Abstract
Objective:
Provide insights into the defective POMC processing and invasive behavior in silent pituitary corticotroph tumors.
Design and methods:
Single cell RNAseq was used to compare the cellular makeup and transcriptome of silent and active corticotroph tumors.
Results:
A series of transcripts related to hormone processing peptidases and genes involved in the structural organization of secretory vesicles were reduced in silent compared to active corticotroph tumors. Most relevant to their invasive behavior, silent corticotroph tumors exhibited several features of epithelial-to-mesenchymal transition (EMT), with increased expression of mesenchymal genes along with the loss of transcripts which regulate hormonal biogenesis and secretion. Silent corticotroph tumor vascular smooth muscle cell and pericyte stromal cell populations also exhibited plasticity in their mesenchymal features.
Conclusions:
Our findings provide novel insights into the mechanisms of impaired POMC processing and invasion in silent corticotroph tumors and suggest that a common transcriptional reprogramming mechanism simultaneously impairs POMC processing and activates tumor invasion.
Keywords: Pituitary tumor, Cushing disease, silent corticotroph tumor, epithelial to mesenchymal transition, de-differentiation
Introduction
Silent corticotroph tumors (SCTs) account for 3–6% of all pituitary tumors, 10–20% of silent pituitary tumors, and ~30% of all corticotroph tumors (1–3). Patients with SCTs typically do not exhibit clinical and/or biochemical features of hypercortisolism. They can have elevated circulating plasma ACTH levels, although the latter peptide is often dysfunctional and incapable of driving ACTH-receptor mediated hypercortisolism (4). Exhibiting a female preponderance, they occur in younger patients than other clinical non-functioning pituitary tumors and are invariably large and invasive of adjacent anatomical structures at presentation, making complete surgical resection almost impossible. Additionally, they exhibit higher rates of recurrence and although pituitary carcinoma is extremely rare, SCTs are disproportionally represented in pituitary tumors that exhibit metastatic spread compared to other pituitary tumor sub-types (5). Mechanisms proposed for their underlying “biochemically” silent nature include disrupted transcriptional activation of the POMC gene, reduced expression of the POMC processing prohormone convertase enzymes such as PCSK1 (aka PC1/3), and/or enhanced lysosomal degradation of generated POMC peptides, leading to impaired ACTH synthesis, processing and maturation. Impaired corticotroph differentiation supported by reduced expression of the lineage restricted corticotroph specific transcription factor TBX19 (aka Tpit) has also been proposed as a mechanism for their silent phenotype (6).
Therapy for this group of tumors typically includes multi-modal approaches with often multiple surgical debulking procedures, radiation therapy and occasionally chemotherapy. Unfortunately, these therapies are not always successful and there is an unmet need for additional treatment options for these rare but extremely challenging tumors. We reasoned that single cell RNA sequencing would allow us to analyze and compare the global transcriptomic landscape and cellular make-up of clinically silent coricotroph tumors (SCT, n=3) and functioning corticotroph tumors (FCTs, n=5) and could provide unique insights into their different behaviors and identify novel disease targets.
Results
Cellular composition of FCTs and SCTs
We used single cell RNAseq (scRNAseq) to analyze 5 clinically functioning corticotroph tumors (FCTs) causing Cushing disease with hypercortisolism (4 microadenomas, 1 macroadenoma), and 3 silent corticotroph macroadenomas (SCTs, clinical details in Table 1). We obtained an average of 249 million reads made up of 24,413 genes per patient with an average of 1,060 genes expressed and 2,877 unique molecular identifiers (UMIs) detected per cell (Supplementary Table 1). According to cell-type specific markers, we identified 7 distinct cell populations from a total of 56,458 cells, namely tumor cells (41,943 cells, 74.3%), stromal cells (7,189 cells, 12.7%, including 4,210 fibroblasts 7.5% and 2,979 endothelial cells 5.2%), immune cells (3,936 cells, 7.0%), progenitor cells (2,054 cells, 3.6%), and a minor population of proliferating cells (146 cells, 0.3%, Fig. 1A, Table 2, and Supplementary Table 2). A cluster of uncharacterized cells (1,190 cells, 2.1%) mainly comprised of erythrocytes-derived hemoglobin genes and background RNA was not included in further analysis.
Table-1.
Patient Characteristics and Sample Information
| Sample ID | F/M | Age | Size (mm) | Pathology | Ki67 | Pre-Surgical Biochemical Test | |||
|---|---|---|---|---|---|---|---|---|---|
| Plasma ACTH (NR: 4 – 48 pg/mL) |
Mor. Ser. Cort. (NR: 8–25 μg/dL) |
L N Sal. Cort. (NR:<0.09 μg/dL) |
24h UFC (NR: < 50 μg/24h) |
||||||
| FCT1 | F | 66 | 5.5 | ACTH+ | <1–2% | 164 | 116 | 0.248 | 1,633 |
| FCT2 | F | 15 | 4 | ACTH+ | 3–6% | 56 | 19 | 0.499 | 196 |
| FCT3 | F | 43 | 8 | ACTH+ | <1% | 73 | 31 | 0.42 | 426 |
| FCT4 | F | 36 | 48 | ACTH+ | <1% | 99 | 22 | ND | 528 |
| FCT5 | F | 34 | 5 | ACTH+ | rare | 61 | 18 | 0.22 | 179 |
| SCT1 | M | 39 | 19 | ACTH+ | <1% | 60 | 15 | 0.05 | ND |
| SCT2 | F | 22 | 22 | ACTH+ | 2–5% | 42 | 10.8 | 0.03 | ND |
| SCT3 | F | 50 | 27 | ACTH-/TBX19+ | 1–3% | 10.7 | 19 | ND | ND |
| SGT | M | 35 | 25 | LH+ FSH+ | 2–3% | ND | ND | ND | ND |
UFC, urinary free cortisol; Mor. Ser. Cort., morning serum cortisol; L N Sal. Cort., late night salivary cortisol; NR, normal range
Figure 1. Single-cell RNA-sequencing (scRNA-seq) analysis depicting the heterogenous cellular composition of FCTs (n=5) and SCTs (n=3).
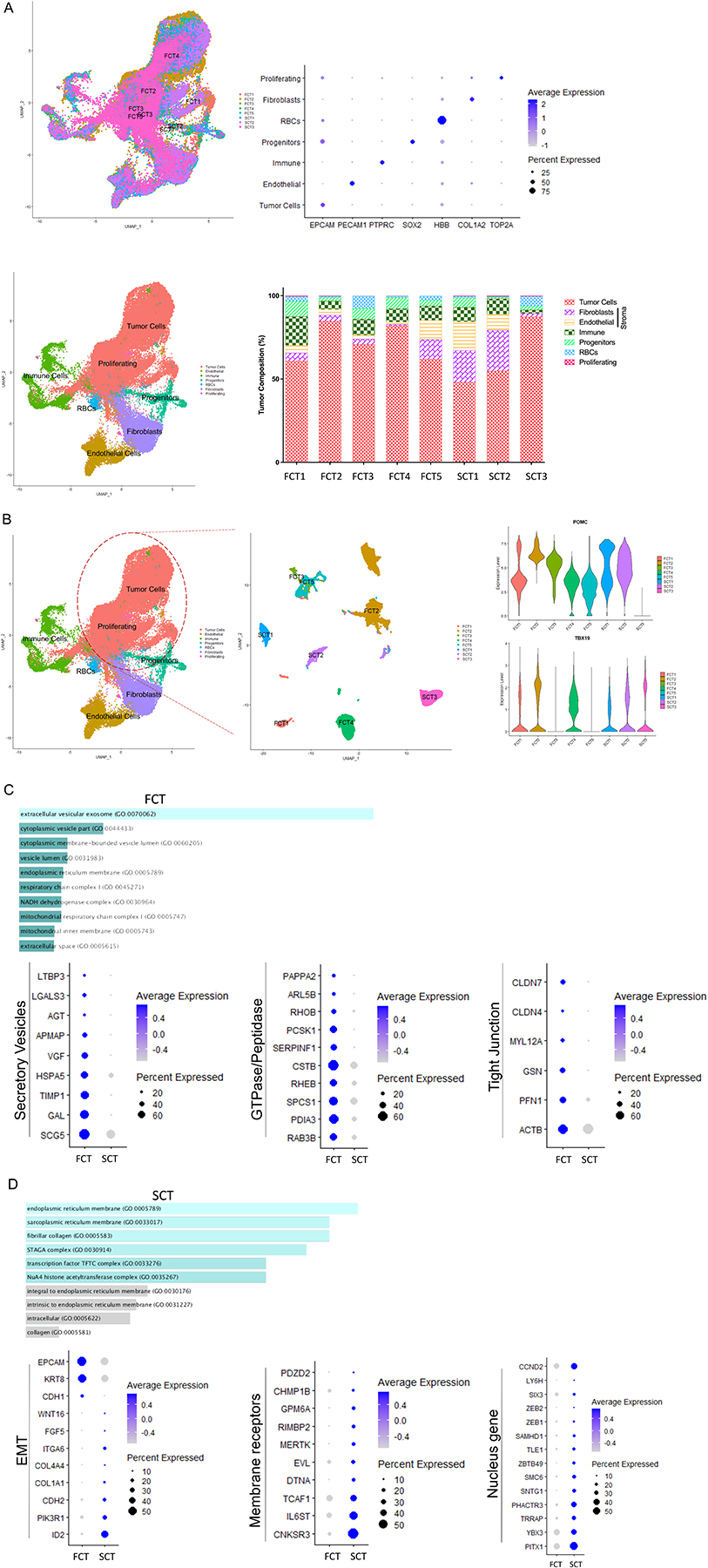
(A) Uniform manifold approximation and projection (UMAP) was used to visualize corticotroph tumor cellular composition and revealed 7 cell types. Each cell type was annotated based on specific marker expression as shown in the dot plot. The percentage of cells detected in the individual cell types is depicted in the lower panel. (B) The corticotroph tumor population was then subclustered and noted to group in a tumor-subtype specific manner, with all tumor samples expressing the corticotroph specific transcription factor, TBX19. (C & D) ENRICHR was then used to analyze GO cellular components of tumor population, and observed that genes involved in membrane bound vesicle formation and prohormonal processing were highly expressed in FCTs (C). In contrast, SCTs predominantly expressed genes involved in epithelial to mesenchymal transition (EMT, D). p<10−4.
Table 2.
The composition of cell populations. Data of the population of individual samples is presented as n (%).
| Tumor Cells | Fibroblasts | Endothelial | Immune | Progenitors | Uncharacterized | Proliferating | Total | |
|---|---|---|---|---|---|---|---|---|
| FCT1 | 2244 (60.9) | 180 (4.88) | 164 (4.45) | 639 (17.34) | 333 (9.04) | 107 (2.9) | 18 (0.49) | 3685 |
| FCT2 | 15142 (84.79) | 653 (3.66) | 617 (3.46) | 889 (4.98) | 388 (2.17) | 118 (0.66) | 51 (0.29) | 17858 |
| FCT3 | 1218 (70.9) | 55 (3.2) | 42 (2.44) | 163 (9.49) | 109 (6.34) | 130 (7.57) | 1 (0.06) | 1718 |
| FCT4 | 5920 (81.88) | 71 (0.98) | 104 (1.44) | 577 (7.98) | 484 (6.69) | 69 (0.95) | 5 (0.07) | 7230 |
| FCT5 | 3662 (62.07) | 695 (11.78) | 675 (11.44) | 512 (8.68) | 199 (3.37) | 157 (2.66) | 0 (0) | 5900 |
| SCT1 | 2277 (48.17) | 903 (19.1) | 812 (17.18) | 413 (8.74) | 267 (5.65) | 39 (0.83) | 16 (0.34) | 4727 |
| SCT2 | 3266 (54.97) | 1454 (24.47) | 565 (9.51) | 551 (9.27) | 59 (0.99) | 19 (0.32) | 27 (0.45) | 5941 |
| SCT3 | 8214 (87.39) | 199 (2.12) | 0 (0) | 192 (2.04) | 215 (2.29) | 551 (5.86) | 28 (0.3) | 9399 |
| Total | 41943 (74.29) | 4210 (7.46) | 2979 (5.28) | 3936 (6.97) | 2054 (3.64) | 1190 (2.11) | 146 (0.26) | 56458 |
Using the subset function of Seurat to further analyze the corticotroph tumor cell population, we observed that they were clustered in an origin-dependent manner on UMAP with 7 samples expressing POMC and 1 sample (SCT3) expressing TBX19 but not POMC, which was consistent with the histopathological findings (Fig. 1B). By profiling differentially expressed genes (DEGs), we identified gene signatures that distinguished the FCT tumor cells from SCT tumor cells (Supplementary Table 3, p<10−4). Using GO cellular component analysis by ENRICHR, we demonstrated that the FCT tumor cells (both micro- and macro-adenomas) exhibited high expression of membrane bound vesicle genes, such as those involved in organization of secretory vesicles (SCG5, GAL, TIMP1, HSPA5, VGF, APMAP, AGT, LGALS3, and LTBP3), small GTPase and peptidases (RAB3B, PDIA3, SPCS1, RHEB, CSTB, SERPINF1, PCSK1, RHOB, ARL5B, and PAPPA2), and genes associated with tight junctions and actin-mediated motility (ACTB, PFN1, GSN, MYL12A, and CLDN4/7, Fig. 1C, Supplementary Table 4, p<10−4).
In marked contrast, the SCT tumor cells exhibited several features of epithelial-to-mesenchymal transition (EMT) such as increased expression of the mesenchymal genes ID2, PIK3R1, CDH2 (aka NCAD), COL1A1, COL4A4, ITGA6, FGF5, and WNT16, all of which regulate cell migration and movement in association with loss of EPCAM, KRT8, and CDH1 (Fig. 1D, Supplementary Table 3, p<10−4). In line with this, several membrane receptors (CNKSR3, IL6ST, TCAF1, DTNA, EVL, MERTK, RIMBP2, GPM6A, CHMP1B, and PDZD2), and various nuclear genes and transcription factors (YBX3, TRRAP, PHACTR3, SNTG1, SMC6, ZBTB49, TLE1, and SAMHD1, Fig. 1D, Supplementary Table 3, p<10−4) were preferentially upregulated in SCT tumor cells. We also noted higher expression of several genes involved in early embryonic pituitary organogenesis, such as PITX1, SIX3, and LY6H and the cell cycle regulator CCND2 in the SCT tumor cells (Fig. 1D, Supplementary Table 3, p<10−4).
Striking differences in tumor stroma between FCTs and SCTs
The stromal cell populations included fibroblasts expressing TAGLN, TPM2, CPE, COL1A2, SOD3, NDUFA4L2, FN1, CARMN, ACTA2, DCN, and TIMP1; and endothelial cells expressing high levels of VWF, PLVAP, CLDN5, PODXL, TM4SF1, HSPG2, IFI17, RAMP2, PTPRB, CD74, INSR, and IGFBP3 (Fig. 2A and Supplementary Table 5, p<10−4). Our stromal analysis became particularly interesting when we further examined the fibroblast population subsets. We observed 3 distinct fibroblast sub-types that comprised myofibroblasts, adventitial fibroblasts and resident stromal fibroblasts. Whereas expression levels of the myofibroblast markers TPM1, TPM2, MYL9, MYL, CAV1, ACTA2, FRZB, LTBP1, SLIT3, SOD3, TAGLN, and MCAM were comparable between FCT and SCT stomal cells (Fig. 2B, p=ns), markers of vascular smooth muscle cells (vSMCs) or adventitial fibroblasts (RGS5, NOTCH3, PTN, PHLDA1, CCND2, MDK, MYH11, EDNRA, COL5A3, FBXO32, and ITGA6) were much more abundant in the SCT stromal cells (Fig. 2B, p<10−4). In contrast, the fibroblasts that predominated in the FCT stromal cells expressed genes involved in ECM organization, ECM receptor interaction and inflammation such as C1S, C1R, C7, LUM, MGP, CST3, CFH, EFEMP1, SDC4, DCN, BGN, and OGN, in keeping with their role in hormone secretion (Fig. 2B, p<10−4).
Figure 2. Striking intratumor stromal cell heterogeneity between FCTs and SCTs.
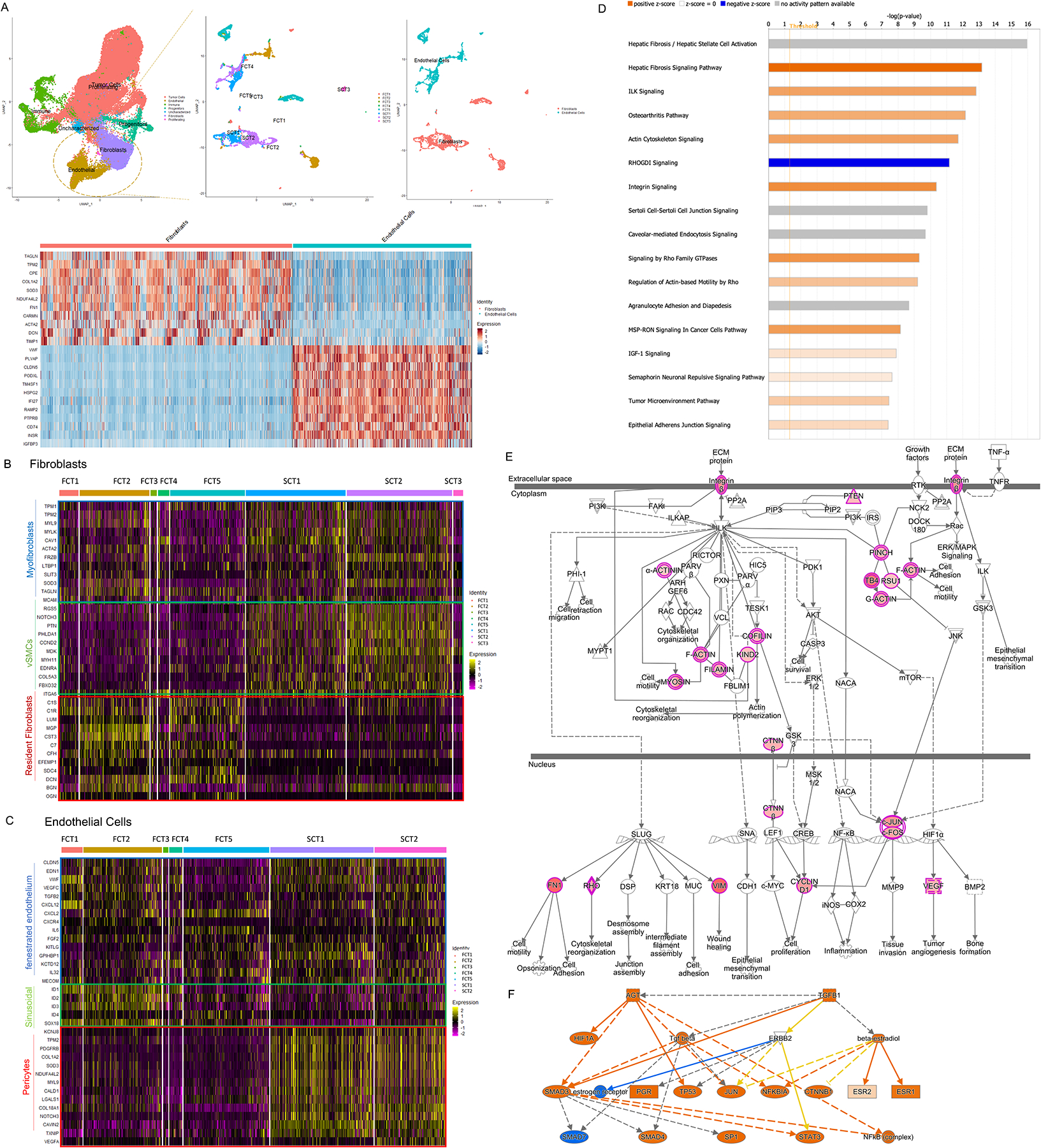
(A) The subset function of Seurat was used to analyze the stromal fibroblast and endothelial cell populations. (B & C) Using canonical transcript markers, the fibroblast and endothelial cell populations were further sub-grouped into myofibroblasts, vascular smooth muscle cells (vSMCs) and resident fibroblasts (B); and fenestrated endothelium, sinusoidal endothelium and pericytes respectively (C). (D-F) Examples of activated pathways in the mural cells (D), signal transduction (E) and their upstream regulators (F) observed in SCTs. p<10−4.
Analysis of the endothelial cells demonstrated that both the FCTs and SCTs exhibited similar expression of markers of fenestrated endothelium such as CLDN5, EDN1, VWF, and classical cytokines, mitogens and inflammatory factors such as VEGFC, TGFB2, CXCL12, CXCL2, CXCR4, IL6, FGF2, KITLG, GPIHBP1, KCTD12, IL32, and MECOM (Fig. 2C, p<10−4). However, whereas the FCT endothelial cells also expressed the sinusoidal endothelial transcription factors ID1-3 and the lymphatic vessel endothelial marker SOX18 (Fig. 2C, p<10−4), the SCT endothelial cells exhibited higher expression of mural cell markers such as the pericyte transcripts (TPM2, PDGFRB, COL1A2, SOD3, NDUFA4L2, MYL9, CALD1, LGALS1, COL18A1, NOTCH3, CAVIN2, TXNIP, and VEGFA, Fig. 2C, p<10−4), reflecting their mesenchymal stromal phenotype.
We also observed that a variety of transcripts involved with fibrosis, actin cytoskeleton activation, integrin-linked kinase (ILK), and integrin signaling were uniquely upregulated in the SCTs and these factors can cross talk and interact to alter cell adhesion, cytoskeletal rearrangement, motility, and tissue invasion (Figs. 2D & 2E, p<10−7). Noting these unique features in the SCT fibroblast and endothelial cell populations, we then examined for upstream activators of these various pathways. Causal network analysis identified TGFB1 and AGT as key upstream regulators (p<10−4), and these in turn can modulate the transcriptional activities of SMADs, HIF1A, NFKB, CTNNB1, STAT3 and several nuclear receptors to cooperatively alter stromal cell function in SCTs (Fig. 2F).
The transcriptomic features of immune cells in FCTs and SCTs
Our single-cell analysis also demonstrated a heterogenous and diverse intratumor immune microenvironment in the corticotroph tumors, comprising 3,936 immune cells in 5 main clusters (Fig. 3A, and Supplementary Table 6). Interestingly, each of the immune clusters contained cells from all the tumor samples, indicating that the tumor immune cell types were broadly consistent and did not vary much in a patient-specific manner (Fig. 3A). The main immune cluster comprised myeloid origin cells (2,435 cells, 62%). Based on the co-expression of CD14 & CD68 in monocytes, HAL-DAQ1 and APOE in macrophages, and LYZ and S100A9 in MDSCs, they were sub-divided into 2 immunosuppressive cell types, namely tumor associated macrophages (1,860 cells, 47%) and myeloid derived suppressor cells (MDSCs, 575 cells, 15%, Fig. 3B). The immunosuppressive chemokines CXCL8/16 and CCL3 were the most highly expressed CXC chemokines in both MDSCs and macrophages (Figs. 3D& 3E, p<10−4). Using a similar approach, we identified 1,501 lymphoid origin immune cells (38%) based on their expression of CD2/3 for T cells (1,173 cells, 30%), MS4A1 (aka CD20)/CD79A for B cells (78 cells, 2%), and GNLY and NKG7 for nature killer cells (NK, 250 cells, 6%, Fig. 3C). CD8+ T cells and NK cells exhibited prominent expression of a variety of cytotoxic genes including IL2RB, KLRG1, GNLY, GZMK, IL32, and RORA (Fig. 3F, p<10−4). Overall, the immune cell transcriptome was quite similar in FCTs and SCTs apart from the inhibitory immunoglobin HAVCR2 (7) which was more highly expressed in SCTs (Fig. 3G, p<10−4).
Figure 3. The transcriptomic features of immune cells in FCTs and SCTs.
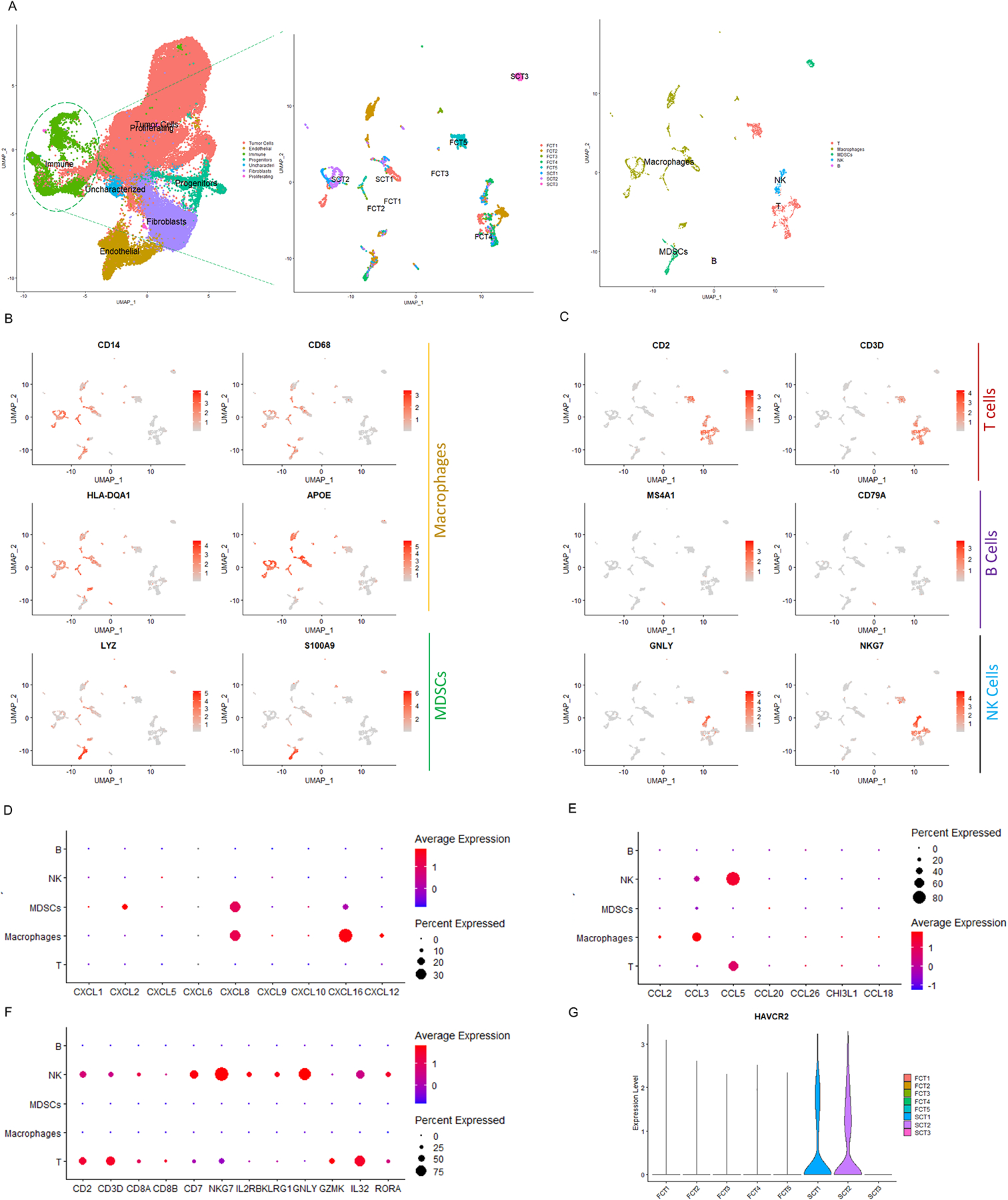
(A) Subset function of Seurat was used to analyze the immune cell population. (B) Monocytes, macrophages and monocytic human myeloid-derived suppressor cells (MDSCs) were identified by CD14 and CD86 expression, HLA-DQA1, apolipoprotein E (APOE) expression and S100 calcium binding protein A8 (S100A8), and LYZ expression respectively. (C) T/NK cells, B cells and NK cells were denoted based on expression of CD2 and CD3E; MS4A1 and CD79A; and GNLY and natural killer cell granule protein 7 (NKG7) expression respectively. (D-F) Depiction using dot plots of various inflammatory cytokines that were present in the various immune cells from the corticotroph tumors. (G) Depiction by violin plot of HAVCR2 expression in the immune cell population of 2 of 3 SCTs. p<10−4.
Different transcriptomic features of progenitor cells in FCTs and SCTs
In addition to the tumor, stromal and immune cell populations, we also characterized a group of epithelial origin progenitor cells, based on their expression of the epithelial markers EPCAM/KRT8 in addition to the archetypical progenitor cell markers SOX2/SOX9 (Fig. 4A). The progenitor population comprised 3 types, firstly endocrine progenitors identified from expression of PITX2, LHX3, PAX6, HEY1, MSX1, and RFX4; secondly mucus secreting cells, that expressed the mucins MUC16/MUC4, several transcripts related to cilia organization (CEACAM6 and DNAH5), and luminal epithelial cytokeratin KRT7/8/17/19; and thirdly posterior pituitary cells which expressed the lineage specific TFs, LHX2, NKX2-1, and the secretory factors LAMA4, CXCL14 and WIF1 (8) (Fig. 4B). The mucus producing cells were primarily derived from one FCT-macro (FCT4) that invaded the entire clivus bone and bilateral cavernous sinuses. It expressed the serous cell markers SLPI/WFDC2 and stem cell markers AGR2, BTG2, KLF2, RFX3, and PROM1, and exhibited some features of ciliated luminal submucosal cells similar to respiratory epithelium (9) (Fig. 4C). Similarly, a spectrum of posterior pituitary derived genes including COL25A1, SCN1A, RAX, ADM, TIMP3, A2M, HMCN1, ATP1A2, CLDN10, FAT3, GRIK3, NTRK2, PCDH9, SLC1A3, LAMA4, CXCL14, and WIF1 (Fig. 4D) were present in one of the 5 FCTs (FCT1). This tumor was noted on pre-operative imaging and at surgical resection to be located deep in the posterior pituitary region. As we suspected these latter 2 progenitor cell populations may have been due to contamination with normal mucosal (FCT4) and posterior pituitary (FCT1) tissues respectively, we only further analyzed the endocrine progenitor cell population. These endocrine progenitor cells expressed folliculo-stellate cell (FSC) hallmark transcripts such as S100B, S100A1/6/10/11, and ANXA1/2 (10), suggesting this regenerative progenitor cell pool may be able to give rise to mesenchymal supportive cells (Fig. 4E) (11).
Figure 4. Different transcriptomic features of progenitor cells in FCTs and SCTs.
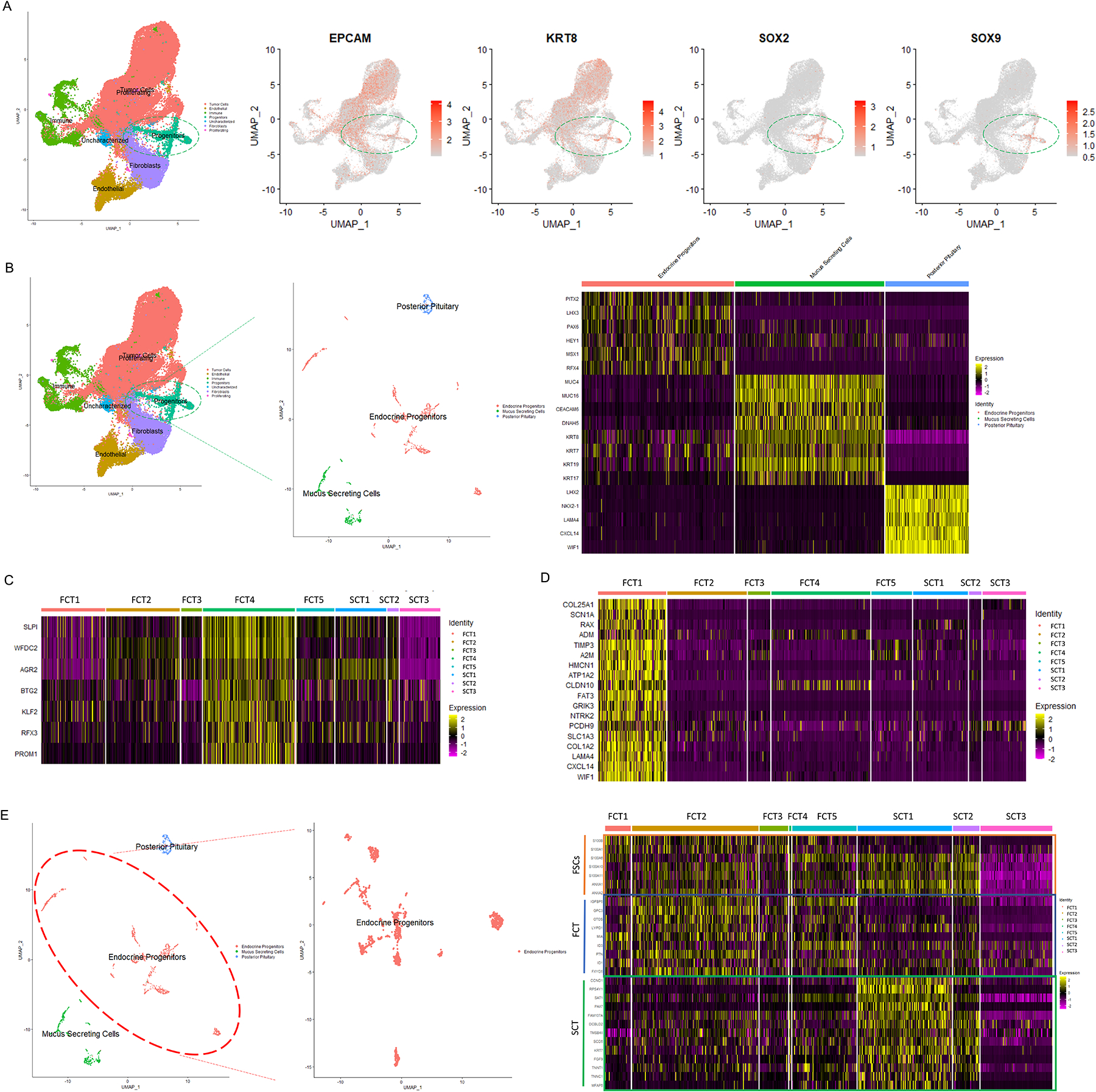
(A-B) Analysis of the progenitor cell population revealed 3 sub-groups, namely endocrine progenitors, mucin producing cells, and posterior pituitary cells. (C-D) The mucin producing and posterior pituitary cells were restricted to two of the FCT samples (FCT4 & FCT1), suggesting these populations may have been due to sample contamination from resected tumor-adjacent normal tissues. (E) Further analysis of endocrine progenitor subpopulation revealed different transcriptional profiles between FCTs and SCTs. p<10−4.
We did note some differences in the progenitor cells in the FCTs and SCTs. Whereas FCT progenitors expressed higher levels of mitogenic factors involved in cranial morphogenesis, survival and senescence (IGFBP5, GPC3, OTOS, MIA, LYPD1, FXYD5, ID1/3) (12–15), the SCT progenitor cells expressed transcripts associated with myogenesis (PAX7, DCBLD2, TMSB4X, MFAP5, TNNT1 and TNNC1) (16–19), metabolism (SCD5, SAT1, and FAM107A) (20, 21) and proliferation (CCND1, Fig. 4E, p<10−4).
The SCT transcriptome is distinct from that of FCT-micro and -macro & silent gonadotroph tumor (SGT)
To address the possibility that the transcriptional changes we observed were simply due to comparison of macro- (SCTs) versus micro-adenoma (4 of 5 FCTs), we directly compared the transcriptome of the single FCT-macro (FCT4, Fig. 5B), a recurrent silent gonadotroph macroadenoma (SGT, Fig. 5A, Table 1, and Supplementary Table 7), the 4 FCT-micro and the 3 SCT-macro (SCT1-3, Fig. 5C). The cellular make-up of the SGT was similar to the corticotroph tumors and comprised tumor (63.25%, 2,985 cells), progenitor (8.9%, 420 cells), stromal (6.87% including endothelial 4.28%, 202 cells; and fibroblasts 2.59%, 122 cells), uncharacterized (5.53%, 261 cells), immune (4.01%, 189 cells), and proliferating cell populations (1.25%, 59 cells, Supplementary Table 7, Fig. 5A). Analysis of the tumor cell populations in both FCT-macro and SGT-macro demonstrated evidence of active hormone biogenesis and secretion with enrichment of secretory vesicle components (SCG2, 3, 5, and CHGB), hormonal processing peptidases and GTPases (PCSK2, SEC11C, CALM, and GNAS, Figs. 5A & 5B). The expression of the majority of these vesicle (VGF, APMAP, LTBP3 & SDC2), peptidase (RAB3B & SERPINF1) and tight junction (CLDN7) transcripts were significantly higher not only in the FCT-micros (blue box), but also in the FCT-macro (black box) and in the SGT-macro samples (red box, Fig 5C). In contrast, the SCTs (yellow box) exhibited higher expression of EMT markers in both tumor cells (CNKSR3, ID2 and CCND2), and stromal cells (SNCG, NSG1, and CALD1) in comparison to the FCT-micros (FCT1-3, blue box), the FCT-macro (FCT4, black box), and the SGT-macro (red box, Fig. 5D). These findings suggest that these EMT changes are not simply associated with tumor size but indicate that SCTs and FCTs (whether micro or macro) are actually quite distinct entities.
Figure 5. The SCT transcriptome is distinct from that of macroadenomas.
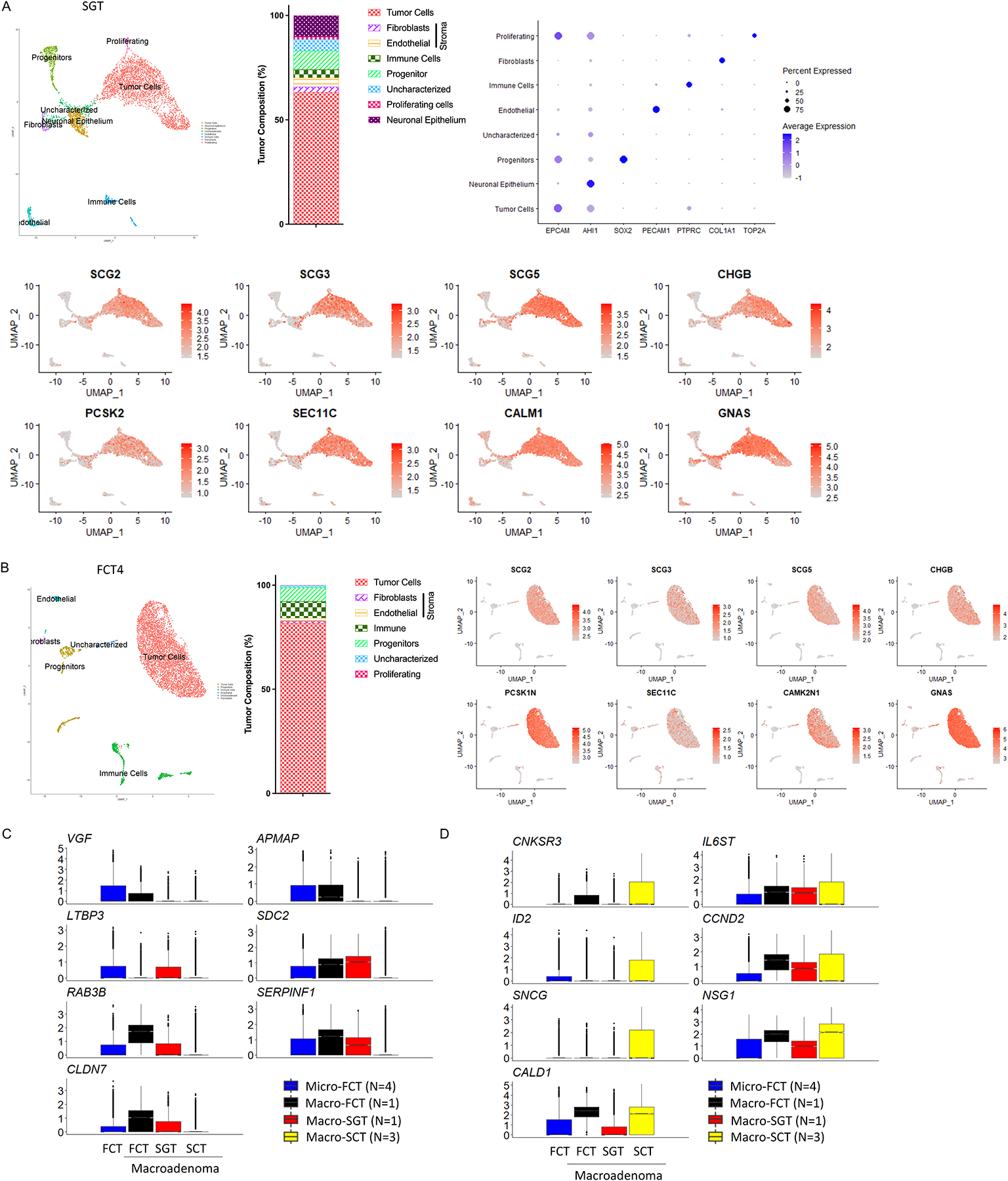
(A-B) Individual analysis of a silent gonadotroph macroadenoma (SGT-macro, A), and a FCT-macro (FCT4, B) demonstrating the cellular composition, and well-differentiated features of the tumor cells with expression of transcripts associated with secretory function. (C-D) Depiction of the relative similarity in expression of a series of secretory (C) and EMT-associated (D) transcripts in 3 FCT-micros (FCT1-3, blue box), a FCT-macro (FCT4, black box), and a SGT-macro (red box), and their striking difference to expression levels of these transcripts in SCTs (yellow box).
Discussion
Clinical management of SCTs can be challenging and sometimes despite multiple surgeries, radiation and even systemic therapies, they exhibit a propensity to recur and cause significant morbidity and even mortality. The molecular mechanisms of this invasive and recurrent behavior are not well understood. Therefore, we used scRNAseq to analyze and compare the global transcriptomic profiling at an individual cellular level from 3 SCTs and 5 FCTs causing hypercortisolism. One of our important findings, as described above, was that the SCTs exhibited a striking difference in their tumor cell populations. This included not just reduced expression of genes involved in POMC processing, but also the organization of secretory vesicles, and absence of tight junctions, the latter essential for hormone cellular storage and release.
Peptide hormone biogenesis and secretion are highly orchestrated spatiotemporal cellular events that involve hormone biosynthesis followed by intracellular trafficking, then sorting into constitutive or regulated secretory vesicles, granule maturation, cargo transportation and ultimately exocytosis. During these processes, immature prohormonal precursors undergo a variety of post-translational modifications, to ultimately reach a bioactive state and thereafter are packaged into vesicles, ready for secretion (22, 23). Our scRNAseq analyses confirm and significantly extend prior findings that key proteases and their regulators involved in prohormonal processing are expressed at much lower levels in SCT compared to FCT tumor cells. These included prohormone convertase (PCSK1), signal peptidase (SPCS1), dipeptidyl peptidase (DPP7), disulfide isomerase (PDIA3), cathepsin (CTSB/D/Z), pappalysin (PAPPA2) and serpin (SERPINF1, Fig. 1C, p<10−4). Our analysis further demonstrated that granin proteins (SCG5 and VGF), the building blocks and regulators of dense core secretory vesicles that process, transport, store and release peptide hormones (24, 25); small GTPases and their partners (RAB3B, RHOB, RHEB, ARL5B and PLD3) and several cytoskeleton components (ACTB, PFN1, GSN and MYL12A) which regulate granule exocytosis (26, 27), were all expressed at much lower levels in SCT compared to FCT tumor cells (Fig. 1C, p<10−4). Together with the tight junction claudins (CLDN4 and CLDN7), these small GTPase and cytoskeleton components are critical to maintaining endocrine cell polarization and may limit cell permeability in FCT tumor cells in contrast to SCTs (depicted schematically in Fig. 6) (28–30).
Figure 6. Schematic representation summarizing the major transcriptomic differences in various cell populations between FCTs and SCTs revealed by scRNAseq.
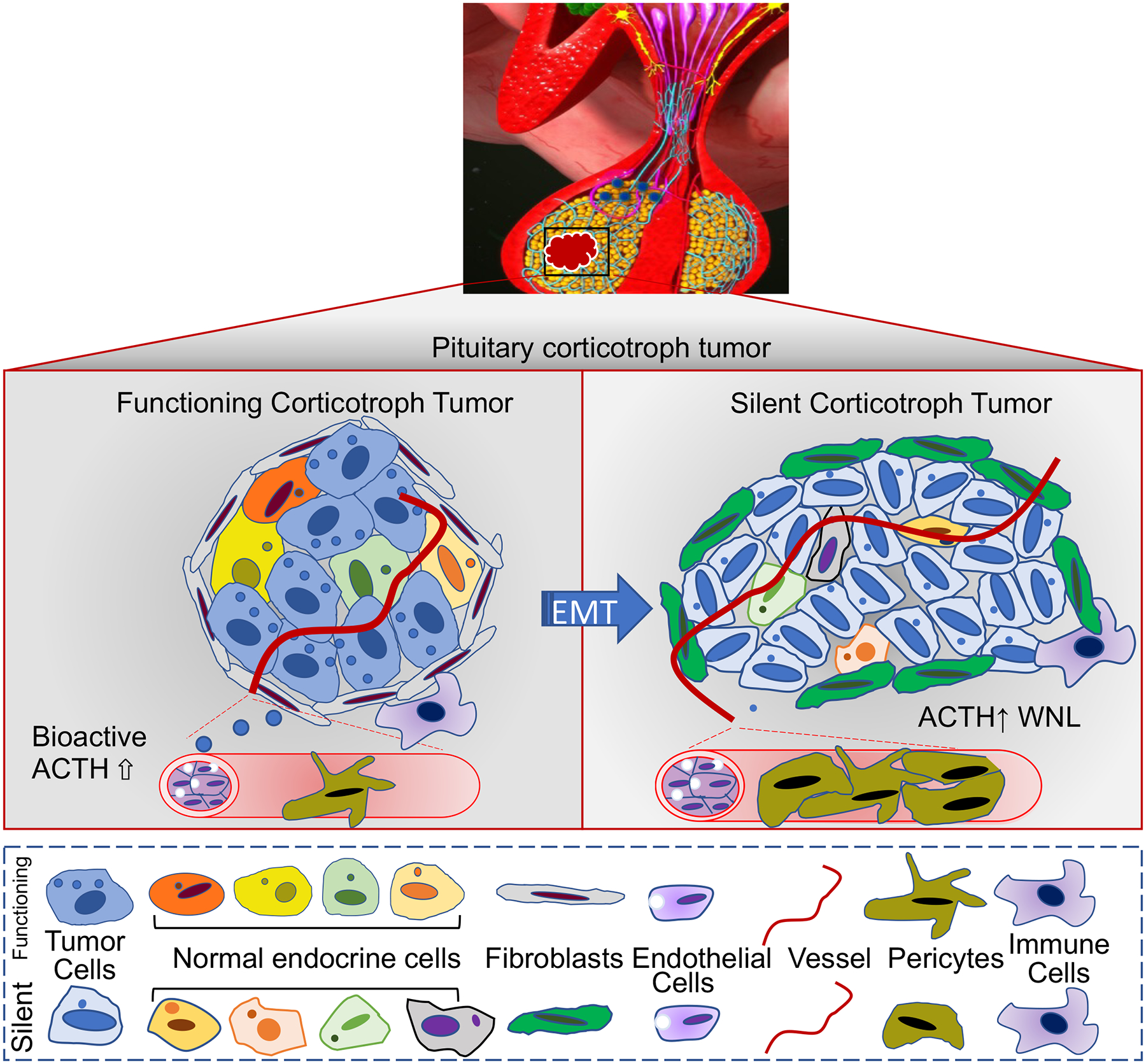
The tumor cells of FCTs (Micro and Macroadenomas) expressed higher levels of genes involved in prohormonal processing, hormone storage and secretion. Fibroblasts in both functioning corticotroph micro- and macro-adenomas exhibited features of residential fibroblasts that participate in ECM organization, ECM receptor interaction and inflammation. In contrast, SCT tumor cells exhibited reduced expression of genes involved in vesicle biogenesis and granule exocytosis, but increased stromal expression of transcripts of vSMCs and increased expression of genes involved in ECM reorganization, cell motility and migration. WNL: Within normal limits.
Our analysis also showed that FCT tumor cells expressed higher levels of several secretory growth factors, such as LTBP3 (TGFB regulator), RSPO3 (WNT pathway ligand), TIMP1 (MMPs inhibitor), galectins (LGALS1/3), galanin (GAL), and angiotensinogen (AGT, Fig. 1C, p<10−4), suggesting that FCT tumor cells exhibit multiple features of a well differentiated phenotype similar to their sibling normal mature corticotroph cells (31). In contrast, we noted higher expression of the pituitary organogenesis genes PITX1, SIX3, and LY6H in SCTs (Fig. 1D, p<10−4), pointing to a degree of de-differentiation in these tumors. Together, these findings may partly explain the not infrequent clinical observation that although some patients may exhibit increased circulating ACTH levels, they are often eucortisolemic (4). As ACTH antibodies employed in commercial ACTH assays recognize only the N-terminal region of the POMC fragment, they cannot distinguish authentic ACTH (1–39aa) from partially processed inactive ACTH-like peptides that harbor an extended C-terminal peptide sequence. These biologically inactive ACTH-like peptides may be caused by defects in components of POMC processing as we and others have described and supported by the finding of high molecular weight ACTH immunoreactive molecules found in many SCTs (Fig. 6) (32).
In addition to deficiency of factors involved in prohormonal processing and secretion, SCT tumor cells exhibited several features of epithelial to mesenchymal transition (EMT). EMT was initially recognized as a dynamic functional biological process during embryogenesis, whereby polarized epithelial cells lose their interaction with the basement membrane and cell-cell tight junctions loosen during cell division. These cells subsequently exhibit increased migratory capability and resistance to cell death which facilitates their movement to local and regional sites (33). Later studies noted that even terminally differentiated mature adult epithelial cells can also undergo trans-differentiation by activation of EMT in response to inflammatory, hypoxic or other pathological stressors that trigger regenerative and/or fibrotic events (34). In addition to these 2 types of restrained EMT, the tumorigenic process can also utilize EMT regulatory circuits in conjunction with epigenetic and genetic changes to drive clonal tumor outgrowth, invasion and metastasis (35).
The features of EMT that we observed in the SCT tumor cells included loss of the hall marker transcripts for EPCAM, KRT8, and E-cadherin (CDH1), along with increased expression of N-cadherin (CDH2) and the mesenchymal matrix markers (COL1A1 and COL4A1), as well as increased expression of the transcription factors ID2, TLE1, and ZEB1/2 (Fig. 1C, p<10−4) (35). Additionally, activation of the WNT pathway (WNT16 and RIMBP2) (36), increased expression of genes involved in growth signaling (IL6ST, FGF5, PI3KR1, and MERTK) (37), cell-cell/ECM modulation (ITGA6, YBX3, and EVL) (38, 39), cell motility and migration (CNKSR3, PHACTR3, GPM6A, DTNA, SNTG1, and PDZD2) (40–48), and DNA replication/cell cycle progression (CCND2, SMC6, ZBTB49, TRRAP, and SAMHD1 (Fig. 1D, p<10−4) (49–52) were also noted in the SCT tumor cells. Of particular relevance, the signaling pathways that can trigger EMT in all of the 3 scenarios above were actively expressed in the SCT tumor cells, and included activation of the WNT, FGFs, BMPs and TGFB pathways, which can in turn modulate neighboring epithelium, stromal and immune cells.
In addition to the changes between SCT and FCT tumor cells themselves, we demonstrated striking differences in the stromal cells observed in SCTs and FCTs. It is clearly established that fibroblasts act to modulate the tumor microenvironment by secretion of a variety of growth factors, in addition to extracellular matrix and these co-operate to modulate tumor architecture (53). Our studies demonstrate that whereas fibroblasts in FCTs exhibited features of residential fibroblasts that participate in ECM organization, ECM receptor interaction and inflammation, markers of vascular SMCs (vSMCs) were much more abundant in the stromal cells observed in the SCTs (Fig. 2B, p<10−4). The pituitary is a highly vascular organ supplied by the anterior and posterior hypophyseal arteries (54–56). Endothelial and mural cells line the inner and encase the outer surface of blood vessels respectively (57). Mural vSMCs encapsulate the larger caliber blood vessels whereas pericytes envelop small branching capillaries (58). The mesenchymal features observed in both the vSMC and pericyte stromal cell populations in the SCTs indicated that active vasculogenesis and remodeling was present (Figs. 2B & 2C, depicted schematically in Fig. 6). Some of these tumor stromal cell factors driving EMT such as TGFB1 blockade could represent potential therapeutic targets (Figs. 2D & 2E) to modify SCT behavior, and small molecule TGFB1 inhibitors such as YL-13027 already exist. Unlike the fibroblast and endothelial cells, which were quite different, the immune cell populations were comparatively similar between the FCTs and SCTs except that the inhibitory HAVCR2 transcript (7) was expressed at much higher levels in SCTs (Fig. 3G, p<10−7). What role, if any, HAVCR2 plays in SCTs is unclear but this certainly warrants further study.
Amongst the endocrine progenitor cell populations in both FCTs and SCTs, we noted the presence of folliculo-stellate cells (FSCs), identified by the hallmark transcripts S100B, S100A1/6/10/11, and ANXA1/2 (Fig. 4E) (10). FSCs are a group of non-hormonal producing cells that make up 3–5% of the anterior pituitary cell population and act to provide sustentacular structural support and facilitate paracrine intracellular communications (59). However, whereas the progenitor cells of FCTs expressed developmental morphogenesis, survival and senescence genes such as IGFBP5, GPC3, OTOS, MIA, LYPD1, FXYD5, ID1/3 (12–15), transcripts associated with myogenesis and myofibrillary contractile genes including PAX7, DCBLD2, TMSB4X, MFAP5, TNNT1 and TNNC1 (16–19) were much more abundant in the progenitor cells in SCTs (Fig. 4E, p<10−4).
In totality, this quite different transcriptional repertoire that we observed in various cell populations including the tumor cells but additionally the stroma, immune and progenitor cells between FCTs and SCTs, emphasizes the importance of cell-cell/ECM interaction, and paracrine/autocrine communication in shaping the heterogenous corticotroph tumor microenvironment (60). We believe the striking differences in the gene signatures of the SCTs versus FCTs which reflect a phenotypic shift towards a mesenchymal-like cell entity in SCTs, potentially afford these tumors increased mobility and provide previously unknown insights into their invasive potential (Figs 2, 4 & 5). These findings also raise the intriguing possibility that a common transcriptional reprogramming mechanism may be at work to control multiple cell types in SCTs driving them simultaneously towards a relatively de-differentiated mesenchymal phenotype (61).
Our scRNAseq analysis provides an unprecedented elucidation of the transcriptomic features of thousands of heterogenous FCT and SCT tumor cells simultaneously and provides novel insights into the mechanism(s) of their different clinical behaviors. We acknowledge that our sample number is small but the high consistency of our findings in these corticotroph tumor sub-types is reassuring. A further challenge with any molecular analysis is the confounding effect of sample contamination from adjacent surgically resected normal tissues. This issue is minimized in pituitary tumors due to their careful removal within a pseudo capsule with no margin co-resection. However, as we observed, entrapped normal tissues or invasive margins can still be a source of contamination, and is an inherent challenge of any analysis. In particular, bulk transcriptional analysis is confounded by this, whereas, single cell transcriptional analysis enables the operator to characterize and identify individual cell populations, thereby allowing targeted transcriptional analysis of these populations and simultaneously reducing any confounding effects of contaminating elements. In so doing, scRNAseq analysis minimizes over- or mis-interpretation of transcriptional readouts in tumor samples compared to bulk tumor transcriptional approaches.
In summary, our findings support a hypothesis that SCTs originate from incompletely differentiated corticotroph cells, which display deficiencies in secretory machinery from early in the tumorigenic process potentially due to a different tumor microenvironment (Fig. 6) (3). Our findings may further support a role for blocking EMT or targeting factors such as TGFB1 in mesenchymal stromal cells as novel treatment strategies in the management of these often challenging tumors.
Materials and Methods:
Patient consent
This study was approved by UCLA Institutional Review Board (IRB#20–002235). Written consent was obtained from each patient after full explanation of the purpose and nature of all procedures used.
Patient Information
Individual patient clinical manifestations and laboratory results are listed in Table-1 and described below. Clinically functioning corticotroph tumors (FCTs) were defined as patients exhibiting typical Cushingoid features (a combination of central fat accumulation, striae, acne, easy bruising, proximal myopathy, diabetes and/or hypertension) along with biochemical evidence of hypercortisolism with failed suppression of total serum cortisol <1.8μg/dL following Dexamethasone (either 1mg overnight or 48h 0.5mg Q 6h testing), elevated late night salivary (LNSC, > 0.112nmol/L on at least 2 measures) and 24-h urinary free cortisol (UFC, > 2-fold upper limit normal on at least 2 measures). Clinically silent corticotroph tumors (SCTs) were diagnosed based on the absence of Cushingoid features and no biochemical evidence of hypercortisolism with normal morning serum cortisol, LNSC, 24h-UFC and either normal or elevated plasma ACTH (>45pg/ml). Four of 5 FCTs (FCT1-3 & FCT5) were microadenomas. The remaining FCT (FCT4) was a macroadenoma that exhibited clival and bilateral cavernous sinus invasion and the 3 SCTs were macroadenomas, two of which (SCT1 & 3) exhibited mass effect on the optic tract. Corticotroph tumor was confirmed histopathologically in all cases, 7 tumors (FCT1-5 & SCT1 & 2) exhibited ACTH immunostaining and one ACTH immunonegative SCT (SCT3) exhibited immunoreactivity for the transcription factor TBX19 (aka Tpit). The gonadotroph tumor was a recurrent pituitary macroadenoma measuring 1.8 × 2.2 × 2.5cm causing severe optic chiasm compression. Histopathology demonstrated LH and FSH immunostaining with a KI-67 labelling index of 2%.
Single-cell RNA-Sequencing
A single cell suspension of surgically resected human pituitary tumors was obtained by mechanical and enzymic digestion using a gentleMACS dissociator, and human tumor dissociation kit (Miltenyi Biotec Inc., Germany, Cat# 130-095-929). Library generation was performed on the 10x Genomics Chromium Controller following the manufacturer’s protocol for the v3 reagent kit (10x Genomics). In brief, cell suspensions were loaded onto a Chromium Single Cell A Chip, aiming for 10,000 cells per channel for generation of single-cell gel bead-in-emulsions (GEMs), following which reverse transcription was performed. The resulting post-GEM reverse transcription product was then cleaned using DynaBeads MyOne silane beads (Thermo Fisher Scientific, Waltham, MA). The cDNA was amplified, cleaned and quantified, then enzymatically fragmented and size selected prior to library construction. Libraries were quantified by KAPA quantitative PCR for Illumina adapters (Roche, Pleasanton, CA) and size was determined by Agilent TapeStation D1000 tapes. Libraries were sequenced on a NextSeq 500 sequencer (Illumina, San Diego, CA).
Bioinformatic analyses of scRNAseq data
Demultiplexed fastq files generated at UCLA Technology Center for Genomics & Bioinformatics (TCGB) were analyzed with the 10x Genomics Cell Ranger 2.1.1. The pipeline aligned the reads to the University of California Santa Cruz (UCSC) human reference (GRCh38) transcriptome using the RNAseq alignment program STAR. Data were imported and analyzed using Seurat package within Rstudio (62). For quality assurance, cells were selected for downstream analysis using the following conservative cut-offs: 1) Cell barcodes associated with the most UMIs were employed by estimating the number of cells captured as 5% of the input beads and retained this number of cell barcodes for downstream analysis; 2) Only cells with >300 and <7,500 unique genes detected, and UMI >1,100 and <50,000 were analyzed; and 3) Only cells with <20% of their counts mapping to MT genes were included; and 4) Only genes detected in >3 cells were included. Cumulatively, from all 8 corticotroph tumor tissue samples, we obtained 56,458 cells for the subsequent analysis. Normalization and variance stabilization was performed to reduce batch effect, and individual Seurat objects from each sample were integrated into one large Seurat object (63–65). For dimension reduction, principal component analysis (PCA) and uniform manifold approximation and projection (UMAP) were performed. Cell clusters were annotated using the “FindNeighbors” function, and the “mast” method (FC. threshold = 1.5) was used for differentially expressed gene (DEG) analyses through the “FindAllMarkers” function in Seurat within Rstudio (62). Raw data of scRNAseq for functioning corticotroph tumors FCT1–4 were deposited as described in our pirior publication (CD1-4) (66). scRNAseq fastq files for the rest samples are deposited in NCBI BioSample database. No customized codes were used.
Statistical analysis:
The “FindAllMarkers” function in the Seurat package was used to identify DEGs based on wilcox testing with Bonferroni correction. Significant DEGs (shown in Supplementary Table 2–7) were selected from genes with adjusted pvalues (p_val_adj) <10−4.
Supplementary Material
Funding:
This work was supported by the National Cancer Institute NIH/NCI R01CA251930 (APH), the Warley Trust (APH), and a Career Enhancement Program from the UCLA SPORE in Brain Cancer NIH/NCI P50CA211015 (DZ).
Footnotes
Declaration of interest: The authors have no conflict of interest to declare.
Reference
- 1.Alahmadi H, Lee D, Wilson JR, Hayhurst C, Mete O, Gentili F, Asa SL, Zadeh G. Clinical features of silent corticotroph adenomas. Acta neurochirurgica. 2012;154(8):1493–8. [DOI] [PubMed] [Google Scholar]
- 2.Saeger W, Lüdecke DK, Buchfelder M, Fahlbusch R, Quabbe HJ, Petersenn S. Pathohistological classification of pituitary tumors: 10 years of experience with the German Pituitary Tumor Registry. European journal of endocrinology. 2007;156(2):203–16. [DOI] [PubMed] [Google Scholar]
- 3.Drummond J, Roncaroli F, Grossman AB, Korbonits M. Clinical and Pathological Aspects of Silent Pituitary Adenomas. The Journal of Clinical Endocrinology & Metabolism. 2018;104(7):2473–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raverot G, Wierinckx A, Jouanneau E, Auger C, Borson-Chazot F, Lachuer J, Pugeat M, Trouillas J. Clinical, hormonal and molecular characterization of pituitary ACTH adenomas without (silent corticotroph adenomas) and with Cushing’s disease. European journal of endocrinology. 2010;163(1):35–43. [DOI] [PubMed] [Google Scholar]
- 5.Flores L, Sleightholm R, Neilsen B, Baine M, Drincic A, Thorell W, Shonka N, Oupicky D, Zhang C. Highly Aggressive and Radiation-Resistant, “Atypical” and Silent Pituitary Corticotrophic Carcinoma: A Case Report and Review of the Literature. Case Reports in Oncology. 2019;12(1):139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen Y, Heaney AP. Pathogenesis and Treatment of Aggressive Corticotroph Pituitary Tumors. In: Geer EB, editor. The Hypothalamic-Pituitary-Adrenal Axis in Health and Disease: Cushing’s Syndrome and Beyond. Cham: Springer International Publishing; 2017. p. 93–110. [Google Scholar]
- 7.Wolf Y, Anderson AC, Kuchroo VK. TIM3 comes of age as an inhibitory receptor. Nat Rev Immunol. 2020;20(3):173–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q, Leshkowitz D, Blechman J, Levkowitz G. Single-Cell Molecular and Cellular Architecture of the Mouse Neurohypophysis. eneuro. 2020;7(1):ENEURO.0345–19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adivitiya Kaushik MS, Chakraborty S, Veleri S, Kateriya S. Mucociliary Respiratory Epithelium Integrity in Molecular Defense and Susceptibility to Pulmonary Viral Infections. Biology. 2021;10(2):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fletcher PA, Smiljanic K, Maso Prévide R, Iben JR, Li T, Rokic MB, Sherman A, Coon SL, Stojilkovic SS. Cell Type- and Sex-Dependent Transcriptome Profiles of Rat Anterior Pituitary Cells. Front Endocrinol (Lausanne). 2019;10:623-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vankelecom H, Chen J. Pituitary stem cells: where do we stand? Mol Cell Endocrinol. 2014;385(1–2):2–17. [DOI] [PubMed] [Google Scholar]
- 12.Mulry E, Parham K. Inner Ear Proteins as Potential Biomarkers. Otology & Neurotology. 2020;41(2):145–52. [DOI] [PubMed] [Google Scholar]
- 13.Miwa JM, Anderson KR, Hoffman KM. Lynx Prototoxins: Roles of Endogenous Mammalian Neurotoxin-Like Proteins in Modulating Nicotinic Acetylcholine Receptor Function to Influence Complex Biological Processes. Front Pharmacol. 2019;10:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolluri A, Ho M. The Role of Glypican-3 in Regulating Wnt, YAP, and Hedgehog in Liver Cancer. Front Oncol. 2019;9:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nogueira JF, de Sousa Lobo Ferreira Querido R, Gonçalves da Silva Leite J, Cabral da Costa T Future of Endoscopic Ear Surgery. Otolaryngol Clin North Am. 2021;54(1):221–31. [DOI] [PubMed] [Google Scholar]
- 16.Kikuta K, Kubota D, Yoshida A, Qiao Z, Morioka H, Nakamura M, Matsumoto M, Chuman H, Kawai A, Kondo T. Discoidin, CUB and LCCL domain-containing protein 2 (DCBLD2) is a novel biomarker of myxofibrosarcoma invasion identified by global protein expression profiling. Biochim Biophys Acta Proteins Proteom. 2017;1865(9):1160–6. [DOI] [PubMed] [Google Scholar]
- 17.Banerji CRS, Zammit PS. Pathomechanisms and biomarkers in facioscapulohumeral muscular dystrophy: roles of DUX4 and PAX7. EMBO Mol Med. 2021;13(8):e13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox MD, Carson VJ, Feng HZ, Lawlor MW, Gray JT, Brigatti KW, Jin JP, Strauss KA. TNNT1 nemaline myopathy: natural history and therapeutic frontier. Hum Mol Genet. 2018;27(18):3272–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao L, Westerhoff M, Hornick JL, Krausz T, Antic T, Xiao SY, Hart J. Loss of microfibril-associated protein 5 (MFAP5) expression in colon cancer stroma. Virchows Arch. 2020;476(3):383–90. [DOI] [PubMed] [Google Scholar]
- 20.Manigandan S, Mukherjee S, Yun JW. Loss of family with sequence similarity 107, member A (FAM107A) induces browning in 3T3-L1 adipocytes. Archives of Biochemistry and Biophysics. 2021;704:108885. [DOI] [PubMed] [Google Scholar]
- 21.Zahedi K, Barone S, Soleimani M. Polyamine Catabolism in Acute Kidney Injury. Int J Mol Sci. 2019;20(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cawley NX, Li Z, Loh YP. 60 YEARS OF POMC: Biosynthesis, trafficking, and secretion of pro-opiomelanocortin-derived peptides. Journal of molecular endocrinology. 2016;56(4):T77–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nillni EA. The Cell Biology Neuropeptide Hormones. In: Nillni EA , editor. Textbook of Energy Balance, Neuropeptide Hormones, and Neuroendocrine Function. Cham: Springer International Publishing; 2018. p. 109–39. [Google Scholar]
- 24.Kim T, Gondré-Lewis MC, Arnaoutova I, Loh YP. Dense-Core Secretory Granule Biogenesis. Physiology. 2006;21(2):124–33. [DOI] [PubMed] [Google Scholar]
- 25.Taupenot L, Harper KL, O’Connor DT. The Chromogranin–Secretogranin Family. New England Journal of Medicine. 2003;348(12):1134–49. [DOI] [PubMed] [Google Scholar]
- 26.Darchen F, Senyshyn J, Brondyk WH, Taatjes DJ, Holz RW, Henry JP, Denizot JP, Macara IG. The GTPase Rab3a is associated with large dense core vesicles in bovine chromaffin cells and rat PC12 cells. Journal of Cell Science. 1995;108(4):1639–49. [DOI] [PubMed] [Google Scholar]
- 27.Streit L, Brunaud L, Vitale N, Ory S, Gasman S. Hormones Secretion and Rho GTPases in Neuroendocrine Tumors. Cancers (Basel). 2020;12(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson JM, Van Itallie CM. Tight junctions. Current Biology. 2008;18(20):R941–R3. [DOI] [PubMed] [Google Scholar]
- 29.Meda P Gap junction proteins are key drivers of endocrine function. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2018;1860(1):124–40. [DOI] [PubMed] [Google Scholar]
- 30.Terry S, Nie M, Matter K, Balda MS. Rho Signaling and Tight Junction Functions. Physiology. 2010;25(1):16–26. [DOI] [PubMed] [Google Scholar]
- 31.Eieland AK, Normann KR, Sundaram AYM, Nyman TA, Øystese KAB, Lekva T, Berg JP, Bollerslev J, Olarescu NC. Distinct Pattern of Endoplasmic Reticulum Protein Processing and Extracellular Matrix Proteins in Functioning and Silent Corticotroph Pituitary Adenomas. Cancers (Basel). 2020;12(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuno A, Okazaki R, Oki Y, Nagashima T. Secretion of high-molecular-weight adrenocorticotropic hormone from a pituitary adenoma in a patient without Cushing stigmata. Case report. J Neurosurg. 2004;101(5):874–7. [DOI] [PubMed] [Google Scholar]
- 33.Hay ED An overview of epithelio-mesenchymal transformation. Acta Anat (Basel). 1995;154(1):8–20. [DOI] [PubMed] [Google Scholar]
- 34.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. The Journal of clinical investigation. 2009;119(6):1420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gil J, Jordà M, Soldevila B, Puig-Domingo M. Epithelial–Mesenchymal Transition in the Resistance to Somatostatin Receptor Ligands in Acromegaly. Front Endocrinol (Lausanne). 2021;12(226). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez M, Torres VI, Vio CP, Inestrosa NC. Canonical Wnt Signaling Modulates the Expression of Pre- and Postsynaptic Components in Different Temporal Patterns. Molecular Neurobiology. 2020;57(3):1389–404. [DOI] [PubMed] [Google Scholar]
- 37.Cummings CT, Deryckere D, Earp HS, Graham DK. Molecular pathways: MERTK signaling in cancer. Clin Cancer Res. 2013;19(19):5275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luciani A, Festa BP, Chen Z, Devuyst O. Defective autophagy degradation and abnormal tight junction-associated signaling drive epithelial dysfunction in cystinosis. Autophagy. 2018;14(7):1157–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwayer C, Shamipour S, Pranjic-Ferscha K, Schauer A, Balda M, Tada M, Matter K, Heisenberg C-P. Mechanosensation of Tight Junctions Depends on ZO-1 Phase Separation and Flow. Cell. 2019;179(4):937–52.e18. [DOI] [PubMed] [Google Scholar]
- 40.Attar MA, Salem JC, Pursel HS, Santy LC. CNK3 and IPCEF1 produce a single protein that is required for HGF dependent Arf6 activation and migration. Experimental Cell Research. 2012;318(3):228–37. [DOI] [PubMed] [Google Scholar]
- 41.Sagara J, Arata T, Taniguchi S. Scapinin, the protein phosphatase 1 binding protein, enhances cell spreading and motility by interacting with the actin cytoskeleton. PLoS One. 2009;4(1):e4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghatnatti V, Vastrad B, Patil S, Vastrad C, Kotturshetti I. Identification of potential and novel target genes in pituitary prolactinoma by bioinformatics analysis. AIMS Neurosci. 2021;8(2):254–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michibata H, Okuno T, Konishi N, Kyono K, Wakimoto K, Aoki K, Kondo Y, Takata K, Kitamura Y, Taniguchi T. Human GPM6A is associated with differentiation and neuronal migration of neurons derived from human embryonic stem cells. Stem Cells Dev. 2009;18(4):629–39. [DOI] [PubMed] [Google Scholar]
- 44.Falch CM, Sundaram AYM, Øystese KA, Normann KR, Lekva T, Silamikelis I, Eieland AK, Andersen M, Bollerslev J, Olarescu NC. Gene expression profiling of fast- and slow-growing non-functioning gonadotroph pituitary adenomas. European journal of endocrinology. 2018;178(3):295–307. [DOI] [PubMed] [Google Scholar]
- 45.Requena T, Cabrera S, Martín-Sierra C, Price SD, Lysakowski A, Lopez-Escamez JA. Identification of two novel mutations in FAM136A and DTNA genes in autosomal-dominant familial Meniere’s disease. Hum Mol Genet. 2014;24(4):1119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hafner A, Obermajer N, Kos J. Gamma-1-Syntrophin Mediates Trafficking of Gamma-Enolase towards the Plasma Membrane and Enhances Its Neurotrophic Activity. Neurosignals. 2010;18(4):246–58. [DOI] [PubMed] [Google Scholar]
- 47.Suen PM, Zou C, Zhang YA, Lau TK, Chan J, Yao KM, Leung PS. PDZ-domain containing-2 (PDZD2) is a novel factor that affects the growth and differentiation of human fetal pancreatic progenitor cells. Int J Biochem Cell Biol. 2008;40(4):789–803. [DOI] [PubMed] [Google Scholar]
- 48.Yanagawa T, Sumiyoshi H, Higashi K, Nakao S, Higashiyama R, Fukumitsu H, Minakawa K, Chiba Y, Suzuki Y, Sumida K, et al. Identification of a Novel Bone Marrow Cell-Derived Accelerator of Fibrotic Liver Regeneration Through Mobilization of Hepatic Progenitor Cells in Mice. Stem cells (Dayton, Ohio). 2019;37(1):89–101. [DOI] [PubMed] [Google Scholar]
- 49.Outwin EA, Irmisch A, Murray JM, O’Connell MJ. Smc5-Smc6-dependent removal of cohesin from mitotic chromosomes. Mol Cell Biol. 2009;29(16):4363–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeon BN, Kim MK, Yoon JH, Kim MY, An H, Noh HJ, Choi WI, Koh DI, Hur MW. Two ZNF509 (ZBTB49) isoforms induce cell-cycle arrest by activating transcription of p21/CDKN1A and RB upon exposure to genotoxic stress. Nucleic Acids Res. 2014;42(18):11447–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daddacha W, Koyen AE, Bastien AJ, Head PE, Dhere VR, Nabeta GN, Connolly EC, Werner E, Madden MZ, Daly MB, et al. SAMHD1 Promotes DNA End Resection to Facilitate DNA Repair by Homologous Recombination. Cell reports. 2017;20(8):1921–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tapias A, Zhou ZW, Shi Y, Chong Z, Wang P, Groth M, Platzer M, Huttner W, Herceg Z, Yang YG, et al. Trrap-dependent histone acetylation specifically regulates cell-cycle gene transcription to control neural progenitor fate decisions. Cell Stem Cell. 2014;14(5):632–43. [DOI] [PubMed] [Google Scholar]
- 53.Kendall RT, Feghali-Bostwick CA. Fibroblasts in fibrosis: novel roles and mediators. Frontiers in Pharmacology. 2014;5(123). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Negm IM. The vascular blood supply of the pituitary and its development. Acta Anat (Basel). 1971;80(4):604–19. [DOI] [PubMed] [Google Scholar]
- 55.Daniel PM. The blood supply of the hypothalamus and pituitary gland. Br Med Bull. 1966;22(3):202–8. [DOI] [PubMed] [Google Scholar]
- 56.Cironi KA, Decater T, Iwanaga J, Dumont AS, Tubbs RS. Arterial Supply to the Pituitary Gland: A Comprehensive Review. World Neurosurgery. 2020;142:206–11. [DOI] [PubMed] [Google Scholar]
- 57.Hill J, Rom S, Ramirez SH, Persidsky Y. Emerging roles of pericytes in the regulation of the neurovascular unit in health and disease. J Neuroimmune Pharmacol. 2014;9(5):591–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu J, Shenoy AK. Epithelial-to-Pericyte Transition in Cancer. Cancers (Basel). 2017;9(7):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horvath E, Kovacs K. Folliculo-stellate cells of the human pituitary: a type of adult stem cell? Ultrastruct Pathol. 2002;26(4):219–28. [DOI] [PubMed] [Google Scholar]
- 60.Roche J The Epithelial-to-Mesenchymal Transition in Cancer. Cancers (Basel). 2018;10(2):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shenoy AK, Jin Y, Luo H, Tang M, Pampo C, Shao R, Siemann DW, Wu L, Heldermon CD, Law BK, et al. Epithelial-to-mesenchymal transition confers pericyte properties on cancer cells. The Journal of Clinical Investigation. 2016;126(11):4174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Molecular Systems Biology. 2019;15(6):e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biology. 2019;20(1):296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahlmann-Eltze C, Huber W. Transformation and Preprocessing of Single-Cell RNA-Seq Data. bioRxiv. 2021:2021.06.24.449781. [Google Scholar]
- 65.Arzalluz-Luque Á, Devailly G, Mantsoki A, Joshi A. Delineating biological and technical variance in single cell expression data. The international journal of biochemistry & cell biology. 2017;90:161–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang D, Hugo W, Redublo P, Miao H, Bergsneider M, Wang MB, Kim W, Yong WH, Heaney AP. A human ACTH-secreting corticotroph tumoroid model: Novel Human ACTH-Secreting Tumor Cell in vitro Model. EBioMedicine. 2021;66:103294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


