Abstract
Colistin, the last resort for multidrug and extensively drug-resistant bacterial infection treatment, was reintroduced after being avoided in clinical settings from the 1970s to the 1990s because of its high toxicity. Colistin is considered a crucial treatment option for Acinetobacter baumannii and Pseudomonas aeruginosa, which are listed as critical priority pathogens for new antibiotics by the World Health Organization. The resistance mechanisms of colistin are considered to be chromosomally encoded, and no horizontal transfer has been reported. Nevertheless, in November 2015, a transmissible resistance mechanism of colistin, called mobile colistin resistance (MCR), was discovered. Up to ten families with MCR and more than 100 variants of Gram-negative bacteria have been reported worldwide. Even though few have been reported from Acinetobacter spp. and Pseudomonas spp., it is important to closely monitor the epidemiology of mcr genes in these pathogens. Therefore, this review focuses on the most recent update on colistin resistance and the epidemiology of mcr genes among non-fermentative Gram-negative bacilli, especially Acinetobacter spp. and P. aeruginosa.
Keywords: colistin resistance, mcr genes, Acinetobacter, Pseudomonas, polymyxin
1 Introduction
Presently, beta-lactams, cephalosporins, carbapenems, fluoroquinolones, aminoglycosides, and macrolides are frequently used to treat bacterial infections. However, the emergence of drug-resistant microorganisms, particularly Gram-negative pathogens, has become a public health threat. In 2017, the World Health Organization classified carbapenem-resistant (CR) Acinetobacter baumannii and Pseudomonas aeruginosa as priority pathogens in critical need of alternative treatment options (World Health Organization, 2017). P. aeruginosa, A. baumannii, Stenotrophomonas maltophilia, and Burkholderia cepacia complex are non-fermentative Gram-negative bacteria that cause significant problems in healthcare settings. Because these bacteria are highly adaptable and have various intrinsic and acquired resistance mechanisms, they are typically resistant to major classes of antimicrobial agents, leaving only a few therapeutic options (Enoch et al., 2007). Among these, P. aeruginosa and A. baumannii are the most common causes of nosocomial infections (Mancuso et al., 2021). P. aeruginosa is the most common pathogen in the Pseudomonas genus. This bacterium is an opportunistic pathogen that causes skin, wound, and lung infections. Respiratory infections caused by P. aeruginosa are often associated with defective respiratory systems or ventilation, such as in cystic fibrosis (Bassetti et al., 2018). In contrast, A. baumannii is one of the most common causes of nosocomial infections, such as bloodstream infections and pneumonia (Garnacho-Montero and Timsit, 2019). This organism is part of what is known as the Acinetobacter calcoaceticus–baumannii complex, which also includes Acinetobacter pittii, Acinetobacter nosocomialis, and Acinetobacter calcoaceticus (Ramirez et al., 2020).
With the limitations of new drug development, many outdated antibiotics have been reintroduced into the clinical setting despite their high toxicity, including the polymyxin drug group (Hermsen et al., 2003). As there are no other options available, this drug group has become crucial to combat antibiotic resistance (Bialvaei and Samadi Kafil, 2015). Modern therapeutic drug monitoring of colistin is prone to have a lower incidence rate of toxicity when compared to the past.
The polymyxin group contains many drugs, but polymyxin E, also known as colistin, is recognized as the main agent (Rhouma et al., 2016b). Colistin is one of the remaining treatment options for life-threatening infections caused by multidrug and extensively drug-resistant A. baumannii and P. aeruginosa (Bialvaei and Samadi Kafil, 2015). Moreover, colistin resistance mechanisms are quite rare and chromosomally encoded, which makes transfer difficult (Olaitan et al., 2014). Therefore, the resistance rate against colistin in Gram-negative pathogens appears to be lower than that of other antibiotic classes. However, the increasing trend of colistin resistance in Enterobacteriaceae led to the discovery of the transmissible resistance mechanism of colistin in 2015 (Liu et al., 2016). Since then, the resistance rate of last-resort drugs has been closely monitored; the more the antibiotic resistance rate increases, the fewer treatment options are available. Transferable polymyxin resistance has been extensively reported worldwide. To date, at least ten variations in mcr genes have been described and are currently ongoing. This problem is critical, especially for pathogens with limited treatment options, such as A. baumannii and P. aeruginosa. Therefore, this review focuses on colistin drug resistance and its epidemiology among the non-fermentative Gram-negative bacilli, Acinetobacter spp., and P. aeruginosa.
2 The Polymyxins
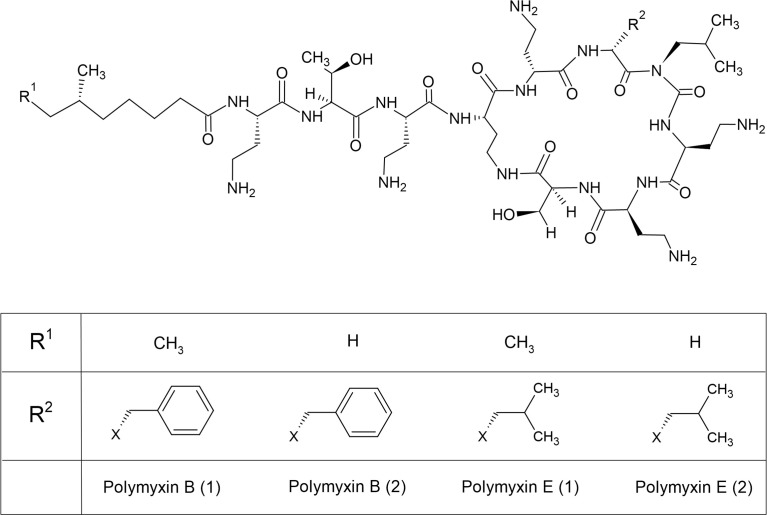
Colistin (polymyxin E) belongs to the polymyxin drug group and appears commercially in two forms as inactive prodrugs: colistin methanesulfonate for parenteral use and colistin sulfate for topical use and use in animal production in some countries (Rhouma et al., 2016b). Another type of polymyxin used in clinical practice is polymyxin B, which is administered in its active form (Tsuji et al., 2019). These antibiotics have been described as old-generation antibiotics, but because of the limitations of antibiotic options, colistin was reintroduced as a last resort for multidrug-resistant (MDR) and extensively drug-resistant (XDR) bacterial infection treatment. Polymyxins were discovered in the 1940s from Bacillus polymyxa, later known as Paenibacillus polymyxa, and were approved by the United States Food and Drug Administration before being used in hospitals in the 1950s (Lim et al., 2010). Polymyxins are polypeptide antibiotic groups that include five different chemical compounds: polymyxins A, B, C, D, and E; however, only polymyxin B and polymyxin E are used in clinical settings (Bialvaei and Samadi Kafil, 2015). Polymyxin B consists of two compounds, polymyxins B1 and B2, whereas colistin contains polymyxins E1 and E2. Colistin differs from polymyxin B in its amino acid composition ( Figure 1 ) (Hermsen et al., 2003; Nation and Li, 2009; Bialvaei and Samadi Kafil, 2015). It has a molecular weight of 1,750 Da and consists of a polycationic cyclic heptapeptide attached to a lipophilic fatty acid side chain (Bialvaei and Samadi Kafil, 2015). The structure of colistin is amphipathic, containing both aqueous and non-aqueous soluble parts (Hermsen et al., 2003).
Figure 1.
Chemical structures of polymyxin B and E.
Colistin has been demonstrated to have a concentration-dependent bactericidal effect, but its mechanism of action is unclear (Nation and Li, 2009; Bialvaei and Samadi Kafil, 2015). The proposed mechanism of action is based on the chemical structure of colistin, which destabilizes lipopolysaccharide (LPS), increases membrane permeability, and leads to bacterial cell leakage (Hermsen et al., 2003; Nation and Li, 2009). The antibiotic spectrum of colistin is narrow, but it is active against many important MDR Gram-negative bacteria, including P. aeruginosa, A. baumannii, Escherichia coli, Klebsiella pneumoniae, Enterobacter spp., and some other bacteria in Enterobacterales (Hermsen et al., 2003; Nation and Li, 2009; Bialvaei and Samadi Kafil, 2015). Colistin is generally not recommended for Gram-positive pathogens because they lack an outer membrane structure.
Its prominent toxicity, including nephrotoxicity and neurotoxicity, is a drawback of colistin use (Nation and Li, 2009). However, toxicity is usually reversible upon discontinuing the medication and is believed to be dose dependent (Li et al., 2006). In the early 2000s, when a resurgence of colistin use occurred, the lack of information on appropriate colistin dosage was the main problem. The International Consensus Guidelines for the Optimal Use of Polymyxins were published in 2020, making colistin safer for use. The recommended PK/PD therapeutic target for efficacy maximization of colistin is a target plasma colistin Css,avg of 2 mg/L, which can provide an area under the plasma concentration–time curve across 24 h at a steady state (AUCss,24 h) of approximately 50 mg h/L (Tsuji et al., 2019). This concentration is considered the maximum tolerable exposure. Higher concentrations can increase the nephrotoxicity incidence and severity (Tsuji et al., 2019). Patients with renal impairment should have their colistin dosage adjusted based on creatinine clearance (Tsuji et al., 2019).
3 The Laboratory Detection of Colistin Resistance
The phenotypic detection of colistin resistance is usually based on antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute (CLSI) guidelines recommend that the broth dilution method be used for colistin because the disc diffusion method is unreliable (Falagas et al., 2010). As a result, the CLSI-EUCAST Working Group recommended a reference method for polymyxin susceptibility testing using broth microdilution without additives (Dafopoulou et al., 2019). The microbe is considered colistin or polymyxin B resistant when the minimum inhibitory concentration (MIC) is equal to or greater than 4 mg/ml in tested organisms, including in Enterobacterales, P. aeruginosa, and Acinetobacter spp. (Clinical and Laboratory Standards Institute, 2022). Colistin resistance in Enterobacterales and Acinetobacter spp. is when the MIC is greater than 2 mg/ml, but resistance in P. aeruginosa is when the MIC is greater than 4 mg/ml, according to the EUCAST breakpoints table (The European Committee on Antimicrobial Susceptibility Testing, 2022). Additionally, sulfate salts of polymyxins must be used instead of colistin methanesulfonate because of their slow breakdown from the inactive prodrug form (Tsuji et al., 2019). Agar dilution is another antimicrobial susceptibility method based on dilution techniques. However, the reliability of the MICs obtained using this method remains inconclusive. Therefore, the CLSI-EUCAST Working Group advised to avoid using the agar dilution method until more data are available (Dafopoulou et al., 2019). Notably, the phenotypic detection method cannot distinguish between colistin resistance mechanisms. To identify the resistance mechanisms, analyses at the genotypic level should be applied subsequent to the antimicrobial susceptibility test. Genotypic detection is based on polymerase chain reaction (PCR) and whole-genome sequencing (WGS) methods. WGS seems to be the most effective strategy for collecting these data because it can identify all targeted antimicrobial resistance genes, including acquired colistin resistance genes (World Health Organization, 2020). The PCR detection method has limitations owing to its selective amplification of only the known sequence. If suspected organisms carry novel mcr genes or mutations, the PCR method alone may not detect that information (World Health Organization, 2021).
Since the presence of mcr genes on transmissible plasmids was reported, the colistin resistance rate has increased significantly, especially in Asia, Africa, and Europe. Therefore, rapid screening methods for mcr-harboring microorganisms are necessary. The only recommended phenotypic detection method is broth dilution, which is laborious compared to the disc diffusion or gradient diffusion methods. Although genetic-based detection methods are the gold standard, they require sophisticated instruments and experienced users. It is also difficult to detect all the responsible colistin resistance genes, especially the acquired genes. Therefore, a more practical method for routine laboratory screening is required (World Health Organization, 2021). Phenotypic detection methods that are still under development have been proposed, such as agar-based screening media (CHROMID® Colistin R agar, Superpolymyxin™, CHROMagar™ COL-APSE), the Rapid Polymyxin NP test, Colispot, and disc prediffusion (Boyen et al., 2010; Nordmann et al., 2016a; Nordmann et al., 2016b; Abdul Momin et al., 2017; Jouy et al., 2017; Garcia-Fernandez et al., 2019).
4 Colistin Resistance Surveillance
Public health awareness of the increasing prevalence of antimicrobial-resistant microorganisms has led to the implementation of antimicrobial stewardship programs worldwide. One strategy is to monitor the resistance of bacteria to slow the spread of resistant microorganisms. Therefore, many surveillance programs have been initiated to monitor antimicrobial resistance in all countries, including the Global Antimicrobial Resistance and Use Surveillance System (GLASS), Central Asian and European Surveillance of Antimicrobial Resistance (CAESAR), Latin American and Caribbean Network for Antimicrobial Resistance Surveillance (ReLAVRA), and the European Antimicrobial Resistance Surveillance Network (EARS-Net). A report of colistin resistance in bloodstream infections from the SENTRY program from 2009 to 2016 showed a resistance rate of less than 1% in P. aeruginosa, 3.1% in A. baumannii, and more than 10% in Enterobacteriaceae (Diekema et al., 2019). In Canada, the CANWARD surveillance study showed that between 2007 and 2016, Enterobacter cloacae, P. aeruginosa, and A. baumannii were the top three microorganisms with the highest colistin resistance rates of approximately 18.1%, 5.0%, and 2.5%, respectively (Zhanel et al., 2019). Similar results were obtained by Bialvaei and Kafil, who also detected a high resistance rate of colistin among Enterobacteriaceae, especially from Enterobacter spp. and K. pneumoniae, in the Asia-Pacific and Latin American regions (Bialvaei and Samadi Kafil, 2015). The abrupt increase in colistin resistance in Asian countries has led to the discovery of mobile colistin resistance (MCR). In Thailand, the National Antimicrobial Resistance Surveillance Center, Thailand (NARST) also monitors colistin resistance in clinically important microorganisms. Fortunately, the resistance rates to colistin in E. coli, K. pneumoniae, P. aeruginosa, and A. baumannii in Thailand in 2019 were less than 5% (National Antimicrobial Resistant Surveillance Center, 2020).
5 The Importance of the Polymyxins in Non-Fermentative Bacteria Treatment
Polymyxins and carbapenems are considered last-resort antibiotics for the treatment of Gram-negative bacteria; however, owing to their misuse, the problem of antibiotic resistance is worsening, particularly in low- to middle-income countries (de Carvalho et al., 2022). A multicenter surveillance study in Taiwan found that the incidence of MDR, XDR, and CR P. aeruginosa infections in hospitalized patients increased from 25.1% to 27.5%, 7.7 to 8.4%, and 19.7% to 27.5%, respectively, between 2016 and 2018 (Jean et al., 2022). In the past decade, hospital-associated P. aeruginosa has showed high MDR/CR numbers in Europe, with prevalence rates of more than 30% (Micek et al., 2015). In 2020, more than half of the countries in Europe showed carbapenem resistance of more than 25% among invasive isolates (European Centre for Disease Prevention and Control, 2022). A meta-analysis found that colistin is the most effective antibiotic for the treatment of Pseudomonas spp. Throughout the study period, colistin was the only antibiotic with a resistance rate of less than 10% (Bonyadi et al., 2022).
In China, Acinetobacter spp. showed a high level of resistance to all carbapenems caused by plasmids carrying various carbapenemase genes (Jean et al., 2022). In a 2022 report from Europe, healthcare-associated isolates of CR-Acinetobacter spp. were >50% in at least 20 countries, especially in southern and eastern Europe (European Centre for Disease Prevention and Control, 2022). Data from many surveillance studies have indicated that carbapenem resistance has been increasing over the last decade, suggesting that carbapenems may not be a suitable standard treatment for MDR, XDR, and CR non-fermentative Gram-negative bacteria. Therefore, polymyxin-based therapy has become the recommended treatment option for CR A. baumannii (CRAB) and XDR P. aeruginosa infections (de Carvalho et al., 2022). In clinical practice, colistin or polymyxin has always been used in combination therapy with at least one additional antibiotic from a different class against CR microorganisms or in patients with risk factors (Bassetti et al., 2018; Doi 2019).
The “Guidelines Recommendations for Evidenced-based Antimicrobial use in Taiwan” (GREAT) working group has launched recommendations and guidelines for the treatment of infections caused by MDR organisms (Sy et al., 2022). In bloodstream infections caused by CRAB, the recommended treatment is colistin 5 mg/kg IV loading dose, followed by IV every 12 h of 2.5 mg × (1.5 × creatine clearance + 30) and/or imipenem/cilastatin 500 mg IV every 6 h or meropenem 2 g IV every 8 h. In pneumonia caused by CRAB, the recommended treatment is colistin 5 mg/kg IV loading dose, then IV every 12 h of 2.5 mg × (1.5 × creatine clearance + 30) and/or imipenem/cilastatin 500 mg IV every 6 h or meropenem 2 g IV every 8 h and adjunctive colistin inhalation 1.25–15 MIU/day in 2–3 divided doses. For any clinical symptoms caused by difficult-to-treat P. aeruginosa, one of the recommended regimens is colistin 5 mg/kg IV loading dose, followed by IV every 12 h at 2.5 mg × (1.5 × creatine clearance + 30) or combination therapy for 5–14 days. Colistin plays a crucial role in MDR microorganism treatment. Therefore, if colistin resistance mechanisms can be transmitted more easily like MCR, it would significantly impact non-fermentative Gram-negative bacterial treatment.
6 Chromosomal Resistance of Colistin
Before the 2000s, reports of resistance to colistin were quite rare, which might have been caused by its low usage over the last 30 years (Nation and Li, 2009). The main mechanism of polymyxin resistance in Gram-negative bacteria is the modification of lipid A, which reduces electrostatic interactions with polymyxins (Cai et al., 2012). Some Gram-negative bacteria, such as Proteus spp. and Burkholderia spp., demonstrated resistance to polymyxins naturally by modifying LPS with 4-amino-4-deoxy-L-arabinose (L-Ara4N) (Olaitan et al., 2014). Chromosomal encoding enzymes (EptA, EptB, and EptC) have been identified in some Gram-negative bacteria, such as Salmonella. EptA, also known as PmrC, is a complex operon. These enzymes, encoded by phosphoethanolamine (pEtN) transferases, can add pEtN to LPS (Zhang et al., 2019; Hamel et al., 2021).
The acquired resistance mechanisms of chromosomally encoded polymyxins are mainly caused by modification of the LPS charge (Olaitan et al., 2014). These resistance mechanisms have been reported in many Gram-negative microorganisms, such as Salmonella enterica, K. pneumoniae, A. baumannii, P. aeruginosa, and E. coli. They are involved in the two-component system genes phoP/phoQ and pmrA/pmrB (Needham and Trent, 2013; Olaitan et al., 2014). PhoQ and PmrB proteins possess tyrosine kinase activity, which phosphorylates the regulator protein (PhoP or PmrA), activates the pmrHFIJKLM operon, and finally modifies the surface of bacteria by adding L-Ara4N or pEtN to lipid A (Ayoub Moubareck, 2020). PhoP/PhoQ is also regulated by the ColR/ColS and CprR/CprS systems. Mutations in these regulatory systems can lead to overexpression of PhoP/PhoQ in P. aeruginosa (Gutu et al., 2013). ParR/ParS is also involved in colistin resistance in P. aeruginosa, with upregulation of the LPS modification operon at sub-inhibitory concentrations of polymyxins (Fernandez et al., 2010). The two-component systems found in P. aeruginosa are PhoP/PhoQ and PmrA/PmrB, but only PmrA/PmrB has been reported in A. baumannii (McPhee et al., 2003; Adams et al., 2009; Beceiro et al., 2011). In addition, in A. baumannii, the insertion of ISAba11 into the biosynthesis genes lpxA, lpxC, and lpxD leads to the complete loss of LPS and colistin resistance (Moffatt et al., 2010; Moffatt et al., 2011).
Additional resistance mechanisms, such as overexpression of efflux pumps, outer membrane remodeling, and lack of LPS formation, have also been reported to be involved in colistin resistance (Olaitan et al., 2014; Ayoub Moubareck, 2020). However, these resistance mechanisms appear to be located on the chromosome. Therefore, the transmission of these mechanisms is difficult, and the horizontal gene transfer of these mechanisms has never been reported (Liu et al., 2016).
7 Transmissible Resistance of Colistin
Although the use of colistin in human clinical settings was reduced in the 1970s and was reintroduced in the late 1990s, colistin is commonly consumed in animal farming to prevent E. coli and Salmonella spp. infections (Kempf et al., 2013; Rhouma et al., 2016a). Prior to the discovery of MCR-1, surveillance of antimicrobial resistance revealed a significant increase in colistin resistance. In 2015, the first mobilized colistin resistance gene, mcr-1, was discovered in E. coli in a Chinese pig farm using a routine antimicrobial resistance surveillance program (Liu et al., 2016). MCR-1 encodes pEtN-lipid A transferase, which can modify the lipid A portion of LPS by the addition of pEtN. MCR-1 also demonstrates transmission and maintenance properties in K. pneumoniae and P. aeruginosa (Liu et al., 2016). Moreover, the microorganisms that harbored MCR-1 showed an increase in the MIC values of colistin. Furthermore, researchers have identified the mcr-1 gene in clinical isolates from inpatients in the same area of a pig farm (Liu et al., 2016). Therefore, awareness of this gene’s transferable properties is of great concern because colistin is currently considered one of the last-resort treatments for XDR microorganisms.
7.1 The Variation of MCR
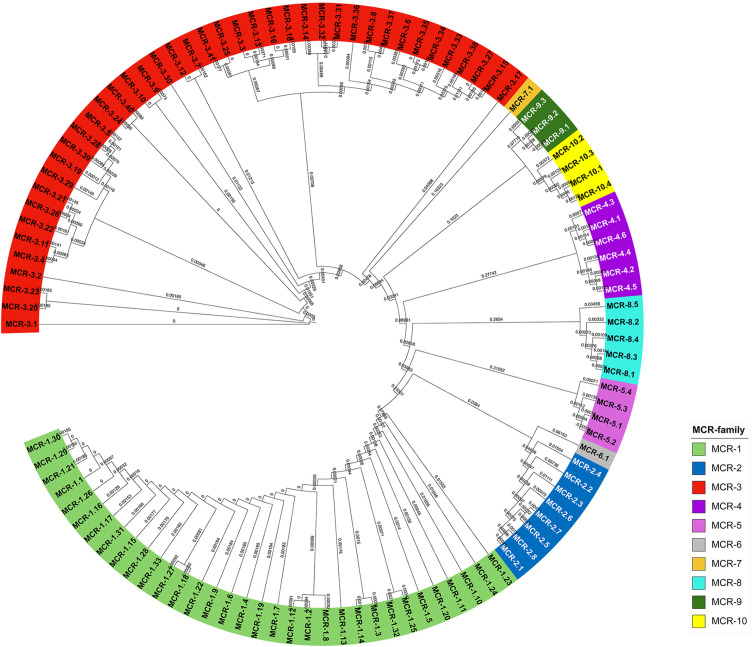
After the MCR-1 discovery, many surveillance programs discovered MCR variation. The nomenclature of mcr genes was proposed in 2018 (Partridge et al., 2018). As of January 2022, ten mcr-gene families with more than 100 variants have been reported in GenBank. The highest number of MCR variants was found in MCR-3 followed by MCR-1, with 42 and 32 variants, respectively (Medicine, 2022). Only MCR-6 and MCR-7 showed one variant each. MCR-5, MCR-8, and MCR-10 all had four variants. The rest of the MCR families, i.e., MCR-2, MCR-4, and MCR-9, have 8, 6, and 3 variants, respectively. The phylogenetic tree of the MCR families is shown in Figure 2 .
Figure 2.
The phylogenetic tree of MCR-gene variants using the Neighbor-Joining method alignment. Multiple sequence alignment was calculated by the clustal omega (Madeira et al., 2019), and the results were illustrated by the Interactive Tree of Life (Letunic and Bork, 2021).
7.2 The Epidemiology of the mcr Gene
The discovery of the mcr gene occurred from a surveillance study in China before it spread around the world (Liu et al., 2016). In 2016, many countries across all continents except Australia reported the discovery of MCR-1 ( Table 1 ). Most MCR-harboring microorganisms belong to the Enterobacterales order, such as E. coli, Salmonella spp., and K. pneumoniae. Apart from Enterobacterales, colistin was also considered a last-resort antibiotic option for non-fermentative Gram-negative bacteria, Acinetobacter spp., and P. aeruginosa. Pseudomonas spp. and Acinetobacter spp. are the most common microorganisms that cause nosocomial infections. They have many intrinsic resistance mechanisms and readily acquire transmissible antibiotic resistance genes, which limit antibiotic treatment options. These organisms belong to ESKAPE (Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacteriaceae), a group of bacteria that are considered an emerging threat in this century (Boucher et al., 2009). Transferable colistin resistance mechanisms in these organisms are a serious problem in the healthcare setting. Therefore, monitoring the mcr gene in non-fermentative Gram-negative bacteria is necessary to combat multidrug resistance.
Table 1.
List of countries that reported the mcr-1 gene in 2016.
| Continent | Country | List of organisms | Source of specimens | Reference |
|---|---|---|---|---|
| Africa | Algeria | E. coli | A, C | (Berrazeg et al., 2016; Olaitan et al., 2016) |
| Egypt | E. coli | A, C | (Elnahriry et al., 2016; Khalifa et al., 2016) | |
| South Africa | E. coli | A, C | (Perreten et al., 2016; Poirel et al., 2016) | |
| Tunisia | E. coli | A | (Grami et al., 2016) | |
| Asia | Bahrain | E. coli | C | (Sonnevend et al., 2016) |
| Cambodia | E. coli | C | (Stoesser et al., 2016) | |
| China |
E. coli
K. pneumoniae E. aerogenes E. cloacae Kluyvera ascorbata S. entirica |
A, C A,C C C E A |
(Li et al., 2016; Liu et al., 2016; Zeng et al., 2016; Zhao and Zong, 2016) | |
| Japan |
E. coli
S. enterica |
A A |
(Kusumoto et al., 2016; Suzuki et al., 2016) | |
| Laos |
E. coli
K. pneumoniae |
A, C C |
(Olaitan et al., 2016) (Rolain et al., 2016) |
|
| Malaysia | E. coli | A, E, C | (Yu et al., 2016) | |
| Pakistan | E. coli | C | (Mohsin et al., 2017) | |
| Singapore |
E. coli
E. aerogenes K. pneumoniae |
C C C |
(Teo et al., 2016a; Teo et al., 2016b) | |
| Saudi Arabia | E. coli | C | (Sonnevend et al., 2016) | |
| South Korea | E. coli | A | (Lim et al., 2016) | |
| Thailand | E. coli | C | (Olaitan et al., 2016) | |
| United Arab Emirates | E. coli | C | (Sonnevend et al., 2016) | |
| Vietnam |
E. coli
Shigella sonnei |
A C |
(Malhotra-Kumar et al., 2016b) (Pham Thanh et al., 2016) |
|
| Australia | – | |||
| Europe | Denmark | E. coli | A, C | (Hasman et al., 2015) |
| Estonia | E. coli | A | (Brauer et al., 2016) | |
| Germany | E. coli | A, C | (Falgenhauer et al., 2016) | |
| France |
Salmonella spp. E. coli K. pneumoniae |
A A C |
(Webb et al., 2016) (Haenni et al., 2016) (Rolain et al., 2016) |
|
| Belgium | E. coli# | A | (Malhotra-Kumar et al., 2016a; Xavier et al., 2016) | |
| Italy |
E. coli
K. pneumoniae* Salmonella spp. |
A, C C A, C |
(Cannatelli et al., 2016; Carnevali et al., 2016; Di Pilato et al., 2016; Zogg et al., 2016) | |
| Lithuania | E. coli | A | (Ruzauskas and Vaskeviciute, 2016) | |
| Netherlands |
E. coli, Salmonella spp. |
A, C A |
(Arcilla et al., 2016; Kluytmans-van den Bergh et al., 2016; Veldman et al., 2016) | |
| Norway | E. coli | C | (Solheim et al., 2016) | |
| Poland | E. coli | C | (Izdebski et al., 2016) | |
| Portugal |
E. coli
Salmonella spp. |
E A, C |
(Campos et al., 2016; Figueiredo et al., 2016; Jones-Dias et al., 2016) | |
| Russia | E. coli | C | (Castanheira et al., 2016) | |
| Spain |
E. coli
Salmonella spp. |
A, C A |
(Prim et al., 2016; Quesada et al., 2016) | |
| Sweden | E. coli | C | (Vading et al., 2016) | |
| Switzerland | E. coli | E, C | (Zurfuh et al., 2016) (Nordmann et al., 2016c) |
|
| United Kingdom |
E. coli
S. enterica |
C A, C |
(Doumith et al., 2016) | |
| North America | Canada | E. coli | A, C | (Mulvey et al., 2016) |
| United States of America | E. coli | A, C | (McGann et al., 2016; Meinersmann et al., 2016) | |
| South America | Argentina | E. coli | A, C | (Liakopoulos et al., 2016; Rapoport et al., 2016) |
| Brazil | E. coli | A | (Fernandes et al., 2016) | |
| Ecuador | E. coli | C | (Ortega-Paredes et al., 2016) | |
| Venezuela | E. coli | A, C | (Delgado-Blas et al., 2016) |
A, animal sources; C, clinical sources; E, environmental sources; *mcr1.2 gene reported; #mcr-1 and mcr-2 gene reported.
7.2.1 The Epidemiology of the mcr Gene in Pseudomonas spp.
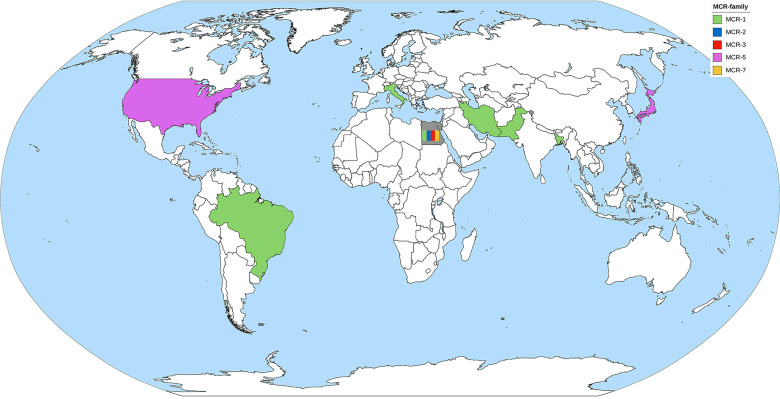
MCR-1 is the major MCR family member found in Pseudomonas spp. ( Table 2 ). Pseudomonas spp. harboring the mcr-gene have been reported by at least one country in all continents, except Australia ( Figure 3 ). P. aeruginosa is a major species of Pseudomonas that harbors the mcr gene. There are also some reports of mcr genes in Pseudomonas putida (Caselli et al., 2018; Ara et al., 2021). PCR is used as the primary detection method for mcr genes in Pseudomonas spp. However, the first report of Pseudomonas spp. carrying the mcr gene by Snesrud et al. used the WGS method combining short-read and long-read sequences (Snesrud et al., 2018). Moreover, they also found that the mcr-5 gene was located within a Tn3-like transposon structure on the chromosome (Snesrud et al., 2018). Considering a health approach, animal and environmental sources may also be reservoirs of mcr genes. Ahmed et al. collected fecal samples from migratory birds in Egypt during the winter season and detected MCR-1 in P. aeruginosa (Ahmed et al., 2019). Some studies have detected mcr genes in cow’s milk, animal meat, and soil (Fujihara et al., 2015; Javed et al., 2020; Tartor et al., 2021). It is noteworthy that the oldest specimen was retrieved from the environment in 1983 but was never recognized until the WGS era (Fujihara et al., 2015). Therefore, dissemination of the mcr gene in the environment via animal hosts is another issue that needs to be considered.
Table 2.
Summary of the mcr gene identified in Pseudomonas spp., specimen description, MCR family, and susceptibility profile.
| Country | Sources of samples [details (if any)] | Detection method | Year of sample collection | Genus species | MCR family | Number of detected samples | Susceptibility profile (μg/ml) (determination method) | Reference |
|---|---|---|---|---|---|---|---|---|
| Bangladesh | Clinical samples (urine) |
PCR | 2017–2018 | P. putida | MCR-1 | 3 | 32–128 (Agar dilution) |
(Ara et al., 2021) |
| Brazil | Clinical sample (urine) |
PCR | 2015–2016 | P. aeruginosa | MCR-1 | 1 | ≥8 (Vitek R 2 Compact) |
(Nitz et al., 2021) |
| Brazil | Animal samples (ear swabs from cat and dog) |
PCR | 2018–2020 | Pseudomonas spp. | MCR-1 | 11 | n/a (Disk diffusion) |
(Martins et al., 2022) |
| Egypt | Clinical samples | PCR | No data | P. aeruginosa | MCR-1 | 8 | 8–256 (Agar dilution) |
(Abd El-Baky et al., 2020) |
| Egypt | Animal samples (bird feces) |
PCR | 2017–2018 | P. aeruginosa | MCR-1 | 6 | n/d | (Ahmed et al., 2019) |
| MCR-2 | 1 | n/d | ||||||
| Egypt | Animal samples (milk from dairy cows) |
PCR | 2018–2020 | P. aeruginosa | MCR-1 | 3 | 32–>128 (Broth microdilution) |
(Tartor et al., 2021) |
| MCR-2 | 1 | 128 (Broth microdilution) |
||||||
| MCR-3 | 3 | 16–64 (Broth microdilution) |
||||||
| MCR-7 | 1 | 128 (Broth microdilution) |
||||||
| Egypt | Clinical sample | PCR | 2019 | P. aeruginosa | MCR-1 | 1 | ≥4 (E-test®) |
(Shabban et al., 2020) |
| Iran | Clinical samples (burn and wound) |
PCR | 2017–2018 | P. aeruginosa | MCR-1 | 3 | >4 (Broth microdilution) |
(Tahmasebi et al., 2020b) |
| Iran | Clinical samples (blood) |
PCR | 2018–2019 | P. aeruginosa | MCR-1 | 10 | ≥4 (E-test®) |
(Tahmasebi et al., 2020a) |
| Italy | Environment sample (hospital surfaces) |
PCR | 2016–2017 | P. aeruginosa | MCR-1 | 1 | 4 (Broth microdilution) |
(Caselli et al., 2018) |
| P. putida | MCR-1 | 1 | 8 (Broth microdilution) |
|||||
| Japan | Environment sample (soil) |
WGS | 1983 | P. aeruginosa | MCR-5 a | 1 | n/a | (Fujihara et al., 2015) |
| Pakistan | Clinical sample (urine) | PCR | 2017–2018 | P. aeruginosa | MCR-1 | 1 | 16 (Broth microdilution) |
(Hameed et al., 2019) |
| Pakistan | Animal sample | PCR | No data (18 months) | P. aeruginosa | MCR-1 | 1 | ≥8 (SensiTest™ Colistin) |
(Javed et al., 2020) |
| Pakistan | Clinical samples (urine, wound) | PCR | No data (6 months) | P. aeruginosa | MCR-1 | 2 | ≥4 (SensiTest™ Colistin) |
(Ejaz et al., 2021) |
| United States of America | Clinical sample (wound) | WGS (short read and long read) | 2012 | P. aeruginosa | MCR-5 | 1 | 4 (Broth microdilution) |
(Snesrud et al., 2018) |
Antimicrobial resistance gene database (NCBI). n/d, not determined; n/a, no data available.
Figure 3.
The worldwide dissemination of the mcr gene in Pseudomonas spp. Countries that reported only one type of mcr gene were colored to represent the mcr gene. The country that reported more than one type of mcr gene was filled with gray background containing color bands of the reported mcr gene.
Because of the low incidence of the mcr gene in Pseudomonas compared to other microorganisms in ESKAPE pathogens, this raises the question of the fitness barrier or transferability properties of the mcr gene among Pseudomonas. The transmissibility of mcr genes in P. aeruginosa was demonstrated by Tartor et al. via conjugation with E. coli J53 (Tartor et al., 2021). Four mcr genes, mcr-1, mcr-2, mcr-3, and mcr-7, were able to transfer into the recipient bacteria and increased the MIC of the recipient cells up to 64 μg/ml (Tartor et al., 2021). Cervoni et al. also demonstrated that MCR-1 increases colistin resistance in recipient cells. Moreover, the expression of MCR-1 in Pseudomonas does not affect bacterial growth or cell envelope homeostasis (Cervoni et al., 2021).
7.2.2 The Epidemiology of the mcr Gene in Acinetobacter spp.
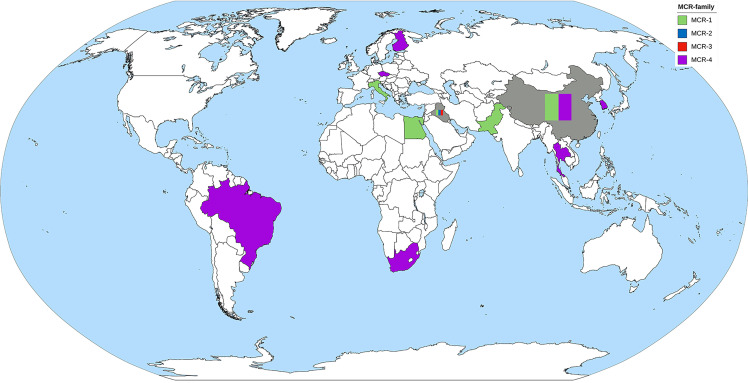
MCR-1 and MCR-4 are the major MCR families reported in Acinetobacter spp. ( Table 3 ). Other mcr genes found in A. baumannii include mcr-2 and mcr-3 (Al-Kadmy et al., 2020). Reports of MCR harboring Acinetobacter spp. have been obtained from all continents, except North America and Australia ( Figure 4 ). The oldest Acinetobacter specimen in which mcr genes were identified was from a stored clinical sample retained from a patient in Brazil in 2008, indicating that the mcr gene circulated for quite a period prior to its discovery by Liu et al. in 2015 (Martins-Sorenson et al., 2020). PCR has been used in Acinetobacter spp. for mcr gene detection, but short- and long-read WGS has been applied in Acinetobacter spp. genome studies and many mobile genetic elements involved in gene transfer have been identified ( Table 4 ). While A. baumannii has been shown to harbor a plasmid carrying the mcr gene, plasmids in A. nosocomialis have also been reported (Cha et al., 2021; Kalova et al., 2021; Snyman et al., 2021). The mcr-4.3 gene in Acinetobacter spp. was found on a plasmid surrounding the transposon Tn3-family and/or insertion sequence. These mobile genetic elements are important for the transfer of many antibiotic resistance genes. Interestingly, some of these plasmids were unable to conjugate and/or were transferable between bacterial species.
Table 3.
Summary of the mcr gene identified in Acinetobacter spp., specimen description, MCR family, and susceptibility profile.
| Country | Sources of samples [details if any] | Detection method | Year of sample collection | Genus species | MCR family | Number of detected samples | Susceptibility profile (μg/ml) (determination method) | Reference |
|---|---|---|---|---|---|---|---|---|
| Brazil | Clinical sample (cerebrospinal fluid) | WGS | 2008 | A. baumannii | MCR-4.3 | 1 | 64 (Broth dilution) |
(Martins-Sorenson et al., 2020) |
| China | Clinical sample | PCR | 2018 | A. baumannii | MCR-1.1 | 1 | 8 (Broth microdilution) |
(Fan et al., 2020) |
| China | Animal sample (pig feces) |
WGS | 2018 | A. baumannii | MCR-4.3 | 1 | 8 (Broth dilution) |
(Hameed et al., 2019) |
| China | Animal sample (pig lung) |
WGS | 2018 a | A. pittii | MCR-1 b | 1 | n/a | (Yang and Zhang, 2018) |
| Czech Republic | Animals sample (imported aquaculture products) |
WGS | 2019 | A. baumannii | MCR-4.3 | 1 | >16 (Broth microdilution) |
(Kalova et al., 2021) |
| A. nosocomialis | MCR-4.3 | 1 | >16 (Broth microdilution) |
|||||
| Czech Republic | Animal sample (imported raw turkey liver) |
WGS | 2017 | A. baumannii | MCR-4.3 | 1 | 16 (Broth microdilution) |
(Bitar et al., 2019) |
| Clinical sample (tracheal) | WGS | 2017 | A. baumannii | MCR-4.3 | 1 | 16 (Broth microdilution) |
||
| Egypt | Clinical samples | PCR | 2019 | A. baumannii | MCR-1 | 2 | ≥4 (E-test®) |
(Shabban et al., 2020) |
| Finland | Environment sample (paper pulp mill) |
WGS | 2020 a | A. baumannii | MCR-4 b | 1 | n/a | (Hamidian et al., 2020) |
| Iraq | Clinical samples | PCR | 2014–2018 | A. baumannii | MCR-1 | 22 | ≥4 (Broth microdilution) |
(Kareem, 2020) |
| Iraq | Clinical and environmental samples | PCR | 2016–2018 | A. baumannii | MCR-1 | 89 | >2 (Broth microdilution) |
(Al-Kadmy et al., 2020) |
| MCR-2 | 78 | |||||||
| MCR-3 | 82 | |||||||
| Italy | Environment samples (hospital surfaces) | PCR | 2016–2017 | A. lwoffii | MCR-1 | 4 | 4-8 (Broth microdilution) |
(Caselli et al., 2018) |
| Pakistan | Clinical sample (blood) | PCR | 2017–2018 | A. baumannii | MCR-1 | 1 | 16 (Agar dilution and broth microdilution) |
(Hameed et al., 2019) |
| Pakistan | Clinical samples (pus, wound, tracheal) |
PCR | No data (6 months) | A. baumannii | MCR-1 | 3 | ≥4 (SensiTest™ colistin) |
(Ejaz et al., 2021) |
| Republic of Korea | Animal sample (imported pork) |
WGS | 2019 | A. nosocomialis | MCR-4.3 | 1 | 16 (Broth microdilution) |
(Cha et al., 2021) |
| South Africa | Clinical sample | PCR, WGS | 2017 | A. nosocomialis | MCR-4.3 | 1 | 16 (Broth microdilution and SensiTest™ Colistin) |
(Snyman et al., 2021) |
| Thailand | Clinical sample | WGS | 2010 | A. nosocomialis | MCR-4.3 b | 1 | n/a | (Kamolvit et al., 2014) |
Year of genome assembly.
Antimicrobial resistance gene database (NCBI); n/a, no data available.
Figure 4.
The worldwide dissemination of the mcr gene in Acinetobacter spp. Countries that reported only one type of mcr gene were colored to represent the mcr gene. Countries that reported more than one type of mcr gene were filled with gray background containing color bands of the reported mcr gene.
Table 4.
Location of the mcr gene in Acinetobacter spp. and surrounding mobile genetic elements.
| Bacteria strain | MCR family | Type of WGS | Gene location | Mobile genetic elements surrounding MCR gene | Year of sample collection | Reference |
|---|---|---|---|---|---|---|
| A. baumannii 597A | MCR-4.3 | Short and long read | pAb-MCR4.3 | -Tn3-family transposon -Insertion sequence ISAba19 |
2008 | (Martins-Sorenson et al., 2020) |
|
A. baumannii
LEV1449/17Ec |
MCR-4.3 | Short and long read | pEC_mcr4.3 (nonconjugative and nontransformable plasmid) | Insertion sequence ISAba19 | 2017 | (Bitar et al., 2019) |
|
A. baumannii
39741 |
MCR-4.3 | Short and long read | pEH _mcr4.3 (nonconjugative and nontransformable plasmid) | Insertion sequence ISAba19 | 2017 | (Bitar et al., 2019) |
| A. nosocomialis CAC13 | MCR-4.3 | Short and long read | Plasmid pCAC13a | -IS3 family transposase (ISAba19) -Tn3 family transposase (ISPsy42) |
2017 | (Snyman et al., 2021) |
| A. baumannii AB18PR065 | MCR-4.3 | Short read | pAB18PR065 (nonconjugative plasmid) |
Tn3 element | 2018 | (Hameed et al., 2019) |
| A. baumannii CT263 | MCR-4.3 | Short and long read | Untypeable plasmid | Tn3 family transpose ISPsy42 | 2019 | (Kalova et al., 2021) |
| A. nosocomialis KUFSE-ACN036 | MCR-4.3 | Short read | Unidentified location due to the limitation of the short-read WGS technique | -Insertion sequence ISAba19
-Transposase |
2019 | (Kalova et al., 2021) |
| A. nosocomialis CT237 | MCR-4.3 | Short and long read | Untypeable plasmid | Tn3 family transpose ISPsy42 | 2019 | (Cha et al., 2021) |
8 Concluding Remarks
Colistin is recognized as a highly toxic old-generation antimicrobial agent. Owing to the shortage of antibiotic options in fighting against MDR Gram-negative bacteria, this drug was reintroduced into the clinical setting and has been recognized as one of the last-resort drugs. Non-fermentative Gram-negative bacteria such as A. baumannii and P. aeruginosa are already recognized as major threats in this century; when combined with resistance to last-resort antibiotics, the severity of the situation is critical. The resistance mechanism against colistin is considered to be chromosomally encoded and difficult to transfer. MCR was discovered in 2015, and several investigations have been subsequently published. To date, ten families of the mcr gene with more than 100 variants have been registered. More efforts are now being made to address this issue, and rapid detection with a high-sensitivity method is essential to track the resistance pattern. A sophisticated approach, such as WGS, is also needed to enhance the knowledge of resistance mechanisms. While waiting for the discovery of a rapid detection technique and more information, a strategy to control the resistant pathogens should be implemented along with the antibiotic stewardship program, which is recognized as a good practice in hospital settings.
Author Contributions
PK wrote the manuscript and conceived the figures. MC and KT reviewed the manuscript draft. All authors contributed to the article and approved the submitted version.
Funding
This work is financially supported by MU-Talents program and Specific League Funds from Mahidol University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors wish to thank the staff from the Department of Microbiology, Faculty of Pharmacy, Mahidol University for suggestions on this work. Their support and collaboration are gratefully acknowledged.
References
- Abd El-Baky R. M., Masoud S. M., Mohamed D. S., Waly N. G., Shafik E. A., Mohareb D. A., et al. (2020). Prevalence and Some Possible Mechanisms of Colistin Resistance Among Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas Aeruginosa . Infect. Drug Resist. 13, 323–332. doi: 10.2147/IDR.S238811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul Momin M. H. F., Bean D. C., Hendriksen R. S., Haenni M., Phee L. M., Wareham D. W. (2017). CHROMagar COL-APSE: A Selective Bacterial Culture Medium for the Isolation and Differentiation of Colistin-Resistant Gram-Negative Pathogens. J. Med. Microbiol. 66 (11), 1554–1561. doi: 10.1099/jmm.0.000602 [DOI] [PubMed] [Google Scholar]
- Adams M. D., Nickel G. C., Bajaksouzian S., Lavender H., Murthy A. R., Jacobs M. R., et al. (2009). Resistance to Colistin in Acinetobacter Baumannii Associated With Mutations in the PmrAB Two-Component System. Antimicrob. Agents Chemother. 53 (9), 3628–3634. doi: 10.1128/AAC.00284-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Z. S., Elshafiee E. A., Khalefa H. S., Kadry M., Hamza D. A. (2019). Evidence of Colistin Resistance Genes (Mcr-1 and Mcr-2) in Wild Birds and its Public Health Implication in Egypt. Antimicrob. Resist. Infect. Control 8, 197. doi: 10.1186/s13756-019-0657-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Kadmy I. M. S., Ibrahim S. A., Al-Saryi N., Aziz S. N., Besinis A., Hetta H. F. (2020). Prevalence of Genes Involved in Colistin Resistance in Acinetobacter Baumannii: First Report From Iraq. Microb. Drug Resist. 26 (6), 616–622. doi: 10.1089/mdr.2019.0243 [DOI] [PubMed] [Google Scholar]
- Ara B., Urmi U. L., Haque T. A., Nahar S., Rumnaz A., Ali T., et al. (2021). Detection of Mobile Colistin-Resistance Gene Variants (Mcr-1 and Mcr-2) in Urinary Tract Pathogens in Bangladesh: The Last Resort of Infectious Disease Management Colistin Efficacy is Under Threat. Expert Rev. Clin. Pharmacol. 14 (4), 513–522. doi: 10.1080/17512433.2021.1901577 [DOI] [PubMed] [Google Scholar]
- Arcilla M. S., van Hattem J. M., Matamoros S., Melles D. C., Penders J., de Jong M. D., et al. (2016). Dissemination of the Mcr-1 Colistin Resistance Gene. Lancet Infect. Dis. 16 (2), 147–149. doi: 10.1016/S1473-3099(15)00541-1 [DOI] [PubMed] [Google Scholar]
- Ayoub Moubareck C. (2020). Polymyxins and Bacterial Membranes: A Review of Antibacterial Activity and Mechanisms of Resistance. Membranes (Basel) 10 (8), 181. doi: 10.3390/membranes10080181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassetti M., Vena A., Croxatto A., Righi E., Guery B. (2018). How to Manage Pseudomonas Aeruginosa Infections. Drugs Context 7, 212527. doi: 10.7573/dic.212527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beceiro A., Llobet E., Aranda J., Bengoechea J. A., Doumith M., Hornsey M., et al. (2011). Phosphoethanolamine Modification of Lipid A in Colistin-Resistant Variants of Acinetobacter Baumannii Mediated by the pmrAB Two-Component Regulatory System. Antimicrob. Agents Chemother. 55 (7), 3370–3379. doi: 10.1128/AAC.00079-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrazeg M., Hadjadj L., Ayad A., Drissi M., Rolain J. M. (2016). First Detected Human Case in Algeria of Mcr-1 Plasmid-Mediated Colistin Resistance in a 2011 Escherichia Coli Isolate. Antimicrob. Agents Chemother. 60 (11), 6996–6997. doi: 10.1128/AAC.01117-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialvaei A. Z., Samadi Kafil H. (2015). Colistin, Mechanisms and Prevalence of Resistance. Curr. Med. Res. Opin. 31 (4), 707–721. doi: 10.1185/03007995.2015.1018989 [DOI] [PubMed] [Google Scholar]
- Bitar I., Medvecky M., Gelbicova T., Jakubu V., Hrabak J., Zemlickova H., et al. (2019). Complete Nucleotide Sequences of Mcr-4.3-Carrying Plasmids in Acinetobacter Baumannii Sequence Type 345 of Human and Food Origin From the Czech Republic, the First Case in Europe. Antimicrob. Agents Chemother. 63 (10), e01166-19. doi: 10.1128/AAC.01166-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonyadi P., Saleh N. T., Dehghani M., Yamini M., Amini K. (2022). Prevalence of Antibiotic Resistance of Pseudomonas Aeruginosa in Cystic Fibrosis Infection: A Systematic Review and Meta-Analysis. Microb. Pathog. 165, 105461. doi: 10.1016/j.micpath.2022.105461 [DOI] [PubMed] [Google Scholar]
- Boucher H. W., Talbot G. H., Bradley J. S., Edwards J. E., Gilbert D., Rice L. B., et al. (2009). Bad Bugs, No Drugs: No ESKAPE! An Update From the Infectious Diseases Society of America. Clin. Infect. Dis. 48 (1), 1–12. doi: 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- Boyen F., Vangroenweghe F., Butaye P., De Graef E., Castryck F., Heylen P., et al. (2010). Disk Prediffusion is a Reliable Method for Testing Colistin Susceptibility in Porcine E. Coli Strains. Vet. Microbiol. 144 (3-4), 359–362. doi: 10.1016/j.vetmic.2010.01.010 [DOI] [PubMed] [Google Scholar]
- Brauer A., Telling K., Laht M., Kalmus P., Lutsar I., Remm M., et al. (2016). Plasmid With Colistin Resistance Gene Mcr-1 in Extended-Spectrum-Beta-Lactamase-Producing Escherichia Coli Strains Isolated From Pig Slurry in Estonia. Antimicrob. Agents Chemother. 60 (11), 6933–6936. doi: 10.1128/AAC.00443-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Chai D., Wang R., Liang B., Bai N. (2012). Colistin Resistance of Acinetobacter Baumannii: Clinical Reports, Mechanisms and Antimicrobial Strategies. J. Antimicrob. Chemother. 67 (7), 1607–1615. doi: 10.1093/jac/dks084 [DOI] [PubMed] [Google Scholar]
- Campos J., Cristino L., Peixe L., Antunes P. (2016). MCR-1 in Multidrug-Resistant and Copper-Tolerant Clinically Relevant Salmonella 1,4,[5],12:I:- and S. Rissen Clones in Portuga to 2015. Euro Surveill 21 (26), pii=30270. doi: 10.2807/1560-7917.ES.2016.21.26.30270 [DOI] [PubMed] [Google Scholar]
- Cannatelli A., Giani T., Antonelli A., Principe L., Luzzaro F., Rossolini G. M. (2016). First Detection of the Mcr-1 Colistin Resistance Gene in Escherichia Coli in Italy. Antimicrob. Agents Chemother. 60 (5), 3257–3258. doi: 10.1128/AAC.00246-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevali C., Morganti M., Scaltriti E., Bolzoni L., Pongolini S., Casadei G. (2016). Occurrence of Mcr-1 in Colistin-Resistant Salmonella Enterica Isolates Recovered From Humans and Animals in Italy 2012 to 2015. Antimicrob. Agents Chemother. 60 (12), 7532–7534. doi: 10.1128/AAC.01803-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli E., D’Accolti M., Soffritti I., Piffanelli M., Mazzacane S. (2018). Spread of Mcr-1-Driven Colistin Resistance on Hospital Surfaces, Italy. Emerg. Infect. Dis. 24 (9), 1752–1753. doi: 10.3201/eid2409.171386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanheira M., Griffin M. A., Deshpande L. M., Mendes R. E., Jones R. N., Flamm R. K. (2016). Detection of Mcr-1 Among Escherichia Coli Clinical Isolates Collected Worldwide as Part of the SENTRY Antimicrobial Surveillance Program in 2014 and 2015. Antimicrob. Agents Chemother. 60 (9), 5623–5624. doi: 10.1128/AAC.01267-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervoni M., Lo Sciuto A., Bianchini C., Mancone C., Imperi F. (2021). Exogenous and Endogenous Phosphoethanolamine Transferases Differently Affect Colistin Resistance and Fitness in Pseudomonas Aeruginosa . Front. Microbiol. 12. doi: 10.3389/fmicb.2021.778968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha M. H., Kim S. H., Kim S., Lee W., Kwak H. S., Chi Y. M., et al. (2021). Antimicrobial Resistance Profile of Acinetobacter Spp. Isolates From Retail Meat Samples Under Campylobacter-Selective Conditions. J. Microbiol. Biotechnol. 31 (5), 733–739. doi: 10.4014/jmb.2102.02027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2022). Performance Standards for Antimicrobial Susceptibility Testing, 32nd Ed - CLSI supplement M100. USA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Dafopoulou K., Vourli S., Tsakris A., Pournaras S. (2019). An Update on Polymyxin Susceptibility Testing Methods for Acinetobacter Baumannii . Expert Rev. Anti Infect. Ther. 17 (9), 699–713. doi: 10.1080/14787210.2019.1667230 [DOI] [PubMed] [Google Scholar]
- de Carvalho F. R. T., Telles J. P., Tuon F. F. B., Rabello Filho R., Caruso P., Correa T. D. (2022). Antimicrobial Stewardship Programs: A Review of Strategies to Avoid Polymyxins and Carbapenems Misuse in Low Middle-Income Countries. Antibiotics (Basel) 11 (3), 378. doi: 10.3390/antibiotics11030378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Blas J. F., Ovejero C. M., Abadia-Patino L., Gonzalez-Zorn B. (2016). Coexistence of Mcr-1 and blaNDM-1 in Escherichia Coli From Venezuela. Antimicrob. Agents Chemother. 60 (10), 6356–6358. doi: 10.1128/AAC.01319-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekema D. J., Hsueh P. R., Mendes R. E., Pfaller M. A., Rolston K. V., Sader H. S., et al. (2019). The Microbiology of Bloodstream Infection: 20-Year Trends From the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 63 (7), e00355-19. doi: 10.1128/AAC.00355-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pilato V., Arena F., Tascini C., Cannatelli A., Henrici De Angelis L., Fortunato S., et al. (2016). Mcr-1.2, A New Mcr Variant Carried on a Transferable Plasmid From a Colistin-Resistant KPC Carbapenemase-Producing Klebsiella Pneumoniae Strain of Sequence Type 512. Antimicrob. Agents Chemother. 60 (9), 5612–5615. doi: 10.1128/AAC.01075-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi Y. (2019). Treatment Options for Carbapenem-Resistant Gram-Negative Bacterial Infections. Clin. Infect. Dis. 69 (Suppl 7), S565–S575. doi: 10.1093/cid/ciz830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumith M., Godbole G., Ashton P., Larkin L., Dallman T., Day M., et al. (2016). Detection of the Plasmid-Mediated Mcr-1 Gene Conferring Colistin Resistance in Human and Food Isolates of Salmonella Enterica and Escherichia Coli in England and Wales. J. Antimicrob. Chemother. 71 (8), 2300–2305. doi: 10.1093/jac/dkw093 [DOI] [PubMed] [Google Scholar]
- Ejaz H., Younas S., Qamar M. U., Junaid K., Abdalla A. E., Abosalif K. O. A., et al. (2021). Molecular Epidemiology of Extensively Drug-Resistant Mcr Encoded Colistin-Resistant Bacterial Strains Co-Expressing Multifarious Beta-Lactamases. Antibiotics (Basel) 10 (4), 467. doi: 10.3390/antibiotics10040467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnahriry S. S., Khalifa H. O., Soliman A. M., Ahmed A. M., Hussein A. M., Shimamoto T., et al. (2016). Emergence of Plasmid-Mediated Colistin Resistance Gene Mcr-1 in a Clinical Escherichia Coli Isolate From Egypt. Antimicrob. Agents Chemother. 60 (5), 3249–3250. doi: 10.1128/AAC.00269-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch D. A., Birkett C. I., Ludlam H. A. (2007). Non-Fermentative Gram-Negative Bacteria. Int. J. Antimicrob. Agents 29 Suppl 3, S33–S41. doi: 10.1016/S0924-8579(07)72176-3 [DOI] [PubMed] [Google Scholar]
- Falagas M. E., Rafailidis P. I., Matthaiou D. K. (2010). Resistance to Polymyxins: Mechanisms, Frequency and Treatment Options. Drug Resist. Update 13 (4-5), 132–138. doi: 10.1016/j.drup.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Falgenhauer L., Waezsada S. E., Yao Y., Imirzalioglu C., Kasbohrer A., Roesler U., et al. (2016). Colistin Resistance Gene Mcr-1 in Extended-Spectrum Beta-Lactamase-Producing and Carbapenemase-Producing Gram-Negative Bacteria in Germany. Lancet Infect. Dis. 16 (3), 282–283. doi: 10.1016/S1473-3099(16)00009-8 [DOI] [PubMed] [Google Scholar]
- Fan R., Li C., Duan R., Qin S., Liang J., Xiao M., et al. (2020). Retrospective Screening and Analysis of Mcr-1 and Bla NDM in Gram-Negative Bacteria in China 2010-2019. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes M. R., Moura Q., Sartori L., Silva K. C., Cunha M. P., Esposito F., et al. (2016). Silent Dissemination of Colistin-Resistant Escherichia Coli in South America Could Contribute to the Global Spread of the Mcr-1 Gene. Euro Surveill 21 (17), 30214. doi: 10.2807/1560-7917.ES.2016.21.17.30214 [DOI] [PubMed] [Google Scholar]
- Fernandez L., Gooderham W. J., Bains M., McPhee J. B., Wiegand I., Hancock R. E. (2010). Adaptive Resistance to the “Last Hope” Antibiotics Polymyxin B and Colistin in Pseudomonas Aeruginosa is Mediated by the Novel Two-Component Regulatory System ParR-ParS. Antimicrob. Agents Chemother. 54 (8), 3372–3382. doi: 10.1128/AAC.00242-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo R., Card R. M., Nunez J., Pomba C., Mendonca N., Anjum M. F., et al. (2016). Detection of an Mcr-1-Encoding Plasmid Mediating Colistin Resistance in Salmonella Enterica From Retail Meat in Portugal. J. Antimicrob. Chemother. 71 (8), 2338–2340. doi: 10.1093/jac/dkw240 [DOI] [PubMed] [Google Scholar]
- Fujihara H., Yamazoe A., Hosoyama A., Suenaga H., Kimura N., Hirose J., et al. (2015). Draft Genome Sequence of Pseudomonas Aeruginosa KF702 (NBRC 110665), A Polychlorinated Biphenyl-Degrading Bacterium Isolated From Biphenyl-Contaminated Soil. Genome Announc 3 (3), e00517-15. doi: 10.1128/genomeA.00517-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernandez S., Garcia-Castillo M., Ruiz-Garbajosa P., Morosini M. I., Bala Y., Zambardi G., et al. (2019). Performance of CHROMID(R) Colistin R Agar, A New Chromogenic Medium for Screening of Colistin-Resistant Enterobacterales. Diagn. Microbiol. Infect. Dis. 93 (1), 1–4. doi: 10.1016/j.diagmicrobio.2018.07.008 [DOI] [PubMed] [Google Scholar]
- Garnacho-Montero J., Timsit J. F. (2019). Managing Acinetobacter Baumannii Infections. Curr. Opin. Infect. Dis. 32 (1), 69–76. doi: 10.1097/QCO.0000000000000518 [DOI] [PubMed] [Google Scholar]
- Grami R., Mansour W., Mehri W., Bouallegue O., Boujaafar N., Madec J. Y., et al. (2016). Impact of Food Animal Trade on the Spread of Mcr-1-Mediated Colistin Resistance, Tunisia, July 2015. Euro Surveill 21 (8), 30144. doi: 10.2807/1560-7917.ES.2016.21.8.30144 [DOI] [PubMed] [Google Scholar]
- Gutu A. D., Sgambati N., Strasbourger P., Brannon M. K., Jacobs M. A., Haugen E., et al. (2013). Polymyxin Resistance of Pseudomonas Aeruginosa phoQ Mutants is Dependent on Additional Two-Component Regulatory Systems. Antimicrob. Agents Chemother. 57 (5), 2204–2215. doi: 10.1128/AAC.02353-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni M., Poirel L., Kieffer N., Chatre P., Saras E., Metayer V., et al. (2016). Co-Occurrence of Extended Spectrum Beta Lactamase and MCR-1 Encoding Genes on Plasmids. Lancet Infect. Dis. 16 (3), 281–282. doi: 10.1016/S1473-3099(16)00007-4 [DOI] [PubMed] [Google Scholar]
- Hameed F., Khan M. A., Muhammad H., Sarwar T., Bilal H., Rehman T. U. (2019). Plasmid-Mediated Mcr-1 Gene in Acinetobacter Baumannii and Pseudomonas Aeruginosa: First Report From Pakistan. Rev. Soc. Bras. Med. Trop. 52, e20190237. doi: 10.1590/0037-8682-0237-2019 [DOI] [PubMed] [Google Scholar]
- Hamel M., Rolain J. M., Baron S. A. (2021). The History of Colistin Resistance Mechanisms in Bacteria: Progress and Challenges. Microorganisms 9 (2), 442. doi: 10.3390/microorganisms9020442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidian M., Maharjan R., Cain A., Faruga D., Paulsen I. (2020). Acinetobacter Baumannii Strain E-072658, Complete Genome.Available at: https://www.ncbi.nlm.nih.gov/nuccore/CP061705.1/ [Accessed January 31, 2022]. [Google Scholar]
- Hasman H., Hammerum A. M., Hansen F., Hendriksen R. S., Olesen B., Agerso Y., et al. (2015). Detection of Mcr-1 Encoding Plasmid-Mediated Colistin-Resistant Escherichia Coli Isolates From Human Bloodstream Infection and Imported Chicken Meat, Denmark 2015. Euro Surveill 20 (49), pii=30085. doi: 10.2807/1560-7917.ES.2015.20.49.30085 [DOI] [PubMed] [Google Scholar]
- Hermsen E. D., Sullivan C. J., Rotschafer J. C. (2003). Polymyxins: Pharmacology, Pharmacokinetics, Pharmacodynamics, and Clinical Applications. Infect. Dis. Clin. North Am. 17 (3), 545–562. doi: 10.1016/s0891-5520(03)00058-8 [DOI] [PubMed] [Google Scholar]
- Izdebski R., Baraniak A., Bojarska K., Urbanowicz P., Fiett J., Pomorska-Wesolowska M., et al. (2016). Mobile MCR-1-Associated Resistance to Colistin in Poland. J. Antimicrob. Chemother. 71 (8), 2331–2333. doi: 10.1093/jac/dkw261 [DOI] [PubMed] [Google Scholar]
- Javed H., Saleem S., Zafar A., Ghafoor A., Shahzad A. B., Ejaz H., et al. (2020). Emergence of Plasmid-Mediated Mcr Genes From Gram-Negative Bacteria at the Human-Animal Interface. Gut Pathog. 12 (1), 54. doi: 10.1186/s13099-020-00392-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean S. S., Harnod D., Hsueh P. R. (2022). Global Threat of Carbapenem-Resistant Gram-Negative Bacteria. Front. Cell Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.823684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Dias D., Manageiro V., Ferreira E., Barreiro P., Vieira L., Moura I. B., et al. (2016). Architecture of Class 1, 2, and 3 Integrons From Gram Negative Bacteria Recovered Among Fruits and Vegetables. Front. Microbiol. 7, 1400. doi: 10.3389/fmicb.2016.01400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouy E., Haenni M., Le Devendec L., Le Roux A., Chatre P., Madec J. Y., et al. (2017). Improvement in Routine Detection of Colistin Resistance in E. Coli Isolated in Veterinary Diagnostic Laboratories. J. Microbiol. Methods 132, 125–127. doi: 10.1016/j.mimet.2016.11.017 [DOI] [PubMed] [Google Scholar]
- Kalova A., Gelbicova T., Overballe-Petersen S., Litrup E., Karpiskova R. (2021). Characterisation of Colistin -Resistant Enterobacterales and Acinetobacter Strains Carrying Mcr Genes From Asian Aquaculture Products. Antibiotics (Basel) 10 (7), 838. doi: 10.3390/antibiotics10070838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamolvit W., Sidjabat H., Kiratisin P., Paterson D. (2014). Acinetobacter Nosocomialis Strain T228, Whole Genome Shortgun Sequencing Project. Available at: https://www.ncbi.nlm.nih.gov/nuccore/JRUA00000000.1/ [Accessed January 31, 2022]. [Google Scholar]
- Kareem S. M. (2020). Emergence of Mcr- and Fosa3-Mediated Colistin and Fosfomycin Resistance Among Carbapenem-Resistant Acinetobacter Baumannii in Iraq. Meta Gene 25, 100708. doi: 10.1016/j.mgene.2020.100708 [DOI] [Google Scholar]
- Kempf I., Fleury M. A., Drider D., Bruneau M., Sanders P., Chauvin C., et al. (2013). What Do We Know About Resistance to Colistin in Enterobacteriaceae in Avian and Pig Production in Europe? Int. J. Antimicrob. Agents 42 (5), 379–383. doi: 10.1016/j.ijantimicag.2013.06.012 [DOI] [PubMed] [Google Scholar]
- Khalifa H. O., Ahmed A. M., Oreiby A. F., Eid A. M., Shimamoto T., Shimamoto T. (2016). Characterisation of the Plasmid-Mediated Colistin Resistance Gene Mcr-1 in Escherichia Coli Isolated From Animals in Egypt. Int. J. Antimicrob. Agents 47 (5), 413–414. doi: 10.1016/j.ijantimicag.2016.02.011 [DOI] [PubMed] [Google Scholar]
- Kluytmans-van den Bergh M. F., Huizinga P., Bonten M. J., Bos M., De Bruyne K., Friedrich A. W., et al. (2016). Presence of Mcr-1-Positive Enterobacteriaceae in Retail Chicken Meat But Not in Humans in the Netherlands Since 2009. Euro Surveill 21 (9), 30149. doi: 10.2807/1560-7917.ES.2016.21.9.30149 [DOI] [PubMed] [Google Scholar]
- Kusumoto M., Ogura Y., Gotoh Y., Iwata T., Hayashi T., Akiba M. (2016). Colistin-Resistant Mcr-1-Positive Pathogenic Escherichia Coli in Swine, Japan 2007-2014. Emerg. Infect. Dis. 22 (7), 1315–1317. doi: 10.3201/eid2207.160234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2021). Interactive Tree Of Life (iTOL) V5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 49 (W1), W293–W296. doi: 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos A., Mevius D. J., Olsen B., Bonnedahl J. (2016). The Colistin Resistance Mcr-1 Gene is Going Wild. J. Antimicrob. Chemother. 71 (8), 2335–2336. doi: 10.1093/jac/dkw262 [DOI] [PubMed] [Google Scholar]
- Li X. P., Fang L. X., Song J. Q., Xia J., Huo W., Fang J. T., et al. (2016). Clonal Spread of Mcr-1 in PMQR-Carrying ST34 Salmonella Isolates From Animals in China. Sci. Rep. 6, 38511. doi: 10.1038/srep38511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S. K., Kang H. Y., Lee K., Moon D. C., Lee H. S., Jung S. C. (2016). First Detection of the Mcr-1 Gene in Escherichia Coli Isolated From Livestock Between 2013 and 2015 in South Korea. Antimicrob. Agents Chemother. 60 (11), 6991–6993. doi: 10.1128/AAC.01472-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L. M., Ly N., Anderson D., Yang J. C., Macander L., Jarkowski A., 3rd, et al. (2010). Resurgence of Colistin: A Review of Resistance, Toxicity, Pharmacodynamics, and Dosing. Pharmacotherapy 30 (12), 1279–1291. doi: 10.1592/phco.30.12.1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Nation R. L., Turnidge J. D., Milne R. W., Coulthard K., Rayner C. R., et al. (2006). Colistin: The Re-Emerging Antibiotic for Multidrug-Resistant Gram-Negative Bacterial Infections. Lancet Infect. Dis. 6 (9), 589–601. doi: 10.1016/S1473-3099(06)70580-1 [DOI] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 16 (2), 161–168. doi: 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Madeira F., Park Y. M., Lee J., Buso N., Gur T., Madhusoodanan N., et al. (2019). The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 47 (W1), W636–W641. doi: 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra-Kumar S., Xavier B. B., Das A. J., Lammens C., Butaye P., Goossens H. (2016. a). Colistin Resistance Gene Mcr-1 Harboured on a Multidrug Resistant Plasmid. Lancet Infect. Dis. 16 (3), 283–284. doi: 10.1016/S1473-3099(16)00012-8 [DOI] [PubMed] [Google Scholar]
- Malhotra-Kumar S., Xavier B. B., Das A. J., Lammens C., Hoang H. T., Pham N. T., et al. (2016. b). Colistin-Resistant Escherichia Coli Harbouring Mcr-1 Isolated From Food Animals in Hanoi, Vietnam. Lancet Infect. Dis. 16 (3), 286–287. doi: 10.1016/S1473-3099(16)00014-1 [DOI] [PubMed] [Google Scholar]
- Mancuso G., Midiri A., Gerace E., Biondo C. (2021). Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 10 (10), 1310. doi: 10.3390/pathogens10101310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins E., Maboni G., Battisti R., da Costa L., Selva H. L., Levitzki E. D., et al. (2022). High Rates of Multidrug Resistance in Bacteria Associated With Small Animal Otitis: A Study of Cumulative Microbiological Culture and Antimicrobial Susceptibility. Microb. Pathog. 165, 105399. doi: 10.1016/j.micpath.2022.105399 [DOI] [PubMed] [Google Scholar]
- Martins-Sorenson N., Snesrud E., Xavier D. E., Cacci L. C., Iavarone A. T., McGann P., et al. (2020). A Novel Plasmid-Encoded Mcr-4.3 Gene in a Colistin-Resistant Acinetobacter Baumannii Clinical Strain. J. Antimicrob. Chemother. 75 (1), 60–64. doi: 10.1093/jac/dkz413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGann P., Snesrud E., Maybank R., Corey B., Ong A. C., Clifford R., et al. (2016). Escherichia Coli Harboring Mcr-1 and blaCTX-M on a Novel IncF Plasmid: First Report of Mcr-1 in the United States. Antimicrob. Agents Chemother. 60 (7), 4420–4421. doi: 10.1128/AAC.01103-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee J. B., Lewenza S., Hancock R. E. (2003). Cationic Antimicrobial Peptides Activate a Two-Component Regulatory System, PmrA-PmrB, That Regulates Resistance to Polymyxin B and Cationic Antimicrobial Peptides in Pseudomonas Aeruginosa . Mol. Microbiol. 50 (1), 205–217. doi: 10.1046/j.1365-2958.2003.03673.x [DOI] [PubMed] [Google Scholar]
- Meinersmann R. J., Ladely S. R., Plumblee J. R., Hall M. C., Simpson S. A., Ballard L. L., et al. (2016). Colistin Resistance Mcr-1-Gene-Bearing Escherichia Coli Strain From the United States. Genome Announc 4 (5), e00 898-16. doi: 10.1128/genomeA.00898-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micek S. T., Wunderink R. G., Kollef M. H., Chen C., Rello J., Chastre J., et al. (2015). An International Multicenter Retrospective Study of Pseudomonas Aeruginosa Nosocomial Pneumonia: Impact of Multidrug Resistance. Crit. Care 19, 219. doi: 10.1186/s13054-015-0926-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt J. H., Harper M., Adler B., Nation R. L., Li J., Boyce J. D. (2011). Insertion Sequence ISAba11 is Involved in Colistin Resistance and Loss of Lipopolysaccharide in Acinetobacter Baumannii . Antimicrob. Agents Chemother. 55 (6), 3022–3024. doi: 10.1128/AAC.01732-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt J. H., Harper M., Harrison P., Hale J. D., Vinogradov E., Seemann T., et al. (2010). Colistin Resistance in Acinetobacter Baumannii is Mediated by Complete Loss of Lipopolysaccharide Production. Antimicrob. Agents Chemother. 54 (12), 4971–4977. doi: 10.1128/AAC.00834-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsin M., Raza S., Roschanski N., Guenther S., Ali A., Schierack P. (2017). Description of the First Escherichia Coli Clinical Isolate Harboring the Colistin Resistance Gene Mcr-1 From the Indian Subcontinent. Antimicrob. Agents Chemother. 61 (1), e01945-16. doi: 10.1128/AAC.01945-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M. R., Mataseje L. F., Robertson J., Nash J. H., Boerlin P., Toye B., et al. (2016). Dissemination of the Mcr-1 Colistin Resistance Gene. Lancet Infect. Dis. 16 (3), 289–290. doi: 10.1016/S1473-3099(16)00067-0 [DOI] [PubMed] [Google Scholar]
- National Antimicrobial Resistant Surveillance Center, T (2020). Antibiogram 2019 (Jan - Dec). Available at: http://narst.dmsc.moph.go.th/antibiograms.html [Accessed January 20, 2022]. [Google Scholar]
- National Library of Medicine (2022). NCBI National Database of Antibiotic Resistant Organisms (NDARO). Available at: https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/ [Accessed January 31, 2022]. [Google Scholar]
- Nation R. L., Li J. (2009). Colistin in the 21st Century. Curr. Opin. Infect. Dis. 22 (6), 535–543. doi: 10.1097/QCO.0b013e328332e672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham B. D., Trent M. S. (2013). Fortifying the Barrier: The Impact of Lipid A Remodelling on Bacterial Pathogenesis. Nat. Rev. Microbiol. 11 (7), 467–481. doi: 10.1038/nrmicro3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitz F., de Melo B. O., da Silva L. C. N., de Souza Monteiro A., Marques S. G., Monteiro-Neto V., et al. (2021). Molecular Detection of Drug-Resistance Genes of blaOXA-23-blaOXA-51 and Mcr-1 in Clinical Isolates of Pseudomonas Aeruginosa . Microorganisms 9 (4), 786. doi: 10.3390/microorganisms9040786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P., Jayol A., Poirel L. (2016. a). Rapid Detection of Polymyxin Resistance in Enterobacteriaceae. Emerg. Infect. Dis. 22 (6), 1038–1043. doi: 10.3201/eid2206.151840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P., Jayol A., Poirel L. (2016. b). A Universal Culture Medium for Screening Polymyxin-Resistant Gram-Negative Isolates. J. Clin. Microbiol. 54 (5), 1395–1399. doi: 10.1128/JCM.00446-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P., Lienhard R., Kieffer N., Clerc O., Poirel L. (2016. c). Plasmid-Mediated Colistin-Resistant Escherichia Coli in Bacteremia in Switzerland. Clin. Infect. Dis. 62 (10), 1322–1323. doi: 10.1093/cid/ciw124 [DOI] [PubMed] [Google Scholar]
- Olaitan A. O., Chabou S., Okdah L., Morand S., Rolain J. M. (2016). Dissemination of the Mcr-1 Colistin Resistance Gene. Lancet Infect. Dis. 16 (2), 147. doi: 10.1016/S1473-3099(15)00540-X [DOI] [PubMed] [Google Scholar]
- Olaitan A. O., Morand S., Rolain J. M. (2014). Mechanisms of Polymyxin Resistance: Acquired and Intrinsic Resistance in Bacteria. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Paredes D., Barba P., Zurita J. (2016). Colistin-Resistant Escherichia Coli Clinical Isolate Harbouring the Mcr-1 Gene in Ecuador. Epidemiol. Infect. 144 (14), 2967–2970. doi: 10.1017/S0950268816001369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge S. R., Di Pilato V., Doi Y., Feldgarden M., Haft D. H., Klimke W., et al. (2018). Proposal for Assignment of Allele Numbers for Mobile Colistin Resistance (Mcr) Genes. J. Antimicrob. Chemother. 73 (10), 2625–2630. doi: 10.1093/jac/dky262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreten V., Strauss C., Collaud A., Gerber D. (2016). Colistin Resistance Gene Mcr-1 in Avian-Pathogenic Escherichia Coli in South Africa. Antimicrob. Agents Chemother. 60 (7), 4414–4415. doi: 10.1128/AAC.00548-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham Thanh D., Thanh Tuyen H., Nguyen Thi Nguyen T., Chung The H., Wick R. R., Thwaites G. E., et al. (2016). Inducible Colistin Resistance via a Disrupted Plasmid-Borne Mcr-1 Gene in a 2008 Vietnamese Shigella Sonnei Isolate. J. Antimicrob. Chemother. 71 (8), 2314–2317. doi: 10.1093/jac/dkw173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Kieffer N., Brink A., Coetze J., Jayol A., Nordmann P. (2016). Genetic Features of MCR-1-Producing Colistin-Resistant Escherichia Coli Isolates in South Africa. Antimicrob. Agents Chemother. 60 (7), 4394–4397. doi: 10.1128/AAC.00444-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prim N., Rivera A., Rodriguez-Navarro J., Espanol M., Turbau M., Coll P., et al. (2016). Detection of Mcr-1 Colistin Resistance Gene in Polyclonal Escherichia Coli Isolates in Barcelona, Spain 2012 to 2015. Euro Surveill 21 (13), pii=30183. doi: 10.2807/1560-7917.ES.2016.21.13.30183 [DOI] [PubMed] [Google Scholar]
- Quesada A., Ugarte-Ruiz M., Iglesias M. R., Porrero M. C., Martinez R., Florez-Cuadrado D., et al. (2016). Detection of Plasmid Mediated Colistin Resistance (MCR-1) in Escherichia Coli and Salmonella Enterica Isolated From Poultry and Swine in Spain. Res. Vet. Sci. 105, 134–135. doi: 10.1016/j.rvsc.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Ramirez M. S., Bonomo R. A., Tolmasky M. E. (2020). Carbapenemases: Transforming Acinetobacter Baumannii Into a Yet More Dangerous Menace. Biomolecules 10 (5), 720. doi: 10.3390/biom10050720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport M., Faccone D., Pasteran F., Ceriana P., Albornoz E., Petroni A., et al. (2016). First Description of Mcr-1-Mediated Colistin Resistance in Human Infections Caused by Escherichia Coli in Latin America. Antimicrob. Agents Chemother. 60 (7), 4412–4413. doi: 10.1128/AAC.00573-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhouma M., Beaudry F., Letellier A. (2016. a). Resistance to Colistin: What is the Fate for This Antibiotic in Pig Production? Int. J. Antimicrob. Agents 48 (2), 119–126. doi: 10.1016/j.ijantimicag.2016.04.008 [DOI] [PubMed] [Google Scholar]
- Rhouma M., Beaudry F., Theriault W., Letellier A. (2016. b). Colistin in Pig Production: Chemistry, Mechanism of Antibacterial Action, Microbial Resistance Emergence, and One Health Perspectives. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolain J. M., Kempf M., Leangapichart T., Chabou S., Olaitan A. O., Le Page S., et al. (2016). Plasmid-Mediated Mcr-1 Gene in Colistin-Resistant Clinical Isolates of Klebsiella Pneumoniae in France and Laos. Antimicrob. Agents Chemother. 60 (11), 6994–6995. doi: 10.1128/AAC.00960-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzauskas M., Vaskeviciute L. (2016). Detection of the Mcr-1 Gene in Escherichia Coli Prevalent in the Migratory Bird Species Larus Argentatus. J. Antimicrob. Chemother. 71 (8), 2333–2334. doi: 10.1093/jac/dkw245 [DOI] [PubMed] [Google Scholar]
- Shabban M., Fahim N. A. E., Montasser K., Abo El Magd N. M. (2020). Resistance to Colistin Mediated by Mcr-1 Among Multidrug Resistant Gram Negative Pathogens at a Tertiary Care Hospital, Egypt. J. Pure Appl. Microbiol. 14 (2), 1125–1132. doi: 10.22207/JPAM.14.2.07 [DOI] [Google Scholar]
- Snesrud E., Maybank R., Kwak Y. I., Jones A. R., Hinkle M. K., McGann P. (2018). Chromosomally Encoded Mcr-5 in Colistin-Nonsusceptible Pseudomonas Aeruginosa . Antimicrob. Agents Chemother. 62 (8), e00679-18. doi: 10.1128/AAC.00679-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyman Y., Reuter S., Whitelaw A. C., Stein L., Maloba M. R. B., Newton-Foot M. (2021). Characterisation of Mcr-4.3 in a Colistin-Resistant Acinetobacter Nosocomialis Clinical Isolate From Cape Town, South Africa. J. Glob Antimicrob. Resist. 25, 102–106. doi: 10.1016/j.jgar.2021.03.002 [DOI] [PubMed] [Google Scholar]
- Solheim M., Bohlin J., Ulstad C. R., Schau Slettemeas J., Naseer U., Dahle U. R., et al. (2016). Plasmid-Mediated Colistin-Resistant Escherichia Coli Detected From 2014 in Norway. Int. J. Antimicrob. Agents 48 (2), 227–228. doi: 10.1016/j.ijantimicag.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Sonnevend A., Ghazawi A., Alqahtani M., Shibl A., Jamal W., Hashmey R., et al. (2016). Plasmid-Mediated Colistin Resistance in Escherichia Coli From the Arabian Peninsula. Int. J. Infect. Dis. 50, 85–90. doi: 10.1016/j.ijid.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Stoesser N., Mathers A. J., Moore C. E., Day N. P., Crook D. W. (2016). Colistin Resistance Gene Mcr-1 and Phnshp45 Plasmid in Human Isolates of Escherichia Coli and Klebsiella Pneumoniae . Lancet Infect. Dis. 16 (3), 285–286. doi: 10.1016/S1473-3099(16)00010-4 [DOI] [PubMed] [Google Scholar]
- Suzuki S., Ohnishi M., Kawanishi M., Akiba M., Kuroda M. (2016). Investigation of a Plasmid Genome Database for Colistin-Resistance Gene Mcr-1 . Lancet Infect. Dis. 16 (3), 284–285. doi: 10.1016/S1473-3099(16)00008-6 [DOI] [PubMed] [Google Scholar]
- Sy C. L., Chen P. Y., Cheng C. W., Huang L. J., Wang C. H., Chang T. H., et al. (2022). Recommendations and Guidelines for the Treatment of Infections Due to Multidrug Resistant Organisms. J. Microbiol. Immunol. Infect. doi: 10.1016/j.jmii.2022.02.001. (Accessed April 20, 2022). [DOI] [PubMed] [Google Scholar]
- Tahmasebi H., Dehbashi S., Arabestani M. R. (2020. a). Co-Harboring of Mcr-1 and Beta-Lactamase Genes in Pseudomonas Aeruginosa by High-Resolution Melting Curve Analysis (HRMA): Molecular Typing of Superbug Strains in Bloodstream Infections (BSI). Infect. Genet. Evol. 85, 104518. doi: 10.1016/j.meegid.2020.104518 [DOI] [PubMed] [Google Scholar]
- Tahmasebi H., Dehbashi S., Arabestani M. R. (2020. b). Prevalence and Molecular Typing of Colistin-Resistant Pseudomonas Aeruginosa (CRPA) Among Beta-Lactamase-Producing Isolates: A Study Based on High-Resolution Melting Curve Analysis Method. Infect. Drug Resist. 13, 2943–2955. doi: 10.2147/IDR.S264796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartor Y. H., Gharieb R. M. A., Abd El-Aziz N. K., El Damaty H. M., Enany S., Khalifa E., et al. (2021). Virulence Determinants and Plasmid-Mediated Colistin Resistance Mcr Genes in Gram-Negative Bacteria Isolated From Bovine Milk. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.761417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo J. W., Chew K. L., Lin R. T. (2016. b). Transmissible Colistin Resistance Encoded by Mcr-1 Detected in Clinical Enterobacteriaceae Isolates in Singapore. Emerg. Microbes Infect. 5 (8), e87. doi: 10.1038/emi.2016.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo J. Q., Ong R. T., Xia E., Koh T. H., Khor C. C., Lee S. J., et al. (2016. a). Mcr-1 in Multidrug-Resistant blaKPC-2-Producing Clinical Enterobacteriaceae Isolates in Singapore. Antimicrob. Agents Chemother. 60 (10), 6435–6437. doi: 10.1128/AAC.00804-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing (2022). Clinical Breakpoints - Breakpoints and Guidance. Available at: https://www.eucast.org/clinical_breakpoints/ [Accessed January 20, 2022]. [Google Scholar]
- Tsuji B. T., Pogue J. M., Zavascki A. P., Paul M., Daikos G. L., Forrest A., et al. (2019). International Consensus Guidelines for the Optimal Use of the Polymyxins: Endorsed by the American College of Clinical Pharmacy (ACCP), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Infectious Diseases Society of America (IDSA), International Society for Anti-Infective Pharmacology (ISAP), Society of Critical Care Medicine (SCCM), and Society of Infectious Diseases Pharmacists (SIDP). Pharmacotherapy 39 (1), 10–39. doi: 10.1002/phar.2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vading M., Kabir M. H., Kalin M., Iversen A., Wiklund S., Naucler P., et al. (2016). Frequent Acquisition of Low-Virulence Strains of ESBL-Producing Escherichia Coli in Travellers. J. Antimicrob. Chemother. 71 (12), 3548–3555. doi: 10.1093/jac/dkw335 [DOI] [PubMed] [Google Scholar]
- Veldman K., van Essen-Zandbergen A., Rapallini M., Wit B., Heymans R., van Pelt W., et al. (2016). Location of Colistin Resistance Gene Mcr-1 in Enterobacteriaceae From Livestock and Meat. J. Antimicrob. Chemother. 71 (8), 2340–2342. doi: 10.1093/jac/dkw181 [DOI] [PubMed] [Google Scholar]
- Webb H. E., Granier S. A., Marault M., Millemann Y., den Bakker H. C., Nightingale K. K., et al. (2016). Dissemination of the Mcr-1 Colistin Resistance Gene. Lancet Infect. Dis. 16 (2), 144–145. doi: 10.1016/S1473-3099(15)00538-1 [DOI] [PubMed] [Google Scholar]
- WHO Regional Office for Europe/European Centre for Disease Prevention and Control (2022). Antimicrobial Resistance Surveillance in Europe 2022 - 2020 data. Copenhagen: WHO Regional Office for Europe. [Google Scholar]
- World Health Organization (2017) WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed. Available at: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (Accessed December 19, 2021).
- World Health Organization (2020). GLASS Whole-Genome Sequencing for Surveillance of Antimicrobial Resistance. Available at: https://www.who.int/publications/i/item/9789240011007 [Accessed January 20, 2022]. [Google Scholar]
- World Health Organization (2021). GLASS: The Detection and Reporting of Colistin Resistance. Available at: https://www.who.int/publications/i/item/glass-the-detection-and-reporting-of-colistin-resistance-2nd-ed [Accessed January 20, 2022]. [Google Scholar]
- Xavier B. B., Lammens C., Ruhal R., Kumar-Singh S., Butaye P., Goossens H., et al. (2016). Identification of a Novel Plasmid-Mediated Colistin-Resistance Gene, Mcr-2, in Escherichia Coli, Belgium, June 2016. Euro Surveill 21 (27), 30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280 [DOI] [PubMed] [Google Scholar]
- Yang Y., Zhang A. (2018). Acinetobacter Pittii Strain SCsI25, Whole Genome Shortgun Sequencing Project. https://www.ncbi.nlm.nih.gov/nuccore/VDIH00000000.1/ [Accessed January 31, 2022]. [Google Scholar]
- Yu C. Y., Ang G. Y., Chin P. S., Ngeow Y. F., Yin W. F., Chan K. G. (2016). Emergence of Mcr-1-Mediated Colistin Resistance in Escherichia Coli in Malaysia. Int. J. Antimicrob. Agents 47 (6), 504–505. doi: 10.1016/j.ijantimicag.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Zeng K. J., Doi Y., Patil S., Huang X., Tian G. B. (2016). Emergence of the Plasmid-Mediated Mcr-1 Gene in Colistin-Resistant Enterobacter Aerogenes and Enterobacter Cloacae . Antimicrob. Agents Chemother. 60 (6), 3862–3863. doi: 10.1128/AAC.00345-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhanel G. G., Adam H. J., Baxter M. R., Fuller J., Nichol K. A., Denisuik A. J., et al. (2019). 42936 Pathogens From Canadian Hospitals: 10 Years of Results,(2017-16) From the CANWARD Surveillance Study. J. Antimicrob. Chemother. 74 (Suppl 4), iv5–iv21. doi: 10.1093/jac/dkz283 [DOI] [PubMed] [Google Scholar]
- Zhang H., Srinivas S., Xu Y., Wei W., Feng Y. (2019). Genetic and Biochemical Mechanisms for Bacterial Lipid A Modifiers Associated With Polymyxin Resistance. Trends Biochem. Sci. 44 (11), 973–988. doi: 10.1016/j.tibs.2019.06.002 [DOI] [PubMed] [Google Scholar]
- Zhao F., Zong Z. (2016). Kluyvera Ascorbata Strain From Hospital Sewage Carrying the Mcr-1 Colistin Resistance Gene. Antimicrob. Agents Chemother. 60 (12), 7498–7501. doi: 10.1128/AAC.01165-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogg A. L., Zurfluh K., Nuesch-Inderbinen M., Stephan R. (2016). Characteristics of ESBL-Producing Enterobacteriaceae and Methicillinresistant Staphylococcus Aureus (MRSA) Isolated From Swiss and Imported Raw Poultry Meat Collected at Retail Level. Schweiz Arch. Tierheilkd 158 (6), 451–456. doi: 10.17236/sat00071 [DOI] [PubMed] [Google Scholar]
- Zurfuh K., Poirel L., Nordmann P., Nuesch-Inderbinen M., Hachler H., Stephan R. (2016). Occurrence of the Plasmid-Borne Mcr-1 Colistin Resistance Gene in Extended-Spectrum-Beta-Lactamase-Producing Enterobacteriaceae in River Water and Imported Vegetable Samples in Switzerland. Antimicrob. Agents Chemother. 60 (4), 2594–2595. doi: 10.1128/AAC.00066-16 [DOI] [PMC free article] [PubMed] [Google Scholar]