Abstract
The oral microbiome is linked to oral and systemic health, but its fluctuation under frequent daily activities remains elusive. Here, we sampled saliva at 10- to 60-min intervals to track the high-resolution microbiome dynamics during the course of human activities. This dense time series data showed that eating activity markedly perturbed the salivary microbiota, with tongue-specific Campylobacter concisus and Oribacterium sinus and dental plaque-specific Lautropia mirabilis, Rothia aeria, and Neisseria oralis increased after every meal in a temporal order. The observation was reproducible in multiple subjects and across an 11-mo period. The microbiome composition showed significant diurnal oscillation patterns at different taxonomy levels with Prevotella/Alloprevotella increased at night and Bergeyella HMT 206/Haemophilus slowly increased during the daytime. We also identified microbial co-occurring patterns in saliva that are associated with the intricate biogeography of the oral microbiome. Microbial source tracking analysis showed that the contributions of distinct oral niches to the salivary microbiome were dynamically affected by daily activities, reflecting the role of saliva in exchanging microbes with other oral sites. Collectively, our study provides insights into the temporal microbiome variation in saliva and highlights the need to consider daily activities and diurnal factors in design of oral microbiome studies.
The oral cavity, the primary gateway to the digestive tract, harbors one of the most diverse and abundant microbial communities within the human body. Usually, the oral microbiome stays in a balanced immunoinflammatory state with the host. Dysbiosis of the microbial ecosystem is closely linked to various oral diseases, including dental caries (Teng et al. 2015; Kim et al. 2020), gingivitis (Nowicki et al. 2018; Kahharova et al. 2020), and periodontitis (Shi et al. 2015). Notably, longitudinal studies reveal that changes in oral microbiota preceded clinical symptoms of early childhood caries (Teng et al. 2015) and gingivitis (Kahharova et al. 2020), implying the potential for microbiome-based diagnosis and microbiome-targeting treatment for oral diseases. Further, oral microbes are found substantially disseminated from oral cavity to distal gastrointestinal tract, and significantly contribute to systemic diseases like gastric cancer (Sung et al. 2020), pancreatic cancer (Fan et al. 2018), inflammatory bowel disease (Gevers et al. 2014), and colorectal cancer (Flemer et al. 2018). Recent studies consistently reported that the proportion of oral bacteria was higher in cancerous samples compared to precancerous samples (Nakatsu et al. 2015; Coker et al. 2018; Gaiser et al. 2019). Besides gastrointestinal diseases, oral microbiota is also associated with other systemic diseases, such as rheumatoid arthritis (Zhang et al. 2015), Alzheimer's disease (Dominy et al. 2019), and coronary artery disease (Chhibber-Goel et al. 2016). Despite the crucial role of oral microbes in human health, its fundamental dynamic nature remains unclear. Elucidating the basic microbial behavior may provide insight into the connections between oral microbes and human diseases.
The oral microbiome can be highly dynamic and show fast growth. The doubling time for oral microbes in an edentulous mouth is estimated to be 1.38 h on average (Dawes 2003). Oral resident Streptococcus mitis can divide in 40 min (Wei et al. 2021). The opposing forces of adherence and shedding imposed by flowing saliva further complicates the dynamic changes of oral microbiome: Saliva flow sweeps away shed or weakly binding bacteria from other oral sites and transmits microbes to other sites for colonization (Mark Welch et al. 2020). The oral cavity is also frequently perturbed by daily activities including food intake, dental hygiene, exercise, and sleep. The exact effects of human activities on the oral microbiome remain elusive to our knowledge. Therefore, high-frequency sampling is required to reveal the precise dynamic changes of microbial communities under daily perturbations.
Here, we used the salivary microbiome as a noninvasive and easily accessible model to characterize its dense time series dynamics. We tracked healthy adults for 6 d continuously (except sleep). Saliva samples were collected every 10–60 min from multiple subjects, with detailed records of daily activities, including diet, teeth brushing, sleep, and exercise. We found significant microbial dynamic patterns in saliva that were consistent in multiple hosts and reproducible over an 11-mo period. This high-frequency time series data unveils the profound dynamics of human salivary microbiota and helps us better understand the complex interactions of the microbiome among different oral sites.
Results
The microbiome overview of densely sampled saliva samples
To collect oral samples with high frequency, we adapted a nondestructive saliva sampling protocol by gently touching the saliva on the tongue with a clean, sterile swab. To validate that this frequent sampling has negligible impact on salivary microbiome, we computed the beta-diversity distances between all sample pairs from the same individual. The intra-individual sample–sample beta distances and differences in alpha-diversity (Faith's phylogenetic diversity [PD]) index were stable, regardless of the length of the sampling time interval (Fig. 1A; Supplemental Fig. S1A) or the number of sampling efforts between two samples (Fig. 1B; Supplemental Fig. S1B), indicating that our high-frequency sampling protocol did not systematically change salivary microbiome composition. The three participants harbored a distinct and relatively stable salivary microbiome along the experiment period of 6 d (Fig. 1C), in line with previous studies (Caporaso et al. 2011; Hall et al. 2017; Johnson et al. 2019). The salivary microbiome composition was dominated by Firmicutes phylum, followed by Proteobacteria, Bacteroidetes, Fusobacteria, Actinobacteria, and others (Fig. 1E; Supplemental Fig. S1E), in agreement with previous research (Segata et al. 2012; David et al. 2014). Within each participant, daily activities, especially eating and sleeping, contribute considerably to the overall composition variation (PERMANOVA for both eat and sleep: P < 0.01) (Fig. 1D; Supplemental Fig. S1D), with the relative abundances of various taxa displaying rapid changes owing to daily activities (Fig. 1E; Supplemental Fig. S1E). We computed the effect size over the covariates that contribute to microbiota variation, showing that host factor explained the largest amount of variation (∼37.93%), followed by eating activity (∼7.75%) (Supplemental Fig. S1C). The effects of other activities like brushing and exercise were marginal, perhaps because of the small sample size for these activities.
Figure 1.
The microbiome overview of densely sampled saliva samples in subject MA. (A,B) The Canberra distance and delta in Faith's PD index between every two salivary samples (measured by time interval in A or sampling efforts in B) showed that our dense sampling protocol does not systematically change microbiome composition. (C) The PCoA plot of three subjects along the sampling time of more than 6 d. Samples were clustered by individuals. (D) The PCoA plot colored by daily activities in MA. (E) Phylum-level taxonomy composition of MA samples along time. (F) Trace plot of alpha diversity in MA. Activity windows are marked in different background colors. Red, blue, green, and purple represent eat, brush, sleep, and exercise activities, respectively.
Eating-responsive microbes
We detected chloroplast, mitochondrial, and other rare non-oral amplicon sequence variants (ASVs) in the salivary samples, which appeared immediately during eating, followed by a rapid monotonic decrease (Supplemental Fig. S2A). These sequences are presumably relic DNA from food sources and were removed from downstream analyses. Yogurt- or cheese-introduced probiotics (e.g., Lactobacillus species) can also be detected. The sudden appearance in the first sample during eating, together with the rapid monotonic decrease and their absence between meals, indicate that these bacteria are also from the consumed food and transient in the oral cavity (Supplemental Fig. S2B).
Food eating activities considerably affected microbial alpha diversity. Faith's PD index increased sharply during eating and decreased soon afterward (Fig. 1F; Supplemental Fig. S1F). To identify the microbes that were associated with eating activity, we developed a Z-score method to evaluate how strongly each ASV reacts to food intake. The Z-score is computed to measure how much the abundance change induced by eating deviates from normal fluctuations for a given ASV (for details, see Methods). Notably, bacteria with the highest Z-scores, including Campylobacter concisus, Lautropia mirabilis, and Neisseria oralis, consistently increased after every meal in all individuals. They maintained at a relatively low baseline abundance in saliva and peaked around eating. Their increases were not associated with diet types or individuality (Fig. 2A). The delayed change of these bacteria compared to chloroplast suggested that their increases were unlikely due to food (Supplemental Fig. S2D). Meanwhile, bacteria with the lowest Z-scores, including Prevotella nanceiensis, SR1 HMT 875, and Prevotella melaninogenica, decreased during eating (Fig. 2B).
Figure 2.
Eating-responsive bacteria. (A) The top three bacteria that increased during eating in the three subjects of MA, ME, and MO. (B) The top three bacteria that decreased during eating. Certain ASVs are missing in some subjects because their prevalences were lower than 10% in those subjects. (C) The bacterial increases can be reproduced in the lunch data set. Lines of distinct colors represent different individuals. (D) Eating-increased bacteria displayed a temporal order after averaging over all the eating time windows, smoothing with moving average, and scaling to the same level. Tongue-specific Campylobacter concisus, Leptotrichia HMT 221, Oribacterium sinus, and Actinomyces HMT 172 increased earlier; teeth-specific Lautropia mirabilis, Neisseria oralis, and Rothia aeria increased later. Blue lines indicate ASVs that were more abundant on tongue dorsum; orange lines indicate ASVs that were more abundant on dental plaque.
To investigate whether these observations are reproducible in a large cohort, we performed a separate experiment in which we collected saliva samples from 19 subjects before and while they ate lunch (lunch data set). We observed similar changes in these eating-increased and eating-decreased bacteria in most subjects, suggesting consistent microbial dynamics associated with eating in populations (Fig. 2C).
The rise of the eating-increased microbes displayed a specific temporal order. Campylobacter concisus, Leptotrichia HMT 221, Oribacterium sinus, and Actinomyces HMT 172 increased earlier, whereas Lautropia mirabilis, Neisseria oralis, and Rothia aeria rose and peaked later (Fig. 2D; Supplemental Fig. S2D). This temporal order was also observed in the lunch data set (Fig. 2C). Additionally, for the lunch data set, the microbiome composition variation increased immediately after lunch (∼0.5 h) and slightly increased again around 76 min, which roughly coincides with the peak time of early- and late-increased bacteria (Supplemental Fig. S2E).
To further test the reproducibility of the microbial patterns from a large time scale, we additionally collected saliva samples from subject MA for another 1.5 d, 11 mo apart (MA validation data set). The microbial eating-increased, eating-decreased, and temporal patterns were all repeated in this time series data set (Supplemental Fig. S2F–H).
Diurnal patterns of the salivary microbiome
Two reports have studied the diurnal oscillations of salivary microbiome. They both sampled every 4 h. Belstrøm et al. (2016) found that the salivary microbiome showed little diurnal variation; another study pointed out that the majority of genera oscillated with the rhythm of 24 h (Takayasu et al. 2017). We also noticed a periodicity pattern in beta distances between intra-individual samples collected 24 h apart (Lomb–Scargle test: false discovery rate [FDR] < 0.01 for MA, ME, and MO), indicating the diurnal oscillation of overall salivary microbiome composition (Fig. 1A; Supplemental Fig. S1A). We therefore performed a statistic test using Lomb–Scargle analysis (Glynn et al. 2006; Wu et al. 2016) on our data set to evaluate periodicity of the salivary microbiome at phylum, genus, and ASV levels.
At the phylum level, the five most abundant phyla in the MA saliva underwent diurnal oscillations (FDR ≤ 0.01 for Bacteroidetes, Firmicutes, Fusobacteria, Actinobacteria, and SR1). The relative abundances of Bacteroidetes and SR1 were higher in the evening (before waking up in the morning), whereas the abundances of Fusobacteria and Actinobacteria were higher in the daytime (Fig. 3A; Supplemental Fig. S3). All oscillation patterns were not homogeneous among individuals. For example, only Bacteroidetes showed statistically significant rhythmicity in subject ME (FDR < 0.01) and none in MO at phylum level (Supplemental Fig. S3).
Figure 3.
Diurnal patterns of the salivary microbiome. (A) The phylum that oscillated diurnally in two or more individuals. (B) The ASVs that oscillated diurnally in two or more individuals. (Left) Trace plots. (Right) Abundances of these bacteria averaged over every 24-h window with wake-up time point set as the 0 h for reference. The black line shows the average abundance, with the band depicting the 95% confidence intervals and green shade representing the average sleep window over the experimental period. Microbes of MA are plotted here; the others are in Supplemental Figures S3–S5.
At the genus level, there were more genera that oscillated significantly in the salivary microbiome of MA than that of ME or MO. In line with the study by Takayasu et al. (2017), genus Streptococcus, Granulicatella, Gemella, Veillonella, Prevotella, and Porphyromonas showed diurnal oscillation in the saliva of one or multiple individuals. In addition, Bergeyella, which has not been reported before, showed strong periodicity in all three individuals (Supplemental Fig. S4).
At ASV level, despite diurnal variations among different subjects (Supplemental Fig. S5), several ASVs showed similar oscillation patterns in multiple subjects (Fig. 3B). Haemophilus and Bergeyella HMT 206 slowly increased in the daytime, remained at high abundance at night, but decreased upon waking. Notably, multiple ASVs within the Prevotella/Alloprevotella genus increased at night. This increasing trend apparently lasts beyond waking, until breakfast when there was a drastic reduction, likely indicating a different reason for the decrease compared to Haemophilus and Bergeyella HMT 206.
The statistically significant oscillating phyla, genera, and ASVs displayed similar oscillation patterns in the MA validation data set (Supplemental Fig. S6), which showed that the diurnal rhythm of the salivary microbiome may not be temporary but a lasting phenomenon.
Microbial co-occurrence network
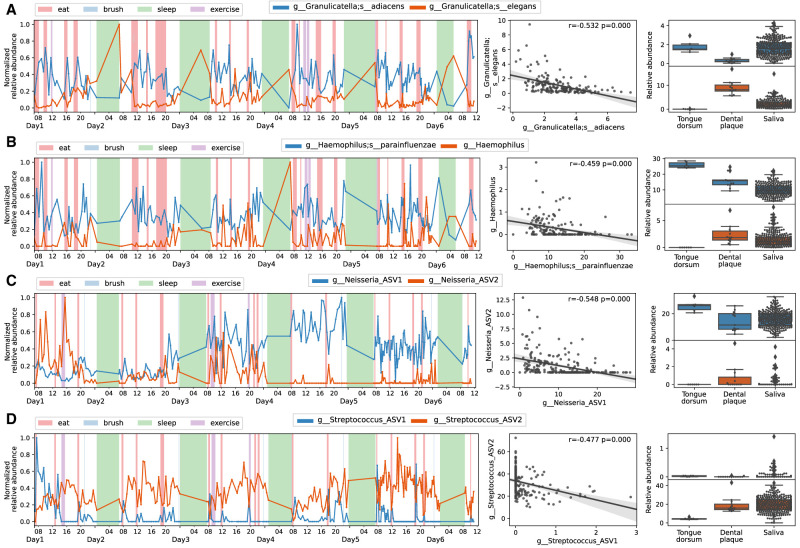
Bioinformatics algorithms like Deblur (Amir et al. 2017), DADA2 (Callahan et al. 2016), and oligotyping (Eren et al. 2013) can distinguish sequences differing in only one single nucleotide. Oral microbiome analyses with these methods infer high-resolution amplicon sequences and reveal that closely related ASVs prefer different habitats in the oral cavity (The Human Microbiome Project Consortium 2012; Eren et al. 2014; Mark Welch et al. 2019; Proctor et al. 2020; Wilbert et al. 2020). To infer the site preference of microbes detected in saliva, we additionally collected samples from tongue dorsum and dental plaque from subject MA—two of the most typical habitats within the oral cavity representing mucosal and nonmucosal surfaces. A permutation-based nonparametric test was applied to identify differential ASVs between these two sites (Xu et al. 2019). Specifically, species within Granulicatella, Haemophilus, Neisseria, and Streptococcus genera were found to be negatively correlated in multiple individuals and tended to be enriched on different oral surfaces (Fig. 4).
Figure 4.
Reverse correlation of ASVs within the same genus. (A) Granulicatella adiacens versus Granulicatella elegans. (B) Haemophilus parainfluenzae versus Haemophilus. (C) Neisseria versus Neisseria. (D) Streptococcus versus Streptococcus. (Left) Trace plots. The relative abundance of each ASV was scaled to [0,1]. (Middle) Scatter plots. The Spearman's correlation coefficient and P-value were shown in the upper right corner. (Right) Box plots displaying the relative abundance of negatively correlated ASV pairs in different oral sites. Tongue-specific ASVs are blue-colored and plaque-specific ASVs are orange-colored in trace and box plots.
We next explored the longitudinal co-occurring relationships between microbes with multiple correlation methods, including CCLasso, which is robust for compositional microbiome data (Fang et al. 2015), and Spearman's, which measures rank relationships (Hirano and Takemoto 2019). Both methods identified several strongly associated pairs in all of the three subjects (Fig. 5; Supplemental Fig. S7): eating early-increased ASVs (Campylobacter concisus and Oribacterium sinus, red box) and late-increased ASVs (two strains of Lautropia mirabilis, Rothia aeria, and Neisseria oralis, orange box) visually clustered separately in the co-occurrence networks. The colors of the nodes in the network represent the oral-site information based on tongue and plaque samples. Notably, early- and late-increased ASVs were enriched in tongue dorsum (blue colored) and dental plaque samples (red colored), respectively. Diurnal oscillating bacteria that increased at night and reduced at breakfast, namely, SR1 HMT 875 and multiple Prevotella/Alloprevotella species (including Prevotella nanceiensis, Prevotella melaninogenica, Prevotella pallens), were correlated together (cyan box), and were found enriched on the tongue surface. In contrast, Bergeyella sp. HMT 206, Haemophilus, Granulicatella elegans, and Streptococcus, displaying an increasing trend in the daytime, were tightly associated with each other (green box) and were found enriched in plaque samples. Porphyrobacter tepidarius, Acidovorax temperans, and Delfia acidovorans were also strongly correlated in multiple subjects.
Figure 5.
Microbial co-occurrence networks computed with CCLasso in the three individuals. Red and green lines represent positive and negative correlations, respectively. A solid line indicates that the correlation of this pair exists in two or more individuals; a dashed line indicates that the significant correlation is only present in this subject. The size of the node is proportional to the average relative abundance. The shape of the node represents different phyla. The color of the node represents the site preference, in which the blue indicates that the ASV is tongue-specific, and the red indicates the ASV is dental plaque-specific. Gray color means that its site information is not available based on our data. The bold fluorescent green–circled nodes indicate diurnal oscillating ASVs. Gray-colored taxonomy labels indicate that the ASVs do not exist in the current subject.
Temporal changes of microbiome functional capacity
The functional capacity of the salivary microbiome in each subject was predicted using PICRUSt2 (Douglas et al. 2020). There was a clear separation among individuals on the principal coordinates analysis (PCoA) plot of predicted functional profile, similar to composition profile, but with a much smaller effect size (Supplemental Fig. S8A; cf. Fig. 1C; Supplemental Fig. S1C). Host factor (∼18.81%) was still the most important variable affecting the functional profile variation, followed by food intake (∼1.74%). We further detected the eating-responsive KEGG pathways using the Z-score method. Pathways changed consistently among individuals upon eating (Supplemental Fig. S8B). Specifically, the proportions of bacterial chemotaxis, biosynthesis of unsaturated fatty acids, flagellar assembly, and chlorocyclohexane and chlorobenzene degradation increased most profoundly in the three subjects (Supplemental Fig. S8C). Twenty-five KEGG pathways showed statistically significant diurnal oscillation in two or more individuals, including apoptosis, glycosaminoglycan degradation, sphingolipid metabolism, steroid biosynthesis, and fatty acid biosynthesis (Supplemental Fig. S8D).
Dynamic site sources of saliva microbiota
Salivary microbiota is often regarded as a sink of bacteria shed from distinct oral surfaces. The intricate topology of the oral cavity creates profoundly different ecological niches (Proctor et al. 2020). Microbiota harbored in separate niches are very dissimilar. Certain microbes primarily localize in only one oral site (Segata et al. 2012; Mark Welch et al. 2019). To understand the dynamic contribution of other oral sites to the saliva microbiome, we estimated the source proportions of our densely sampled saliva microbiome with SourceTracker2 (Knights et al. 2011) by integrating the Human Microbiome Project (HMP) data with the closed-reference OTU picking strategy, because the HMP data set, which includes the comprehensive profiling of oral sites, is not directly compatible on the ASV analysis with our sequencing data.
The buccal mucosa and tongue dorsum were the predominant contributing sources, accounting for half of the total origins, followed by attached keratinized gingiva, supragingival plaque, palatine tonsils, subgingival plaque, hard palate, and throat (Supplemental Fig. S9A). The composition of these sources was similar among the three subjects. On average, 12%–17% of reads came from unknown sources, which may arise from the fact that the source (our samples) and the sink (HMP data) were from different individuals. Alternatively, it might be ascribable to other heterogeneous niches besides the nine oral sites that were studied in HMP (Segata et al. 2012).
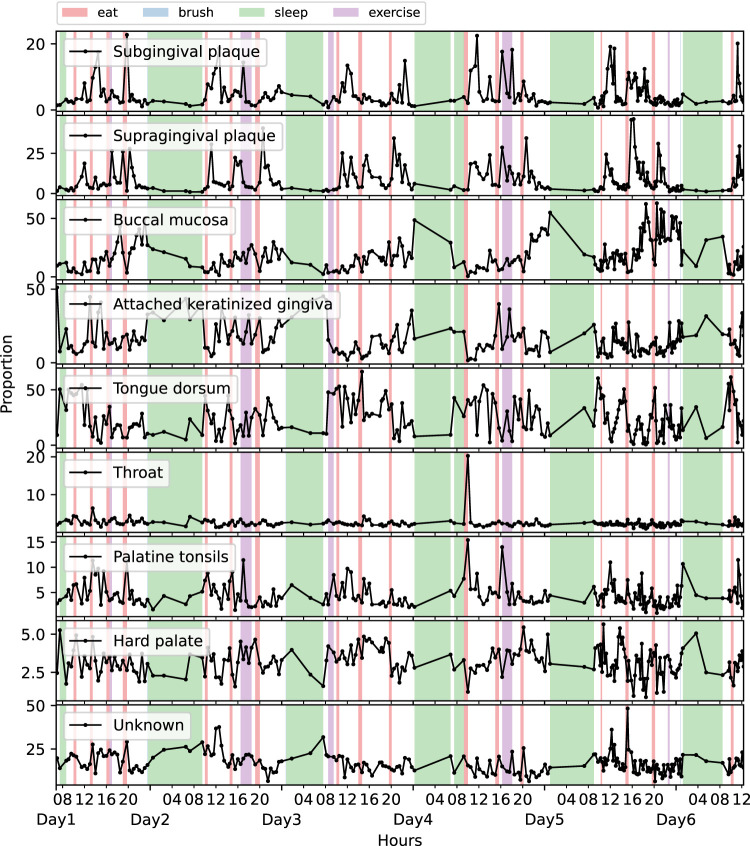
Daily activities affected the temporal source contributions (Fig. 6; Supplemental Fig. S9B,C). Both supragingival and subgingival plaque sources surged after eating, and they were usually low proportion at baseline level. The proportion of buccal mucosa source accumulated during the daytime and reached its lowest point in the morning. The attached keratinized gingiva source showed a similar trend with buccal mucosa, while it also seemed to increase during sleep. The proportion of tongue dorsum source rose immediately after waking up; throat, palatine tonsils, and unknown sources primarily increased at eating time points. Collectively, the source tracking analysis suggests that the microbes dislodged from various oral surfaces may significantly change the composition of the salivary microbiota. Daily activities may affect the extent and timing of this dislodging, thus probably contributing to the salivary microbiome patterns observed in the previous sections.
Figure 6.
Source tracker analysis in MA. The trace plots showing the proportional contributions from different oral sites at each time point.
Discussion
In this study, we tracked dynamic changes of the human salivary microbiome with nondestructive high-frequency sampling. Our dense time series data set revealed that, despite the relative stability within each individual over time, the salivary microbiome is highly dynamic and affected by daily activities. Preeminently, eating and sleep drive the microbial variability. We identified that specific microbes increased after every meal in a temporal order, which was reproducible in multiple subjects. Diurnal patterns of the salivary microbiome were unveiled at phylum, genus, and ASV levels. We also observed interesting microbial co-occurrences shared across three individuals, with inverse correlation existing between species within the same genus. Notably, similar results of these microbes were also identified when using the normalization method in the study by David et al. (2014) to account for the compositional effect in microbiome data, indicating the robustness of the results (Supplemental Fig. S10).
Longitudinal microbiome studies to date are sampled in months, days, or most frequently, hours (Caporaso et al. 2011; David et al. 2014; Hall et al. 2017; Marotz et al. 2021). These studies reported that the oral microbiota was relatively stable within individuals in the short-term (within days) or long-term (within years) (Caporaso et al. 2011; David et al. 2014; Hall et al. 2017; Marotz et al. 2021). Despite the stability in the overall structure, only a small fraction of operational taxonomic units (OTUs) were present across all the sampling time points (Caporaso et al. 2011; Hall et al. 2017), suggesting that the exact microorganisms in the community can fluctuate over time. The most intensive human microbiome research before this study took samples every 2 h (Marotz et al. 2021), in which they discovered that the oral microbiome composition was disturbed after an acute intervention trial using mouthwash and returned back to normal level within 2 h. These studies indicate that high-frequency sampling strategies are necessary to explore the precise dynamic changes of the oral microbiome.
We found that eating activity profoundly changed salivary microbiome composition. Alpha diversity increased during eating time windows, and several bacteria changed consistently upon every meal in multiple subjects. We could think of two potential explanations for these dynamic patterns: (1) mastication causes the microbes on other surfaces within the oral cavity to drop into the saliva; and (2) food-introduced nutrients promote the growth of the bacteria. The data in the soda intervention experiment conducted by Marotz et al. (2021) may provide some insights into this question. In that experiment, saliva samples were collected from seven individuals before each of them consumed a 12-ounce can of Coca-Cola soda as well as 15 min and 2 h afterward (Marotz et al. 2021). We reanalyzed this data set and found that drinking soda, which presumably provided carbohydrate without chewing movement, did not affect the abundance of those eating-responsive bacteria (Supplemental Fig. S11), indicating that mastication, rather than carbohydrate, gave rise to these microbial changes. However, it is worth noting that the increase of these bacteria in saliva lagged after meal in a temporal order, suggesting that these bacteria were not passively shed from other oral sites to saliva. Additionally, KEGG pathways associated with cell motility (e.g., bacterial chemotaxis and flagella assembly) congruently increased during eating. There could exist other mechanisms yet to be discovered by which oral bacteria dislodge themselves from biofilms. It warrants further investigation to mechanistically understand the contributions of all the factors to the rapid dynamics of these bacteria.
The eating-increased microbes rose in abundance in a temporal order. The early-increased bacteria were enriched on the tongue dorsum, and the late-increased bacteria were enriched on dental plaque. This site preference results are consistent with literature: Campylobacter concisus, Oribacterium spp., and Leptotrichia sp.HMT 221 are regarded as tongue specialists (Segata et al. 2012; Mark Welch et al. 2019), and Lautropia mirabilis and Rothia aeria are supragingival plaque specialists (Mark Welch et al. 2016, 2019; Utter et al. 2020). The source tracking analysis also showed that the proportional contributions of bacteria from supragingival plaque and subgingival plaque sources increased after each meal. Campylobacter concisus normally grows under anaerobic conditions (Lee et al. 2014). Ovesen et al. (2019) revealed that its motility, which was negatively correlated with biofilm forming capacity, was significantly increased under microaerophilic conditions compared to anaerobic environments. Speculatively, the increase of Campylobacter concisus in saliva might be related to fluctuations in oxygen exposure during eating, leading to its high motility and dislodging from oral surfaces. Besides, Campylobacter concisus is reported to be associated with inflammatory bowel disease (Zhang et al. 2014; Liu et al. 2018) and detected active in gastric fluid samples (von Rosenvinge et al. 2013). It would be interesting to study if Campylobacter concisus could transmit to the gastrointestinal tract and contribute to inflammatory bowel disease. Rothia aeria and Lautropia mirabilis were abundant in dental plaque and were reported as health-associated bacteria, as they were depleted in gingivitis and periodontitis populations (Huang et al. 2021). Their dynamics in saliva might reflect their shedding from dental plaque.
Many microbes in saliva showed diurnal oscillation. We detected more oscillating taxa in subject MA than in ME or MO, which is probably because more samples were collected during sleep from MA than ME and MO, leading to higher statistical power. Despite the diurnal variations among different individuals, some bacteria were found to be universally rhythmic. We observed an increase in the abundance of Prevotella and Alloprevotella at night, and they decreased sharply at breakfast. Prevotella and Alloprevotella were found positively correlated with the concentrations of volatile sulfur compound gases, such as H2S and CH3SH (Ye et al. 2020), and were enriched in individuals with halitosis (Takeshita et al. 2012; Yang et al. 2013; Ye et al. 2020). The increase of Prevotella and Alloprevotella at night might be linked to bad breath in the morning. These bacteria were more abundant in tongue samples compared to dental plaque samples, agreeing with the study by Mark Welch et al. (2019). Wilbert et al. (2020) visualized microbial spatial structure lining on the tongue surface using fluorescence spectral imaging, showing that Prevotella was more often observed as free floating or sparsely epithelial-bound bacteria than embedded in the densely structured microbial consortia that were organized around the keratinized epithelial cells. The result implies that Prevotella might attach loosely to the tongue surface. Hence, their dynamics may be closely associated with saliva flow. Their decrease during every meal, especially at breakfast, may result from the washing effects of saliva secretion. The low salivary flow at night in turn favors their increase (Pedersen et al. 2018).
Other diurnal oscillating bacteria include Bergeyella sp. HMT 206, Haemophilus, Granulicatella elegans, and Streptococcus. They increased from the late afternoon, stayed abundant in the evening, and decreased after waking up. They were more abundant in dental plaque samples compared to tongue dorsum samples, but our data only includes two oral sites, which may obscure the precise site preference. An oral microbiome research involving nine HMP oral sites suggested that Granulicatella elegans was enriched in keratinized gingiva/buccal mucosa (KG/BM); Streptococcus and Haemophilus are generalists that are abundant at most oral sites (Mark Welch et al. 2019). Our source tracking results showed that the proportion of KG and BM source showed a similar diurnal oscillation pattern as the dynamics of Granulicatella elegans. Research showed that the concentration of secretory immunoglobulin A (sIgA), which protects mucosal tissue and teeth from opportunistic pathogens, oscillates diurnally and was elevated at night (Shirakawa et al. 2004). Streptococcus and Haemophilus species possess the IgA1 protease activity (Cole et al. 1994; Spahich and St. Geme 2011; Huus et al. 2021), which might have connection to their high abundances at night.
Common co-occurring patterns were observed among different individuals. The positive correlations were mostly between the microbes from the same oral site, whereas the ASVs that tended to be enriched in different oral surfaces were largely negatively correlated in the networks. In our data set, two ASVs of Lautropia mirabilis were closely associated in all of three subjects. Only one species of Lautropia, L. mirabilis, has been identified, until recent research isolated a novel species named Lautropia dentalis from the subgingival dental plaque of a gingivitis patient (Lim et al. 2019). One Lautropia mirabilis ASV is 100% identical to this Lautropia dentalis strain KCOM 2505 16S ribosomal RNA; the other ASV sequence, with two bases differing from the first one, is 100% identical to L. mirabilis genome. The relative abundance of these two sequences did not shadow each other (Supplemental Fig. S12); thus, we believe that they are not different copies from the same strain but rather represent two unique strains present in the samples.
Our study provides insight into the baseline dynamics of the human salivary microbiome in healthy adults and deepens our understanding of the general rules governing the microbial variation. Our findings highlight the need to consider the influence of daily activities and diurnal oscillations in oral microbiome studies.
Methods
Recruitment criteria and participant demographics
Volunteers were laboratory researchers recruited from the University of Colorado at Boulder. Inclusion criteria included the following: healthy adults without usage of antibiotics or other medications in the last 3 mo and no oral diseases in the last 3 mo. Participant demographics are summarized in Supplemental Table S1. The time series experiment included three volunteers: one female and two males, aged 20–45 yr old; and the lunch experiment included 19 volunteers: 6 females and 13 males, aged 20–45 yr old.
Saliva sampling and metadata
Time series data set
Three healthy subjects were tracked for 6 consecutive days. Their saliva samples were collected every 10–60 min, except when sleeping, by gently touching saliva on tongue surfaces (without scraping) using sterile swabs (BD BBL CultureSwab 220145). The first sample within a day was taken right after waking up, and the last sample was taken before sleep. Self-reported daily activities, including sleep, food intake, physical exercise, and tooth brushing, were recorded at the sampling time. Samples were collected and frozen at −20°C. After 6 d of experiments, samples were transported collectively to the laboratory on ice and stored at −20°C until further processing. In total, 614 samples were collected (subject MA: 212 samples; subject ME: 221 samples; subject MO: 181 samples).
Lunch data set
Nineteen volunteers were recruited to have lunch together. Saliva samples were collected at 12 fixed time points for all the volunteers: −46, −36, −24, 25, 37, 47, 65, 76, 87, 98, 109, and 121 min, with time of 0 min indicating the start of a meal. All the individuals had finished eating at time of 25 min. In total, 222 samples were collected.
MA validation data set
MA was sampled for another 1.5 d, 11 mo apart from the time series data set mentioned above. Additionally, an extra 13 dental plaque samples and six tongue dorsum samples were scraped from subject MA. Dental plaque samples were collected by repeated swabbing (30 sec) of the facial supragingival surfaces of the upper and lower incisors, canines, and premolars. Tongue dorsum samples were collected by repeated swabbing (30 sec) of the entire dorsal tongue surface, as described in Costello et al. (2009).
16S rRNA gene amplicon sequencing
DNA purification and 16S rRNA amplicon sequencing was performed using Earth Microbiome Project standard protocols (https://earthmicrobiome.org/protocols-and-standards/). In brief, DNA was extracted using the MoBio PowerSoil DNA Isolation kit (Qiagen) as described in Marotz et al. (2017). Amplicon library was constructed using universal primers (F515/R806) targeted across the V4 region of the 16S rRNA (Caporaso et al. 2012) and sequenced on the Illumina HiSeq 4000 sequencing platform. Blank negative controls were used and went through all the sample processing steps together with the saliva samples (Supplemental Fig. S13A). During processing, all samples were randomized across the 96-well plates to reduce systematic bias in contaminations and well-to-well effects (Minich et al. 2019).
Microbial data analysis
We used Quantitative Insights Into Microbial Ecology version 2 (QIIME2, version 2019.4) tool to process raw sequencing data into an ASV table (Bolyen et al. 2019). Raw reads were first filtered based on sequence quality scores using qiime quality-filter q-score command with default settings (Bokulich et al. 2013), and next processed using the Deblur algorithm, resulting in high-quality 150-bp sequence variant data with single-nucleotide resolution. Deblur pipeline did sequence quality control by removing singleton and artifactual reads, followed by de novo chimera removal (Amir et al. 2017). A median was obtained of 8484 reads per sample (Supplemental Fig. S13B). Samples were next rarefied to 2533 reads according to the rarefaction curve (Supplemental Fig. S13C).
The taxonomy assignment for the oral ASVs was improved by matching against the Human Oral Microbiome Database (HOMD, 16S rRNA RefSeq V15.2) (Chen et al. 2010) compared to Greengenes (Supplemental Fig. S1G). The ASVs with <97% similarity against HOMD were defined as non-oral ASVs, which includes 19 chloroplast, 23 mitochondrial, and 538 other non-oral ASVs. Their abundances in all samples were very low (on average 0.849%, 0.135%, and 0.560%, respectively). These sequences were mostly from food and discarded from downstream analyses. One sample in MO contained high abundant non-oral bacteria, which was likely contaminated, and was also discarded (Supplemental Fig. S2B).
Alpha and beta diversity metrics were computed by q2-diversity plugin. PCoA plots were generated using Emperor (Vázquez-Baeza et al. 2013). Effect sizes of variables on microbiome composition were calculated as previously described (Dimitriu et al. 2019).
Besides rarefaction, we also used another normalization method as described previously (David et al. 2014) to mitigate the compositionality problem in microbiome analysis.
Eating-responsive bacteria analysis
We developed a Z-score method to measure how much the abundance change induced by eating deviates from normal fluctuations for a given ASV. Z-score is frequently used in bioinformatic analysis to quantify the deviation of the observed value from the random expected distribution (Rouskin et al. 2014; Mathys et al. 2019). Specifically, for a given ASV in a subject, we computed a vector D consisting of the abundance differences between every two samples that were collected within 1 h. The vector D represents the distribution of normal fluctuations for this ASV. For each eating time window, we computed the Z-score as , where d is the abundance fluctuation induced by eating, is the mean of D, and σ (D) is the standard deviation of D. The mean of the Z-scores of all the eating windows was the final measure on how much this ASV responded to eating.
A rooted phylogenetic tree was generated by aligning sequences with MAFFT (Katoh and Standley 2013) and building a tree with FastTree (using the QIIME2 phylogeny plugin) (Price et al. 2010). The heatmap of Z-scores of ASVs was created using an online tool, Interactive Tree of Life (iTOL) (Ciccarelli et al. 2006; Letunic and Bork 2019).
Eight ASVs with the highest Z-scores were further analyzed by integrating and averaging all the eating time windows to reveal the temporal order of these bacteria. These ASVs were further scaled to the same numeric scale and smoothed with moving average for visualization.
Diurnal oscillation detection
We applied the Lomb–Scargle (LS) method in the R MetaCycle package to detect the rhythmic pattern of 24 h (±2 h) in our 6-d longitudinal study (Glynn et al. 2006; Wu et al. 2016). The Meta3d function, which integrates analysis results from multiple individuals, was used to detect the periodic signal from the time series data. FDR ≤ 0.01 was considered statistically significant. Statistically significant phyla, genera, and ASVs in the salivary microbiome were plotted in Supplemental Figures S3–S5.
Microbial co-occurrence network
We applied CCLasso and Spearman's correlation to estimate microbial co-occurrence. Benchmark showed that they performed well in inferring relationships based on the compositional microbiome data (Hirano and Takemoto 2019). CCLasso was run with default settings.
The network was visualized via Cytoscape (version 3.7.2) for each subject (Shannon et al. 2003). To identify the general co-occurring relationships among multiple individuals, an identical subset of nodes was included. The nodes were selected if they were strongly associated (|correlation| ≥ 0.6 and FDR ≤ 0.01) in any of the three individuals. Significant associations with FDR ≤ 0.01 were displayed in the visualized networks.
An extra set of 12 samples from dental plaque and six samples from tongue dorsum were collected from subject MA. A permutation-based nonparametric test followed by FDR correction (calour.analysis.diff_abundance) was applied to detect bacterial site preference (Xu et al. 2019).
Prediction of metagenomic functions
Potential metagenome functions were predicted using PICRUSt2 (Douglas et al. 2020). Kyoto Encyclopedia of Gene and Genomes (KEGG) pathways abundances were inferred by mapping file of KEGG orthologs to pathways. The distribution of the NSTI scores for ASVs in the time series data set was shown in Supplemental Figure S14. Only 0.02% ASVs scored greater than 2, which is the default maximum NSTI cutoff in PICRUSt2. The mean value of the NSTI scores was 0.05. These results indicate a reasonable reliability of the PICRUSt2 prediction. Downstream analyses were performed as same as microbiome data.
SourceTracker analysis
We downloaded the 16S v3-v5 OTU table from HMP (https://hmpdacc.org/hmp/HMQCP/). To match up HMP sequences and our time series data, we used closed-reference clustering on both sets of the sequences using q2-vsearch plugin (Rognes et al. 2016) against HOMD (16S rRNA RefSeq V15.2) based on 97% identity. We then applied SourceTracker (v2) to estimate the potential sources of oral microbial community (Knights et al. 2011). HMP samples covering eight oral sites (supragingival plaque, subgingival plaque, buccal mucosa, keratinized gingiva, hard palate, tongue dorsum, throat, and palatine tonsils) were used as sources; our saliva samples were used as sinks.
Soda intervention data set
A previous study investigated the effect of four acute treatments (including soda) on saliva microbial composition (Marotz et al. 2021). The data set was downloaded from Qiita (Gonzalez et al. 2018) under study ID 11899. Non-oral ASVs were removed as described previously. Our discovered top eating-responsive ASVs in the time series experiment were visualized before and after the soda intervention with Matplotlib.
Ethics approval and consent to participate
Volunteers were recruited under IRB number 150275 approved by the UCSD Human Research Protections Program.
Data access
The raw sequencing data and metadata from this study have been submitted to Qiita (https://qiita.ucsd.edu) under study IDs 14062 (time series), 14057 (lunch), 14058 (MA validation), and 14059 (tongue and plaque samples), and the European Nucleotide Archive (https://www.ebi.ac.uk/ena/browser/home) under accession numbers ERP133012 (time series), ERP133010 (lunch), ERP133009 (MA validation), and ERP133011 (tongue and plaque samples). The ASV table obtained after removing non-oral sequences is available as Supplemental Table S2. The code is available as Supplemental Code and at GitHub (https://github.com/maque4004/saliva-time-series-analyses).
Supplementary Material
Acknowledgments
This research was supported by the National Natural Science Foundation of China (grant no. 31970088) and the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (grant no. 2020YFA0509600).
Author contributions: A.A., Z.Z.X., and R.K. conceived and designed the study. A.A. and Z.Z.X. ran the experiment. A.A., Y.H., Y.L., and Z.Z.X. analyzed and interpreted the data. Z.Z.X. and Y.H. wrote the manuscript, and all authors edited the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276482.121.
Competing interest statement
The authors declare no competing interests.
References
- Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, et al. 2017. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2: e00191-16. 10.1128/mSystems.00191-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belstrøm D, Holmstrup P, Bardow A, Kokaras A, Fiehn NE, Paster BJ. 2016. Temporal stability of the salivary microbiota in oral health. PLoS One 11: e0147472. 10.1371/journal.pone.0147472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10: 57–59. 10.1038/nmeth.2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37: 852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13: 581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, et al. 2011. Moving pictures of the human microbiome. Genome Biol 12: R50. 10.1186/gb-2011-12-5-r50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6: 1621–1624. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010: baq013. 10.1093/database/baq013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhibber-Goel J, Singhal V, Bhowmik D, Vivek R, Parakh N, Bhargava B, Sharma A. 2016. Linkages between oral commensal bacteria and atherosclerotic plaques in coronary artery disease patients. NPJ Biofilms Microbiomes 2: 7. 10.1038/s41522-016-0009-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287. 10.1126/science.1123061 [DOI] [PubMed] [Google Scholar]
- Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, Wu WK, Wong SH, Chen Z, Sung JJY, et al. 2018. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 67: 1024–1032. 10.1136/gutjnl-2017-314281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MF, Evans M, Fitzsimmons S, Johnson J, Pearce C, Sheridan MJ, Wientzen R, Bowden G. 1994. Pioneer oral streptococci produce immunoglobulin A1 protease. Infect Immun 62: 2165–2168. 10.1128/iai.62.6.2165-2168.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697. 10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. 2014. Host lifestyle affects human microbiota on daily timescales. Genome Biol 15: R89. 10.1186/gb-2014-15-7-r89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes C. 2003. Estimates, from salivary analyses, of the turnover time of the oral mucosal epithelium in humans and the number of bacteria in an edentulous mouth. Arch Oral Biol 48: 329–336. 10.1016/S0003-9969(03)00014-1 [DOI] [PubMed] [Google Scholar]
- Dimitriu PA, Iker B, Malik K, Leung H, Mohn WW, Hillebrand GG. 2019. New insights into the intrinsic and extrinsic factors that shape the human skin microbiome. mBio 10: e00839-19. 10.1128/mBio.00839-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A, Nguyen M, Haditsch U, Raha D, Griffin C, et al. 2019. Porphyromonas gingivalis in Alzheimer's disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv 5: eaau3333. 10.1126/sciadv.aau3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI. 2020. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38: 685–688. 10.1038/s41587-020-0548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML. 2013. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4: 1111–1119. 10.1111/2041-210X.12114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Borisy GG, Huse SM, Mark Welch JL. 2014. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci 111: E2875–E2884. 10.1073/pnas.1409644111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, Purdue MP, Abnet CC, Stolzenberg-Solomon R, Miller G, et al. 2018. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 67: 120–127. 10.1136/gutjnl-2016-312580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Huang C, Zhao H, Deng M. 2015. CCLasso: correlation inference for compositional data through Lasso. Bioinformatics 31: 3172–3180. 10.1093/bioinformatics/btv349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, Hurley E, O'Riordain M, Shanahan F, O'Toole PW. 2018. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 67: 1454–1463. 10.1136/gutjnl-2017-314814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiser RA, Halimi A, Alkharaan H, Lu L, Davanian H, Healy K, Hugerth LW, Ateeb Z, Valente R, Moro CF, et al. 2019. Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut 68: 2186–2194. 10.1136/gutjnl-2018-317458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. 2014. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15: 382–392. 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn EF, Chen J, Mushegian AR. 2006. Detecting periodic patterns in unevenly spaced gene expression time series using Lomb–Scargle periodograms. Bioinformatics 22: 310–316. 10.1093/bioinformatics/bti789 [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Navas-Molina JA, Kosciolek T, McDonald D, Vázquez-Baeza Y, Ackermann G, DeReus J, Janssen S, Swafford AD, Orchanian SB, et al. 2018. Qiita: rapid, web-enabled microbiome meta-analysis. Nat Methods 15: 796–798. 10.1038/s41592-018-0141-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MW, Singh N, Ng KF, Lam DK, Goldberg MB, Tenenbaum HC, Neufeld JD, Beiko RG, Senadheera DB. 2017. Inter-personal diversity and temporal dynamics of dental, tongue, and salivary microbiota in the healthy oral cavity. NPJ Biofilms Microbiomes 3: 2. 10.1038/s41522-016-0011-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano H, Takemoto K. 2019. Difficulty in inferring microbial community structure based on co-occurrence network approaches. BMC Bioinformatics 20: 329. 10.1186/s12859-019-2915-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, He T, Yue F, Xu V, Wang S, Zhu P, Teng F, Sun Z, Liu X, Jing G, et al. 2021. Longitudinal multi-omics and microbiome meta-analysis identify an asymptomatic gingival state that links gingivitis, periodontitis, and aging. mBio 12: e03281-20. 10.1128/mBio.03281-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huus KE, Petersen C, Finlay BB. 2021. Diversity and dynamism of IgA−microbiota interactions. Nat Rev Immunol 21: 514–525. 10.1038/s41577-021-00506-1 [DOI] [PubMed] [Google Scholar]
- Johnson AJ, Vangay P, Al-Ghalith GA, Hillmann BM, Ward TL, Shields-Cutler RR, Kim AD, Shmagel AK, Syed AN, Walter J, et al. 2019. Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe 25: 789–802.e5. 10.1016/j.chom.2019.05.005 [DOI] [PubMed] [Google Scholar]
- Kahharova D, Brandt BW, Buijs MJ, Peters M, Jackson R, Eckert G, Katz B, Keels MA, Levy SM, Fontana M, et al. 2020. Maturation of the oral microbiome in caries-free toddlers: a longitudinal study. J Dent Res 99: 159–167. 10.1177/0022034519889015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Barraza JP, Arthur RA, Hara A, Lewis K, Liu Y, Scisci EL, Hajishengallis E, Whiteley M, Koo H. 2020. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc Natl Acad Sci 117: 12375–12386. 10.1073/pnas.1919099117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. 2011. Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8: 761–763. 10.1038/nmeth.1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Ma R, Grimm MC, Riordan SM, Lan R, Zhong L, Raftery M, Zhang L. 2014. Examination of the anaerobic growth of Campylobacter concisus strains. Int J Microbiol 2014: 476047. 10.1155/2014/476047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2019. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47: W256–W259. 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YK, Park SN, Lee WP, Shin JH, Jo E, Shin Y, Paek J, Chang YH, Kim H, Kook JK. 2019. Lautropia dentalis sp. nov., isolated from human dental plaque of a gingivitis lesion. Curr Microbiol 76: 1369–1373. 10.1007/s00284-019-01761-1 [DOI] [PubMed] [Google Scholar]
- Liu F, Ma R, Wang Y, Zhang L. 2018. The clinical importance of Campylobacter concisus and other human hosted Campylobacter species. Front Cell Infect Microbiol 8: 243. 10.3389/fcimb.2018.00243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. 2016. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci 113: E791–E800. 10.1073/pnas.1522149113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch JL, Dewhirst FE, Borisy GG. 2019. Biogeography of the oral microbiome: the site-specialist hypothesis. Annu Rev Microbiol 73: 335–358. 10.1146/annurev-micro-090817-062503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch JL, Ramírez-Puebla ST, Borisy GG. 2020. Oral microbiome geography: micron-scale habitat and niche. Cell Host Microbe 28: 160–168. 10.1016/j.chom.2020.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotz C, Amir A, Humphrey G, Gaffney J, Gogul G, Knight R. 2017. DNA extraction for streamlined metagenomics of diverse environmental samples. BioTechniques 62: 290–293. 10.2144/000114559 [DOI] [PubMed] [Google Scholar]
- Marotz C, Morton JT, Navarro P, Coker J, Belda-Ferre P, Knight R, Zengler K. 2021. Quantifying live microbial load in human saliva samples over time reveals stable composition and dynamic load. mSystems 6: e01182-20. 10.1128/mSystems.01182-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, Menon M, He L, Abdurrob F, Jiang X, et al. 2019. Single-cell transcriptomic analysis of Alzheimer's disease. Nature 570: 332–337. 10.1038/s41586-019-1195-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minich JJ, Sanders JG, Amir A, Humphrey G, Gilbert JA, Knight R. 2019. Quantifying and understanding well-to-well contamination in microbiome research. mSystems 4: e00186-19. 10.1128/mSystems.00186-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WKK, Ng SC, Tsoi H, Dong Y, Zhang N, et al. 2015. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun 6: 8727. 10.1038/ncomms9727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki EM, Shroff R, Singleton JA, Renaud DE, Wallace D, Drury J, Zirnheld J, Colleti B, Ellington AD, Lamont RJ, et al. 2018. Microbiota and metatranscriptome changes accompanying the onset of gingivitis. mBio 9: e00575-18. 10.1128/mBio.00575-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovesen S, Durack J, Kirk KF, Nielsen HL, Nielsen H, Lynch SV. 2019. Motility and biofilm formation of the emerging gastrointestinal pathogen Campylobacter concisus differs under microaerophilic and anaerobic environments. Gut Microbes 10: 34–44. 10.1080/19490976.2018.1472201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen A, Sørensen C, Proctor G, Carpenter G. 2018. Salivary functions in mastication, taste and textural perception, swallowing and initial digestion. Oral Dis 24: 1399–1416. 10.1111/odi.12867 [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor DM, Shelef KM, Gonzalez A, Davis CL, Dethlefsen L, Burns AR, Loomer PM, Armitage GC, Ryder MI, Millman ME, et al. 2020. Microbial biogeography and ecology of the mouth and implications for periodontal diseases. Periodontol 2000 82: 26–41. 10.1111/prd.12268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4: e2584. 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. 2014. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505: 701–705. 10.1038/nature12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J. 2012. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol 13: R42. 10.1186/gb-2012-13-6-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B, Chang M, Martin J, Mitreva M, Lux R, Klokkevold P, Sodergren E, Weinstock GM, Haake SK, Li H. 2015. Dynamic changes in the subgingival microbiome and their potential for diagnosis and prognosis of periodontitis. mBio 6: e01926-14. 10.1128/mBio.01926-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa T, Mitome M, Oguchi H. 2004. Circadian rhythms of S-IgA and cortisol in whole saliva—compensatory mechanism of oral immune system for nocturnal fall of saliva secretion—. Pediatr Dent J 14: 115–120. 10.1016/S0917-2394(04)70017-8 [DOI] [Google Scholar]
- Spahich NA, St. Geme JW III. 2011. Structure and function of the Haemophilus influenzae autotransporters. Front Cell Infect Microbiol 1: 5. 10.3389/fcimb.2011.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung JJY, Coker OO, Chu E, Szeto CH, Luk STY, Lau HCH, Yu J. 2020. Gastric microbes associated with gastric inflammation, atrophy and intestinal metaplasia 1 year after Helicobacter pylori eradication. Gut 69: 1572–1581. 10.1136/gutjnl-2019-319826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayasu L, Suda W, Takanashi K, Iioka E, Kurokawa R, Shindo C, Hattori Y, Yamashita N, Nishijima S, Oshima K, et al. 2017. Circadian oscillations of microbial and functional composition in the human salivary microbiome. DNA Res 24: 261–270. 10.1093/dnares/dsx001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita T, Suzuki N, Nakano Y, Yasui M, Yoneda M, Shimazaki Y, Hirofuji T, Yamashita Y. 2012. Discrimination of the oral microbiota associated with high hydrogen sulfide and methyl mercaptan production. Sci Rep 2: 215. 10.1038/srep00215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Yang F, Huang S, Bo C, Xu ZZ, Amir A, Knight R, Ling J, Xu J. 2015. Prediction of early childhood caries via spatial-temporal variations of oral microbiota. Cell Host Microbe 18: 296–306. 10.1016/j.chom.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Utter DR, Borisy GG, Eren AM, Cavanaugh CM, Mark Welch JL. 2020. Metapangenomics of the oral microbiome provides insights into habitat adaptation and cultivar diversity. Genome Biol 21: 293. 10.1186/s13059-020-02200-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. 2013. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2: 16. 10.1186/2047-217X-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Rosenvinge EC, Song Y, White JR, Maddox C, Blanchard T, Fricke WF. 2013. Immune status, antibiotic medication and pH are associated with changes in the stomach fluid microbiota. ISME J 7: 1354–1366. 10.1038/ismej.2013.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Joyce LR, Wall AM, Guan Z, Palmer KL. 2021. Streptococcus pneumoniae, S. mitis, and S. oralis produce a phosphatidylglycerol-dependent, ltaS-independent glycerophosphate-linked glycolipid. mSphere 6: e01099-20. 10.1128/mSphere.01099-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbert SA, Mark Welch JL, Borisy GG. 2020. Spatial ecology of the human tongue dorsum microbiome. Cell Rep 30: 4003–4015.e3. 10.1016/j.celrep.2020.02.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB. 2016. MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics 32: 3351–3353. 10.1093/bioinformatics/btw405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZZ, Amir A, Sanders J, Zhu Q, Morton JT, Bletz MC, Tripathi A, Huang S, McDonald D, Jiang L, et al. 2019. Calour: an interactive, microbe-centric analysis tool. mSystems 4: e00269-18. 10.1128/mSystems.00269-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Huang S, He T, Catrenich C, Teng F, Bo C, Chen J, Liu J, Li J, Song Y, et al. 2013. Microbial basis of oral malodor development in humans. J Dent Res 92: 1106–1112. 10.1177/0022034513507065 [DOI] [PubMed] [Google Scholar]
- Ye W, Zhang Y, He M, Zhu C, Feng XP. 2020. Relationship of tongue coating microbiome on volatile sulfur compounds in healthy and halitosis adults. J Breath Res 14: 016005. 10.1088/1752-7163/ab47b4 [DOI] [PubMed] [Google Scholar]
- Zhang L, Lee H, Grimm MC, Riordan SM, Day AS, Lemberg DA. 2014. Campylobacter concisus and inflammatory bowel disease. World J Gastroenterol 20: 1259–1267. 10.3748/wjg.v20.i5.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, et al. 2015. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 21: 895–905. 10.1038/nm.3914 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.