Short abstract
Content available: Audio Recording
Abbreviations
- MAFLD
metabolic associated fatty liver disease
- NAFLD
non‐alcoholic fatty liver disease
- PDFF
proton density fat fraction
Metabolic associated fatty liver disease (MAFLD) encompasses a broad disease spectrum that impacts an estimated one billion people worldwide. 1 Initially described as non‐alcoholic fatty liver disease (NAFLD) in 1980, there is now increasing recognition that the absence of excess alcohol use may be insufficient to characterize this disease process, and interaction between several metabolic risk factors leads to an array of dynamic phenotypes. Although this paradigm change could ultimately help guide new therapeutic strategies, multiple barriers continue to pose clinical care and discovery challenges, including disease heterogeneity, natural history variability, imperfect nomenclature, and suboptimal diagnostic and surveillance tools (Table 1). The application of precision medicine may hold promise for meaningful progress in the future.
TABLE 1.
Challenges in Diagnosis, Prognostication, and Therapeutics Development for MAFLD
| Process | Challenges |
|---|---|
| Screening/Diagnosis | Imperfect/evolving nomenclature |
| Disease heterogeneity | |
| Coexistance of alternative chronic liver diseases | |
| Lack of early and sensitive testing for at‐risk individuals | |
| Cost and availability of testing modalities | |
| Staging/surveillance | Lack of accurate modalities to detect steatohepatitis and early/intermediate fibrosis |
| Inefficient surveillance tools to monitor therapeutic response | |
| Treatment | Limited pharmacotherapy |
| Lack of tools to predict therapeutic response | |
| Clinical trial enrollment and retention limitations | |
| Prognostication | Cardiovascular and oncologic comorbidities |
| Long latency period prior to the development of liver‐related outcomes | |
| Variability in disease progression |
MAFLD Clinical Phenotypes
Phenotypes for MAFLD occur in the context of multiple metabolic risk factors that affect hepatic lipid accumulation, inflammation, and fibrosis. They include demographic traits (age, sex, and ethnicity), lifestyle characteristics (diet, tobacco and alcohol use, and weight), medical comorbidities (glucose intolerance, diabetes mellitus, and other endocrinopathies), surgical interventions (cholecystectomy), intestinal microbiomic composition, genetics, epigenetics, and metabolomics. 1 , 2 These elements contribute to both established and evolving phenotypes.
Among the established MAFLD phenotypes, the traditional phenotype includes individuals with excess weight, a history of minimal‐to‐moderate alcohol use and, and comorbidities such as diabetes mellitus, hypertension, dyslipidemia, and vascular disease. It is particularly common among Caucasians and captures approximately 80% of patients with MAFLD. 3 Disease progression among patients with this phenotype is typically limited. Only a small fraction develops cirrhosis, and the average rate of progression is several years to decades between fibrosis stages. 2 , 4 Alternatively, lean MAFLD is observed more frequently in the East Asian population, occurring in non‐obese individuals with possible genetic predisposition, intestinal dysbiosis, and endocrinopathies; it portends an increased risk for advanced liver disease. 5 Finally, Hispanic patients suffer excess burden from MAFLD, with higher disease prevalence and relatively faster progression; although this phenotype is likely multifactorial in etiology, common genetic variants have been identified. 6 , 7 Overall, among patients with MAFLD, the risks of hepatic decompensation, cardiovascular events, and malignancy are well‐described (Fig. 1). 8 , 9 , 10 , 11 , 12
FIG 1.
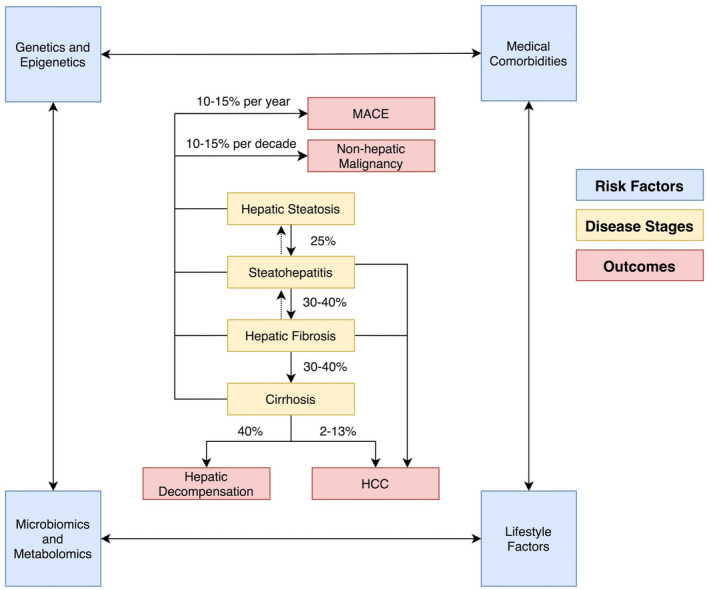
Natural history of MAFLD. Abbreviation: MACE, major adverse cardiovascular events.
Although these phenotypes have become increasingly recognized in the medical community, they lack the necessary granularity for clinical practice and research. Through the use of precision medicine tools, diverse genetic, epigenetic, and metabolomic signatures can ultimately be used to identify specific and targetable phenotypes. The building blocks for this approach currently exist, but future studies are necessary to determine how different types of molecular data can be synthesized in ways that are applicable for clinicians and researchers. Before this can be accomplished, barriers in diagnosis and staging will likely need to be overcome.
Challenges in Diagnosis and Staging
The diagnosis of MAFLD currently depends on a combination of clinical, laboratory, and radiographic assessments that include individuals’ risk factor profiles, basic labs and liver chemistries, serological testing to exclude alternative forms of liver disease, and relevant imaging findings, including features of hepatic steatosis and/or fibrosis. In conjunction with the diagnostic evaluation, an early determination of the presence and severity of fibrosis is critical given its association with all‐cause and cardiovascular mortality. 13
Non‐invasive tools, including clinical scoring systems, plasma biomarkers, and elastography, are currently used to identify those with MAFLD, MAFLD with steatohepatitis, and MAFLD with advanced fibrosis. In particular, transient elastography (FibroScan), shear wave elastography, and MR elastography are well‐established tools that allow for non‐invasive assessment of hepatic fibrosis. 14 Unfortunately, a number of these modalities lack the diagnostic discrimination for intermediate fibrosis stages, as well as identification of those with high steatohepatitis inflammatory activity. Thus, these diagnostic tests are likely not adequately sensitive to identify high‐risk patients, particularly those seeking enrollment in clinical trials. However, recent multi‐national validation of the FibroScan‐AST is one example of a risk score developed to identify those individuals with MAFLD, high inflammatory activity, and moderate‐to‐advanced fibrosis for clinical trial enrollment. 15
Finally, a lack of consistent nomenclature also continues to pose challenges, limiting the transition of phenotypic concepts to the clinical medicine. The term “MAFLD” is potentially more representative and inclusive than “NAFLD,” highlighting the premise that metabolic stress dictates phenotypes (and not alcohol use) and emphasizes that MAFLD is not a diagnosis of exclusion. However, it remains imperfect largely due to lack of specificity. Replacing the phrase “non‐alcoholic” with a general term such as “metabolic associated” can create ambiguity among providers and researchers since metabolic dysregulation plays a role in a multitude of disease processes that impact the liver. Frequent changes in nomenclature may also lead to confusion among non‐hepatologists and lead to barriers in interdisciplinary practices.
Future clinical care and research in MAFLD, therefore, is heavily contingent on appropriate host identification, specific and consistent nomenclature, and the application of non‐invasive, widely available, effective, and dynamic diagnostic and staging tools that incorporate precision medicine techniques.
Emerging Phenotyping Tools
Genetic and Epigenetic Biomarkers
Genome‐ and phenome‐wide association studies in obese and non‐obese patients of different ethnic backgrounds have identified single nucleotide polymorphisms in candidate genes that impact processes such as lipid remodeling, lipid metabolism, glycogen storage, and/or lipophagy (Table 2). 16 , 17 , 18 The effects of PNPLA3 polymorphisms have been evaluated in multiple epidemiologic studies, and findings suggest that specific variants can impact disease severity, progression, and responses to intervention. In particular, the PNPLA3 G risk allele is associated with an earlier age of diagnosis, especially among Hispanic patients, and M‐variants have been associated with an increased risk for adverse outcomes, including hepatic decompensation, hepatocellular carcinoma, and death. 6 , 7 Patients with the I148M polymorphism have been shown to have limited responses to statin use and may derive significant benefits from dietary modification. 19
TABLE 2.
Genetic, Epigenetic, Metabolomic, and Microbiomic Markers in MAFLD
| Genetic Polymorphisms | Risk Modification | Outcomes |
|---|---|---|
| PNPLA3 (G allele, M variants) | ↑ | Steatosis, steatohepatitis, fibrosis, decompensation, hepatocellular cancer, death |
| GCRK (P446L) | ↑ | Steatosis, steatohepatitis, fibrosis, hepatocellular cancer; synergistic effect with PNPLA3 I148M |
| HSD17B13 (inactivating variants) | ↓ | Steatohepatitis, fibrosis, hepatocellular cancer; mitigates risk in patients with PNPLA3 I148M |
| TM6SF2 (E167K) | ↑ | Steatosis, steatohepatitis, fibrosis, hepatocellular cancer |
| MBOAT7 (rs641738) | ↑ | Steatosis, steatohepatitis, fibrosis, hepatocellular cancer |
| Genes with epigenetic changes | ||
| AQP1 (overexpression) | ↑ | Fibrosis |
| FGFR2 (overexpression) | ↑ | Fibrosis |
| MicroRNAs | ||
| miR‐34a (overexpression) | ↑ | Steatosis, steatohepatitis |
| miR‐122 (underexpression) | ↑ | Steatosis, steatohepatitis; in human studies (potentially differing effects in mice) |
| Metabolites | ||
| Branched‐chain amino acids | ↑ or ↓ (depending on disease stage) | Steatosis, steatohepatitis, fibrosis |
| Lipids (triglycerides and fatty acids) | ↑ or ↓ (depending on molecular subtype) | Steatosis, steatohepatitis, fibrosis |
| Carbohydrates (glycolytic products) | ↑ | Steatosis, steatohepatitis |
| Bile acids (total) | ↑ (predominantly) | Steatosis, steatohepatitis, fibrosis; some bile acids reduce risk for steatosis and steatohepatitis |
| Gut microbiome | ||
| Proteobacteria, Enterobacteria | ↑ | Steatosis, steatohepatitis, fibrosis |
| Firmicutes | ↑ or ↓ (depending on bacterial species and disease stage) | Steatosis, steatohepatitis, fibrosis; Firmicutes concentrations may decrease with fibrosis progression |
| Bacteroidetes | ↑ or ↓ (depending on bacterial species and disease stage) | Steatosis, steatohepatitis, fibrosis |
Factors that impact gene regulation such as differential DNA methylation and miRNA expression have also been implicated in the pathogenesis and progression of MAFLD. Epigenome‐wide association studies and microarrays have identified a subset of genes and miRNA sequences that impact lipid metabolism and inflammation in MAFLD (Table 2). 20 , 21
Metabolomics
The addition of metabolomics to genetic and epigenetic data, microbiomics, and additional surrogate markers may enable targeted clinical research in MAFLD. Studies have already highlighted its potential impact by demonstrating that metabolic profiles incorporating lipid, carbohydrate, amino acid, bacterial, and/or bile acid markers from plasma, urine, or stool samples can be used to identify disease subtypes, monitor for disease progression, assess the risk for outcomes such as cardiovascular disease and cancer, and even discriminate between MAFLD fibrosis stages, in some cases outperforming standard scoring systems for the detection of advanced fibrosis (Table 2). 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 In particular, changes in the levels of particular amino acids (branched chain and glutathione metabolites) and alterations in fatty acid and bile salt composition have been studied. Increased levels of branched‐chain amino acids, increased frequency of fatty acids with low carbon number and double bonds, and a preferential increase in primary bile acids may signify metabolically active MAFLD, whereas with disease progression, one may expect decreased levels of branched‐chain amino acids and significant reductions in glutathione precursors. 22 Biomarker panels, which can be used to rapidly measure multiple metabolite levels using spectroscopy or chromatography, offer clinicians, patients, and researchers the possibility of trending disease activity and monitoring therapeutic responses in a much more robust manner, overcoming many of the limitations posed by conventional tools.
Radiographic Biomarkers
Finally, a number of MR‐based biomarkers and methods, including proton density fat fraction (PDFF), spectroscopy, T1 mapping, gadoxetate, and multiparametric imaging, have been studied in the detection of steatosis, steatohepatitis, fibrosis, and hepatocyte function in MAFLD. 31 Unfortunately, the application of some of these techniques are limited by technological constraints (availability of specialized scanners). However, PDFF is readily available and has been applied longitudinally to track disease activity. Studies have demonstrated that changes in liver fat content measured via PDFF correlate with changes histologic disease activity, including fibrosis. 32 , 33
Future Directions: The Path to Precision Medicine
MAFLD is a heterogeneous disease with diverse phenotypes that incorporate a variety of risk factors. The spectrum of disease activity is vast, and outcomes can differ markedly among patients. However, our current diagnostic and therapeutic approaches are homogenous and rely largely on insensitive tools that are insufficient to identify the varied phenotypes in MAFLD. The rise of precision medicine in the form of genetic, epigenetic, metabolomic, and microbiomic techniques will help overcome these challenges as the burden of MAFLD continues to increase globally.
In the future, it may become possible to screen high‐risk patients with a combination of genetic testing and metabolomic assays that augment conventional modalities, such as elastography and serological studies. The results of these assays can be used to phenotype patients using specific terminology, readily monitor the impact of conventional and experimental treatments, serve as the basis for new highly targeted molecular therapies, and inform prognosis (Fig. 2). The application of machine learning can potentially further improve the efficacy and efficiency of these precision medicine tools.
FIG 2.
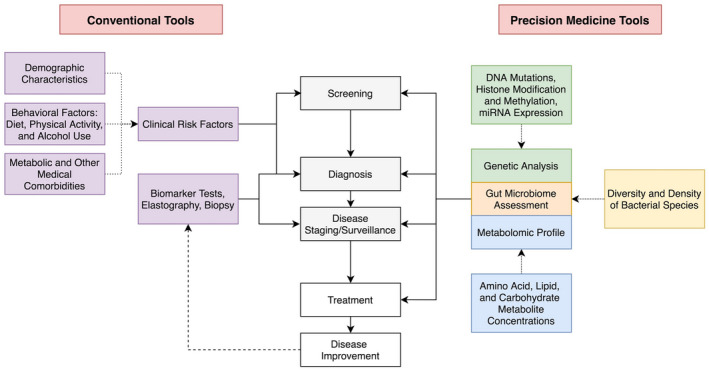
Precision medicine tools in diagnosis and management of MAFLD.
Although additional research will be required to understand how different types of data can be synthesized to develop more holistic diagnostic and treatment models, precision medicine will ultimately change the landscape of MAFLD. Initiatives, such as the Liver Investigation: Testing Marker Utility in Steatohepatitis (LITMUS) project, which aim to accomplish this goal, have been established. Armed with new tools, researchers and clinicians will soon be able to apply molecular techniques to accurately identify and monitor patients and tailor therapies based on personalized molecular signatures.
Potential conflict of interest: Nothing to report.
References
- 1. Eslam M, Sanyal AJ, George J, et al. International consensus panel. MAFLD: a consensus‐driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology 2020;158:1999‐2014.e1. [DOI] [PubMed] [Google Scholar]
- 2. Kechagias S, Nasr P, Blomdahl J, Ekstedt M. Established and emerging factors affecting the progression of nonalcoholic fatty liver disease. Metabolism 2020;111S:154183. [DOI] [PubMed] [Google Scholar]
- 3. Albhaisi S, Chowdhury A, Sanyal AJ. Non‐alcoholic fatty liver disease in lean individuals. JHEP Rep 2019;1:329‐341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Singh S, Allen AM, Wang Z, Prokop LJ, Murad MH, Loomba R. Fibrosis progression in nonalcoholic fatty liver vs nonalcoholic steatohepatitis: a systematic review and meta‐analysis of paired‐biopsy studies. Clin Gastroenterol Hepatol 2015;13:643‐654.e1‐9; quiz e39–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hagström H, Nasr P, Ekstedt M, et al. Risk for development of severe liver disease in lean patients with nonalcoholic fatty liver disease: a long‐term follow‐up study. Hepatol Commun 2017;2:48‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yuan L, Terrrault NA. PNPLA3 and nonalcoholic fatty liver disease: towards personalized medicine for fatty liver. Hepatobiliary Surg Nutr 2020;9:353‐356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walker RW, Belbin GM, Sorokin EP, et al. A common variant in PNPLA3 is associated with age at diagnosis of NAFLD in patients from a multi‐ethnic biobank. J Hepatol 2020;72:1070‐1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Loomba R, Wong R, Fraysse J, et al. Nonalcoholic fatty liver disease progression rates to cirrhosis and progression of cirrhosis to decompensation and mortality: a real world analysis of Medicare data. Aliment Pharmacol Ther 2020;51:1149‐1159. [DOI] [PubMed] [Google Scholar]
- 9. White DL, Kanwal F, El‐Serag HB. Association between nonalcoholic fatty liver disease and risk for hepatocellular cancer, based on systematic review. Clin Gastroenterol Hepatol 2012;10:1342‐1359.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simon TG, Roelstraete B, Khalili H, Hagström H, Ludvigsson JF. Mortality in biopsy‐confirmed nonalcoholic fatty liver disease: results from a nationwide cohort. Gut 2020. 10.1136/gutjnl-2020-322786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Abeles RD, Mullish BH, Forlano R, et al. Derivation and validation of a cardiovascular risk score for prediction of major acute cardiovascular events in non‐alcoholic fatty liver disease; the importance of an elevated mean platelet volume. Aliment Pharmacol Ther 2019;49:1077‐1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Allen AM, Hicks SB, Mara KC, Larson JJ, Therneau TM. The risk of incident extrahepatic cancers is higher in non‐alcoholic fatty liver disease than obesity ‐ a longitudinal cohort study. J Hepatol 2019;71:1229‐1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ekstedt M, Hagström H, Nasr P, et al. Fibrosis stage is the strongest predictor for disease‐specific mortality in NAFLD after up to 33 years of follow‐up. Hepatology 2015;61:1547‐1554. [DOI] [PubMed] [Google Scholar]
- 14. Xiao G, Zhu S, Xiao X, Yan L, Yang J, Wu G. Comparison of laboratory tests, ultrasound, or magnetic resonance elastography to detect fibrosis in patients with nonalcoholic fatty liver disease: a meta‐analysis. Hepatology 2017;66:1486‐1501. [DOI] [PubMed] [Google Scholar]
- 15. Newsome PN, Sasso M, Deeks JJ, et al. FibroScan‐AST (FAST) score for the non‐invasive identification of patients with non‐alcoholic steatohepatitis with significant activity and fibrosis: a prospective derivation and global validation study. Lancet Gastroenterol Hepatol 2020;5:362‐373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jonas W, Schürmann A. Genetic and epigenetic factors determining NAFLD risk. Mol Metab 2020. 10.1016/j.molmet.2020.101111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anstee QM, Darlay R, Cockell S, et al. EPoS Consortium Investigators. Genome‐wide association study of non‐alcoholic fatty liver and steatohepatitis in a histologically characterised cohort. J Hepatol 2020;73:505‐515. [DOI] [PubMed] [Google Scholar]
- 18. Yoshida K, Yokota K, Kutsuwada Y, et al. Genome‐wide association study of lean nonalcoholic fatty liver disease suggests human leukocyte antigen as a novel candidate locus. Hepatol Commun 2020;4:1124‐1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grimaudo S, Pipitone RM, Pennisi G, et al. Association between PNPLA3 rs738409 C>G variant and liver‐related outcomes in patients with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol 2020;18:935‐944.e3. [DOI] [PubMed] [Google Scholar]
- 20. Gerhard GS, Malenica I, Llaci L, et al. Differentially methylated loci in NAFLD cirrhosis are associated with key signaling pathways. Clin Epigenetics 2018;10:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Torres J‐L, Novo‐Veleiro I, Manzanedo L, et al. Role of microRNAs in alcohol‐induced liver disorders and non‐alcoholic fatty liver disease. World J Gastroenterol 2018;24:4104‐4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Masoodi M, Gastaldelli A, Hyötyläinen T, et al. Metabolomics and lipidomics in NAFLD: biomarkers and non‐invasive diagnostic tests. Nat Rev Gastroenterol Hepatol 2021;18(12):835‐856. 10.1038/s41575-021-00502-9. [DOI] [PubMed] [Google Scholar]
- 23. Gitto S, Schepis F, Andreone P, Villa E. Study of the serum metabolomic profile in nonalcoholic fatty liver disease: research and clinical perspectives. Metabolites 2018;8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Masarone M, Troisi J, Aglitti A, et al. Untargeted metabolomics as a diagnostic tool in NAFLD: discrimination of steatosis, steatohepatitis and cirrhosis. Metabolomics 2021;17:12. [DOI] [PubMed] [Google Scholar]
- 25. Mayo R, Crespo J, Martínez‐Arranz I, et al. Metabolomic‐based noninvasive serum test to diagnose nonalcoholic steatohepatitis: results from discovery and validation cohorts. Hepatol Commun 2018;2:807‐820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ioannou GN, Nagana Gowda GA, Djukovic D, Raftery D. Distinguishing NASH Histological Severity Using a Multiplatform Metabolomics Approach. Metabolites 2020;10:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mazzini F, Cook F & Gounarides J et al. Plasma and stool metabolomic biomarkers of non‐alcoholic fatty liver disease in Argentina. 10.1101/2020.07.30.20165308. [DOI]
- 28. Caussy C, Ajmera VH, Puri P, et al. Serum metabolites detect the presence of advanced fibrosis in derivation and validation cohorts of patients with non‐alcoholic fatty liver disease. Gut 2019;68:1884‐1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Loomba R, Seguritan V, Li W, et al. Gut microbiome‐based metagenomic signature for non‐invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab 2017;25:1054‐1062.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jayakumar S, Loomba R. Review article: emerging role of the gut microbiome in the progression of nonalcoholic fatty liver disease and potential therapeutic implications. Aliment Pharmacol Ther 2019;50:144‐158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Caussy C, Johansson L. Magnetic resonance‐based biomarkers in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Endocrinol Diabetes Metab 2020;3:e00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patel J, Bettencourt R, Cui J, et al. Association of noninvasive quantitative decline in liver fat content on MRI with histologic response in nonalcoholic steatohepatitis. Therap Adv Gastroenterol 2016;9:692‐701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ajmera V, Park CC, Caussy C, et al. Magnetic resonance imaging proton density fat fraction associates with progression of fibrosis in patients with nonalcoholic fatty liver disease. Gastroenterology 2018;155:307‐310.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]


