Abstract
The identification of Cryptosporidium oocysts in environmental samples is largely made by the use of an immunofluorescent assay. In this study, we have used a small-subunit rRNA-based PCR-restriction fragment length polymorphism technique to identify species and sources of Cryptosporidium oocysts present in 29 storm water samples collected from a stream in New York. A total of 12 genotypes were found in 27 positive samples; for 4 the species and probable origins were identified by sequence analysis, whereas the rest represent new genotypes from wildlife. Thus, this technique provides an alternative method for the detection and differentiation of Cryptosporidium parasites in environmental samples.
Cryptosporidiosis is a significant cause of waterborne outbreaks of diarrheal diseases. Molecular typing tools have shown that two genotypes of Cryptosporidium parvum are responsible for these outbreaks (28, 35). The human genotype (genotype 1) parasites have so far been found only in humans, whereas the bovine genotype (genotype 2) parasites have been found in farm animals and some humans (3–5, 23, 24, 28, 31, 33, 35, 36, 39, 40). Although a high prevalence of Cryptosporidium oocysts has been found in water (18, 19, 21, 30), a direct laboratory linkage between oocysts found in water and parasites in outbreak cases has not been made. This could be due to the lack of sensitivity and specificity of oocyst detection in environmental samples. More sensitive identification and differentiation of Cryptosporidium parasites in water samples might be invaluable in the evaluation of sources of environmental contamination.
Attempts have been made to use molecular techniques for the analysis of environmental samples (7, 9, 11–14, 16, 29, 41). Although earlier molecular diagnostic techniques did not have the capability to differentiate Cryptosporidium oocysts at the species and strain levels, recent advances in the molecular characterization of Cryptosporidium parasites now make this possible. Three small-subunit rRNA (SSU rRNA) gene-based PCR-restriction fragment length polymorphism (RFLP) techniques have been developed to differentiate C. parvum, Cryptosporidium muris, and Cryptosporidium baileyi (2, 15, 20). Many protocols have also been described to differentiate the human and bovine genotypes of C. parvum (3–5, 23, 24, 28, 31–33, 35–37, 39, 40). Although most of these techniques perform satisfactorily for the analysis of fecal samples or purified oocysts, the usefulness of some techniques for the analysis of environmental samples has been questioned recently (29, 41). For example, the species-differentiating techniques use primers that cross-react with other apicomplexan parasites or other eukaryotic organisms, which leads to reduced specificity due to interference from other organisms present in clinical and environmental samples (34). Most of the C. parvum genotyping tools fail to amplify genomic DNA from other human-pathogenic Cryptosporidium parasites (the C. parvum dog genotype, C. meleagridis, and C. felis) and therefore may lead to underestimation of the hazardous potential of oocysts found in waters. The single-step PCR format used for most genotyping methods may also lack the sensitivity required for the analysis of environmental samples. The potential of genotyping tools in the analysis of environmental samples, however, has been demonstrated recently. Six types of 70-kDa heat shock protein (HSP70) nucleotide sequences from the C. parvum bovine, mouse, and human genotypes have been found in cell cultures inoculated with oocysts isolated from environmental samples (8, 37).
We and others have recently found the presence of various host-adapted Cryptosporidium species and strains (25, 42, 44). An SSU rRNA-based nested PCR-RFLP technique was developed for the differentiation of Cryptosporidium species and C. parvum strains in fecal samples from humans and animals (42, 44). In the present study, we evaluated the performance of this technique in the analysis of Cryptosporidium parasites in storm water samples collected from a catchment area of the New York City water system.
Water samples and sample processing.
Most storm water samples were collected from Ashokan Brook, which drains the Ashokan Brook basin located in the Eastern Catskill Mountains in New York State and contributes to the New York City water supply, except for two samples which were collected from Johnson Hollow Brook. The drainage basin for Ashokan Brook is mostly undeveloped and forested, consisting of 84% forested areas, 10% grassy land, 2% wetland, and approximately 4% impervious surface (homes and roads). Storm flow in the stream during collections at times exceeded 363 ft3/s, and turbidity during storms usually ranged from 7 to 100 nephelometric turbidity units. The water pH was fairly stable during storms, and dissolved oxygen ranged between 9 and 11 mg/liter.
Most storm samples used in this study were collected after seven storm events between May 1999 and March 2000, with the exception of one sample (sample N), which was taken in September 1998 (Table 1). Several water samples were taken from the same site during each storm, at intervals of 0.5 to 1 h. One-hour composites, with an average sample volume of 59 gal, or 50-gal grab samples of stream water were collected for each sample.
TABLE 1.
Storm water samples used in this study
| Collection date (mo/day/yr) | Collection sitea | No. of samples | Genotype name(s) (no. of positive samples) |
|---|---|---|---|
| 9/22/98 | E13i | 1 | W5 (1) |
| 5/19/99 | E13i | 6 | W1 (2), W4 (5), W7 (1), W11 (1) |
| 8/14/99 | E13i | 4 | W6 (4), W7 (1), W11 (3) |
| 9/7/99 | E13i | 4 | W1 (2), W3 (1), W8 (2), W10 (3) |
| 9/16/99 | FB4 | 2 | W3 (1)b |
| 11/2/99 | E13i | 4 | W1 (1), W2 (2), W7 (1), W9 (1) |
| 1/10/00 | E13i | 4 | W2 (1), W4 (2), W7 (2) |
| 3/16/00 | E13i | 4 | W7 (3), W12 (1)b |
E13i, Ashokan Brook; FB4, Johnson Hollow Brook.
One sample was negative for the given genotype.
The samples used to determine the number of oocysts by microscopy were prepared using capture by filtration, concentration by centrifugation, and purification by sucrose-Percoll flotation using the Information Collection Rule (ICR) method recommended by the U.S. Environmental Protection Agency (38). Final sample concentrates were filtered through cellulose acetate filters on a Hoefer manifold, stained using a Hydrofluor Combo immunofluorescent detection procedure, rinsed, and placed on 75- by 38-mm slides as described for the ICR method. Slides were examined for the presence of Cryptosporidium oocysts under an epifluorescent microscope. Cryptosporidium oocysts were diagnosed by fluorescence characteristics, size, and shape and confirmed by the presence of internal structures under differential interference contrast microscopy. Usually, only 0.5 ml of the original ICR pellet was examined by microscopy for each sample, which was the equivalent of 1.5 to 20 gal of storm water. The subsample concentrates used for the PCR-RFLP and sequencing analysis were prepared using pellet material remaining from the original centrifugation procedure completed for the ICR method (total pellet size was between 1.5 and 20 ml, depending on the water turbidity during sampling). Multiple (usually two) 0.5-ml pellet portions (each equivalent to 3 to 40 gal of storm water) were separated from the remaining pellet and were purified as separate aliquots using sucrose-Percoll flotation. Each float suspension was reduced to 5 ml, vortexed, and combined with the other floated subsample of the original pellet to maximize possible oocyst recovery. After tubes were rinsed with a small amount of eluting solution, the final subsample concentrate volume submitted for PCR-RFLP and sequencing analysis was approximately 10 ml.
DNA extraction.
Cryptosporidium oocysts present in water samples concentrated by filtration and sucrose-Percoll flotation were further purified by immunomagnetic separation (IMS), using magnetic beads coated with an anti-Cryptosporidium monoclonal antibody (Dynal, Lake Success, N.Y.) and following the manufacturer's recommended procedures. The IMS-purified oocysts were then subjected to five freeze-thaw cycles, incubated with 1 mg of proteinase K (Sigma, St. Louis, Mo.) per ml at 56°C for at least 1 h, and diluted with an equal volume of pure ethanol. Oocyst DNA was extracted by passing the oocyst-ethanol suspension through QIAamp DNA Mini isolate columns (Qiagen, Valencia, Calif.).
PCR-RFLP analysis.
Cryptosporidium oocysts in water samples were identified to the species and genotype levels by a previously described PCR-RFLP technique (42, 44), except that a correction (change of TAA to ATT) was made in the sequence of the reverse primer for primary PCR (the corrected primary reverse primer is 5′-CCCATTTCCTTCGAAACAGGA-3′). Each sample was analyzed at least three times by PCR-RFLP, using different volumes of DNA preparation (0.25, 0.5, and 1 μl) for the PCR.
Sequence analysis.
For confirmation, the secondary PCR products were sequenced using an ABI377 autosequencer. Sequence accuracy was confirmed by two-directional sequencing and by sequencing of a second PCR product. Nucleotide sequences generated were aligned with each other and with known Cryptosporidium species and a C. parvum genotype previously obtained by us, using the computer software Wisconsin Package, version 9.0 (Genetics Computer Group, Madison, Wis.) and manual adjustment. Phylogenetic analysis was used for the aligned sequences to assess the relationship among isolates (42, 44). Neighbor-joining trees were constructed with the program Treecon W, based on the evolutionary distances between different isolates calculated by Kimura two-parameter analysis and using C. muris and Cryptosporidium serpentis as the out-group to assess the relatedness of isolates from water samples. Tree reliability was assessed by the bootstrap method with 1,000 pseudoreplicates, and only values above 50% are reported (42, 44).
Detection of heterogeneous genotypes of Cryptosporidium parasites in storm water samples by PCR-RFLP.
The SSU rRNA-based PCR-RFLP technique was initially used for the analysis of DNA directly extracted from 11 water pellets prepared by the ICR method, using the traditional phenol-chloroform extraction technique (42, 44). No positive amplification was achieved from these samples, although eight samples were positive for Cryptosporidium oocysts by microscopic examinations, and the corresponding volume of water examined for these samples was similar to that for the samples processed further by IMS (see below). Even after secondary purification of the extracted DNA through passage in Qiagen columns, none of the samples produced positive results. Analysis with spiked Cryptosporidium DNA had shown that the DNA extracted from water pellets was inhibitory to PCR (data not shown).
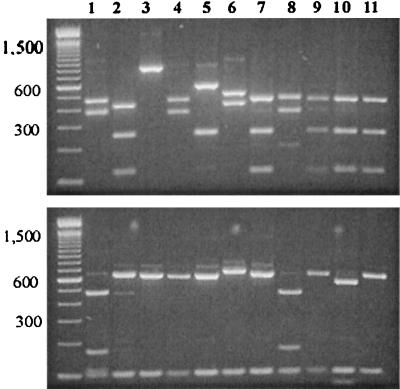
Subsequently, the PCR-RFLP technique was used for the analysis of DNA extracted by Qiagen columns from oocysts purified by IMS. Twenty-seven of the 29 storm water samples analyzed by this technique produced positive PCR amplification, including 12 of 13 samples that were negative by microscopy (Table 1). RFLP analysis of the secondary PCR products revealed extensive differences in the band patterns between different samples (Fig. 1). At least seven RFLP band patterns were seen. Only four of the seven RFLP patterns showed similarity to RFLP patterns of known Cryptosporidium parasites from animals genetically characterized by us (Fig. 1, lanes 3, 5, 6, 7, and 9). Some samples apparently had mixed Cryptosporidium parasites, as shown by the presence of extra RFLP bands (data not shown).
FIG. 1.
Differentiation of the Cryptosporidium parasites in storm water samples by SSU rRNA-based PCR-RFLP. Lanes 1, 2, 4, 8, 10, and 11, unknown Cryptosporidium spp.; lane 3, Cryptosporidium from snakes; lane 5, C. baileyi; lane 6, Cryptosporidium opossum genotype 2; lanes 7 and 9, C. parvum bovine-like genotype.
Nucleotide sequence characterization of Cryptosporidium parasites from storm water.
To confirm the identification of Cryptosporidium parasites, all secondary PCR products were sequenced. Twelve major sequence types were obtained, and these were named W1 to W12 (Fig. 2). Four of the genotypes showed 100% homology to nucleotide sequences we previously obtained for various animals; W2 was identical to Cryptosporidium opossum genotype 1, W8 was identical to Cryptosporidium opossum genotype 2, W10 was identical to C. baileyi, and W11 was identical to an unnamed Cryptosporidium parasite from snakes. PCR products from 12 samples had multiple genotypes, as shown by the underlying signals in the electropherogram of the autosequencing results. Sequencing of multiple PCR products confirmed the presence of different genotypes in these samples (Table 1 and Fig. 1). However, there were usually only one or two genotypes dominating each sampling time. For example, W6 was found in all samples taken on 14 August 1999, and W4 was seen in five of six samples taken on 19 May 1999 (Table 1).
FIG. 2.
Nucleotide sequence diversity among Cryptosporidium water genotypes in one of the polymorphic regions of the SSU rRNA gene. Dots denote nucleotides identical to those from the C. parvum bovine (Bov) genotype sequence, and dashes indicate deletions. Hum, C. parvum human genotype.
Genetic relationship among Cryptosporidium genotypes found in storm water.
The genetic differences among the Cryptosporidium genotypes found in water were relatively large. The genetic differences between the genotypes found in water and the C. parvum bovine or human genotype were 1.55 to 6.25%. With the exception of genetic distance between the W3 and W4 genotypes, which had a nucleotide difference of 1.28 changes per 100 bp, the genetic differences among various water genotypes were 2.07 to 8.19% (Table 2).
TABLE 2.
Genetic distances among various Cryptosporidium water genotypesa
| Genotype | Genetic distance (nucleotide changes/100 bp) from:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bov | Hum | W1 | W2 | W3 | W4 | W5 | W6 | W7 | W8 | W9 | W10 | W11 | W12 | |
| Bovb | 0.00 | 0.60 | 1.68 | 2.07 | 2.19 | 2.86 | 3.81 | 2.09 | 6.25 | 5.13 | 4.23 | 4.71 | 4.57 | 3.65 |
| Humb | 0.00 | 1.55 | 2.20 | 2.06 | 2.60 | 3.81 | 2.09 | 6.11 | 5.66 | 4.38 | 4.71 | 4.71 | 3.52 | |
| W1 | 0.00 | 2.07 | 2.86 | 3.28 | 4.72 | 2.75 | 6.27 | 5.14 | 5.14 | 5.39 | 5.38 | 4.08 | ||
| W2 | 0.00 | 2.99 | 3.13 | 4.00 | 2.09 | 5.85 | 5.02 | 3.92 | 4.44 | 4.29 | 4.20 | |||
| W3 | 0.00 | 1.28 | 4.38 | 2.35 | 5.14 | 5.26 | 4.69 | 4.99 | 4.98 | 2.32 | ||||
| W4 | 0.00 | 4.95 | 2.89 | 4.61 | 4.47 | 5.32 | 5.28 | 5.26 | 2.33 | |||||
| W5 | 0.00 | 2.17 | 8.19 | 7.19 | 4.63 | 5.38 | 4.99 | 5.89 | ||||||
| W6 | 0.00 | 6.18 | 5.33 | 3.46 | 3.75 | 3.61 | 3.96 | |||||||
| W7 | 0.00 | 6.03 | 7.70 | 7.81 | 7.36 | 4.32 | ||||||||
| W8 | 0.00 | 6.24 | 6.36 | 6.35 | 4.45 | |||||||||
| W9 | 0.00 | 3.74 | 4.04 | 5.48 | ||||||||||
| W10 | 0.00 | 2.91 | 5.55 | |||||||||||
| W11 | 0.00 | 5.26 | ||||||||||||
| W12 | 0.00 | |||||||||||||
Based on Kimura two-parameter analysis.
Bov, C. parvum bovine genotype; Hum, C. parvum human genotype.
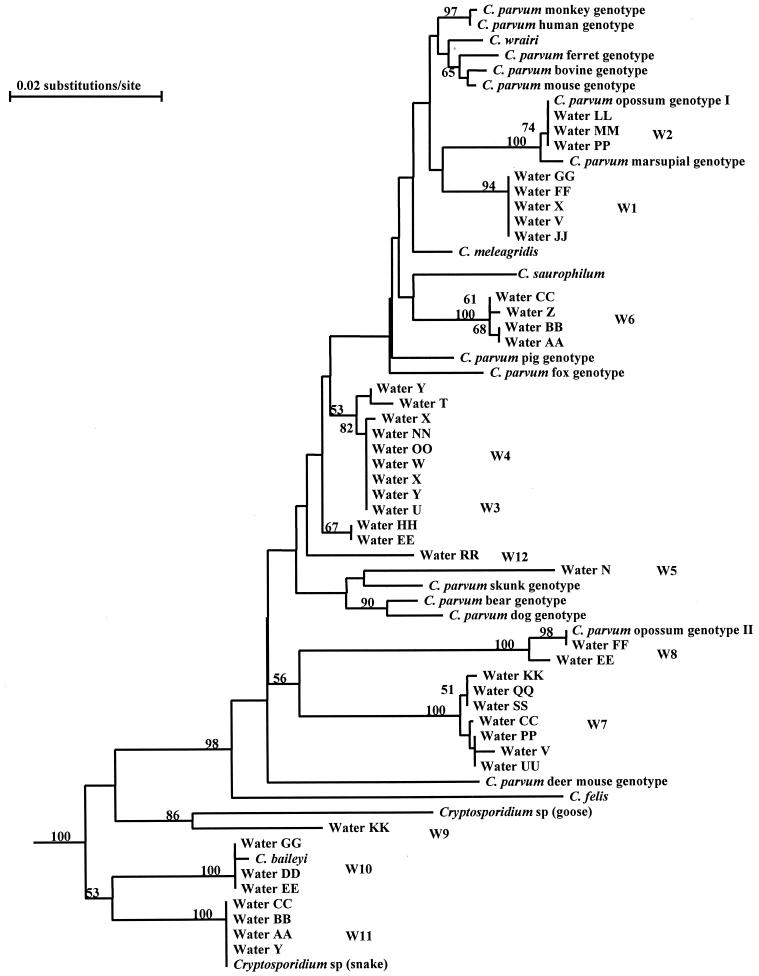
To assess the genetic relatedness of the water genotypes to known Cryptosporidium parasites, phylogenetic analysis was conducted with the water genotype sequences and SSU rRNA sequences from various Cryptosporidium spp. (C. muris, C. andersoni, C. serpentis, C. baileyi, C. felis, C. saurophilum, C. meleagridis, C. wrairi, and several unnamed species) and C. parvum genotypes (bovine, human, mouse, monkey, ferret, pig, dog, bear, skunk, marsupial, and opossum). All 12 water genotypes clustered in the group containing the intestinal Cryptosporidium parasites (C. baileyi, C. felis, C. saurophilum, C. meleagridis, C. wrairi, and C. parvum), with full statistical reliability. Furthermore, genotypes W1 to W8 and W12 were placed in the previously defined broad C. parvum group (containing C. parvum, C. wrairi, C. meleagridis, C. saurophilum, and C. felis) (42), with a bootstrap value of 98% (Fig. 3).
FIG. 3.
Phylogenetic relationship among various Cryptosporidium water genotypes as inferred by neighbor-joining analysis. Bootstrap values above 50% from 1,000 pseudoreplicates are shown on the branches.
Humans, farm animals, pets, and wildlife have all been proposed as sources of Cryptosporidium oocyst contamination in the environment (1, 6, 26, 27, 30). The contribution of humans and specific animals to the oocyst contamination of source water, however, is difficult to assess due to the lack of laboratory tools to differentiate Cryptosporidium oocysts from various sources. The recent finding of the existence of host-adapted Cryptosporidium parasites and the development of new molecular tools that can differentiate Cryptosporidium parasites at the strain level will now allow the determination of the relative contributions of particular groups of animals to Cryptosporidium oocyst contamination in the environment. In this study, we have evaluated the usefulness of an SSU rRNA-based PCR-RFLP technique for the analysis of storm water samples. This technique was chosen because it has the advantage over other molecular techniques of detecting and differentiating all known Cryptosporidium spp. and divergent C. parvum parasites from various animals (34, 42, 44).
Twelve Cryptosporidium genotypes were found in 27 of 29 storm water samples collected for this study. Only four of these genotypes matched sequences from known Cryptosporidium parasites: C. baileyi, Cryptosporidium from snakes, and Cryptosporidium genotypes 1 and 2 from opossums. None of the genotypes found in the storm samples matched those from humans, farm animals, or companion animals (C. felis, C. meleagridis, C. andersoni, and the human, bovine, dog, and pig genotypes of C. parvum), indicating that the genotypes in storm water were probably from wildlife. This conclusion is also consistent with the environmental settings of the sampling sites and the presence of the four genotypes with known animal sources. The presence of multiple genotypes in some samples is expected, considering the runoff nature of storm water and the likely presence of multiple animal species in the catchment area. The higher prevalence of Cryptosporidium oocysts in storm waters is also not surprising, since runoff from storms can release fecal material from the land cover and cause elevated Cryptosporidium oocyst concentrations in the stream samples compared with base flow samples.
The public health importance of the genotypes we identified in water is not yet known. None of these genotypes belong to the five types of Cryptosporidium parasites found in humans (C. parvum human, bovine, and dog genotypes, C. meleagridis, and C. felis) (43). Although 9 of the 12 genotypes (W1 to W8 and W12) clustered within the clade containing the five known human-pathogenic Cryptosporidium parasites, the genetic differences among the genotypes in storm water are quite large, with most of them exhibiting 2.07 to 8.19 nucleotide changes per 100 bp. These genetic distances are much larger than those between C. parvum and C. wrairi (0.4%), C. parvum and C. meleagridis (0.87%), or C. andersoni and C. muris (0.87%), indicating that some of the genotypes may represent different Cryptosporidium species. Sequence diversity was also found between some sequences categorized as the same genotype, such as members of W4, W7, and W8 (Fig. 1). Because it is known that Cryptosporidium parasites have heterogeneous copies of the SSU rRNA gene that have minor sequence differences from the dominant copies (17, 45), these related sequences were likely from the same genotypes.
Results of this study also confirmed the need for a better technique for oocyst isolation and DNA extraction from environmental samples (41). Due to the presence of PCR inhibitors, the PCR-RFLP tool failed to detect Cryptosporidium parasites in DNA prepared directly from ICR water concentrates. Secondary purification of oocysts by IMS improved the quality of DNA extracted. More positivity was revealed by PCR-RFLP analysis of the IMS-purified ICR pellets than by microscopic examination of the ICR pellets. This is expected, because only a small proportion of the ICR concentrates was examined under the microscope, whereas composite samples were used in IMS purification of oocysts. The PCR-RFLP technique needs only a fraction of the DNA from one sporozoite (one oocyst has four sporozoites, and one sporozoite has enough DNA for five PCR templates). It is conceivable that the sensitivity of detection can be further improved by the direct processing of water filtrates with IMS.
In summary, the SSU rRNA-based PCR-RFLP technique has the potential to differentiate among Cryptosporidium parasites and to assess the sources of Cryptosporidium parasites in environmental samples. Results of the present study suggest that wildlife is a major source of Cryptosporidium oocyst contamination in storm water samples. Further studies are needed to characterize the genetic nature of Cryptosporidium parasites from humans and animals so that enough knowledge is accumulated to determine the exact nature and contamination sources of Cryptosporidium oocysts in water. A complete understanding of the source of human infection and environmental contamination would contribute to the scientific management of watersheds.
Nucleotide sequence accession numbers.
The nucleotide sequences of the SSU rRNA gene of Cryptosporidium parasites have been deposited in GenBank under accession no. AF262324 to AF262334 and AY007254.
Acknowledgments
This work was supported in part by an interagency agreement between the Centers for Disease Control and Prevention and the U.S. Environmental Protection Agency (DW 75937984-01-1).
We thank the New York City Department of Environmental Protection staff, Lisa Blancero, William Kuhne, and Charles Lundy for the microscopy work and subsample preparation and the Pathogen Field Monitoring Program for providing stream storm data.
REFERENCES
- 1.Atwill E R, Sweitzer R A, Pereira M D C, Gardner I A, Vanvuren D, Boyce W M. Prevalence of and associated risk factors for shedding Cryptosporidium parvum oocysts and Giardia cysts within feral pig populations in California. Appl Environ Microbiol. 1997;63:3946–3949. doi: 10.1128/aem.63.10.3946-3949.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awad-el-Kariem F M, Warhurst D C, McDonald V. Detection and species identification of Cryptosporidium oocysts using a system based on PCR and endonuclease restriction. Parasitology. 1994;109:19–22. doi: 10.1017/s0031182000077714. [DOI] [PubMed] [Google Scholar]
- 3.Bonnin A, Fourmaux M N, Dubremetz J F, Nelson R G, Gobet P, Harly G, Buisson M, Puygauthier-Toubas D, Gabriel-Pospisil G, Naciri M, Camerlynck P. Genotyping human and bovine isolates of Cryptosporidium parvum by polymerase chain reaction-restriction fragment length polymorphism analysis of a repetitive DNA sequence. FEMS Microbiol Lett. 1996;137:207–211. doi: 10.1111/j.1574-6968.1996.tb08107.x. [DOI] [PubMed] [Google Scholar]
- 4.Carraway M, Tzipori S, Widmer G. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl Environ Microbiol. 1996;62:712–716. doi: 10.1128/aem.62.2.712-716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carraway M, Tzipori S, Widmer G. A new restriction fragment length polymorphism from Cryptosporidium parvum identifies genetically heterogeneous parasite populations and genotypic changes following transmission from bovine to human hosts. Infect Immun. 1997;65:3958–3960. doi: 10.1128/iai.65.9.3958-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalmers R M, Sturdee A P, Mellors P, Nicholson V, Lawlor F, Kenny F, Timpson P. Cryptosporidium parvum in environmental samples in the Sligo area, Republic of Ireland: a preliminary report. Lett Appl Microbiol. 1997;25:380–384. doi: 10.1046/j.1472-765x.1997.00248.x. [DOI] [PubMed] [Google Scholar]
- 7.Chung E, Aldom J E, Chagla A H, Kostrzynska M, Lee H, Palmateer G, Trevors J T, Unger S, Degrandis S. Detection of Cryptosporidium parvum oocysts in municipal water samples by the polymerase chain reaction. J Microbiol Methods. 1998;33:171–180. [Google Scholar]
- 8.Di Giovanni G D, Hashemi F H, Shaw N J, Abrams F A, LeChevallier M W, Abbaszadegan M. Detection of infectious Cryptosporidium parvum oocysts in surface and filter backwash water samples by immunomagnetic separation and integrated cell culture-PCR. Appl Environ Microbiol. 1999;65:3427–3432. doi: 10.1128/aem.65.8.3427-3432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fricker C R, Crabb J H. Water-borne cryptosporidiosis: detection methods and treatment options. Adv Parasitol. 1998;40:241–278. doi: 10.1016/s0065-308x(08)60123-2. [DOI] [PubMed] [Google Scholar]
- 10.Gibbons C L, Gazzard B G G, Ibrahim M, Morris-Jones S, Ong C S L, Awad-El-Kariem F M. Correlation between markers of strain variation in Cryptosporidium parvum: evidence of clonality. Parasitol Int. 1998;47:139–147. [Google Scholar]
- 11.Gibbons C L, Rigi F M, Awad-El-Kariem F M. Detection of Cryptosporidium parvum and C. muris oocysts in spiked backwash water using three PCR-based protocols. Protist. 1998;149:127–134. doi: 10.1016/S1434-4610(98)70017-3. [DOI] [PubMed] [Google Scholar]
- 12.Hallier-Soulier S, Guillot E. An immunomagnetic separation polymerase chain reaction assay for rapid and ultra-sensitive detection of Cryptosporidium parvum in drinking water. FEMS Microbiol Lett. 1999;176:285–289. doi: 10.1111/j.1574-6968.1999.tb13674.x. [DOI] [PubMed] [Google Scholar]
- 13.Johnson D W, Pieniazek N J, Griffin D W, Misener L, Rose J B. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl Environ Microbiol. 1995;61:3849–3855. doi: 10.1128/aem.61.11.3849-3855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaucner C, Stinear T. Sensitive and rapid detection of viable Giardia cysts and Cryptosporidium parvum oocysts in large-volume water samples with wound fiberglass cartridge filters and reverse transcription PCR. Appl Environ Microbiol. 1998;64:1743–1749. doi: 10.1128/aem.64.5.1743-1749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimbell L M, III, Miller D L, Chavez W, Altman N. Molecular analysis of the 18S rRNA gene of Cryptosporidium serpentis in a wild-caught corn snake (Elaphe guttata guttata) and a five-species restriction fragment length polymorphism-based assay that can additionally discern C. parvum from C. wrairi. Appl Environ Microbiol. 1999;65:5345–5349. doi: 10.1128/aem.65.12.5345-5349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kostrzynska M, Sankey M, Haack E, Power C, Aldom J E, Chagla A H, Unger S, Palmateer G, Lee H, Trevors J T, De Grandis S A. Three sample preparation protocols for polymerase chain reaction based detection of Cryptosporidium parvum in environmental samples. J Microbiol Methods. 1999;35:65–71. doi: 10.1016/s0167-7012(98)00106-7. [DOI] [PubMed] [Google Scholar]
- 17.Le Blancq S M, Khramtsov N V, Zamani F, Upton S J, Wu T W. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol Biochem Parasitol. 1997;90:463–478. doi: 10.1016/s0166-6851(97)00181-3. [DOI] [PubMed] [Google Scholar]
- 18.LeChevallier M W, Norton W D, Lee R G. Giardia and Cryptosporidium spp. in filtered drinking water supplies. Appl Environ Microbiol. 1991;57:2617–2621. doi: 10.1128/aem.57.9.2617-2621.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeChevallier M W, Norton W D, Lee R G. Occurrence of Giardia and Cryptosporidium spp. in surface water supplies. Appl Environ Microbiol. 1991;57:2610–2616. doi: 10.1128/aem.57.9.2610-2616.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leng X, Mosier D A, Oberst R D. Differentiation of Cryptosporidium parvum, C. muris, and C. baileyi by PCR-RFLP analysis of the 18S rRNA gene. Vet Parasitol. 1996;62:1–7. doi: 10.1016/0304-4017(95)00863-2. [DOI] [PubMed] [Google Scholar]
- 21.Madore M S, Rose J B, Gerba C P, Arrowood M J, Sterling C R. Occurrence of Cryptosporidium oocysts in sewage effluents and selected surface waters. J Parasitol. 1987;73:702–705. [PubMed] [Google Scholar]
- 22.Mayer C L, Palmer C J. Evaluation of PCR, nested PCR, and fluorescent antibodies for detection of Giardia and Cryptosporidium species in wastewater. Appl Environ Microbiol. 1996;62:2081–2085. doi: 10.1128/aem.62.6.2081-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan U M, Constantine C C, Forbes D A, Thompson R C A. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J Parasitol. 1997;83:825–830. [PubMed] [Google Scholar]
- 24.Morgan U M, Constantine C C, O'Donahue P, Meloni B P, O'Brien P A, Thompson R C A. Molecular characterization of Cryptosporidium isolates from humans and other animals using random amplified polymorphic DNA analysis. Am J Trop Med Hyg. 1995;52:559–564. doi: 10.4269/ajtmh.1995.52.559. [DOI] [PubMed] [Google Scholar]
- 25.Morgan U M, Monis P T, Fayer R, Deplazes P, Thompson R C A. Phylogenetic relationships among isolates of Cryptosporidium: evidence for several new species. J Parasitol. 1999;85:1126–1133. [PubMed] [Google Scholar]
- 26.Newman R D, Wuhib T, Lima A A, Guerrant R L, Sears C L. Environmental sources of Cryptosporidium in an urban slum in northeastern Brazil. Am J Trop Med Hyg. 1993;49:270–275. doi: 10.4269/ajtmh.1993.49.270. [DOI] [PubMed] [Google Scholar]
- 27.Pell A N. Manure and microbes: public and animal health problem? J Dairy Sci. 1997;80:2673–2681. doi: 10.3168/jds.S0022-0302(97)76227-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng M M, Xiao L, Freeman A R, Arrowood M J, Escalante A A, Weltman A C, Ong C S, Mac Kenzie W R, Lal A A, Beard C B. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–573. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rochelle P A, De Leon R, Stewart M H, Wolfe R L. Comparison of primers and optimization of PCR conditions for detection of Cryptosporidium parvum and Giardia lamblia in water. Appl Environ Microbiol. 1997;63:106–114. doi: 10.1128/aem.63.1.106-114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose J B. Environmental ecology of Cryptosporidium and public health implications. Annu Rev Public Health. 1997;18:135–161. doi: 10.1146/annurev.publhealth.18.1.135. [DOI] [PubMed] [Google Scholar]
- 31.Spano F, Putignani L, Crisanti A, Sallicandro P, Morgan U M, Leblancq S M, Tchack L, Tzipori S, Widmer G. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J Clin Microbiol. 1998;36:3255–3259. doi: 10.1128/jcm.36.11.3255-3259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spano F, Putignani L, Guida S, Crisanti A. Cryptosporidium parvum—PCR-RFLP analysis of the TRAP-C1 (thrombospondin-related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Exp Parasitol. 1998;90:195–198. doi: 10.1006/expr.1998.4324. [DOI] [PubMed] [Google Scholar]
- 33.Spano F, Putignani L, McLauchlin J, Casemore D P, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 34.Sulaiman I M, Xiao L, Lal A A. Evaluation of Crypytosporidium parvum genotyping techniques. Appl Environ Microbiol. 1999;65:4431–4435. doi: 10.1128/aem.65.10.4431-4435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sulaiman I M, Xiao L, Yang C, Escalante L, Moore A, Beard C B, Arrowood M J, Lal A A. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg Infect Dis. 1998;4:681–685. doi: 10.3201/eid0404.980424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sulaiman I M, Lal A A, Arrowood M J, Xiao L H. Biallelic polymorphism in the intron region of beta-tubulin gene of Cryptosporidium parasites. J Parasitol. 1999;85:154–157. [PubMed] [Google Scholar]
- 37.Sulaiman I M, Morgan U M, Thompson R C A, Lal A A, Xiao L. Phylogenetic relationships of Crypytosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl Environ Microbiol. 2000;66:2385–2391. doi: 10.1128/aem.66.6.2385-2391.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.U.S. Environmental Protection Agency. ICR microbial laboratory manual. Washington, D.C.: Office of Research and Development, U.S. Government Printing Office; 1996. [Google Scholar]
- 39.Widmer G, Tchack L, Chappell C L, Tzipori S. Sequence polymorphism in the beta-tubulin gene reveals heterogeneous and variable population structures in Cryptosporidium parvum. Appl Environ Microbiol. 1998;64:4477–4481. doi: 10.1128/aem.64.11.4477-4481.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Widmer G, Tzipori S, Fichtenbaum C J, Griffiths J K. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J Infect Dis. 1998;178:834–840. doi: 10.1086/515373. [DOI] [PubMed] [Google Scholar]
- 41.Wiedenmann A, Kruger P, Botzenhart K. PCR detection of Cryptosporidium parvum in environmental samples—a review of published protocols and current developments. J Ind Microbiol Biotechnol. 1998;21:150–166. [Google Scholar]
- 42.Xiao L, Morgan U M, Limor J, Escalante A, Arrowood M, Shulaw W, Thompson R C A, Fayer R, Lal A A. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. 1999;65:3386–3391. doi: 10.1128/aem.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao L, Morgan U M, Fayer R, Thompson R C A, Lal A A. Cryptosporidium systematics and implications for public health. Parasitol Today. 2000;16:287–292. doi: 10.1016/s0169-4758(00)01699-9. [DOI] [PubMed] [Google Scholar]
- 44.Xiao L H, Escalante L, Yang C F, Sulaiman I, Escalante A A, Montali R J, Fayer R, Lal A A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao L H, Limor J R, Li L X, Morgan U, Thompson R C A, Lal A A. Presence of heterogeneous copies of the small subunit rRNA gene in Cryptosporidium parvum human and marsupial genotypes and Cryptosporidium felis. J Eukaryot Microbiol. 1999;46:44S–45S. [PubMed] [Google Scholar]