limbs. This opens up the exciting possibility that some positional codes can interact, although other combinations are not detected or ignored. In several tadpole species, treating amputated tails with retinoids causes them to regenerate as hindlimbs (Mohanty-Hejmadi et al. 1992; Mahapatra and Hejmadi 1994). Retinoids likely anteriorize the amputated tail field to a hindlimb field by respecifying axial tissue identity, for example through Hox gene expression (Morioka et al. 2018). However, it is tempting to speculate that the precision of this homeotic transformation (from tail to hindlimb) is made possible by an underlying similarity in positional codes.
Are Positional Codes Conserved?
Prod1 encodes proximal limb identity in Notophthalmus and axolotls but does not have homologs outside of salamanders (da Silva et al. 2002; Oliveira et al. 2021). agr2, a homolog of nAG (Prod1 ligand), is ∼1.8× proximally enriched in zebrafish caudal fin (Rabinowitz et al. 2017), but these cells do not obey the rule of distal transformation (Shibata et al. 2018). Thus, the degree of functional conservation between zebrafish and salamanders is unclear. One approach to testing whether positional codes are conserved is to perform interspecies grafts. 1 mm3 implants from the northern leopard frog (Rana pipiens) into Notophthalmus or Ambystoma limbs revealed that tissues including kidney or lung, but not brain or liver, could induce accessory limbs (Ruben 1955; Carlson 1968, 1971). Adult frog kidney induced accessory limbs in salamanders more efficiently than tadpole kidney, perhaps reflecting the acquisition of an “adult” code (Carlson 1968). Frog-to-salamander grafts maintained their capacity to induce supernumerary structures after treatments including lyophilization, indicating that living cells are not necessary, although this morphogenetic activity was abrogated by boiling (Bodemer 1959; Stevens et al. 1965; Carlson and Morgan 1967 and summarized in Carlson 1971). Extracellular matrix from posterior mouse limb induced skeletal elements (albeit with simple patterning) from nerve-deviated anterior wounds in axolotls (Phan et al. 2015). Interestingly, these structures contained bone, whereas “native” accessory limbs typically form cartilage. Thus, some combinations of tissues can collaborate across species to generate a limb-like pattern. As similar results are obtained after intraspecies organ grafts (see above), this could suggest that aspects of adult body coding are similar/conserved between species.
What Are the Implications of Understanding Positional Memory?
The accessory limb model is an exciting demonstration that patterned limb can be created by leveraging positional memory in an engineering setup. Recently, knowledge of positional codes was used to “improve” tail regeneration in the mourning gecko (Lepidodactylus lugubris). Lizards, unlike salamanders, regenerate imperfect tails—they lack roof plate structures, the skeleton encapsulates the spinal cord and is unsegmented. These patterning defects correlate with an adult-specific loss of Pax7-expressing ependymal cells in the dorsal spinal cord (Sun et al. 2018). Transplanting ventralization-resistant Pax7 cells from embryo to adult restored an axolotl-like spinal cord organization (dorsal Pax7 and ventral FoxA2/Shh) in adult lizards. Remarkably, these tails regenerated roof plate structures and the skeletal rod was displaced ventrally, as in axolotls (Lozito et al. 2021). Thus, an engineering approach inspired by axolotl positional coding improved regenerative patterning. Methods to induce adult pattern, guided by rational design principles, have obvious applications in regenerative therapies.
Positional Memory in the Human Body?
Transcriptional profiling indicates that at least some aspects of a positional code are present in adult humans: several cell types express HOX genes in anatomy-specific manners, including fibroblasts (Chang et al. 2002; Rinn et al. 2006), mesenchymal stem/stromal cells (MSCs) (Onizuka et al. 2020), muscle cells, and muscle stem cells (Yoshioka et al. 2021). Interestingly, when Hox-negative mandible MSCs in mice were transplanted to a HoxA11-expressing injury site in the tibia, they initiated HoxA11 expression, differentiated into bone, and repaired the injury (Leucht et al. 2008). In contrast, HoxA11-expressing tibia MSCs transplanted to Hox-negative mandible sustained HoxA11 expression and failed to differentiate or repair tissue. Thus, even though they are not as regeneration-competent as salamanders, mammalian cells might have unique compatibilities during tissue repair based on expression/lack of expression of HOX genes, and/or the specific HOX genes expressed. This would have implications for selecting donor tissues for transplantation-based therapies. Consistent with a link between positional identity and repair in mammals, HoxA11-expressing cells in the developing mouse limb are retained in the adult mouse limb as HoxA11-expressing periosteal cells and bone marrow MSCs, and these cells proliferate after fracture injury to give rise to new skeletal cells (Rux et al. 2016, 2017; Pineault et al. 2019). HoxA11/HoxD11 double deletion in adult mouse limb MSCs compromised differentiation and bone structure (Song et al. 2020), whereas HoxA10 deletion reduced limb muscle stem cell proliferation and regenerative capacity (Yoshioka et al. 2021), indicating roles for these positionally expressed genes in proliferation and differentiation. Many other genes show position-specific expression in adult humans, suggesting that the HOX code is one manifestation of a broader positional memory (Chang et al. 2002; Rinn et al. 2006; Onizuka et al. 2020).
The implications of inducing and manipulating positional memory in human cells could extend to tissue engineering. Huge strides have been made in generating organoids aiming to replicate human tissues like brain or intestine in vitro. If human organs harbor positional codes, it might be necessary to replicate these codes in organoids to achieve true functionality and seamless integration into patient tissues after transplantation. An untested possibility is that organoid tissue is less likely to be rejected if its positional codes are arranged harmoniously with surrounding patient tissue. Conversely, if human organs harbor positional codes, a salient question is why these tissues do not regenerate as well as their counterparts in salamanders. Could parts of the positional code be missing in humans, or is there some epigenetic or genetic block to manifesting the positional code (e.g., distalization) following injury? Could differences in the immune system and the prevalence of scarring in mammals compared with salamanders inhibit the activation or manifestation of the positional memory program in humans? The human liver, which “regenerates” size but not shape after resection, would be an interesting system to delineate the necessity and sufficiency of positional memory molecules for patterning human organs and organoids.
CONCLUDING REMARKS
No model has yielded as many insights into vertebrate positional memory as the salamander limb. The success of a model is dictated by the range and interpretability of phenotypes that can be generated. The salamander limb has a complex yet quantifiable structure, and no shortage of impressive phenotypes. The next step is to connect these phenotypes to genetic and epigenetic mechanisms using recently available technologies. A fascinating take-home is that the adult body is a mosaic of invisible territories with characteristic morphogenetic properties. To show the goal of mapping and interrogating these territories body wide, we have represented them with different colors in our representation of a “patchwork” axolotl (Fig. 6).
Figure 6.
The patchwork axolotl of positional memories. Lewis and Wolpert proposed that the adult body is a mosaic of invisible territories with different morphogenetic properties dictated by developmental history (Lewis and Wolpert 1976). Inspired by their description, we have shown a “patchwork axolotl” in which adult positional memories are represented in different colors. Positional memory is best understood in salamander limbs. To date, only proximodistal memory molecules have been discovered. These are expressed in graded distributions from shoulder to hand (brown gradient, right arm). The left arm represents anteroposterior memory. In the axolotl, posterior memory (purple) encodes more patterning information than anterior memory (green). The hindlimbs represent dorsoventral memory (yellow and blue, respectively). Other tissues in salamanders are also likely to encode memory. The spinal cord (in the tail) expresses dorsoventral patterning genes (purple, yellow, and green stripes; color similarities with limb memory genes do not represent functional interactions). Future work will color in the memory “domains” in other tissues, the functional interactions between these domains, and the degree of evolutionary conservation between species. (Illustration by Moritz Wegscheider.)
Understanding the functions, layout, and compatibility rules of positional memory could have implications for human therapies. The ability to precisely alter positional values and assemble adult cells in in vivo–like configurations could enable the construction of correctly functioning patient-specific organoids compatible with transplantation. Genes encoding positional memory, and molecules that alter these memories, will act as entry points to achieving these goals. An exciting prospect is that these molecules, or similar, could eventually be appropriated to induce or modify positional values directly in patient tissues in vivo, creating a regeneration-stimulating environment for endogenous cells and removing the need for transplantation. Encouraging steps in this direction—for example, the induction of accessory limbs in salamanders using pharmacological agents (McCusker et al. 2014)—make this prospect a realistic possibility.
ACKNOWLEDGMENTS
We thank Moritz Wegscheider for his wonderful illustration of the patchwork axolotl and Yuka Taniguchi-Sugiura for providing the image of the axolotl limb in Figure 1B. L.O. is supported by Human Frontier Science Program (HFSP) fellowship LT000785/2019-L; E.M.T. acknowledges support from ERC Advanced Grant 742046 and Institutional Support from the Vienna Biocenter, Austria.
Footnotes
Editors: Kenneth D. Poss and Donald T. Fox
Additional Perspectives on Regeneration available at www.cshperspectives.org
REFERENCES
- Balinsky BI. 1925. Transplantation des Ohrbläschens bei Triton [Transplantation of the ear vesicle in Triturus]. W Roux’ Archiv f Entwicklungsmechanik 105: 718–731. 10.1007/BF02080664 [DOI] [PubMed] [Google Scholar]
- Bodemer C. 1958. The development of nerve-induced supernumerary limbs in the adult newt, Triturus viridescens. J Morphol 102: 555–581. 10.1002/jmor.1051020304 [DOI] [Google Scholar]
- Bodemer CW. 1959. Observations on the mechanism of induction of supernumerary limbs in adult Triturus viridescens. J Exp Zool 140: 79–99. 10.1002/jez.1401400105 [DOI] [PubMed] [Google Scholar]
- Bryant SV. 1976. Regenerative failure of double half limbs in Notophthalmus viridescens. Nature 263: 676–679. 10.1038/263676a0 [DOI] [PubMed] [Google Scholar]
- Bryant SV, Baca BA. 1978. Regenerative ability of double-half and half upper arms in the newt, Notophthalmus viridescens. J Exp Zool 204: 307–323. 10.1002/jez.1402040302 [DOI] [PubMed] [Google Scholar]
- Bryant SV, French V, Bryant PJ. 1981. Distal regeneration and symmetry. Science 212: 993–1002. 10.1126/science.212.4498.993 [DOI] [PubMed] [Google Scholar]
- Burton R, Holder N, Jesani P. 1986. The regeneration of double dorsal and double ventral limbs in the axolotl. J Embryol Exp Morphol 94: 29–46. [PubMed] [Google Scholar]
- Butler EG. 1955. Regeneration of the urodele forelimb after reversal of its proximo-distal axis. J Morphol 96: 265–281. 10.1002/jmor.1050960204 [DOI] [Google Scholar]
- Capdevila J, Tsukui T, Rodríquez Esteban C, Zappavigna V, Izpisúa Belmonte JC. 1999. Control of vertebrate limb outgrowth by the proximal factor Meis2 and distal antagonism of BMPs by gremlin. Mol Cell 4: 839–849. 10.1016/S1097-2765(00)80393-7 [DOI] [PubMed] [Google Scholar]
- Carlson BM. 1968. Ontogenic development and anatomical distribution of a supernumerary limb-inducing factor. Experientia 24: 1064–1066. 10.1007/BF02138753 [DOI] [PubMed] [Google Scholar]
- Carlson BM. 1971. The distribution of supernumerary limb-inducing capacity in tissues of Rana pipiens. Oncology 25: 365–371. 10.1159/000224586 [DOI] [PubMed] [Google Scholar]
- Carlson BM. 1974. Morphogenetic interactions between rotated skin cuffs and underlying stump tissues in regenerating axolotl forelimbs. Dev Biol 39: 263–285. 10.1016/0012-1606(74)90239-5 [DOI] [PubMed] [Google Scholar]
- Carlson BM. 1975a. Multiple regeneration from axolotl limb stumps bearing cross-transplanted minced muscle regenerates. Dev Biol 45: 203–208. 10.1016/0012-1606(75)90255-9 [DOI] [PubMed] [Google Scholar]
- Carlson BM. 1975b. The effects of rotation and positional change of stump tissues upon morphogenesis of the regenerating axolotl limb. Dev Biol 47: 269–291. 10.1016/0012-1606(75)90282-1 [DOI] [PubMed] [Google Scholar]
- Carlson BM, Morgan CF. 1967. Studies on the mechanism of implant-induced supernumerary limb formation in Urodeles. II: The effect of heat-treatment, lyophilization and homogenization on the inductive capacity of frog kidney. J Exp Zool 164: 243–249. 10.1002/jez.1401640208 [DOI] [PubMed] [Google Scholar]
- Chang HY, Chi JT, Dudoit S, Bondre C, van de Rijn M, Botstein D, Brown PO. 2002. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci 99: 12877–12882. 10.1073/pnas.162488599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen RN, Weinstein M, Tassava RA. 2002. Expression of fibroblast growth factors 4, 8, and 10 in limbs, flanks, and blastemas of Ambystoma. Dev Dyn 223: 193–203. 10.1002/dvdy.10049 [DOI] [PubMed] [Google Scholar]
- Crawford K, Stocum DL. 1988. Retinoic acid coordinately proximalizes regenerate pattern and blastema differential affinity in axolotl limbs. Development 102: 687–698. 10.1242/dev.102.4.687 [DOI] [PubMed] [Google Scholar]
- Cura Costa E, Otsuki L, Albors AR, Tanaka EM, Chara O. 2021. Spatiotemporal control of cell cycle acceleration during axolotl spinal cord regeneration. eLife 10: e55665. 10.7554/eLife.55665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daane JM, Lanni J, Rothenberg I, Seebohm G, Higdon CW, Johnson SL, Harris MP. 2018. Bioelectric-calcineurin signaling module regulates allometric growth and size of the zebrafish fin. Sci Rep 8: 10391–10399. 10.1038/s41598-018-28450-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva SM, Gates PB, Brockes JP. 2002. The newt ortholog of CD59 is implicated in proximodistal identity during amphibian limb regeneration. Dev Cell 3: 547–555. 10.1016/S1534-5807(02)00288-5 [DOI] [PubMed] [Google Scholar]
- Dent JN. 1954. A study of regenerates emanating from limb transplants with reversed proximodistal polarity in the adult newt. Anat Rec 118: 841–856. 10.1002/ar.1091180410 [DOI] [PubMed] [Google Scholar]
- Droin A. 1959. Potentialités morphogènes dans la peau du Triton en régénération [Morphogenetic potentials in the regenerating skin of Triturus]. Rev Suisse Zool 66: 641–709. [Google Scholar]
- Duerr TJ, Jeon EK, Wells KM, Villanueva A, Seifert AW, McCusker CD, Monaghan JR. 2021. A constitutively expressed fluorescence ubiquitin cell cycle indicator (FUCCI) in axolotls for studying tissue regeneration. bioRxiv 10.1101/2021.03.30.437716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri K, Tanaka EM. 2005. Proximodistal patterning during limb regeneration. Dev Biol 279: 391–401. 10.1016/j.ydbio.2004.12.029 [DOI] [PubMed] [Google Scholar]
- Elewa A, Wang H, Talavera-López C, Joven A, Brito G, Kumar A, Hameed LS, Penrad-Mobayed M, Yao Z, Zamani N, et al. 2017. Reading and editing the Pleurodeles waltl genome reveals novel features of tetrapod regeneration. Nat Commun 8: 2286. 10.1038/s41467-017-01964-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Bryant SV, Gardiner DM. 2004. A stepwise model system for limb regeneration. Dev Biol 270: 135–145. 10.1016/j.ydbio.2004.02.016 [DOI] [PubMed] [Google Scholar]
- Evano B, Gill D, Hernando-Herraez I, Comai G, Stubbs TM, Commere P-H, Reik W, Tajbakhsh S. 2020. Transcriptome and epigenome diversity and plasticity of muscle stem cells following transplantation. PLoS Genet 16: e1009022. 10.1371/journal.pgen.1009022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French V, Bryant PJ, Bryant SV. 1976. Pattern regulation in epimorphic fields. Science 193: 969–981. 10.1126/science.948762 [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Bryant SV. 1989. Organization of positional information in the axolotl limb. J Exp Zool 251: 47–55. 10.1002/jez.1402510107 [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Blumberg B, Komine Y, Bryant SV. 1995. Regulation of HoxA expression in developing and regenerating axolotl limbs. Development 121: 1731–1741. 10.1242/dev.121.6.1731 [DOI] [PubMed] [Google Scholar]
- Garza-Garcia A, Harris R, Esposito D, Gates PB, Driscoll PC. 2009. Solution structure and phylogenetics of Prod1, a member of the three-finger protein superfamily implicated in salamander limb regeneration. PLoS ONE 4: e7123. 10.1371/journal.pone.0007123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber T, Murawala P, Knapp D, Masselink W, Schuez M, Hermann S, Gac-Santel M, Nowoshilow S, Kageyama J, Khattak S, et al. 2018. Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362: eaaq0681. 10.1126/science.aaq0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazale H, Ripoll C, Leventoux N, Jacob L, Azar S, Mamaeva D, Glasson Y, Calvo C-F, Thomas JL, Meneceur S, et al. 2019. RNA profiling of the human and mouse spinal cord stem cell niches reveals an embryonic-like regionalization with MSX1+ roof-plate-derived cells. Stem Cell Reports 12: 1159–1177. 10.1016/j.stemcr.2019.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glade RW. 1957. The effects of tail tissue on limb regeneration in Triturus viridescens. J Morphol 101: 477–521. 10.1002/jmor.1051010305 [DOI] [Google Scholar]
- Glick B. 1931. The induction of supernumerary limbs in amblystoma. Anat Rec 48: 407–414. 10.1002/ar.1090480216 [DOI] [Google Scholar]
- Grassme KS, Garza-Garcia A, Delgado JP, Godwin JW, Kumar A, Gates PB, Driscoll PC, Brockes JP. 2016. Mechanism of action of secreted newt anterior gradient protein. PLoS ONE 11: e0154176. 10.1371/journal.pone.0154176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MJ, An JY, Kim WS. 2001. Expression patterns of Fgf-8 during development and limb regeneration of the axolotl. Dev Dyn 220: 40–48. [DOI] [PubMed] [Google Scholar]
- Holder N, Tank PW, Bryant SV. 1980. Regeneration of symmetrical forelimbs in the axolotl, Ambystoma mexicanum. Dev Biol 74: 302–314. 10.1016/0012-1606(80)90432-7 [DOI] [PubMed] [Google Scholar]
- Imokawa Y, Yoshizato K. 1997. Expression of Sonic hedgehog gene in regenerating newt limb blastemas recapitulates that in developing limb buds. Proc Natl Acad Sci 94: 9159–9164. 10.1073/pnas.94.17.9159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iten LE, Bryant SV. 1975. The interaction between the blastema and stump in the establishment of the anterior-posterior and proximal-distal organization of the limb regenerate. Dev Biol 44: 119–147. 10.1016/0012-1606(75)90381-4 [DOI] [PubMed] [Google Scholar]
- Joven A, Elewa A, Simon A. 2019. Model systems for regeneration: salamanders. Development 146. 10.1242/dev.167700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WS, Stocum DL. 1986. Retinoic acid modifies positional memory in the anteroposterior axis of regenerating axolotl limbs. Dev Biol 114: 170–179. 10.1016/0012-1606(86)90393-3 [DOI] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. 2009. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460: 60–65. 10.1038/nature08152 [DOI] [PubMed] [Google Scholar]
- Kujawski S, Lin W, Kitte F, Börmel M, Fuchs S, Arulmozhivarman G, Vogt S, Theil D, Zhang Y, Antos CL. 2014. Calcineurin regulates coordinated outgrowth of zebrafish regenerating fins. Dev Cell 28: 573–587. 10.1016/j.devcel.2014.01.019 [DOI] [PubMed] [Google Scholar]
- Kumar A, Gates PB, Brockes JP. 2007a. Positional identity of adult stem cells in salamander limb regeneration. C R Biol 330: 485–490. 10.1016/j.crvi.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Kumar A, Godwin JW, Gates PB, Garza-Garcia AA, Brockes JP. 2007b. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318: 772–777. 10.1126/science.1147710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Gates PB, Czarkwiani A, Brockes JP. 2015. An orphan gene is necessary for preaxial digit formation during salamander limb development. Nat Commun 6: 8684–8688. 10.1038/ncomms9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucht P, Kim JB, Amasha R, James AW, Girod S, Helms JA. 2008. Embryonic origin and Hox status determine progenitor cell fate during adult bone regeneration. Development 135: 2845–2854. 10.1242/dev.023788 [DOI] [PubMed] [Google Scholar]
- Lewis JH, Wolpert L. 1976. The principle of non-equivalence in development. J Theoret Biol 62: 479–490. 10.1016/0022-5193(76)90132-6 [DOI] [PubMed] [Google Scholar]
- Lheureux E. 1972. Contribution à l'étude du rôle de la peau et des tissus axiaux du membre dans le déclenchement de morphogenèses régénératrices anormales chez le triton Pleurodeles waltlii [Contribution to the study of the role of the skin and axial tissues of the limb in triggering abnormal regenerative morphogenesis in the newt Pleurodeles waltl]. Ann Embryol Morphog 5: 165–172. [Google Scholar]
- Lheureux E. 1975. Régénération des membres irradiés de Pleurodeles waltlii Michah (Urodèle). Influence des qualités et orientations des greffons non irradies [Regeneration of irradiated limbs of Pleurodeles waltl Michah (Urodele). Influence of the qualities and orientations of non-irradiated grafts]. W Roux' Archiv Entwickllungsmechanik 176: 303–327. 10.1007/BF00575323 [DOI] [PubMed] [Google Scholar]
- Lheureux E. 1977. Importance des associations de tissus du membre dans le développement des membres surnuméraires induits par déviation de nerf chez le Triton Pleurodeles waltlii Michah [Importance of limb tissue associations in the development of supernumerary limbs induced by nerve deflection in the newt Pleurodeles waltl Michah]. Development 38: 151–173. 10.1242/dev.38.1.151 [DOI] [PubMed] [Google Scholar]
- Lozito T, Londono R, Sun A, Hudnall M. 2021. Perfecting the imperfect: introducing dorsoventral patterning into adult regenerating lizard tails with gene-edited embryonic neural stem cells. In Review 10.21203/rs.3.rs-142271/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludolph DC, Cameron JA, Stocum DL. 1990. The effect of retinoic acid on positional memory in the dorsoventral axis of regenerating axolotl limbs. Dev Biol 140: 41–52. 10.1016/0012-1606(90)90051-J [DOI] [PubMed] [Google Scholar]
- Maden M. 1977. The regeneration of positional information in the amphibian limb. J Theor Biol 69: 735–753. 10.1016/0022-5193(77)90379-4 [DOI] [PubMed] [Google Scholar]
- Maden M. 1979a. Neurotrophic and x-ray blocks in the blastemal cell cycle. J Embryol Exp Morphol 50: 169–173. [PubMed] [Google Scholar]
- Maden M. 1979b. Regulation and limb regeneration: the effect of partial irradiation. J Embryol Exp Morphol 52: 183–192. [PubMed] [Google Scholar]
- Maden M. 1980. Intercalary regeneration in the amphibian limb and the rule of distal transformation. J Embryol Exp Morphol 56: 201–209. [PubMed] [Google Scholar]
- Maden M. 1982. Vitamin A and pattern formation in the regenerating limb. Nature 295: 672–675. 10.1038/295672a0 [DOI] [PubMed] [Google Scholar]
- Maden M. 1983. The effect of vitamin A on the regenerating axolotl limb. J Embryol Exp Morphol 77: 273–295. [PubMed] [Google Scholar]
- Maden M, Wallace H. 1976. How x-rays inhibit amphibian limb regeneration. J Exp Zool 197: 105–113. 10.1002/jez.1401970112 [DOI] [PubMed] [Google Scholar]
- Maden M, Keeble S, Cox RA. 1985. The characteristics of local application of retinoic acid to the regenerating axolotl limb. Wilhelm Roux Arch Dev Biol 194: 228–235. 10.1007/BF00848251 [DOI] [Google Scholar]
- Mahapatra PK, Hejmadi PM. 1994. Vitamin A-mediated homeotic transformation of tail to limbs, limb suppression and abnormal tail regeneration in the Indian jumping frog Polypedates maculatus. Dev Growth Differ 36: 307–317. 10.1111/j.1440-169X.1994.00307.x [DOI] [PubMed] [Google Scholar]
- McCusker CD, Gardiner DM. 2013. Positional information is reprogrammed in blastema cells of the regenerating limb of the axolotl (Ambystoma mexicanum). PLoS ONE 8: e77064. 10.1371/journal.pone.0077064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker C, Lehrberg J, Gardiner D. 2014. Position-specific induction of ectopic limbs in non-regenerating blastemas on axolotl forelimbs. Regeneration (Oxf) 1: 27–34. 10.1002/reg2.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker CD, Athippozhy A, Diaz-Castillo C, Fowlkes C, Gardiner DM, Voss SR. 2015. Positional plasticity in regenerating Amybstoma mexicanum limbs is associated with cell proliferation and pathways of cellular differentiation. BMC Dev Biol 15: 1–17. 10.1186/s12861-015-0095-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt H. 1983a. A bootstrap model for the proximodistal pattern formation in vertebrate limbs. J Embryol Exp Morphol 76: 139–146. [PubMed] [Google Scholar]
- Meinhardt H. 1983b. A boundary model for pattern formation in vertebrate limbs. J Embryol Exp Morphol 76: 115–137. [PubMed] [Google Scholar]
- Mercader N, Leonardo E, Azpiazu N, Serrano A, Morata G, Martínez-A C, Torres M. 1999. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature 402: 425–429. 10.1038/46580 [DOI] [PubMed] [Google Scholar]
- Mercader N, Leonardo E, Piedra ME, Martínez-A C, Ros MA, Torres M. 2000. Opposing RA and FGF signals control proximodistal vertebrate limb development through regulation of Meis genes. Development 127: 3961–3970. 10.1242/dev.127.18.3961 [DOI] [PubMed] [Google Scholar]
- Mercader N, Tanaka EM, Torres M. 2005. Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development 132: 4131–4142. 10.1242/dev.01976 [DOI] [PubMed] [Google Scholar]
- Mohanty-Hejmadi P, Dutta SK, Mahapatra P. 1992. Limbs generated at site of tail amputation in marbled balloon frog after vitamin A treatment. Nature 355: 352–353. 10.1038/355352a0 [DOI] [PubMed] [Google Scholar]
- Monaghan JR, Maden M. 2012. Visualization of retinoic acid signaling in transgenic axolotls during limb development and regeneration. Dev Biol 368: 63–75. 10.1016/j.ydbio.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morioka S, Hejmadi PM, Yaoita Y, Tazawa I. 2018. Homeotic transformation of tails into limbs in anurans. Dev Growth Differ 60: 365–376. 10.1111/dgd.12547 [DOI] [PubMed] [Google Scholar]
- Muneoka K, Fox WF, Bryant SV. 1986. Cellular contribution from dermis and cartilage to the regenerating limb blastema in axolotls. Dev Biol 116: 256–260. 10.1016/0012-1606(86)90062-X [DOI] [PubMed] [Google Scholar]
- Nachtrab G, Kikuchi K, Tornini VA, Poss KD. 2013. Transcriptional components of anteroposterior positional information during zebrafish fin regeneration. Development 140: 3754–3764. 10.1242/dev.098798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacu E, Glausch M, Le HQ, Damanik FFR, Schuez M, Knapp D, Khattak S, Richter T, Tanaka EM. 2013. Connective tissue cells, but not muscle cells, are involved in establishing the proximo-distal outcome of limb regeneration in the axolotl. Development 140: 513–518. 10.1242/dev.081752 [DOI] [PubMed] [Google Scholar]
- Nacu E, Gromberg E, Oliveira CR, Drechsel D, Tanaka EM. 2016. FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration. Nature 533: 407–410. 10.1038/nature17972 [DOI] [PubMed] [Google Scholar]
- Nardi JB, Stocum DL. 1984. Surface properties of regenerating limb cells: evidence for gradation along the proximodistal axis. Differentiation 25: 27–31. 10.1111/j.1432-0436.1984.tb01334.x [DOI] [Google Scholar]
- Niazi IA, Saxena S. 1978. Abnormal hind limb regeneration in tadpoles of the toad, Bufo andersoni, exposed to excess vitamin A. Folia Biol (Krakow) 26: 3–8. [PubMed] [Google Scholar]
- Niazi IA, Pescitelli MJ, Stocum DL. 1985. Stage-dependent effects of retinoic acid on regenerating urodele limbs. Wilhelm Roux Arch Dev Biol 194: 355–363. 10.1007/BF00877373 [DOI] [Google Scholar]
- Nowoshilow S, Schloissnig S, Fei JF, Dahl A, Pang AWC, Pippel M, Winkler S, Hastie AR, Young G, Roscito JG, et al. 2018. The axolotl genome and the evolution of key tissue formation regulators. Nature 554: 50–55. 10.1038/nature25458 [DOI] [PubMed] [Google Scholar]
- Oliveira CR, Knapp D, Elewa A, Malagon SGG, Gates PB, Petzhold A, Arce H, Cordoba RC, Chara O, Tanaka EM, et al. 2021. Tig1 regulates proximo-distal identity during salamander limb regeneration. bioRxiv 10.1101/2021.02.03.428234. [DOI] [Google Scholar]
- Onizuka S, Yamazaki Y, Park SJ, Sugimoto T, Sone Y, Sjöqvist S, Usui M, Takeda A, Nakai K, Nakashima K, et al. 2020. RNA-sequencing reveals positional memory of multipotent mesenchymal stromal cells from oral and maxillofacial tissue transcriptomes. BMC Genomics 21: 417–413. 10.1186/s12864-020-06825-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescitelli MJ, Stocum DL. 1980. The origin of skeletal structures during intercalary regeneration of larval Ambystoma limbs. Dev Biol 79: 255–275. 10.1016/0012-1606(80)90115-3 [DOI] [PubMed] [Google Scholar]
- Phan AQ, Lee J, Oei M, Flath C, Hwe C, Mariano R, Vu T, Shu C, Dinh A, Simkin J, et al. 2015. Positional information in axolotl and mouse limb extracellular matrix is mediated via heparan sulfate and fibroblast growth factor during limb regeneration in the axolotl (Ambystoma mexicanum). Regeneration (Oxf) 2: 182–201. 10.1002/reg2.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineault KM, Song JY, Kozloff KM, Lucas D, Wellik DM. 2019. Hox11 expressing regional skeletal stem cells are progenitors for osteoblasts, chondrocytes and adipocytes throughout life. Nat Commun 10: 1–15. 10.1038/s41467-019-11100-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JS, Robitaille AM, Wang Y, Ray CA, Thummel R, Gu H, Djukovic D, Raftery D, Berndt JD, Moon RT. 2017. Transcriptomic, proteomic, and metabolomic landscape of positional memory in the caudal fin of zebrafish. Proc Natl Acad Sci 114: E717–E726. 10.1073/pnas.1620755114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani T. 1960. Conflit de potentialités morphogènes et duplicature [Conflicts of morphogenetic potentials and duplication]. Rev Suisse Zool 67: 589–675. 10.5962/bhl.part.75283 [DOI] [Google Scholar]
- Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY. 2006. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet 2: e119. 10.1371/journal.pgen.0020119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben LN. 1955. The effects of implanting anuran cancer into non-regenerating and regenerating larval urodele limbs. J Exp Zool 128: 29–51. 10.1002/jez.1401280104 [DOI] [Google Scholar]
- Rux DR, Song JY, Swinehart IT, Pineault KM, Schlientz AJ, Trulik KG, Goldstein SA, Kozloff KM, Lucas D, Wellik DM. 2016. Regionally restricted Hox function in adult bone marrow multipotent mesenchymal stem/stromal cells. Dev Cell 39: 653–666. 10.1016/j.devcel.2016.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rux DR, Song JY, Pineault KM, Mandair GS, Swinehart IT, Schlientz AJ, Garthus KN, Goldstein SA, Kozloff KM, Wellik DM. 2017. Hox11 function is required for region-specific fracture repair. J Bone Miner Res 32: 1750–1760. 10.1002/jbmr.3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A, Makanae A. 2014. Conservation of position-specific gene expression in axolotl limb skin. Zoolog Sci 31: 6–13. 10.2108/zsj.31.6 [DOI] [PubMed] [Google Scholar]
- Schloissnig S, Kawaguchi A, Nowoshilow S, Falcon F, Otsuki L, Tardivo P, Timoshevskaya N, Keinath MC, Smith JJ, Voss SR, et al. 2021. The giant axolotl genome uncovers the evolution, scaling, and transcriptional control of complex gene loci. Proc Natl Acad Sci 118: e2017176118. 10.1073/pnas.2017176118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp E, Kragl M, Rubin L, Tanaka EM. 2005. Hedgehog signaling controls dorsoventral patterning, blastema cell proliferation and cartilage induction during axolotl tail regeneration. Development 132: 3243–3253. 10.1242/dev.01906 [DOI] [PubMed] [Google Scholar]
- Settles HE. 1967. “Supernumerary regeneration caused by ninety-degree skin rotation in the adult newt, Triturus viridescens.” PhD thesis, Tulane University, New Orleans. [Google Scholar]
- Shaikh N, Gates PB, Brockes JP. 2011. The Meis homeoprotein regulates the axolotl prod 1 promoter during limb regeneration. Gene 484: 69–74. 10.1016/j.gene.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata E, Liu Z, Kawasaki T, Sakai N, Kawakami A. 2018. Robust and local positional information within a fin ray directs fin length during zebrafish regeneration. Dev Growth Differ 60: 354–364. 10.1111/dgd.12558 [DOI] [PubMed] [Google Scholar]
- Shimokawa T, Yasutaka S, Kominami R, Shinohara H. 2013. Lmx-1b and Wnt-7a expression in axolotl limb during development and regeneration. Okajimas Folia Anat Jpn 89: 119–124. 10.2535/ofaj.89.119 [DOI] [PubMed] [Google Scholar]
- Shimozono S, Iimura T, Kitaguchi T, Higashijima SI, Miyawaki A. 2013. Visualization of an endogenous retinoic acid gradient across embryonic development. Nature 496: 363–366. 10.1038/nature12037 [DOI] [PubMed] [Google Scholar]
- Slack JMW. 1980a. A serial threshold theory of regeneration. J Theor Biol 82: 105–140. 10.1016/0022-5193(80)90092-2 [DOI] [PubMed] [Google Scholar]
- Slack MJ. 1980b. Morphogenetic properties of the skin in axolotl limb regeneration. J Embryol Exp Morphol 58: 265–288. [PubMed] [Google Scholar]
- Song JY, Pineault KM, Dones JM, Raines RT, Wellik DM. 2020. Hox genes maintain critical roles in the adult skeleton. Proc Natl Acad Sci 117: 7296–7304. 10.1073/pnas.1920860117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, Ruben LN, Lockwood D, Rose H. 1965. Implant-induced accessory limbs in urodeles: fresh, frozen, and boiled tissues. J Morphol 117: 213–227. 10.1002/jmor.1051170207 [DOI] [PubMed] [Google Scholar]
- Stocum DL. 1975. Regulation after proximal or distal transposition of limb regeneration blastemas and determination of the proximal boundary of the regenerate. Dev Biol 45: 112–136. 10.1016/0012-1606(75)90246-8 [DOI] [PubMed] [Google Scholar]
- Stocum DL. 1978. Regeneration of symmetrical hindlimbs in larval salamanders. Science 200: 790–793. 10.1126/science.644323 [DOI] [PubMed] [Google Scholar]
- Sun AX, Londono R, Hudnall ML, Tuan RS, Lozito TP. 2018. Differences in neural stem cell identity and differentiation capacity drive divergent regenerative outcomes in lizards and salamanders. Proc Natl Acad Sci 115: E8256–E8265. 10.1073/pnas.1803780115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank PW. 1978. The failure of double-half forelimbs to undergo distal transformation following amputation in the axolotl, Ambystoma mexicanum. J Exp Zool 204: 325–336. 10.1002/jez.1402040303 [DOI] [PubMed] [Google Scholar]
- Tank PW, Holder N. 1978. The effect of healing time on the proximodistal organization of double-half forelimb regenerates in the axolotl, Ambystoma mexicanum. Dev Biol 66: 72–85. 10.1016/0012-1606(78)90274-9 [DOI] [PubMed] [Google Scholar]
- Tank PW, Holder N. 1979. The distribution of cells in the upper forelimb of the axolotl, Ambystoma mexicanum. J Exp Zool 209: 435–442. 10.1002/jez.1402090309 [DOI] [Google Scholar]
- Thoms SD, Stocum DL. 1984. Retinoic acid-induced pattern duplication in regenerating urodele limbs. Dev Biol 103: 319–328. 10.1016/0012-1606(84)90320-8 [DOI] [PubMed] [Google Scholar]
- Torok MA, Gardiner DM, Izpisúa Belmonte JC, Bryant SV. 1999. Sonic Hedgehog (shh) expression in developing and regenerating axolotl limbs. J Exp Zool 284: 197–206. [PubMed] [Google Scholar]
- Vieira WA, Wells KM, McCusker CD. 2020. Advancements to the axolotl model for regeneration and aging. Gerontology 66: 212–222. 10.1159/000504294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira WA, Goren S, McCusker CD. 2021. ECM-mediated positional cues are able to induce pattern, but not new positional information, during axolotl limb regeneration. PLoS ONE 16: e0248051. 10.1371/journal.pone.0248051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YT, Tseng TL, Kuo YC, Yu JK, Su YH, Poss KD, Chen CH. 2019. Genetic reprogramming of positional memory in a regenerating appendage. Curr Biol 29: 4193–4207.e4. 10.1016/j.cub.2019.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Li H, Guo Y, Zhao X, Liu Y, Zou X, Zhou L, Yuan Y, Qin Y, Mao C, et al. 2021. An ATAC-seq dataset uncovers the regulatory landscape during axolotl limb regeneration. Front Cell Dev Biol 9: 651145. 10.3389/fcell.2021.651145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigmore P, Holder N. 1985. Regeneration from isolated half limbs in the upper arm of the axolotl. J Embryol Exp Morphol 89: 333–347. [PubMed] [Google Scholar]
- Wolpert L. 1969. Positional information and the spatial pattern of cellular differentiation. J Theor Biol 25: 1–47. 10.1016/S0022-5193(69)80016-0 [DOI] [PubMed] [Google Scholar]
- Yoshioka K, Nagahisa H, Miura F, Araki H, Kamei Y, Kitajima Y, Seko D, Nogami J, Tsuchiya Y, Okazaki N, et al. 2021. Hoxa10 mediates positional memory to govern stem cell function in adult skeletal muscle. Sci Adv 7: eabd7924. 10.1126/sciadv.abd7924 [DOI] [PMC free article] [PubMed] [Google Scholar]